Abstract
Cytokine polymorphisms are associated with disease outcome and interferon (IFN) treatment response in hepatitis C virus (HCV) infection. We genotyped eight SNPs spanning the entire IFN-γ gene in two cohorts and assessed the association between those polymorphisms and treatment response or spontaneous viral clearance. The first cohort was composed of 284 chronically HCV-infected patients who had received IFN-α-based therapy and the second was 251 i.v. drug users who had either spontaneously cleared HCV or become chronically infected. A SNP variant located in the proximal IFN-γ promoter region next to the binding motif of heat shock transcription factor (HSF), −764G, was significantly associated with sustained virological response [P = 0.04, odds ratio (OR) = 3.51 (confidence interval 1.0–12.5)]. The association was independently significant in multiple logistic regression (P = 0.04) along with race, viral titer, and genotype. This variant was also significantly associated with spontaneous recovery [P = 0.04, OR = 3.51 (1.0–12.5)] in the second cohort. Functional analyses show that the G allele confers a two- to three-fold higher promoter activity and stronger binding affinity to HSF1 than the C allele. Our study suggests that the IFN-γ promoter SNP −764G/C is functionally important in determining viral clearance and treatment response in HCV-infected patients and may be used as a genetic marker to predict sustained virological response in HCV-infected patients.
Keywords: genetics, human study, cytokine, viral clearance, antiviral treatment
HCV is a major cause of chronic liver disease worldwide. Its clinical course is highly variable: only a fraction of infected persons exhibit spontaneous self-limited infection with clearance of the virus, whereas the majority (70%) develop chronic infection (1). The approved anti-HCV therapy, polyethylene-glycol-conjugated IFN-α (Peg-IFN-α) in combination with ribavirin, is effective in about half of patients (2, 3). The factors responsible for these different outcomes and response to IFN therapy involve both viral and host factors. Given the polymorphic genetic compositions of human populations, genetic variations likely play a role. Many recent studies have focused on the genetic polymorphisms of cytokines, chemokines, and their receptors (4, 5). However, these studies are limited with respect to study size and polymorphism functionality.
IFN-γ is a multifunctional cytokine, which is produced by effector T and natural killer cells. It defines the development of T helper 1 (Th1) cells and is critical for host defense against a variety of intracellular pathogens, including HCV. IFN-γ efficiently inhibits HCV replication in the replicon system in vitro (6) and the intrahepatic level of IFN-γ appears to be associated with viral clearance in the chimpanzee model (7). The human IFN-γ gene on chromosome 12q24.1 spans ≈5.4 kb and contains four exons that encode a 146-aa protein. Like other cytokines, the IFN-γ coding region is invariant, with no reported polymorphisms (8). Several polymorphisms within the IFN-γ noncoding regions, such as +874A/T, CA repeat microsatellite, and −179T/G, have been implicated in several autoimmune and chronic inflammatory conditions (9, 10). However, none of them has been shown to be related to HCV infection.
The aim of the present study was to identify and validate potential markers of the IFN-γ gene associated with response to IFN-α-based therapy and spontaneous recovery in HCV infection. To this end, we performed comprehensive genotyping of IFN-γ gene polymorphisms in two large cohorts of HCV-positive patients. The allele and carrier frequencies of SNPs were analyzed for association with either sustained response to IFN-α-based therapies or spontaneous viral clearance. The biological relevance of the most significantly associated SNP was studied by various functional assays in vitro.
Results
Genotypes Associated with Response to IFN Therapy.
We performed comprehensive genotyping of IFN-γ gene polymorphisms in two large cohorts of HCV-positive patients. Cohort 1 contained 284 HCV-infected patients, who received IFN-α-based therapy (Table 1). Treatment outcomes were grouped into sustained virological response (SVR) and nonresponse (NR); NR includes relapse after stopping treatment and no response during treatment, according to the standard definition (1). Univariate analyses of demographic and viral factors showed that race, gender, age, viral load, and viral genotype were significantly associated with response to IFN-α therapy (P < 0.05). Among the clinical factors evaluated, both histological grade and fibrosis stage were significantly associated with treatment response. The 251 patients, 92% African Americans (AA) and 8% Caucasian Americans (CA), of cohort 2 were participants in a well characterized study of i.v. drug users, of whom 85 (34%) had spontaneously cleared HCV infection and 166 (66%) had chronic HCV infection (11).
Table 1.
Demographic, virological, and clinical features of IFN-α-treated patients (cohort 1)
| Variable | All | SVR (%) | NR (%)* | P | |
|---|---|---|---|---|---|
| No. of patients | 284 | 98 (34.5) | 186 (65.5) | ||
| Race, no. | CA | 213 | 90 (42.3) | 123 (57.7) | <0.001 |
| AA | 71 | 8 (11.3) | 63 (88.7) | ||
| Sex, no. | Female | 103 | 44 (42.7) | 59 (57.3) | 0.028 |
| Male | 181 | 54 (29.8) | 127 (70.2) | ||
| Age, years† | 48.8 ± 8.5 | 47.2 ± 8.9 | 49.7 ± 8.2 | 0.017 | |
| ALT, units/liter† | 112.8 ± 83.9 | 122.8 ± 99.9 | 107.3 ± 73.4 | 0.222 | |
| Viral titer, log10(copies/ml)† | 6.2 ± 0.6 | 6.1 ± 0.7 | 6.3 ± 0.5 | 0.016 | |
| Virus genotype, no. | 1 | 223 | 53 (23.8) | 170 (76.2) | |
| Other | 53 | 40 (75.5) | 13 (24.5) | <0.001 | |
| Nonclassified | 8 | 5 (62.5) | 3 (37.5) | ||
| Histological grade, no. | Minimal to mild (≤7) | 57 | 27 (47.4) | 30 (52.6) | |
| Moderate to severe (>7) | 178 | 58 (32.6) | 120 (67.4) | 0.044 | |
| No biopsy | 49 | 13 (26.5) | 36 (73.5) | ||
| Fibrosis stage, no. | Minimal to mild (0, 1) | 122 | 57 (46.7) | 65 (53.3) | |
| Moderate to severe (3, 4) | 113 | 28 (24.8) | 85 (75.2) | 0.001 | |
| No biopsy | 49 | 13 (26.5) | 36 (73.5) |
P values in boldface are significant. ALT, alanine aminotransferase.
*Includes relapse after stopping treatment and no response during treatment.
†Data are expressed as mean ± SD.
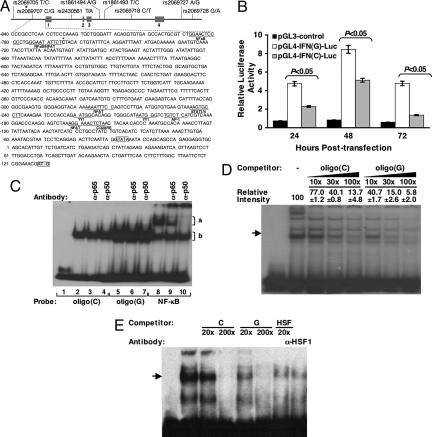
In both cohorts, we genotyped eight SNPs (two in the promoter region, four in the intron, and two downstream of the 3′ UTR) covering the entire IFN-γ gene as shown in Fig. 1A. The genotype distributions for all SNPs were consistent with those expected under Hardy–Weinberg equilibrium. Because allele frequency distribution differs by race, we evaluated the SNPs for association with treatment response by adjusting for race (reflected in P values for the combined group) and presented allele frequencies separately for each racial group (Table 2). One SNP (rs2069707), a C-to-G change located in the proximal promoter region at position −764, was found to be significantly associated with IFN treatment response in cohort 1. The frequencies of G allele differed significantly between those with SVR (3.9%) and NR (2.8%) (P = 0.01). The G carrier was significantly more frequent in those with SVR [P = 0.008, odds ratio (OR) = 2.66 with a confidence interval (CI) of 1.27–5.57]. Under a more restricted analysis comparing all SVRs to those NRs who received the optimal standard therapy (Peg-IFN-α plus ribavirin), the association between G carrier status and SVR was still significant, with a greater OR [P = 0.02, OR = 3.37 (1.15–9.83)]. Consistent with the correlation to IFN response, carrier of the allele −764G was also significantly associated with spontaneous recovery from HCV infection in cohort 2 [P = 0.04, OR = 3.51 (0.98–12.49)]. Haplotype analyses of these eight SNPs demonstrated that only the haplotype containing the G allele of SNP rs2069707 was significantly associated with IFN treatment response [supporting information (SI) Table 5].
Fig. 1.
Functional analyses of SNP −764C/G in vitro. (A) The eight SNPs in the human IFN-γ gene genotyped in this study are shown with their IDs and the corresponding polymorphic bases. The four exons are represented as boxes. Sequence of the promoter and 5′ UTR region is presented with the position number relative to the transcription site as predicted by Gray and Goeddel (35). The open and shaded boxes in the sequence represent the transcription initiative site (TATA box) and the translation start codon (AUG), respectively. The previously reported transcription factor binding sites are underlined. Asterisk indicates the SNP rs2069707. (B) Jurkat cells were cotransfected with individual firefly luciferase (FLuc)-expressing plasmid (pGL3-control, pGL4-IFN(C)-Luc, or pGL4-IFN(G)-Luc) and pRL-CMV as described in Materials and Methods. Cells were harvested after incubation for indicated periods and relative luciferase activity (FLuc/Renilla luciferase) was determined. Bars and error bars indicate mean ± SD of three experiments. (C) The 32P-labeled −764C, −764G, or NF-κB DNA oligonucleotide probe was used in the EMSA as indicated in the presence (lanes 2–10) of a HeLa cell nuclear extract in comparison with the absence of nuclear protein (lane 1). Antibodies against p65 (lanes 3, 6, and 9) and p50 (lanes 4, 7, and 10) were used for supershift assay. (D) EMSA was performed by using 32P-labeled probe containing −764C with HeLa cell nuclear extract, with or without competition from unlabeled oligonucleotides containing −764C and −764G as indicated. Relative densities of bands are shown as mean ± SD. The first lane represents the absence of nuclear extract. (E) EMSA was performed by using biotin-labeled probe containing HSF binding motif with Jurkat cell nuclear extract, with or without competition from unlabeled oligonucleotides containing −764C, −764G, HSF probe itself, or anti-HSF1 antibody as indicated. The first lane represents the absence of nuclear extract.
Table 2.
Allele and carrier frequency analyses of genotyped IFN-γ SNPs
| SNP ID and alleles | Allele frequency, %* |
Carrier† |
||||||
|---|---|---|---|---|---|---|---|---|
| CA | AA | P‡ | OR (95% CI) | P‡ | ||||
| Cohort 1 (IFN-α-treated patients) | ||||||||
| Treatment response | SVR | NR | SVR | NR | ||||
| No. of alleles§ | 180 | 246 | 16 | 126 | ||||
| rs2069705 T/C | T | 66.9 | 33.6 | 81.3 | 54 | 0.398 | 0.81 (0.48–1.35) | 0.407 |
| C | 33.1 | 66.4 | 18.7 | 46 | ||||
| rs2069707 C/G | C | 88.3 | 94.7 | 93.8 | 97.6 | 0.011 | 2.66 (1.27–5.57) | 0.008 |
| G | 11.7 | 5.3 | 6.2 | 2.4 | ||||
| rs2430561 T/A | T | 56.1 | 57 | 62.5 | 80.2 | 0.513 | 1.12 (0.64–1.94) | 0.691 |
| A | 43.9 | 43 | 37.5 | 19.8 | ||||
| rs1861494 A/G | A | 70.8 | 71.1 | 93.8 | 79.8 | 0.729 | 0.89 (0.53–1.50) | 0.663 |
| G | 29.2 | 28.9 | 6.2 | 20.2 | ||||
| rs1861493 T/C | T | 71.3 | 72.1 | 93.8 | 86.5 | 0.963 | 0.92 (0.55–1.56) | 0.771 |
| C | 28.7 | 27.9 | 6.2 | 13.5 | ||||
| rs2069718 C/T | C | 60.1 | 56.1 | 62.5 | 40.3 | 0.174 | 0.72 (0.42–1.24) | 0.23 |
| T | 39.9 | 43.9 | 37.5 | 59.7 | ||||
| rs2069727 A/G | A | 56.7 | 57.8 | 56.3 | 77.8 | 0.434 | 1.16 (0.67–2.01) | 0.591 |
| G | 43.3 | 42.2 | 43.7 | 22.2 | ||||
| rs2069728 G/A | G | 92.1 | 89.3 | 75 | 73.4 | 0.363 | 0.72 (0.37–1.37) | 0.315 |
| A | 7.9 | 10.7 | 25 | 26.6 | ||||
| Cohort 2 (intravenous drug users)¶ | ||||||||
| Viral clearance | C | P | C | P | ||||
| No. of alleles | 16 | 20 | 150 | 302 | ||||
| rs2069707 C/G | C | 87.5 | 100 | 96.7 | 98.3 | 0.086 | 3.51 (0.98–12.49) | 0.044 |
| G | 12.5 | 0 | 3.3 | 1.7 | ||||
P values in boldface are significant.
*Allele frequency is presented as percentage of the column variable: SVR or NR for cohort 1 and spontaneous recovery (C) or persistent infection (P) for cohort 2.
†Carrier refers to carrier of the rarer allele (shown second) according to the National Center for Biotechnology Information dbSNP database.
‡P value is calculated with Cochran Mantel Haenszel (CMH) statistics adjusting for race for comparison between SVR and NR in cohort 1 and C and P in cohort 2.
§In cohort 1, the number of alleles contributing to each SNP analysis depends upon the rate of success in genotyping the particular SNP and varies by no more than ±2 from the number listed.
¶Only the data of SNP rs2069707 are shown for cohort 2. The data for the other SNPs (not significant) are not shown.
To further focus on this apparent association, we evaluated the −764C/G SNP in each of the ethnic groups in cohort 1 (Table 3). Interestingly, the G allele was more common in CA than in AA (8.0% vs. 3.0%, P = 0.03). This finding is consistent with the general observation that CA ethnicity is more closely linked with spontaneous viral clearance and response to IFN therapy than is AA ethnicity. Within the CA population, the G carrier status was significantly associated with SVR [P = 0.01, OR = 2.64 (1.22–5.74)]. The same trend was also observed in the AA group, but the association did not attain statistical significance, which is likely due to the small AA sample size.
Table 3.
Allele, genotype, and carrier frequencies of SNP rs2069707 for IFN-α-treated patients (cohort 1), separately by race
| CA |
AA |
CMH race-adjusted | |||
|---|---|---|---|---|---|
| SVR (%) | NR (%) | SVR (%) | NR (%) | ||
| Allele frequency | |||||
| G | 21 (11.7) | 13 (5.3) | 1 (6.2) | 3 (2.4) | |
| C | 159 (88.3) | 233 (94.7) | 15 (93.8) | 123 (97.6) | |
| P | 0.016 | 0.384 | 0.011 | ||
| Genotype frequency | |||||
| GG | 1 (1.1) | 1 (0.8) | 0 | 0 | |
| CG | 19 (21.1) | 11 (8.9) | 1 (12.5) | 3 (4.8) | |
| CC | 70 (77.8) | 111 (90.3) | 7 (87.5) | 60 (95.2) | |
| P | 0.04 | NC | 0.028 | ||
| G carrier frequency | |||||
| GG or CG | 20 (22.2) | 12 (9.7) | 1 (12.5) | 3 (4.8) | |
| CC | 70 (77.8) | 111 (90.3) | 7 (87.5) | 60 (95.2) | |
| P | 0.012 | 0.387 | 0.008 | ||
| OR (95% CI) | 2.64 (1.22–5.74) | 2.86 (0.26–31.33) | 2.66 (1.27–5.57) | ||
P values in boldface are significant. CMH, Cochran Mantel Haenszel statistics; NC, not calculated due to zero observation for GG genotype.
Multiple logistic regression analyses of this −764 SNP and the clinical variables identified in the univariate analyses indicated that the G carrier status was independently associated with SVR [P = 0.04, OR = 2.92 (1.07–7.94)] (Table 4). Race, viral titer, and virus genotype were also independently significant, whereas gender, age, histological grade, and fibrosis stage were no longer significant in the multiple logistic regression model. This −764 SNP was also evaluated for its association with disease severity and progression, including alanine aminotransferase (ALT), histological grade, and fibrosis stage, but no significant correlation was observed. All these data support the hypothesis that the SNP variant −764G located in the IFN-γ promoter is an important genetic marker for treatment response and spontaneous recovery in HCV infection.
Table 4.
Multiple logistic regression analyses of SNP rs2069707 and clinical factors associated with IFN treatment response
| Variable | Univariate analysis |
Multiple logistic regression analysis |
||
|---|---|---|---|---|
| P | OR (95% CI) | P | OR (95% CI) | |
| rs2069707G | 0.001* | 3.11 (1.52–6.35)* | 0.036 | 2.92 (1.07–7.94) |
| Race | <0.001 | 5.76 (2.63–12.6) | 0.024 | 3.88 (1.19–12.6) |
| Sex | 0.028 | 1.75 (1.06–2.90) | 0.079 | 1.88 (0.93–3.80) |
| Age | 0.019 | 0.96 (0.94–0.99) | 0.486 | 0.99 (0.98–1.03) |
| Viral titer | 0.010 | 0.55 (0.35–0.87) | 0.001 | 0.36 (0.20–0.64) |
| Virus genotype | <0.001 | 9.87 (4.91–19.8) | <0.0001 | 11.8 (4.94–28.3) |
| Histological grade | 0.044 | 1.86 (1.02–3.42) | 0.754 | 0.87 (0.38–2.03) |
| Fibrosis stage | 0.001 | 2.66 (1.53–4.64) | 0.329 | 1.44 (0.69–3.00) |
IFN-γ −764G/C Polymorphism Affects Promoter Activity.
To further understand the significance of this novel SNP, we performed biological assays to study the functionality of the two different alleles. We first measured the promoter activities of different allele constructs by using luciferase reporter gene assay. Jurkat cells transfected with the variant −764G construct had a 2- to 3-fold higher luciferase activity than the common −764C construct, suggesting that the G allele conferred a higher promoter activity (Fig. 1B).
IFN-γ −764G/C Alleles and Promoter Binding Affinity.
The regulation of IFN-γ gene transcription has been shown to involve a large number of transcription factors and multiple regions of the gene (12–18), including a NF-κB binding motif (16) next to −764C/G polymorphism region. Therefore, we performed an EMSA with oligo(C) and oligo(G) containing the −764C and −764G variants as well as NF-κB probe, respectively (Fig. 1C). The oligo(G) had a higher binding activity than the oligo(C) (comparing lanes 2–4 to lanes 5–7), an observation that was confirmed with a competitive EMSA (Fig. 1D). These data suggest that the G oligonucleotide represented a 2- to 3-fold higher binding affinity than the C oligonucleotide, which is consistent with the luciferase reporter assay. Supershift analysis using monoclonal antibodies against the p50 and p65 subunits of NF-κB showed that the oligo(C)– and oligo(G)–protein complex (region b) were not supershifted, whereas the previously reported NF-κB binding complex (region a) was (Fig. 1C) (13).
To identify the putative nuclear factor interacting with this polymorphism, we compared the sequence of IFN-γ promoter from −774 to −764 (GGAATATTCTC) with the binding sites of previously reported transcriptional factors and observed a high level of homology with the HSF binding motif (NGAANNTTCN) (19). When the HSF probe was used in EMSA with Jurkat cell nuclear extract, both unlabeled oligo(C) and oligo(G) could compete against the HSF probe binding in a dose-dependent manner, with oligo(G) appearing to have a higher binding affinity than oligo(C) (Fig. 1E). More importantly, the positive band intensity was diminished in the presence of anti-HSF1 antibody. Thus, our data indicate that the SNP −764C/G is located in an HSF1 binding region of IFN-γ promoter and determines the binding affinity to HSF1.
Discussion
This study presents compelling evidence that a previously undescribed functional variant in the proximal promoter region of IFN-γ gene affects IFN-induced or spontaneous recovery from HCV infection. This relationship is in accordance with the well documented role of IFN-γ in the immune response of hepatitis C and supports the hypothesis that host genetic factors play a major role in disease outcome and treatment response of HCV infection.
We have conducted a comprehensive analysis of IFN-γ gene polymorphisms for association with response to IFN-α-based therapy in a large cohort of chronic hepatitis C patients. The overall response rate to therapy and its association with race, gender, viral titer, viral genotype, histological grade, and fibrosis stage in this cohort are in agreement with previous findings (3, 5, 20, 21). The relationship between several IFN-γ gene polymorphism and outcomes of HCV infection has been studied previously. One paper reported the correlation of the +874T/A polymorphism with early recurrent hepatitis C after liver transplantation (22), but many other groups did not demonstrate any association with either viral clearance or treatment response (23–26). Although our study did not demonstrate a significant association of the +874T/A SNP (rs2430561), we did identify the SNP variant −764G of the IFN-γ gene as an independent marker predicting SVR to IFN-α-based therapy. We also demonstrated that there was a significant association between G carrier status and viral clearance in a well characterized cohort of i.v. drug users. Although this association of the −764G alleles with spontaneous viral clearance was not as strong as with treatment response, results of both cohorts together support an important connection between this IFN-γ polymorphism and the outcome of HCV infection.
To further understand the significance of this SNP, we performed biological assays to study the functionality of the two different alleles. Because the −764C/G was within the IFN-γ promoter region, we first measured promoter activities with different allele constructs by using a luciferase reporter gene assay. A 2- to 3-fold higher level of luciferase was detected with the variant −764G/FLuc IFN-γ promoter compared with −764C/FLuc construct. These constructs could not be induced by either individual or combined cytokine stimuli, e.g., IL-2, IL-2/IL-12, IL-2/IL-18, CD3/CD28, and phorbol 12-myristate 13-acetate (PMA)/ionomycin (data not shown), which have been used to induce IFN-γ production of susceptible cell lines in vitro (27, 28). Therefore we could not assess the role of the −764C/G alleles in IFN-γ induction. Because the regulation of IFN-γ gene transcription involves a large number of transcription factors and multiple regions of the gene (12–18), the promoter region used in this assay may not contain the appropriate sequence response to these stimuli.
To investigate the molecular mechanisms responsible for the effect of the −764C/G promoter polymorphism on transcription, binding of transcription factors to this polymorphism region was further analyzed. We found that binding of a factor involved in stress response, HSF, appeared to be affected by either the C or G nucleotide at position −764. Consistent with luciferase reporter assay, the G oligonucleotide represented a 2- to 3-fold higher binding affinity than the C oligonucleotide. HSF is known to be the transcriptional activator responsible for the induction of heat shock proteins (HSPs). Virus proliferation depends on the successful recruitment of host cellular components for their own replication, protein synthesis, and viral particle assembly. Increase of Hsp70 chaperones, as the central components of the cellular chaperone network, has been widely observed after viral infection (29). The induction can occur indirectly through the production of a large number of viral proteins in an unfolded, aggregation-prone state, which then compete with the HSF for binding to Hsp70 and lead to an increase of free and active HSF and transcription of heat shock genes. The induction of BiP/Grp78 and Grp94, the Hsp70 and Hsp90 homologs resident in the endoplasmic reticulum (ER), has been reported during HCV infection (30, 31). The HCV envelope proteins are retained in a pre-Golgi compartment and bound by BiP in a misfolded state, which activates the unfolded protein response and leads to the induction of ER-resident chaperones, including BiP and Grp94 (32). We reason that during this process, the IFN-γ promoter could be the target of HSF1. The G allele at −764 would confer a stronger induction of the IFN-γ gene and favor viral clearance and response to exogenous IFN-α-based therapy.
In conclusion, we report a polymorphism variant in the IFN-γ promoter, −764C/G, that is functionally important in regulating IFN-γ gene expression in vitro. The G allele is significantly associated with viral clearance and treatment response and may be used as a genetic marker to predict the response of IFN-based therapies in HCV-infected patients. In addition, the less frequent occurrence of the G allele in AA may explain the commonly observed racial difference in persistent infection and response to IFN-α-based therapy.
Patients and Methods
Patients.
The 284 chronic hepatitis C patients (cohort 1) were recruited between July 1999 and December 2005 from four sources: (i) National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health (NIH); (ii) Department of Transfusion Medicine, Clinical Center, NIH; (iii) University of Maryland; and (iv) Portland (OR) Veterans Affairs Medical Center. The study protocols were approved under the institutional review board guidelines of the respective institutions, and all patients signed an informed consent before participation in the study. Complete laboratory evaluations of all patients were obtained before treatment. Liver biopsy specimens collected at time of presentation were scored by using Knodell histology activity index (HAI) for grading of inflammation and fibrosis (33).
Patients enrolled in the study were treated in various clinical trials over a period of more than 15 years. The response to various IFN-α-based therapies was evaluated in patients who had received treatment for at least 3 months. A SVR was defined as the loss of detectable HCV RNA (viremia) during treatment and its continued absence for at least 6 months after stopping therapy. Nonresponders (NVR) were defined as patients with continuous viremia after 6 months of therapy. Relapsers (RL) were those patients with undetectable viremia during the course of treatment and detectable viremia after the end of treatment. Patients who responded initially but could not complete the full course, because of side effects or other reasons, and later relapsed were not included in the analysis. Details of the treatment regimens and the outcomes of therapy of cohort 1 are shown in SI Table 6.
The 251 patients of cohort 2 were participants in a well characterized study of i.v. drug users from Baltimore, MD, of whom 85 (34%) had spontaneously cleared HCV infection and 166 (66%) had chronic HCV infection (11). In this cohort, 231 (92%) are AA and 20 (8%) are CA.
Diagnosis of HCV Infection.
All patients in cohort 1 were initially screened for anti-HCV antibodies by using HCV EIA (Abbott Laboratories, Abbott Park, IL). A strip immunoblot assay (RIBA 3.0, Chiron/Ortho-Clinical Diagnostics, Emeryville, CA) was used for confirmation. Serial serum samples were tested for HCV RNA by using the COBAS AMPLICOR HCV Test, version 2.0 (Roche Diagnostics, Branchburg, NJ), which has a detection limit of 100 international units/ml (270 copies/ml). The viral genotypes of all samples were determined by using INNO-LiPA (Bayer Diagnostics, Tarrytown, NY).
In cohort 2, prior infection was substantiated by detection of HCV antibody (anti-HCV) by using enzyme immunoassay (EIA) and RIBA. Individuals designated spontaneously cleared had anti-HCV (confirmed by RIBA) and undetectable HCV RNA in serum for a minimum of 6 months. Persistently infected individuals had detectable anti-HCV and HCV RNA in serum for a minimum of 6 months.
DNA Extraction and Polymorphism Genotyping.
For cohort 1, genomic DNA was extracted from whole blood by using the FlexiGene DNA kit (Qiagen, Valencia, CA). In cohort 2, Epstein–Barr virus-transformed cell lines were established for each individual, and genomic DNA was extracted from these cell lines by using phenol/chloroform extraction. SNP markers were selected to cover the entire IFN-γ gene, including the 5′ regulatory region and 3′ downstream region at 1- to 2-kb intervals as shown in Fig. 1A. Only those SNPs with a minor allele frequency ≥5% as indicated in the National Center for Biotechnology Information dbSNP database were genotyped. SNPs were genotyped by using ABI TaqMan allelic discrimination method and an ABI7000 Sequence Detection System (Applied Biosystems, Foster City, CA). The probes and primers (SI Table 7) for selected SNPs were designed with ABI's Primer Expression version 2.0 software. Genotyping assay was performed in a reaction mixture of 10 μl, containing 1× Universal PCR Master Mix, 10 ng of DNA, 0.2 μM each probe, and 0.9 μM each of forward and reverse primer.
Statistical Analysis.
The ordinal variables of histological grade and fibrosis stage were dichotomized according to clinically relevant designations of mild vs. moderate to severe: ≤7 (minimal to mild) or >7 (moderate to severe) for histological grade and 0 + 1 (minimal to mild) or 3 + 4 (moderate to severe) for fibrosis stage. Because of the difficulty in determination of the exact age at time of infection for most patients, we used the age at the time of enrollment in the protocol for analysis. Unless otherwise stated, we compared all SVR patients (regardless of the treatment) vs. nonresponders (NVR) and relapsing patients (designated “NR group”).
Genotype distributions in AA and CA populations were tested for Hardy–Weinberg equilibrium by using χ2 analyses. Demographic, clinical, and virological features were evaluated by χ2 tests for association with SVR. Allele, genotype, and carrier frequencies were evaluated separately for AA and CA by using χ2 or Fisher exact tests, whenever appropriate. Cochran Mantel Haenszel (CMH) statistics were used to assess the race-adjusted association between a polymorphism/haplotype and treatment response or spontaneous recovery. We considered two-tailed P values of <0.05 to be statistically significant in χ2 tests and calculated ORs and 95% confidence intervals (CI) when appropriate. Multiple logistic regression was performed to evaluate the association between the IFN-γ markers and SVR in the presence of demographic, clinical, and virological features. All ORs were calculated for the predictor category potentially favorable to IFN response, including race (CA vs. AA), sex (female vs. male), age (in years), viral titer (low vs. high), viral genotype (non-1 type vs. type 1), histological grade (mild vs. moderate to severe), and fibrosis stage (mild vs. moderate to severe). Haplotypes were constructed by using PHASE 2.02 (34). All analyses were performed by using Statistical Analysis System (SAS) version 8.02 (SAS Institute, Cary, NC).
Luciferase Reporter Assay.
The −1058 to +128 regions with both −764C and −764G alleles from the human IFN-γ gene were amplified by PCR using relevant patient genomic DNA as template. PCR primers were designed to create 5′ BglII and 3′ HindIII restriction enzyme digestion sites in the PCR products. The amplified fragments were purified and further subcloned into the BglII and HindIII sites of a promoterless firefly luciferase (FLuc) expression vector, pGL4.10[Luc2] (Promega, Madison, WI) to create two plasmids: pGL4-IFN(G)-Luc and pGL4-IFN(C)-Luc. pGL3-control and pRL-CMV are two control vectors expressing FLuc and Renilla luciferase (RLuc) genes under the simian virus 40 and cytomegalovirus promoters, respectively. Jurkat cells (clone E6-1) were purchased from American Type Culture Collection (Manassas, VA) and maintained with RPMI medium 1640 supplemented with 10% FCS, 1% penicillin/streptomycin, and 1% l-glutamine (Invitrogen, Carlsbad, CA). Cells were cotransfected with FLuc expression plasmid (pGL3-control, pGL4-IFN(G)-Luc or pGL4-IFN(C)-Luc) and pRL-CMV vector by using the Nucleofector Device (Amaxa, Gaithersburg, MD). Cells were harvested at 24, 48, and 72 h after transfection and followed by Dual-Luciferase Assay (Promega) to measure FLuc and RLuc activities. The FLuc activity in each sample was normalized with RLuc activity.
EMSA Analysis.
Complementary single-stranded oligonucleotides were synthesized (Operon Biotechnologies, Huntsville, AL) to span ≈10 bp on either side of the variant nucleotide (C/G) at promoter −764 of IFN-γ gene (Fig. 1A), 5′-CCTGGGAATATTCT(C/G)TACACTGTATT-3′, and annealed to form a double-stranded probe. The NF-κB DNA probe (Promega) is a 28-bp double-stranded commercial probe specifically binding to the NF-κB protein. All three probes were end-labeled by using T4 polynucleotide kinase (Promega) with [γ-32P]dATP (3,000 Ci/mmol, 10 mCi/ml; PerkinElmer, Wellesley, MA; 1 Ci = 37 GBq). The EMSA reaction was performed in a 10-μl reaction mixture consisting of 2 μl of 5× binding buffer, 5 μg of HeLa cell nuclear extract (Promega), and 50 fmol of labeled oligonucleotide probes. In competition experiments, excess unlabeled probes were added 10 min before adding the labeled probes. The reaction mixture was then incubated for 20 min at room temperature and analyzed on the gel. For supershift, antibodies against the p50 and p65 subunits of NF-κB (Santa Cruz Biotechnology, Santa Cruz, CA) were used. The gel images were photographed and the band densities were quantified with a public analysis program, ImageJ (http://rsb.info.nih.gov/ij/). Complementary single-stranded oligonucleotides containing HSF binding motif (CTGGAATATTCCCGACCTGGCAGCCTCATC) with 5′-end-labeled with biotin were synthesized by Sigma–Genosys (Woodlands, TX). For the EMSA shown in Fig. 1E, the oligonucleotides containing −764C or −764G were 5′-end-labeled with biotin and the binding assay was performed with Lightshift Chemiluminescent EMSA Kit (Pierce, Rockford, IL). Antibody against HSF1 was from Santa Cruz Biotechnology. Jurkat cell nuclear extract was purchased from GeneTex (San Antonio, TX).
Supplementary Material
Acknowledgments
We thank Rivkah Gonsky for technical suggestions on EMSA analysis and Jay Hoofnagle, Theo Heller, and Marc Ghany for advice and assistance. This work was supported by grants from National Institutes of Health (U01 DK60341 to H.Y., X.S., S.L.R., K.Y., X.T., and G.T.; DA00441, DA04334, and DA13324 to C.L.T. and D.L.T.) and in part by the Intramural Research Program of the National Institute of Diabetes and Digestive and Kidney Diseases. C.L.T. was supported in part by a grant from the Burroughs Wellcome Fund for Pathogenesis of Infectious Diseases.
Abbreviations
- AA
African Americans
- CA
Caucasian Americans
- CI
confidence interval
- FLuc
firefly luciferase
- HCV
hepatitis C virus
- HSF
heat shock transcription factor
- NR
nonresponse
- OR
odds ratio
- SVR
sustained virological response.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0609954104/DC1.
References
- 1.Liang TJ, Rehermann B, Seeff LB, Hoofnagle JH. Ann Intern Med. 2000;132:296–305. doi: 10.7326/0003-4819-132-4-200002150-00008. [DOI] [PubMed] [Google Scholar]
- 2.Tan SL, He Y, Huang Y, Gale M., Jr Curr Opin Pharmacol. 2004;4:465–470. doi: 10.1016/j.coph.2004.07.003. [DOI] [PubMed] [Google Scholar]
- 3.Feld JJ, Hoofnagle JH. Nature. 2005;436:967–972. doi: 10.1038/nature04082. [DOI] [PubMed] [Google Scholar]
- 4.Promrat K, Liang TJ. Hepatology. 2003;38:1359–1362. doi: 10.1016/j.hep.2003.10.008. [DOI] [PubMed] [Google Scholar]
- 5.Gao B, Hong F, Radaeva S. Hepatology. 2004;39:880–890. doi: 10.1002/hep.20139. [DOI] [PubMed] [Google Scholar]
- 6.Frese M, Schwarzle V, Barth K, Krieger N, Lohmann V, Mihm S, Haller O, Bartenschlager R. Hepatology. 2002;35:694–703. doi: 10.1053/jhep.2002.31770. [DOI] [PubMed] [Google Scholar]
- 7.Woollard DJ, Grakoui A, Shoukry NH, Murthy KK, Campbell KJ, Walker CM. Hepatology. 2003;38:1297–1306. doi: 10.1053/jhep.2003.50478. [DOI] [PubMed] [Google Scholar]
- 8.Hayden C, Pereira E, Rye P, Palmer L, Gibson N, Palenque M, Hagel I, Lynch N, Goldblatt J, Lesouef P. Clin Exp Allergy. 1997;27:1412–1416. doi: 10.1046/j.1365-2222.1997.1800979.x. [DOI] [PubMed] [Google Scholar]
- 9.Pravica V, Asderakis A, Perrey C, Hajeer A, Sinnott PJ, Hutchinson IV. Eur J Immunogenet. 1999;26:1–3. doi: 10.1046/j.1365-2370.1999.00122.x. [DOI] [PubMed] [Google Scholar]
- 10.Bream JH, Carrington M, O'Toole S, Dean M, Gerrard B, Shin HD, Kosack D, Modi W, Young HA, Smith MW. Immunogenetics. 2000;51:50–58. doi: 10.1007/s002510050008. [DOI] [PubMed] [Google Scholar]
- 11.Thomas DL, Astemborski J, Rai RM, Anania FA, Schaeffer M, Galai N, Nolt K, Nelson KE, Strathdee SA, Johnson L, et al. J Am Med Assoc. 2000;284:450–456. doi: 10.1001/jama.284.4.450. [DOI] [PubMed] [Google Scholar]
- 12.Ye J, Cippitelli M, Dorman L, Ortaldo JR, Young HA. Mol Cell Biol. 1996;16:4744–4753. doi: 10.1128/mcb.16.9.4744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sica A, Dorman L, Viggiano V, Cippitelli M, Ghosh P, Rice N, Young HA. J Biol Chem. 1997;272:30412–30420. doi: 10.1074/jbc.272.48.30412. [DOI] [PubMed] [Google Scholar]
- 14.Cippitelli M, Sica A, Viggiano V, Ye J, Ghosh P, Birrer MJ, Young HA. J Biol Chem. 1995;270:12548–12556. doi: 10.1074/jbc.270.21.12548. [DOI] [PubMed] [Google Scholar]
- 15.Penix L, Weaver WM, Pang Y, Young HA, Wilson CB. J Exp Med. 1993;178:1483–1496. doi: 10.1084/jem.178.5.1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sica A, Tan TH, Rice N, Kretzschmar M, Ghosh P, Young HA. Proc Natl Acad Sci USA. 1992;89:1740–1744. doi: 10.1073/pnas.89.5.1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu X, Sun YL, Hoey T. Science. 1996;273:794–797. doi: 10.1126/science.273.5276.794. [DOI] [PubMed] [Google Scholar]
- 18.Sweetser MT, Hoey T, Sun YL, Weaver WM, Price GA, Wilson CB. J Biol Chem. 1998;273:34775–34783. doi: 10.1074/jbc.273.52.34775. [DOI] [PubMed] [Google Scholar]
- 19.Kroeger PE, Morimoto RI. Mol Cell Biol. 1994;14:7592–7603. doi: 10.1128/mcb.14.11.7592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Manns MP, McHutchison JG, Gordon SC, Rustgi VK, Shiffman M, Reindollar R, Goodman ZD, Koury K, Ling M, Albrecht JK. Lancet. 2001;358:958–965. doi: 10.1016/s0140-6736(01)06102-5. [DOI] [PubMed] [Google Scholar]
- 21.Fried MW, Shiffman ML, Reddy KR, Smith C, Marinos G, Goncales FL, Jr, Haussinger D, Diago M, Carosi G, Dhumeaux D, et al. N Engl J Med. 2002;347:975–982. doi: 10.1056/NEJMoa020047. [DOI] [PubMed] [Google Scholar]
- 22.Ben-Ari Z, Pappo O, Druzd T, Sulkes J, Klein T, Samra Z, Gadba R, Tambur AR, Tur-Kaspa R, Mor E. Cytokine. 2004;27:7–14. doi: 10.1016/j.cyto.2004.03.009. [DOI] [PubMed] [Google Scholar]
- 23.Abbott WG, Rigopoulou E, Haigh P, Cooksley H, Mullerova I, Novelli M, Winstanley A, Williams R, Naoumov NV. Liver Int. 2004;24:90–97. doi: 10.1111/j.1478-3231.2004.00904.x. [DOI] [PubMed] [Google Scholar]
- 24.Mangia A, Santoro R, Piattelli M, Pazienza V, Grifa G, Iacobellis A, Andriulli A. Cytokine. 2004;25:103–109. doi: 10.1016/j.cyto.2003.10.005. [DOI] [PubMed] [Google Scholar]
- 25.Barrett S, Collins M, Kenny C, Ryan E, Keane CO, Crowe J. J Med Virol. 2003;71:212–218. doi: 10.1002/jmv.10472. [DOI] [PubMed] [Google Scholar]
- 26.Dai CY, Chuang WL, Chang WY, Chen SC, Lee LP, Hsieh MY, Hou NJ, Lin ZY, Wang LY, Yu ML. Antiviral Res. 2005;67:93–97. doi: 10.1016/j.antiviral.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 27.Rodriguez-Galan MC, Bream JH, Farr A, Young HA. J Immunol. 2005;174:2796–2804. doi: 10.4049/jimmunol.174.5.2796. [DOI] [PubMed] [Google Scholar]
- 28.Young HA. J Interferon Cytokine Res. 1996;16:563–568. doi: 10.1089/jir.1996.16.563. [DOI] [PubMed] [Google Scholar]
- 29.Mayer MP. Rev Physiol Biochem Pharmacol. 2005;153:1–46. doi: 10.1007/s10254-004-0025-5. [DOI] [PubMed] [Google Scholar]
- 30.Choukhi A, Ung S, Wychowski C, Dubuisson J. J Virol. 1998;72:3851–3858. doi: 10.1128/jvi.72.5.3851-3858.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liberman E, Fong YL, Selby MJ, Choo QL, Cousens L, Houghton M, Yen TS. J Virol. 1999;73:3718–3722. doi: 10.1128/jvi.73.5.3718-3722.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kozutsumi Y, Segal M, Normington K, Gething MJ, Sambrook J. Nature. 1988;332:462–464. doi: 10.1038/332462a0. [DOI] [PubMed] [Google Scholar]
- 33.Brunt EM. Hepatology. 2000;31:241–246. doi: 10.1002/hep.510310136. [DOI] [PubMed] [Google Scholar]
- 34.Stephens M, Smith NJ, Donnelly P. Am J Hum Genet. 2001;68:978–989. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gray PW, Goeddel DV. Nature. 1982;298:859–863. doi: 10.1038/298859a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.