Abstract
The Saccharomyces cerevisiae Yor112wp, which we named Cex1p, was identified using a yeast tRNA three-hybrid interaction approach and an in vivo nuclear tRNA export assay as a cytoplasmic component of the nuclear tRNA export machinery. Cex1p binds tRNA saturably, and associates with the nuclear pore complex by interacting directly with Nup116p. Cex1p co-purifies with the nuclear tRNA export receptors Los1p and Msn5p, the eukaryotic elongation factor eEF-1A, which delivers aminoacylated tRNAs to the ribosome, and the RanGTPase Gsp1p, but not with Cca1p, a tRNA maturation enzyme that facilitates translocation of non-aminoacylated tRNAs across the nuclear pore complex. Depletion of Cex1p and eEF-1A or Los1p significantly reduced the efficiency of nuclear tRNA export. Cex1p interacts with Los1p but not with eEF-1A in vitro. These findings suggest that Cex1p is a component of the nuclear aminoacylation-dependent tRNA export pathway in S. cerevisiae. They also suggest that Cex1p collects aminoacyl-tRNAs from the nuclear export receptors at the cytoplasmic side of the nuclear pore complex, and transfers them to eEF-1A using a channelling mechanism.
Keywords: channelling mechanism, nuclear tRNA aminoacylation, nuclear tRNA export, tRNA export components, tRNA three-hybrid
Introduction
In eukaryotes, the nuclear transcription and cytoplasmic translation machineries are separated by the nuclear envelope (NE). Large heteromeric protein structures called nuclear pore complexes (NPC) perforate the NE and allow the passage of macromolecules such as proteins and RNAs between the nucleus and cytoplasm. Translocation of macromolecules across the NPC requires components of the NPC known as nucleoporins (Nups), as well as soluble transport proteins known as nuclear import/export receptors. In most cases, the import/export receptors are members of a family of transporters called β-Karyopherins (β-Kaps). β-Kaps interact directly with a subset of Nups during translocation across the NPC, and their function in the import–export cycle is regulated by the small GTPase Ran (for reviews see Gorlich and Kutay, 1999; Grosshans et al, 2000b; Rout and Aitchison, 2001; Rodriguez et al, 2004). Thus, the NPC, Ran and the nuclear import/export receptors work together to establish the directionality and specificity of cargo translocation across the NE, allowing cells to control growth, gene expression and cellular division (Kaffman and O'Shea, 1999).
Studies in mammalian cells and Saccharomyces cerevisiae established that functional, mature tRNAs are preferentially selected for export to the cytoplasm (Zasloff et al, 1982; Arts et al, 1998b; Lipowsky et al, 1999). In these organisms and other eukaryotes, tRNA genes are transcribed by RNA polymerase III to produce pre-tRNAs. The pre-tRNA transcripts undergo trimming at the 5′ and 3′ ends, base modification, addition of C, C and A to their 3′ ends, and in a small percentage of the transcripts, removal of an intron (Hopper and Phizicky, 2003). For intronless pre-tRNAs, this multistep maturation process occurs in the nucleus. Maturation of intron-containing pre-tRNAs also takes place in the nucleus. However, for removal of the intron, the pre-tRNAs are exported to the cytoplasm and then imported back to the nucleus, most likely for further maturation (Shaheen and Hopper, 2005; Takano et al, 2005). Mature tRNAs are then subjected to a quality assurance step to ensure that they are functional before export to the cytoplasm.
Studies conducted in Xenopus laevis and S. cerevisiae led to the suggestion that aminoacylation is used to select functional tRNAs for export to the cytoplasm (Lund and Dahlberg, 1998; Sarkar et al, 1999; Grosshans et al, 2000a; Azad et al, 2001). This proof-reading step appears to occur in the nucleolus (Steiner-Mosonyi and Mangroo, 2004). However, nuclear tRNA aminoacylation is not the only mechanism used to inspect the functionality of tRNA, as this step is not absolutely required for tRNA export in both X. laevis and S. cerevisiae (Arts et al, 1998b; Lipowsky et al, 1999; Azad et al, 2001). Consequently, nuclear tRNA export is thought to occur by two pathways referred to as aminoacylation-dependent and aminoacylation-independent pathways. Recent evidence, however, suggests that the nuclear aminoacylation-dependent pathway has the primary responsibility for exporting tRNAs from the nucleus in S. cerevisiae (Steiner-Mosonyi and Mangroo, 2004).
Utp8p is a nucleolar tRNA-binding protein that plays a critical role in nuclear tRNA export in S. cerevisiae (Steiner-Mosonyi et al, 2003). It is required for nuclear export of both aminoacylated and non-aminoacylated tRNAs derived from intron-containing and intronless pre-tRNAs, and was proposed to act at a step between tRNA maturation/aminoacylation and translocation of the tRNA across the NPC. Utp8p most likely directs tRNAs that have been deemed functional to the nuclear tRNA export receptors in S. cerevisiae. Whether such a protein is required in mammalian cells is not known.
Los1p and Msn5p are the nuclear export receptors responsible for translocating tRNAs across the NPC in S. cerevisiae (Hellmuth et al, 1998; Takano et al, 2005). This step in mammalian cells is facilitated by exportin-t and exportin-5, the orthologues of Los1p and Msn5p, respectively (Arts et al, 1998a; Kutay et al, 1998; Bohnsack et al, 2002; Calado et al, 2002). These proteins are β-Kaps and bind the tRNA cargo directly in an RanGTP-dependent manner. Los1p, Msn5p and exportin-5 are thought to be receptors of the nuclear aminoacylation-dependent export pathway (Grosshans et al, 2000a; Bohnsack et al, 2002; Calado et al, 2002; Steiner-Mosonyi and Mangroo, 2004). However, Los1p may also facilitate nuclear export of non-aminoacylated tRNAs, as exportin-t has been shown to transport a non-aminoacylated tRNA to the cytoplasm (Arts et al, 1998b). The ATP (CTP):nucleotidyltransferase (Cca1p), an essential enzyme that prepares tRNAs for aminoacylation in the nucleus, cytoplasm and mitochondrion by adding the nucleotides C, C and A to the 3′ ends of tRNAs, has also been implicated in nuclear export of some tRNAs in S. cerevisiae (Feng and Hopper, 2002). Cca1p is thought to function as a tRNA export receptor or an adaptor in a nuclear aminoacylation-independent pathway that permits export of tRNAs arising from intronless pre-tRNAs (Feng and Hopper, 2002). Furthermore, evidence reported suggests that another unidentified nuclear aminoacylation-independent pathway facilitates the nuclear export of tRNAs derived from intron-containing pre-tRNAs (Steiner-Mosonyi et al, 2003).
The genetic and biochemical studies reported indicate that nuclear tRNA export in S. cerevisiae involves multiple redundant pathways. The details of these pathways, however, are poorly understood, and only some of the proteins that facilitate nuclear tRNA export are known. We described previously the use of a yeast tRNA three-hybrid approach and an in vivo nuclear tRNA export assay based on amber suppression to identify proteins that play a role in nuclear tRNA export in S. cerevisiae (Steiner-Mosonyi et al, 2003). This combined strategy resulted in the identification of Utp8p, several known tRNA-binding proteins (Steiner-Mosonyi et al, 2003) and an uncharacterized 85-kDa cytoplasmic protein (encoded by YOR112W), which we named Cex1p. We report that Cex1p, which is similar to a protein found in higher eukaryotes including humans, is a cytoplasmic component of the nuclear aminoacylation-dependent export pathway. The data suggest that Cex1p collects aminoacyl-tRNAs from the nuclear export receptors at the cytoplasmic side of the NPC, and delivers them to eEF-1A in the cytoplasm using a channelling mechanism. Cex1p, therefore, provides a link between the nuclear aminoacylation-dependent export pathway and the protein synthesis apparatus. This may, in part, be necessary to maintain the high translation rate needed to allow cells to reach the critical size required to trigger cell division. Finally, this study underscores the importance of specific hypothesis-driven, directed proteomic experiments, as several genome-scale protein–protein interaction studies in S. cerevisiae did not identify the proteins that we have found to interact with Cex1p (Gavin et al, 2006; Krogan et al, 2006).
Results
Cex1p interacts genetically with Arc1p
We have previously used a yeast tRNA three-hybrid selection method to identify the genes of proteins that interact with tRNA, and a phenotypic in vivo nuclear tRNA export assay based on suppression of amber codons in metabolic reporter genes to test whether the identified proteins participate in nuclear tRNA export in S. cerevisiae (Steiner-Mosonyi et al, 2003). This strategy resulted in the identification of Utp8p, a nucleolar tRNA-binding protein that plays an essential intranuclear role in nuclear tRNA export (Steiner-Mosonyi et al, 2003). The gene YOR112W encoding an 85-kDa protein of unknown function was also identified by the tRNA three-hybrid selection screen and the in vivo nuclear tRNA export assay (Supplementary Figure S1). The function of the Yor112w protein is not essential (Giaever et al, 2002), and it was localized to the cytoplasm by yeast protein localization studies (Huh et al, 2003). PSI-BLAST indicated that Yor112wp is structurally homologous to an uncharacterized ORF in higher eukaryotes, including the human SCYL1/NTKL, Caenorahbditis elegans W07G4.3 and Arabidopsis thaliana At2g40730. These proteins have a characteristic protein kinase-like domain at their N-termini. This domain in Yor112wp is found between amino acids 1 and 280. However, Yor112wp and the orthologues lack characteristic catalytic residues, suggesting that they may not be protein kinases. This is consistent with the finding that SCYL1/NTKL does not possess kinase activity (Kato et al, 2002). Yor112wp and its orthologues also contain a HEAT repeat domain, which is commonly involved in protein–protein interactions. Yor112wp was named Cex1p for cytoplasmic export protein, as findings described below indicate that it has a cytoplasmic role in nuclear tRNA export.
The cytoplasmic Arc1p influences the efficiency of nuclear tRNA export by gathering certain species of non-aminoacylated tRNA exiting the nucleus, and delivering them to their cognate aminoacyl-tRNA synthetases (Simos et al, 1996). However, the function of Arc1p is not essential, suggesting that there is at least one S. cerevisiae protein with function similar to Arc1p (Simos et al, 1996). The cytoplasmic location of Cex1p suggests that it could be functionally equivalent to Arc1p. To investigate this possibility, a haploid cex1 arc1 strain harboring pCEN-URA-GAL1-CEX1 was prepared by tetrad dissection of a heterozygous diploid strain with the pCEN-URA-GAL1-CEX1 plasmid. The cex1 arc1 strain expressing Cex1p showed no growth defect at 30°C (Figure 1A, upper left-hand panel). Glucose repression of Cex1p expression significantly reduced the growth of cex1 arc1 (upper right-hand panel), but had no effect on the growth of the arc1 and cex1 strains (Figure 1A, lower panels). The use of 5-fluoroorotic acid (5-FOA) to select for cells lacking pCEN-URA-GAL1-CEX1 and harboring pUN100-ARC1 (Figure 1B, upper left-hand panel) or pUN100 (upper right-hand panel) indicated that Arc1p provided in trans rescued the growth defect of the cex1 arc1 strain. Growth of the arc1 and cex1 strains (lower panels) was not affected by the loss of pCEN-URA-GAL1-CEX1. In contrast, Los1p provided in trans could not rescue the growth defect of the cex1 arc1 strain (Figure 1C). The data suggest that the function of Cex1p is distinct from that of Los1p, and that it could be a functional homologue of Arc1p.
Figure 1.
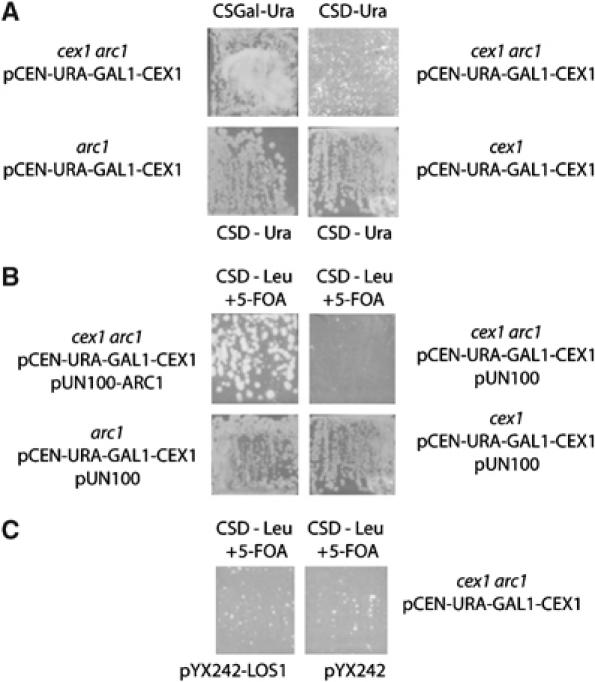
Disruption of the CEX1 and ARC1 genes reduced cell growth, which could be rescued by Arc1p but not by Los1p. The cex1 arc1, arc1 and cex1 strains containing pCEN-URA-GAL1-CEX1 were streaked on CSD-Ura medium and incubated at 30°C. The growth of the cex1 arc1 strain harboring the pCEN-URA-GAL1-CEX1 plasmid on CS medium containing galactose was also monitored (A). The cex1 arc1 pCEN-URA-GAL1-CEX1 strain containing pUN100 without or with the ARC1 gene, and the arc1 pCEN-URA-GAL1-CEX1 and cex1 pCEN-URA-GAL1-CEX1 strains containing pUN100 were streaked on CSD medium lacking Leu and containing 0.1% 5-FOA and incubated at 30°C (B). The cex1 arc1 pCEN-URA-GAL1-CEX1 strain containing pYX242 without or with the LOS1 gene was streaked on CSD medium lacking Leu and containing 0.1% 5-FOA and incubated at 30°C (C).
Depletion of Cex1p and Los1p significantly reduced the efficiency of nuclear tRNA export
Previous studies have shown that disruption of both LOS1 and ARC1 was lethal (Simos et al, 1996). To investigate further whether Cex1p is functionally equivalent to Arc1p, the effect of disruption of CEX1 and LOS1 on cell growth was assessed. A cex1 los1 haploid strain harboring pCEN-URA-GAL1-CEX1 was obtained by tetrad dissection of a heterozygous diploid strain with pCEN-URA-GAL1-CEX1. Growth of cex1 los1 strain was not affected when the pCEN-URA-GAL1-CEX1 plasmid was eliminated by counter-selection using 5-FOA (data not shown). Furthermore, growth of the cex1 los1 strain harboring pYX242 was comparable to the cex1 and los1 strains with pYX242 on YPD and CSD-Leu medium at 24, 30 and 37°C (Figure 2). This finding suggests that Cex1p may not be performing the same function as Arc1p. This is consistent with the observation that overexpression of Cex1p is unable to complement a los1 arc1 strain (data not shown).
Figure 2.
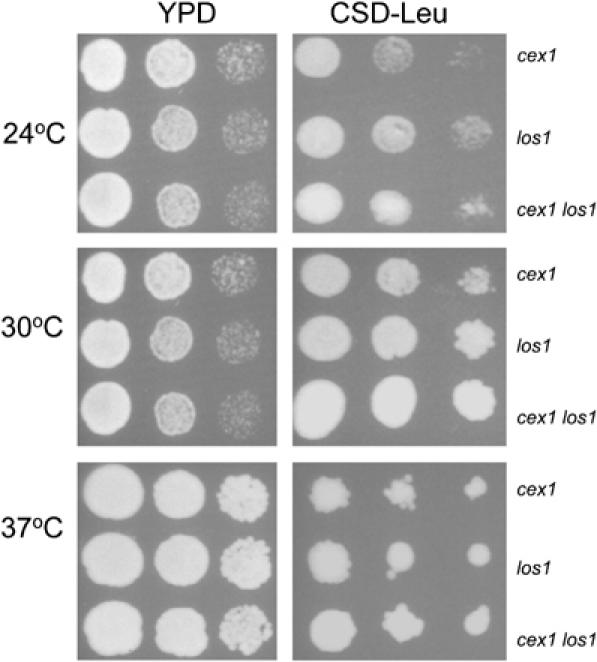
Disruption of the CEX1 and LOS1 genes did not affect cell growth. The cex1, los1 and cex1 los1 strains containing pYX242 were serially diluted and spotted on YPD or CSD medium lacking Leu and incubated at 24, 30 and 37°C.
Previous studies have shown that disruption of LOS1 and MSN5 did not affect cell growth (Takano et al, 2005). Loss of the function of both Los1p and Msn5p, however, reduced the efficiency of nuclear tRNA export (Takano et al, 2005). Consequently, FISH was used to ascertain whether tRNAs accumulate in the nucleus of the cex1 los1 strain by monitoring the cellular location of tRNATyr (Figure 3). The U18 small nucleolar RNA (U18 snoRNA) was used as a nuclear marker. In the wild-type and cex1 strains, tRNATyr was found mainly in the cytoplasm. However, nuclear retention of tRNATyr could be observed in approximately 5% of the cex1 cells. Disruption of the LOS1 gene has been shown to cause nuclear accumulation of several tRNAs, including tRNATyr (Grosshans et al, 2000a; Feng and Hopper, 2002), and we also detect nuclear accumulation of tRNATyr in the los1 strain. This was observed in about 22% of the los1 cells, which is comparable to values reported earlier (Grosshans et al, 2000a). Moreover, retention of tRNATyr was detected in the nucleus of about 48% of the cex1 los1 cells. These findings suggest that the function of Cex1p has an impact on the efficiency of nuclear tRNA export. As Los1p is a receptor of the aminoacylation-dependent nuclear tRNA export pathway (Steiner-Mosonyi and Mangroo, 2004), this suggests that Cex1p plays a role in this pathway. This suggestion is consistent with data presented below.
Figure 3.
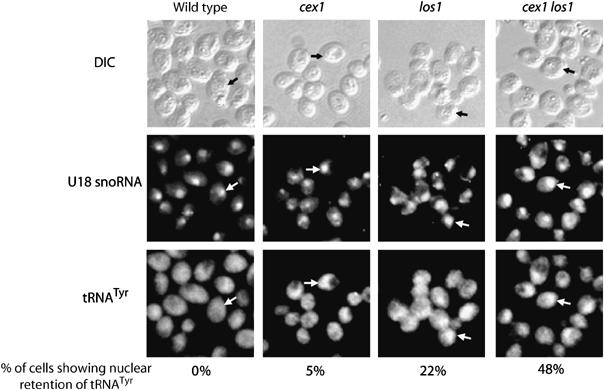
Depletion of both Cex1p and Los1p significantly reduced the efficiency of nuclear tRNA export. The location of the U18 snoRNA and the mature form of tRNATyr in each strain was detected by FISH. Cells showing nuclear retention of tRNATyr, as determined by U18 snoRNA localization, are indicated by arrows. The number of cells showing nuclear retention of tRNATyr was expressed as a percentage of total cells. For each strain, a total of 200 cells were inspected for nuclear accumulation of tRNATyr.
Cex1p binds tRNA directly and saturably in vitro
To understand how Cex1p affects nuclear tRNA export, substrate-induced intrinsic fluorescence quenching of tryptophan residues was used to ascertain whether Cex1p can interact with tRNA. Cex1p containing an N-terminal (His)9 tag was overproduced in Escherichia coli and purified by a combination of affinity and ion-exchange chromatography. The purity of Cex1p is shown in Figure 4A. The interaction analysis showed that recombinant Cex1p binds mature yeast tRNA directly and saturably (Figure 4B), indicating that it has a tRNA-binding site. Cex1p also interacts with a 90-bp DNA oligonucleotide, but to a lower extent compared with that observed for tRNA binding (Figure 4B). Moreover, saturable binding to the oligonucleotide could not be achieved using the same concentrations employed for tRNA binding. Non-saturable binding was also observed when 5S rRNA was used (data not shown). Eadie-Hofstee and Hanes-Woolf analyses of the data indicate that the affinity of Cex1p for tRNA, the DNA oligonucleotide and 5S rRNA is 2, 98 and 100 μM, respectively. This suggests that Cex1p has an RNA-binding site that is specific for tRNA. However, Cex1p is able to bind nucleic acid nonspecifically in vitro. The nonspecific binding property of Cex1p is not unusual, as it is well established that bona fide prokaryotic and eukaryotic tRNA-binding proteins, including Utp8p and Arc1p, interact nonspecifically with non-cognate RNAs in vitro (Gite and RajBhandary, 1997; Wang and Schimmel, 1999; Steiner-Mosonyi et al, 2003). Thus, the data strongly suggest that in vivo tRNA is the substrate for Cex1p.
Figure 4.
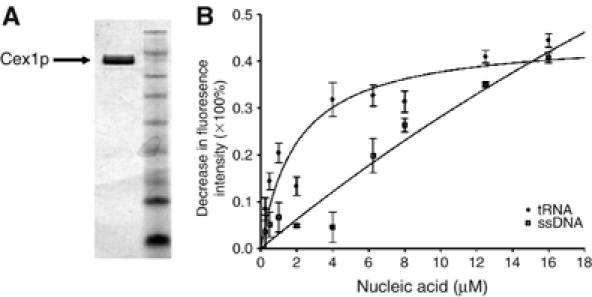
Cex1p binds tRNA directly and saturably. Substrate-induced intrinsic fluorescence quenching of Trp residues was used to determine whether purified Cex1p (A) interacts with tRNA or DNA (B).
Cex1p associates with the nuclear pore complex by interacting directly with Nup116p
Yor112wp/Cex1p was found to interact with Nup116p by a large-scale two-hybrid interaction study (Ito et al, 2001). However, Yor112wp was reported to be cytoplasmic based on direct fluorescence microscopy (Huh et al, 2003). Furthermore, our localization studies using a Cex1-GFP fusion protein verified that Cex1p is a cytoplasmic protein (data not shown). Therefore, to test whether Cex1p associates with the NPC, immunofluorescence confocal scanning microscopy was used to assess whether Cex1p containing a C-terminal Myc epitope colocalizes with the nucleoporin Nup116p or Nup2p in cex1 cells (Figure 5). Overlay analysis indicates that Cex1p colocalizes with Nup116p (A) and Nup2p (B). The data show that, the majority of Cex1p is located in the cytoplasm, whereas a small amount of the protein associates with the NPC (Figure 5A and B).
Figure 5.
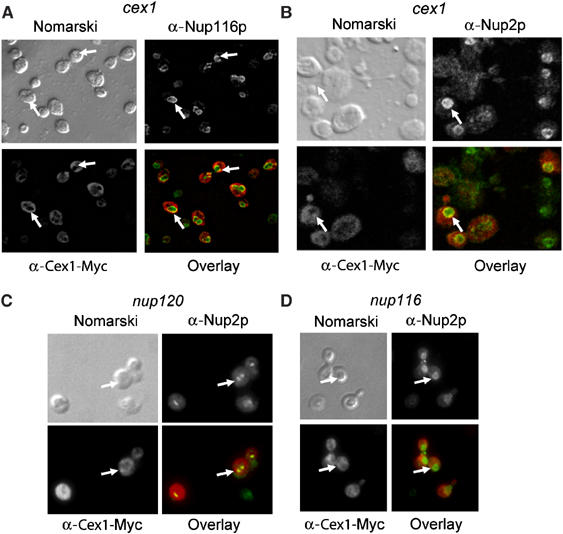
Cex1p associates with the NPC. The location of the Cex1-Myc fusion protein and Nup116p (A) or Nup2p (B) in cex1 harboring pRS416-ADH1-CEX1-MYC was determined by immunofluorescence confocal microscopy using a mouse monoclonal anti-Myc antibody and rabbit polyclonal anti-Nup2p and anti-Nup116p. Colocalization of Cex1p with Nup116p or Nup2p (arrow) was determined by overlay analysis of the images. The location of Cex1-Myc and Nup2p in nup120 (C) and nup116 (D) with pRS416-ADH1-CEX1-MYC was determined as above and overlay analyses of the images were performed to ascertain whether Cex1p colocalizes with Nup2p.
To further test whether the association of Cex1p with the NPC is authentic, colocalization of Cex1-Myc with Nup2p was investigated in nup116 and nup120, which exhibit clustered NPC (Aitchison et al, 1995). Clustering of the NPC in nup120 does not affect nuclear import processes, but blocks nuclear export processes including nuclear tRNA export (Aitchison et al, 1995; Sarkar and Hopper, 1998). Immunofluorescence shows that Nup2p is clustered in the nuclear envelope of nup120 (Figure 5C), but not in nup116, at the permissive temperature (Figure 5D) or in cex1 cells (Figure 5B). As in cex1 cells (Figure 5A and B), Cex1p is distributed throughout nup120 and nup116 cells (Figure 5C and D). Overlay analyses indicate that Cex1p colocalizes with Nup2p in the clustered NPC of nup120 (Figure 5C). However, Cex1p no longer colocalizes with Nup2p in nup116 (Figure 5D). These findings strongly suggest that Cex1p associates with the NPC by interacting with Nup116p.
To investigate whether Cex1p associates with Nup116p in vivo, a tandem affinity purification (TAP) strategy (Rigaut et al, 1999; Puig et al, 2001) was used to determine whether Cex1p co-purifies with Nup116p. TAP was also used to test whether Cex1p co-purifies with other Nups such as Nup2p and Nup57p. For this analysis, total cell extract was prepared from CEX1-TAP, NUP116-TAP, NUP2-TAP or NUP57-TAP and subjected to TAP using IgG–Sepharose and calmodulin–Sepharose. The proteins in the final eluate were separated by SDS–PAGE and transferred to membranes. The blots were stained with Sypro-Ruby to detect the co-purifying proteins (Figure 6A), or subjected to Western blot analyses to detect Cex1p or Nup116p (Figure 6B). Nup116p was found to co-purify with Cex1-TAP (Figure 6B, top panel). Conversely, Cex1p co-purifies with Nup116-TAP (Figure 6B, lower panel, top row). However, Cex1p did not co-purify with Nup2-TAP (Figure 6B, lower panel, middle row) or Nup57-TAP (Figure 6B, lower panel, bottom row), suggesting that Cex1p co-purified specifically with Nup116p.
Figure 6.
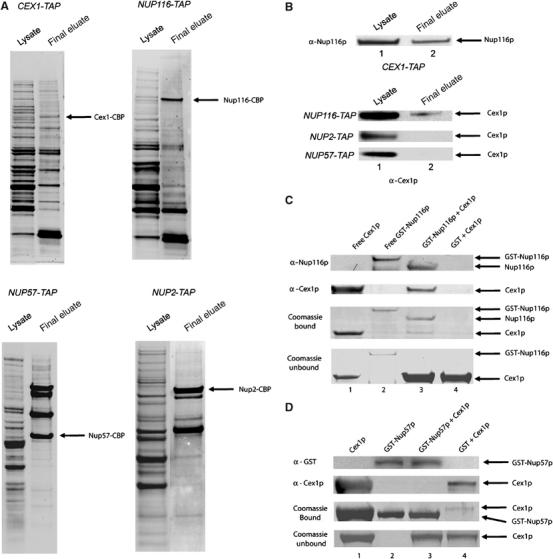
Cex1p associates with the NPC by interacting directly and specifically with Nup116p. Total cell extract was prepared from the CEX1-TAP, NUP116-TAP, NUP2-TAP or NUP57-TAP strain and subjected to TAP. The proteins in the total cell lysate (lane 1) and the final TAP eluate (lane 2) were on separated a 4–12% Novex gel and transferred to nitrocellulose membrane. The blot was stained with Sypro-Ruby to detect co-purifying proteins (A), or probed with anti-Cex1p or anti-Nup116p (B). To investigate whether Cex1p interacts with Nup116p directly, GST-Nup116p (200 μg, 1.72 nmol) (lanes 2 and 3) or GST was bound to glutathione resin (lane 4) (C) and incubated with a two-fold molar excess of Cex1p (291 μg, 3.44 nmol). After the resin was washed, Nup116p was released from GST using TEV. Coomassie blue staining of an SDS–PAGE gel (third row) or Western blot analysis was used to detect Nup116p (first row) and Cex1p (second row) in an aliquot of the eluate. Purified Cex1p (lane 1) and GST-Nup116p (lane 2) were used as size markers. The unbound Cex1p in an aliquot of the wash eluate was detected by Coomassie blue staining of an SDS–PAGE gel (fourth row). The specificity of the interaction between Cex1p and Nup116p was tested by investigating whether Cex1p interacts with Nup57p (D). The analysis was conducted as described above except the amount of GST-Nup57p (200 μg, 3.44 nmol) and Cex1p (590 μg, 6.88 nmol) was increased by two-fold. Bound GST-Nup57p was released by boiling the resin in the SDS–PAGE buffer, and an aliquot was subjected to SDS–PAGE. Coomassie blue staining (third row) or Western blot analysis was used to detect Nup57p (first row) and Cex1p (second row). Purified Cex1p (lane 1) and GST-Nup57p (lane 2) were used as size markers. Unbound Cex1p in an aliquot of the wash eluate was detected by Coomassie blue staining of an SDS–PAGE gel (fourth row).
In vitro binding was used to ascertain whether Cex1p interacts directly with Nup116p. GST-Nup116p bound to glutathione resin was incubated with a two-fold molar excess of Cex1p (lane 3). As a control, Cex1p was incubated with bound GST (lane 4). After washing the matrix, Nup116p was released from the GST tag by cleavage with the TEV protease. The eluate and purified GST-Nup116p and Cex1p were subjected to SDS–PAGE, followed by Coomassie blue staining (Figure 6C, third row), or Western blot analysis to detect Nup116p (Figure 6C, top row) and Cex1p (Figure 6C, second row). Unbound Cex1p in the wash eluate (Figure 6C, fourth row) was detected by Coomassie blue staining of an SDS–PAGE gel. Nup116p was released from the resin owing to cleavage of the TEV site found between GST and Nup116p in the fusion protein (Figure 6C, compare lanes 2 and 3). Western blot analysis and Coomassie blue staining showed that Cex1p interacts with Nup116p (Figure 6C, second and third rows, lane 3). In contrast, Cex1p could not be detected in the eluate from the bound GST control (Figure 6C, second and third rows, lane 4). These findings show that Cex1p interacts directly with Nup116p.
To determine whether the interaction between Cex1p and Nup116p in vitro is specific, we tested whether Cex1p could interact with Nup57p (Figure 6D). Bound GST-Nup57p was incubated with a two-fold molar excess of Cex1p. The resin was washed and boiled in SDS–PAGE buffer to release GST-Nup57p. The eluate and purified GST-Nup57p and Cex1p were subjected to SDS–PAGE, followed by Western blot analysis using anti-GST to detect Nup57p (Figure 6D, top row) and anti-Cex1p to detect Cex1p (Figure 6D, second row). The interaction was also monitored by staining an SDS–PAGE gel with Coomassie blue (Figure 6D, third row). Unbound Cex1p in the wash eluate was detected by Coomassie blue staining of an SDS–PAGE gel (Figure 6D, bottom row). Both Coomassie blue staining and Western blot analysis showed that Cex1p did not interact with Nup57p (Figure 6D, second and third row, lane 3). A very small amount of Cex1p could be detected in the eluate obtained from the sample containing bound GST (Figure 6D, second and third rows, lane 4). Collectively, the data indicate that Cex1p associates with the NPC by interacting directly and specifically with Nup116p. The data also suggest that Cex1p may be playing a direct role in nuclear tRNA export by facilitating a step after the tRNA–export receptor complex has reached the cytoplasmic side of the NPC.
Cex1p may collect tRNA from the nuclear tRNA export receptors of the aminoacylation-dependent export pathway at the NPC
To gain insight into the role of Cex1p at the NPC, TAP was used to determine whether Cex1p co-purifies with the export receptors Los1p and Msn5p of the nuclear aminoacylation-dependent pathway and Cca1p of the aminoacylation-independent pathway (Figure 7). Co-purification of Cex1p with Cse1p, which is an unrelated nuclear export receptor responsible for protein export, was also investigated (Kunzler and Hurt, 1998). The final eluates were separated by SDS–PAGE and transferred to membranes. The blots were stained with Sypro-Ruby to detect the co-purifying proteins (Figure 7A), or subjected to Western blot analysis to detect Cex1p or Los1p (Figure 7B). The analyses show that Cex1p co-purifies with Los1-TAP (Figure 7B, top panel, first row, lane 2) and Los1p co-purifies with Cex1-TAP (lower panel, top row, lane 2). Furthermore, Cex1p co-purifies with Msn5-TAP (top panel, second row, lane 2) but not with Cca1-TAP (top panel, fourth row, lane 2) or Cse1-TAP (top panel, third row, lane 2). These findings suggest that Cex1p associates specifically with receptors of the nuclear tRNA aminoacylation-dependent export pathway. In addition, the RanGTPase Gsp1p co-purified with Cex1-TAP (Figure 7B, lower panel, bottom row, lane 2). As tRNA is translocated across the NPC in a complex with the export receptor and Gsp1p, these results suggest that Cex1p may associate with the export receptor–tRNA–Gsp1p complex in vivo. Interestingly, the S. cerevisiae genome-scale protein–protein interaction studies reported did not detect an association between Cex1p, Los1p, Msn5p, Nup116p and Gsp1p (Gavin et al, 2006; Krogan et al, 2006).
Figure 7.
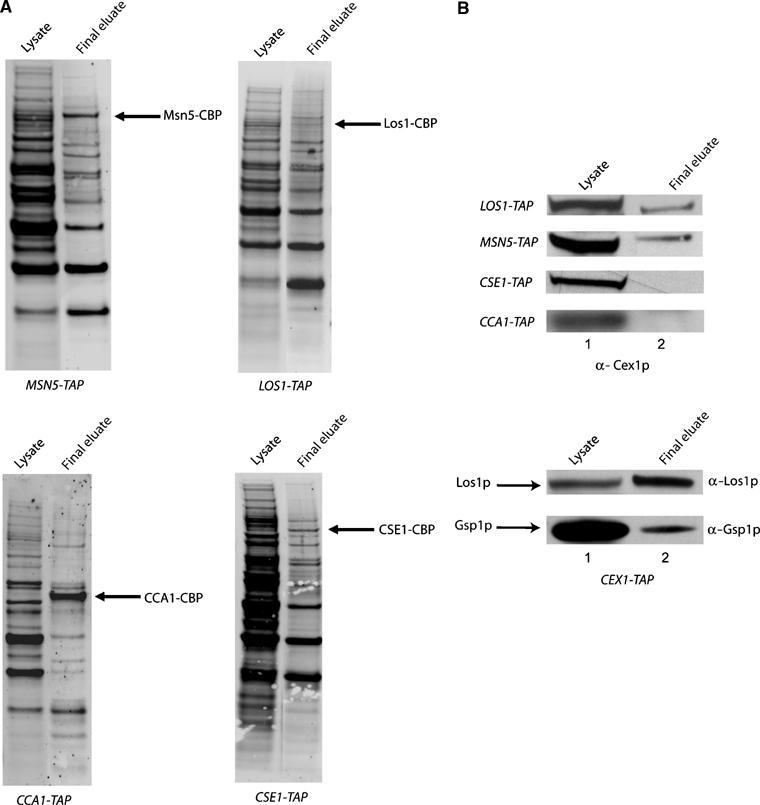
Cex1p co-purifies with Los1p, Msn5p and Gsp1p but not with Cse1p or Cca1p by TAP. TAP was performed using total cell extract prepared from CEX1-TAP, LOS1-TAP, MSN5-TAP, CSE1-TAP, GSP1-TAP and CCA1-TAP. Total cell extract (lane 1) and the final eluate (lane2) were subjected to SDS–PAGE and the separated proteins were transferred to nitrocellulose membranes. The membranes were stained with Sypro-Ruby to detect the co-purifying proteins (A), or used for Western blot analysis (B) to detect Cex1p, Los1p or Gsp1p.
In vitro binding was used to determine whether Cex1p interacts directly with the nuclear tRNA export receptors. This analysis was conducted with Los1p (Figure 8), as its function in nuclear tRNA export is better characterized than that of Msn5p. GST-Los1p bound to glutathione resin was incubated with (lane 4) or without tRNA (26 μM) (Figure 8A, lanes 3 and 5). Substrate-induced intrinsic fluorescence quenching of trytophan residues indicated that under the binding conditions used, the GST-Los1 fusion protein binds tRNA (data not shown); the apparent affinity of Los1p for tRNA is approximately 20 μM. The resin was washed and a two-fold molar excess of Cex1p was incubated with bound GST-Los1p loaded with tRNA (lane 4) or free of tRNA (lane 5). Buffer was also added to bound GST-Los1p free of tRNA (lane 3). As a control, Cex1p was incubated with bound GST (lane 6). Los1p was released from bound GST by cleavage with TEV. The eluate and purified GST-Los1p and Cex1p were subjected to SDS–PAGE, followed by Coomassie blue staining (Figure 8B, top row) or Western blot analysis to detect Los1p (Figure 8A, top row) and Cex1p (Figure 8A, bottom row). Unbound Cex1p in the wash eluate was detected by Coomassie blue staining of an SDS–PAGE gel (Figure 8B, bottom row). Los1p is released from the resin owing to cleavage of the TEV site found between GST and Los1p in the GST-Los1 fusion protein (Figure 8A and B, compare lane 2 with lanes 3–5). Both Western blot analysis and Coomassie blue staining show that Cex1p interacts with Los1p loaded with tRNA (Figure 8A and B, lanes 4 and 3, respectively) or free of tRNA (Figure 8A and B, lanes 5 and 4, respectively). Furthermore, Cex1p could not be detected in the eluate obtained from the sample containing bound GST (Figure 8A and B, lanes 6 and 5, respectively). The ratio of the intensities of Cex1p to Los1p, determined by densitometric analysis, indicates that Los1p loaded with tRNA or free of tRNA bound approximately the same amount of Cex1p (compare lanes 3 and 4 of Figure 8B, top row). Qualitative analysis of the Coomassie blue-stained proteins in the blot indicates that the interaction between Cex1p and Los1p in the presence (lane 3) or absence of tRNA (lane 4) is nearly stoichiometric (Figure 8B, top row).
Figure 8.
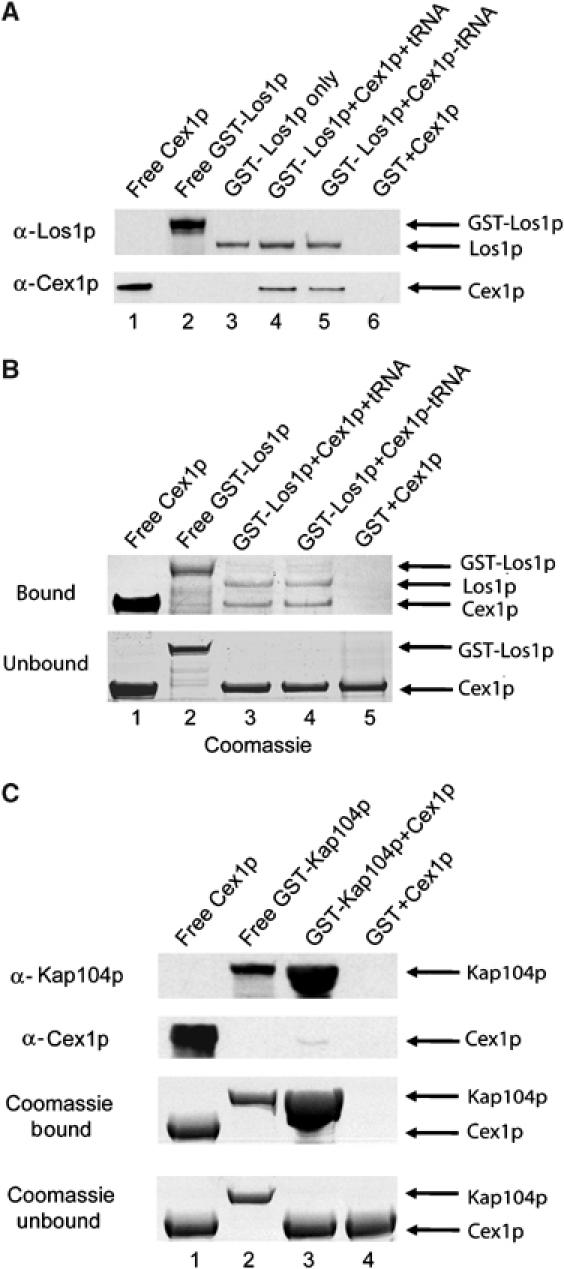
Cex1p interacts directly and specifically with Los1p in a tRNA-independent manner in vitro. GST-Los1p (200 μg, 1.57 nmol) was bound to glutathione resin with (lane 4) or without tRNA (lanes 3 and 5), and incubated with buffer with (lanes 4 and 5) or without (lane 3) a two-fold molar excess of Cex1p (266 μg, 3.14 nmol). Cex1p was also incubated with bound GST (lane 6). The resin was washed and Los1p was released from bound GST using TEV. Western blot analysis was used to detect Los1p (A, top row) and Cex1p (A, bottom row) in an aliquot of the eluate. Purified Cex1p (lane 1) and GST-Los1p (Lane 2) were used as size markers. Coomassie blue staining of an SDS–PAGE gel was also used to monitor the extent of protein binding to Los1p with (lane 3) or without tRNA (lane 4) and GST (lane 5) (B, top row). Unbound Cex1p in an aliquot of the wash eluate was detected by Coomassie blue staining of an SDS–PAGE gel (B, bottom row). The specificity of the interaction of Cex1p with Los1p was tested by investigating whether Cex1p interacts with Kap104p (C). The analysis was performed as above except 1.92 nmol (200 μg) of GST-Kap104p and 3.84 nmol (327 μg) of Cex1p were used. The resin was boiled in SDS–PAGE sample buffer to release GST-Kap104p. Coomassie blue staining of an SDS–PAGE gel (third row) or Western blot analysis was used to detect Kap104p (top row) and Cex1p (second row) in an aliquot of the eluate. Purified Cex1p (lane 1) and GST-Kap104p (lane 2) were used as size markers. Unbound Cex1p in an aliquot of the wash eluate was detected by Coomassie blue staining of an SDS–PAGE gel (bottom panel).
The specificity of the interaction between Cex1p and Los1p was investigated by testing whether Cex1p interacts with Kap104p, which is a β-Kap involved in nuclear protein import (Figure 8C). Bound GST-Kap104p or GST was incubated with a two-fold molar excess of Cex1p. The resin was washed and boiled in SDS–PAGE buffer to release GST-Kap104p. The eluate and purified GST-Kap104p and Cex1p were subjected to SDS–PAGE followed by Coomassie blue staining (Figure 8C, third row) or Western blot analysis to detect Kap104p (Figure 8C, top row) and Cex1p (Figure 8C, second row). The amount of Cex1p in the wash eluate was monitored by Coomassie blue staining of an SDS–PAGE gel (Figure 8D, bottom row). By comparison to Los1p (Figure 8A and B), an extremely small amount of Cex1p was found to interact with Kap104p by Western blot analysis (Figure 8C, second row, lane 3) and Coomassie blue staining (Figure 8C, third row, lane 3). Cex1p could not be detected in the eluate from the bound GST control (Figure 8C, lane 4). The data suggest that Cex1p is interacting specifically with Los1p. Collectively, the data obtained suggest that in vivo Cex1p interacts directly with the nuclear tRNA export receptors at the cytoplasmic side of the NPC to collect tRNA.
Cex1p may deliver aminoacylated tRNAs to the translation machinery
Cex1p may also be required to deliver aminoacyl-tRNAs to eEF-1A, which is necessary for efficient export of tRNAs from the nucleus to the cytoplasm by Los1p (Grosshans et al, 2000a). To investigate this possibility, the effect of depletion of Cex1p and eEF-1A on cell growth was assessed. The cex1 strain with a disruption in one of the two eEF-1A genes was prepared from a heterozygous diploid strain harboring pCEN-URA-GAL-CEX1 by tetrad dissection. The cex1 tef2 strain harboring pYX242 with or without the TEF2 gene grew at 30°C at a rate comparable to cex1 and tef2 with pYX242 when the plasmid containing the CEX1 gene was eliminated by counter-selection using 5-FOA (data not shown). The cex1 tef2 strain containing pYX242 grew at a comparable rate to the tef2 and cex1 strains containing pYX242 on CSD or rich medium at 30°C (Figure 9A, bottom panel). In contrast, the cex1 tef2 strain grew slower than the tef2 and cex1 strains on CSD medium at 24°C (Figure 9A, top right panel). However, this growth was significantly slower on rich medium at 24°C (Figure 9A, top left panel). A los1 tef2 strain was also reported to grow significantly slower than each parental strain on rich medium at 24°C (Grosshans et al, 2000a).
Figure 9.
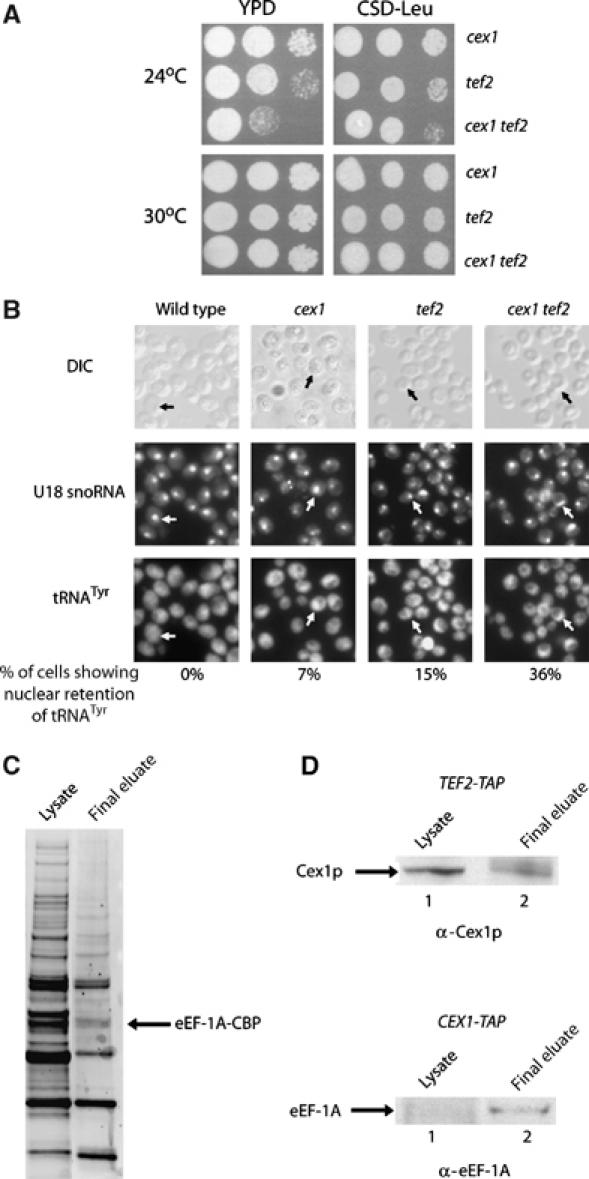
Disruption of the CEX1 and TEF2 genes affected cell growth and the efficiency of nuclear tRNA export and TAP resulted in the co-isolation of Cex1p and eEF-1A. The cex1, tef2 and cex1 tef2 strains containing pYX242 were serially diluted and spotted on YPD (left panels) or CSD medium lacking Leu (right panels) and incubated at 24 and 30°C (A). The location of the U18 snoRNA and the mature form of tRNATyr in the cex1, tef2 and cex1 tef2 strains was detected by FISH. Cells showing nuclear retention of tRNATyr are indicated by arrows as determined by U18 snoRNA localization. The number of cells showing nuclear retention of tRNATyr was expressed as a percentage of total cells. For each strain, a total of 200 cells were inspected for nuclear accumulation of tRNATyr (B). TAP was performed using total cell extract prepared from TEF2-TAP and CEX1-TAP. The proteins in total cell lysate (lane 1) and the final eluate (lane 2) were separated by SDS–PAGE and transferred to nitrocellulose membrane. The membrane was stained with Sypro-Ruby to detect the proteins (C) or used for Western blot analysis to detect Cex1p (D, top row) or eEF-1A (D, bottom row).
FISH was used to ascertain whether nuclear tRNA export was also affected in the cex1 tef2 strain (Figure 9B). Nuclear retention of tRNATyr was observed in approximately 7% of the cex1 cells, whereas 15% of the tef2 cells showed nuclear accumulation of tRNATyr. Studies reported previously also found that a similar percentage of tef2 cells show nuclear retention of tRNALeu (Grosshans et al, 2000a). A higher percentage (36%) of the cex1 tef2 cells exhibited nuclear retention of tRNATyr. Taken together, these results suggest that the growth defect of the cex1 tef2 strain was most likely caused by a reduction in the efficiency of nuclear tRNA export.
TAP was used to investigate whether Cex1p co-purifies with eEF-1A (Tef2-TAP). Proteins that co-purified with Tef2-TAP were separated by SDS–PAGE and transferred to membranes. The bolt was stained with Sypro-Ruby to detect the proteins (Figure 9C) or subjected to Western blot analysis to detect Cex1p or eEF-1A (Figure 9D). Cex1p was found to co-purify with Tef2-TAP (Figure 9D, top row, lane 2), and eEF-1A co-purified with Cex1-TAP (Figure 9D, bottom row, lane 2). These findings also support the suggestion that Cex1p is an extranuclear component of the nuclear aminoacylation-dependent export pathway, as eEF-1A delivers aminoacyl-tRNAs to ribosomes. However, in vitro, Cex1p did not interact with eEF-1A-GTP in the presence or absence of tRNA (data not shown).
Discussion
We show that Cex1p is a novel component of the nuclear tRNA export apparatus of S. cerevisiae. Cex1p, encoded by the YOR112W gene, was originally identified using a yeast tRNA three-hybrid interaction approach and an in vivo nuclear tRNA export assay (Figure S1) to identify proteins associated with the nuclear tRNA export process in S. cerevisiae (Steiner-Mosonyi et al, 2003). Here we have shown that Cex1p is located primarily in the cytoplasm (Figure 5A) and has a tRNA-binding site (Figure 4). Furthermore, Cex1p interacts genetically with Arc1p, a cytoplasmic component that influences the efficiency of the nuclear aminoacylation-independent export pathway (Figure 1A and B). However, disruption of the Cex1p and Los1p genes had no effect on cell growth (Figure 2), whereas previous studies have shown that combining the arc1 and los1 alleles resulted in synthetic lethality (Simos et al, 1996). Overexpression of Cex1p could not restore growth of the los1 arc1 strain (data not shown). Similarly, overproduction of Los1p did not alleviate the growth defect of the cex1 arc1 strain (Figure 1C). These data suggest that the function of Cex1p is distinct from that of Arc1p, but may play an equivalent role in the nuclear aminoacylation-dependent export pathway, which is the principal route used to export tRNA from the nucleus in S. cerevisiae (Steiner-Mosonyi and Mangroo, 2004).
Disruption of the Los1p and eEF-1A genes has been shown to reduce both cell growth and the efficiency of nuclear tRNA export (Grosshans et al, 2000a). Furthermore, tRNAs that have been shown to be exported by Los1p are found in the aminoacylated form in the nucleus of a los1 strain (Steiner-Mosonyi and Mangroo, 2004). These observations suggest that Los1p is responsible for exporting aminoacyl-tRNAs. Msn5p is also a nuclear tRNA export receptor of the nuclear aminoacylation-dependent pathway based on the findings that depletion of both Msn5p and Los1p reduced the efficiency of nuclear export of tRNA, and the Msn5p orthologue exportin-5 associates with aminoacyl-tRNAs in mammalian cells (Bohnsack et al, 2002; Calado et al, 2002; Takano et al, 2005). This is consistent with data showing that the major tRNA species of 19 families are in the aminoacylated form in the nucleus of wild-type S. cerevisiae cells at steady state (Steiner-Mosonyi and Mangroo, 2004). Although the loss of function of Cex1p and Los1p did not affect cell growth, the efficiency of nuclear tRNA export was reduced significantly (Figures 2 and 3). In addition, disruption of Cex1p and eEF-1A genes was found to impair cell growth and the efficiency of nuclear tRNA export (Figure 9A and B). Taken together, the data are consistent with the supposition that Cex1p plays a role in the nuclear aminoacylation-dependent tRNA export pathway. This is further supported by the finding that TAP resulted in the co-purification of Cex1p, Los1p, Msn5p and eEF-1A but not Cca1p (Figures 7 and 9D), which plays a role in facilitating translocation of non-aminoacylated tRNAs across the NPC in S. cerevisiae (Feng and Hopper, 2002), or Cse1p, a nuclear export receptor, that does not play a role in nuclear tRNA export (Kunzler and Hurt, 1998). Although the biochemical and genetic characterizations strongly suggest that Cex1p functions in the nuclear aminoacylation-dependent export pathway, the data however do not rule out the possibility that Cex1p may also participate in the aminoacylation-independent export pathway.
Colocalization studies showed that, although the majority of Cex1p is in the cytoplasm, a small percentage of the protein associates with the NPC (Figure 5A–C). This association is dependent on the presence of Nup116p (Figure 5D). Nup116p is located on the nucleoplasmic, but more so, on the cytoplasmic side of the NPC. Disruption of the NUP116 gene was shown to cause a defect in nuclear tRNA export (Sarkar and Hopper, 1998). Cex1p was found to interact with Nup116p in vivo by a two-hybrid interaction study (Ito et al, 2001) and by our TAP analyses (Figure 6B). Moreover, Cex1p and Nup116p interact directly and specifically in vitro (Figure 6C and D). These results combined with the finding that Cex1p associates with both Los1p and Msn5p but not with Cse1p (Figure 7B) in vivo suggest that Cex1p locates at the NPC by interacting with Nup116p, and facilitates a step after the nuclear export receptor–tRNA complex has reached the cytoplasmic side of the NPC. Although the role of Cex1p in nuclear tRNA export is not fully characterized at this stage, the observation that Cex1p interacts directly with tRNA and Los1p in vitro (Figures 4 and 8) strongly suggests that in vivo it interacts with the export receptors at the cytoplasmic side of the NPC to pick up aminoacyl-tRNAs. However, further studies are required to understand fully how Cex1p interfaces with the nuclear tRNA export receptor such as Los1p at the NPC to retrieve the aminoacylated tRNAs.
The GTPase Ran/Gsp1p plays an essential role in both nuclear import and export processes facilitated by β-Kaps. For nuclear export, interaction of the export receptor with the GTP-bound form of Ran/Gsp1p facilitates binding of the cargo to the receptor. The resulting ternary complex moves across the NPC and, once in the cytoplasm, the GTPase activity of Ran/Gsp1p is activated by the RanGTPase-activating protein RanGAP/Rna1p. Hydrolysis of GTP to GDP by Ran/Gsp1p facilitates dissociation of the receptor–cargo–Ran/Gsp1p complex (Gorlich and Kutay, 1999; Rodriguez et al, 2004). Los1p has been shown to co-purify with Gsp1p-GTP and to interact with Gsp1p-GTP in a tRNA-dependent manner in vitro (Hellmuth et al, 1998). This finding led to the suggestion that loading of Los1p with tRNA is dependent on Gsp1p-GTP in vivo. This is also the case for exportin-t and exportin-5 (Arts et al, 1998a; Kutay et al, 1998; Bohnsack et al, 2002; Calado et al, 2002). Moreover, in vitro and in vivo studies in S. cerevisiae and X. laevis suggest that unloading of the tRNA cargo from Los1p and exportin-t after translocation across the NPC requires GTP hydrolysis by Gsp1p/Ran (Arts et al, 1998a; Hellmuth et al, 1998; Kutay et al, 1998; Sarkar and Hopper, 1998). Cex1p was found to co-purify with Los1p and Gsp1p by TAP (Figure 7B). This combined with the observation that Cex1p binds Los1p directly and specifically in vitro (Figure 8) suggests that in vivo Cex1p interacts with Los1p before activation of the GTPase activity of Gsp1p. A model consistent with the finding that in vitro Cex1p interaction with Los1p is not dependent on tRNA (Figure 8A and B, compare lanes 4 and 5, and 3 and 4, respectively) suggests that in vivo Cex1p interacts first with the export receptor and then removes the aminoacyl-tRNA from the export receptor once the GTPase activity of Gsp1p is activated by the RanGAP Rna1p. This interpretation is consistent with the finding that in vitro Cex1p remained associated with Los1p loaded with tRNA (Figure 8). However, a thorough analysis is required to ascertain whether Gsp1p GTPase activity is required for Cex1p to facilitate the unloading of aminoacyl-tRNAs from the nuclear tRNA export receptors. Nevertheless, the data taken together support the conclusion that Cex1p is directly involved in the nuclear tRNA export process in S. cerevisiae.
Channelling is a mechanism used to spatially compartmentalize biochemical processes. This is achieved by directly transferring a substrate from one component to another within a multistep biochemical pathway. A channelling mechanism is used in mRNA and ribosome biogenesis and export, tRNA maturation, delivery of cytoplasmic tRNAs to aminoacyl-tRNA synthetases and transfer of aminoacyl-tRNAs from aminoacyl-tRNA synthetases to ribosomes (Stapulionis and Deutscher, 1995; Simos et al, 1996; Wolin and Matera, 1999; Grosshans et al, 2000a; Milkereit et al, 2001; Strasser et al, 2002). Aminoacyl-tRNAs exiting the nucleus are also channelled to the cytoplasmic translation machinery through eEF-1A (Grosshans et al, 2000a). This is, in part, based on the finding that a los1 tef2 strain shows a growth defect owing to nuclear accumulation of tRNA. The growth and nuclear tRNA export defect observed in the cex1 tef2 strain also support channelling of tRNAs exiting the nucleus to the translation machinery (Figure 9A and B). Moreover, the finding that Cex1p and eEF-1A co-purify with each other suggests that after Cex1p picks up aminoacyl-tRNAs from the export receptors, it delivers them to eEF-1A (Figure 9D). However, no interaction was detected between the two proteins in the presence or absence of tRNA in vitro. A possible explanation is that eEF-1A binds the tRNA and rapidly dissociates from Cex1p. This is unlikely to be the case, as an association between Cex1p and eEF-1A in vivo could be detected by TAP (Figure 9D). Furthermore, no tRNA was isolated from eEF-1A incubated with GST-Cex1p complexed with tRNA (data not shown). Therefore, a more likely explanation for the lack of interaction between the two proteins in vitro is that an unidentified protein is mediating the interaction between Cex1p and eEF-1A in vivo. Although further studies are required to test this possibility, the genetic and biochemical evidence is consistent with Cex1p delivering aminoacyl-tRNAs to eEF-1A.
We have shown that Cex1p is a cytoplasmic component of the nuclear aminoacylation-dependent tRNA export pathway in S. cerevisiae. The function of Cex1p is conserved, as a similar protein is found in higher eukaryotes, including humans. The biochemical and genetic characterizations suggest that Cex1p picks up aminoacyl-tRNAs from the nuclear export receptors at the cytoplasmic side of the NPC and delivers them to eEF-1A using a channelling mechanism. The data also suggest that Cex1p interacts directly with the export receptors at the NPC and then removes the tRNA from the export receptor, most likely after GTP hydrolysis by Gsp1p. Cex1p then dissociates from the export receptor and delivers the tRNA to eEF-1A, presumably in the cytoplasm. Transfer of the aminoacyl-tRNA from Cex1p to eEF-1A appears to be mediated by an unidentified protein, as no interaction was detected between the two proteins in the presence or absence of tRNA. Cex1p, therefore, provides a link between the nuclear aminoacylation-dependent tRNA export pathway and the protein translation apparatus.
Materials and methods
Strains and plasmids
The S. cerevisiae strains used in this study are listed in Table I. Plasmid construction, and overexpression and purification of the fusion proteins are provided as Supplementary data.
Table 1.
List of strains
| Strain | Genotype | Source |
|---|---|---|
| HEY301-129 | MATa, met8-1, trp1-1, his4-580, leu2-3,112, ura3-1, ade1 | Cleary and Mangroo (2000) |
| los1 | MATα los1∷HIS3, trp1, leu2, ade2, ura3, lys1, his3 | Dr E Hurt, University of Heidelberg (Simos et al, 1996) |
| cex1 | MATα cex1∷KanMX, his3, leu2, lys2, ura3 | Research Genetics |
| tef2 | MATa tef2∷HIS3, ade 2–1, trp 1–1, leu 2–3, 112, his 3–11, 15, ura 3–52, lys+, can 1–100, GAL+ | Dr E Hurt, University of Heidelberg (Grosshans et al, 2000a) |
| arc1 | MATa arc1∷HIS3, ade2, leu2, ura3, his3, trp1 | Dr E Hurt, University of Heidelberg (Simos et al, 1996) |
| CEX1-TAP | MATa CEX1∷TAP-K.I. URA3, his3, leu2, met15, ura3 | Open Biosystems |
| LOS1-TAP | MATa LOS1∷TAP-K.I. URA3, his3, leu2, met15, ura3 | Open Biosystems |
| TEF2-TAP | MATa TEF2∷TAP-K.I. URA3, his3, leu2, met15, ura3 | Open Biosystems |
| cex1 tef2 | MATα cex1∷KanMX, tef2∷HIS3, his3, leu2, ura3 | This study |
| cex1 arc1 | MATα cex1∷KanMX, arc1∷HIS3, his3, leu2, ura3, pCEN-URA-GAL1-CEX1 | This study |
| arc1 los1 | MATα los1∷HIS3, arc1∷HIS3, ade2, leu2, ura3, his3, trp, pHT4467-URA3-ADE3-LOS1 | Dr E Hurt, University of Heidelberg (Simos et al, 1996) |
| cex1 los1 | MATα cex1∷KanMX, los1∷HIS3, his3, leu2, ura3 | This study |
| NUP116-TAP | MATa NUP116∷TAP-K.I. URA3, ade2, arg4, leu2-3,112, trp1-289, ura3-52 | Euroscarf |
| CCA1-TAP | MATa CCA1∷TAP-K.I. URA3, his3, leu2, met15, ura3 | Open Biosystems |
| CSE1-TAP | MATa CSE1∷TAP-K.I. URA3, his3, leu2, met15, ura3 | Open Biosystems |
| MSN5-TAP | MATa MSN5∷TAP-K.I. URA3, his3, leu2, met15, ura3 | Open Biosystems |
| nup116 | MATa nup116∷KanMX, ura3, his3, leu2 | This study |
| NUP2-TAP | MATa NUP2∷TAP-K.I. URA3, his3, leu2, met15, ura3 | Open Biosystems |
| NUP57-TAP | MATa NUP57∷TAP-K.I. URA3, his3, leu2, met15, ura3 | Open Biosystems |
| nup120 | MATα ura3-52, his3-Δ200, trp1-1, leu2-3,112, lys2-8, nup120∷URA3 | Dr JD Aitchison, Institute for Systems Biology (Aitchison et al, 1995) |
Cex1p–tRNA interaction
Substrate-induced intrinsic fluorescence quenching was used to determine whether Cex1p binds tRNA. The reaction mixtures, containing 20 mM HEPES, pH 7.4 buffer containing 100 mM NaCl, 0.25 μM Cex1p, and varying amounts of total yeast tRNA (Sigma), or a single-stranded 90-mer DNA oligonucleotide (1, 2, 4, 6.25, 8, 12.5 and 16 μM), were incubated for 1 h at 4°C. Control reactions lacking Cex1p were prepared as above. Tryptophan fluorescence was measured using a Photon Technology International spectroflourimeter with excitation and emission slits set to 4 nm, and excitation and emission wavelengths of 295 and 334 nm, respectively. The fluorescence intensity of each control containing only tRNA was subtracted from that of the appropriate reaction and expressed as a percent reduction in the fluorescence intensity obtained with Cex1p alone.
Tandem affinity purification and Western blot analysis
TAP strains were grown in 2 l of YPD medium to an A600 of 2.0 at 30°C. The cells were harvested by centrifugation, resuspended in 50 ml NP-40 buffer (15 mM Na2HPO4, pH 7.2 buffer containing 10 mM NaH2PO4, 2% N P-40 (v/v), 150 mM NaCl, 2 mM EDTA, 50 mM NaF, 0.1 mM Na3VO4 and protease inhibitors) and lysed at 20 000 psi using the Emulsiflex-C3 High Pressure Homogenizer (Avestin Inc.). The lysate was clarified by ultracentrifugation at 142 000 g for 1.25 h at 4°C and subjected to tandem affinity purification using IgG–Sepharose 6 (Amersham Biosciences) and calmodulin–Sepharose (Stratagene), as described (Rigaut et al, 1999; Puig et al, 2001). The proteins in the final eluate were precipitated using 25% trichloroacetic acid. The protein precipitate was washed with ice-cold acetone containing 0.05 N HCl, followed by acetone and dried at room temperature. The protein precipitates were solubilized in LDS sample buffer (Invitrogen) and separated on 4–12% Novex bis-tris gels (Invitrogen). The separated proteins were transferred electrophoretically to Protran nitrocellulose membrane and stained with Sypro-Ruby, or probed with rabbit anti-Cex1p, anti-Los1p, anti-Nup116p and anti-Gsp1p, and mouse anti-human eEF-1A (Upstate Cell Signalling).
In vitro protein binding assays
The GST fusion proteins Los1p, Cex1p, Nup116p, Nup57p and Kap104p, as well as GST alone (Sigma), were added to 16 μl of glutathione–Sepharose 4B (Amersham Biosciences) in the presence or absence of a molar excess of total S. cerevisiae tRNA (26 μM) (Sigma) and the volume was adjusted to 1.5 ml with GST binding buffer (IPP150 buffer (25 mM Tris–HCl, pH 8.0, 150 mM NaCl and 0.1% NP-40 (w/v)) containing 1 mM DTT and a protease inhibitor cocktail (complete, EDTA-free)) and incubated for 2 h at 4°C. The resins were washed 3 × with GST binding buffer and incubated with a two-fold molar excess of the interacting protein in a total volume of 1.5 ml of GST binding buffer at 4°C for 3 h. The resins were washed 4 × with GST binding buffer and incubated with TEV protease to release the proteins from bound GST. The eluates were subjected to electrophoresis using a 4–12% Novex bis-tris gel and the proteins were detected by Coomassie blue staining or Western blot analysis. The unbound protein in the wash eluate was detected by Coomassie blue staining of SDS–PAGE gels.
Supplementary Material
Supplementary Information
Acknowledgments
We thank H Dehghani, JD Cleary and JD Steels for their initial participation in the project. We also thank Dr E Hurt, Dr S Wente, Dr D Heinrichs and Dr J Aitchison for their generous gifts of strains and antibodies, and B Strub for the pGEX-GST-TEV-LOS1 plasmid. We also thank the anonymous reviewers for their constructive and insightful suggestions. This work was supported by an operating grant from the Canadian Institutes of Health Research.
References
- Aitchison JD, Blobel G, Rout MP (1995) Nup120p: a yeast nucleoporin required for NPC distribution and mRNA transport. J Cell Biol 131: 1659–1675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arts GJ, Fornerod M, Mattaj IW (1998a) Identification of a nuclear export receptor for tRNA. Curr Biol 8: 305–314 [DOI] [PubMed] [Google Scholar]
- Arts GJ, Kuersten S, Romby P, Ehresmann B, Mattaj IW (1998b) The role of exportin-t in selective nuclear export of mature tRNAs. EMBO J 17: 7430–7441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azad AK, Stanford DR, Sarkar S, Hopper AK (2001) Role of nuclear pools of aminoacyl-tRNA synthetases in tRNA nuclear export. Mol Biol Cell 12: 1381–1392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohnsack MT, Regener K, Schwappach B, Saffrich R, Paraskeva E, Hartmann E, Gorlich D (2002) Exp5 exports eEF1A via tRNA from nuclei and synergizes with other transport pathways to confine translation to the cytoplasm. EMBO J 21: 6205–6215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calado A, Treichel N, Muller EC, Otto A, Kutay U (2002) Exportin-5-mediated nuclear export of eukaryotic elongation factor 1A and tRNA. EMBO J 21: 6216–6224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleary JD, Mangroo D (2000) Nucleotides of the tRNA D-stem that play an important role in nuclear-tRNA export in Saccharomyces cerevisiae. Biochem J 347 (Part 1): 115–122 [PMC free article] [PubMed] [Google Scholar]
- Feng W, Hopper AK (2002) A Los1p-independent pathway for nuclear export of intronless tRNAs in Saccharomycescerevisiae. Proc Natl Acad Sci USA 99: 5412–5417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavin AC, Aloy P, Grandi P, Krause R, Boesche M, Marzioch M, Rau C, Jensen LJ, Bastuck S, Dumpelfeld B, Edelmann A, Heurtier MA, Hoffman V, Hoefert C, Klein K, Hudak M, Michon AM, Schelder M, Schirle M, Remor M, Rudi T, Hooper S, Bauer A, Bouwmeester T, Casari G, Drewes G, Neubauer G, Rick JM, Kuster B, Bork P, Russell RB, Superti-Furga G (2006) Proteome survey reveals modularity of the yeast cell machinery. Nature 440: 631–636 [DOI] [PubMed] [Google Scholar]
- Giaever G, Chu AM, Ni L, Connelly C, Riles L, Veronneau S, Dow S, Lucau-Danila A, Anderson K, Andre B, Arkin AP, Astromoff A, El Bakkoury M, Bangham R, Benito R, Brachat S, Campanaro S, Curtiss M, Davis K, Deutschbauer A, Entian KD, Flaherty P, Foury F, Garfinkel DJ, Gerstein M, Gotte D, Guldener U, Hegemann JH, Hempel S, Herman Z, Jaramillo DF, Kelly DE, Kelly SL, Kotter P, LaBonte D, Lamb DC, Lan N, Liang H, Liao H, Liu L, Luo CY, Lussier M, Mao R, Menard P, Ooi SL, Revuelta JL, Roberts CJ, Rose M, Ross-Macdonald P, Scherens B, Schimmack G, Shafer B, Shoemaker DD, Sookhai-Mahadeo S, Storms RK, Strathern JN, Valle G, Voet M, Volckaert G, Wang CY, Ward TR, Wilhelmy J, Winzeler EA, Yang YH, Yen G, Youngman E, Yu KX, Bussey H, Boeke JD, Snyder M, Philippsen P, Davis RW, Johnston M (2002) Functional profiling of the Saccharomyces cerevisiae genome. Nature 418: 387–391 [DOI] [PubMed] [Google Scholar]
- Gite S, RajBhandary UL (1997) Lysine 207 as the site of cross-linking between the 3′-end of Escherichia coli initiator tRNA and methionyl-tRNA formyltransferase. J Biol Chem 272: 5305–5312 [DOI] [PubMed] [Google Scholar]
- Gorlich D, Kutay U (1999) Transport between the cell nucleus and the cytoplasm. Annu Rev Cell Dev Biol 15: 607–660 [DOI] [PubMed] [Google Scholar]
- Grosshans H, Hurt E, Simos G (2000a) An aminoacylation-dependent nuclear tRNA export pathway in yeast. Genes Dev 14: 830–840 [PMC free article] [PubMed] [Google Scholar]
- Grosshans H, Simos G, Hurt E (2000b) Review: transport of tRNA out of the nucleus–direct channeling to the ribosome? J Struct Biol 129: 288–294 [DOI] [PubMed] [Google Scholar]
- Hellmuth K, Lau DM, Bischoff FR, Kunzler M, Hurt E, Simos G (1998) Yeast Los1p has properties of an exportin-like nucleocytoplasmic transport factor for tRNA. Mol Cell Biol 18: 6374–6386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopper AK, Phizicky EM (2003) tRNA transfers to the limelight. Genes Dev 17: 162–180 [DOI] [PubMed] [Google Scholar]
- Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O'Shea EK (2003) Global analysis of protein localization in budding yeast. Nature 425: 686–691 [DOI] [PubMed] [Google Scholar]
- Ito T, Chiba T, Ozawa R, Yoshida M, Hattori M, Sakaki Y (2001) A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc Natl Acad Sci USA 98: 4569–4574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaffman A, O'Shea EK (1999) Regulation of nuclear localization: a key to a door. Annu Rev Cell Dev Biol 15: 291–339 [DOI] [PubMed] [Google Scholar]
- Kato M, Yano K, Morotomi-Yano K, Saito H, Miki Y (2002) Identification and characterization of the human protein kinase-like gene NTKL: mitosis-specific centrosomal localization of an alternatively spliced isoform. Genomics 79: 760–767 [DOI] [PubMed] [Google Scholar]
- Krogan NJ, Cagney G, Yu HY, Zhong GQ, Guo XH, Ignatchenko A, Li J, Pu SY, Datta N, Tikuisis AP, Punna T, Peregrin-Alvarez JM, Shales M, Zhang X, Davey M, Robinson MD, Paccanaro A, Bray JE, Sheung A, Beattie B, Richards DP, Canadien V, Lalev A, Mena F, Wong P, Starostine A, Canete MM, Vlasblom J, Wu S, Orsi C, Collins SR, Chandran S, Haw R, Rilstone JJ, Gandi K, Thompson NJ, Musso G, St Onge P, Ghanny S, Lam MHY, Butland G, taf-Ui AM, Kanaya S, Shilatifard A, O'Shea E, Weissman JS, Ingles CJ, Hughes TR, Parkinson J, Gerstein M, Wodak SJ, Emili A, Greenblatt JF (2006) Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature 440: 637–643 [DOI] [PubMed] [Google Scholar]
- Kunzler M, Hurt EC (1998) Cse1p functions as the nuclear export receptor for importin alpha in yeast. FEBS Lett 433: 185–190 [DOI] [PubMed] [Google Scholar]
- Kutay U, Lipowsky G, Izaurralde E, Bischoff FR, Schwarzmaier P, Hartmann E, Gorlich D (1998) Identification of a tRNA-specific nuclear export receptor. Mol Cell 1: 359–369 [DOI] [PubMed] [Google Scholar]
- Lipowsky G, Bischoff FR, Izaurralde E, Kutay U, Schafer S, Gross HJ, Beier H, Gorlich D (1999) Coordination of tRNA nuclear export with processing of tRNA. RNA 5: 539–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund E, Dahlberg JE (1998) Proofreading and aminoacylation of tRNAs before export from the nucleus. Science 282: 2082–2085 [DOI] [PubMed] [Google Scholar]
- Milkereit P, Gadal O, Podtelejnikov A, Trumtel S, Gas N, Petfalski E, Tollervey D, Mann M, Hurt E, Tschochner H (2001) Maturation and intranuclear transport of pre-ribosomes requires Noc proteins. Cell 105: 499–509 [DOI] [PubMed] [Google Scholar]
- Puig O, Caspary F, Rigaut G, Rutz B, Bouveret E, Bragado-Nilsson E, Wilm M, Seraphin B (2001) The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods 24: 218–229 [DOI] [PubMed] [Google Scholar]
- Rigaut G, Shevchenko A, Rutz B, Wilm M, Mann M, Seraphin B (1999) A generic protein purification method for protein complex characterization and proteome exploration. Nat Biotechnol 17: 1030–1032 [DOI] [PubMed] [Google Scholar]
- Rodriguez MS, Dargemont C, Stutz F (2004) Nuclear export of RNA. Biol Cell 96: 639–655 [DOI] [PubMed] [Google Scholar]
- Rout MP, Aitchison JD (2001) The nuclear pore complex as a transport machine. J Biol Chem 276: 16593–16596 [DOI] [PubMed] [Google Scholar]
- Sarkar S, Hopper AK (1998) tRNA nuclear export in Saccharomyces cerevisiae: in situ hybridization analysis. Mol Biol Cell 9: 3041–3055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar S, Azad AK, Hopper AK (1999) Nuclear tRNA aminoacylation and its role in nuclear export of endogenous tRNAs in Saccharomyces cerevisiae. Proc Natl Acad Sci USA 96: 14366–14371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaheen HH, Hopper AK (2005) Retrograde movement of tRNAs from the cytoplasm to the nucleus in Saccharomyces cerevisiae. Proc Natl Acad of Sci USA 102: 11290–11295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simos G, Segref A, Fasiolo F, Hellmuth K, Shevchenko A, Mann M, Hurt EC (1996) The yeast protein Arc1p binds to tRNA and functions as a cofactor for the methionyl- and glutamyl-tRNA synthetases. EMBO J 15: 5437–5448 [PMC free article] [PubMed] [Google Scholar]
- Stapulionis R, Deutscher MP (1995) A channeled tRNA cycle during mammalian protein synthesis. Proc Natl Acad Sci USA 92: 7158–7161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner-Mosonyi M, Mangroo D (2004) The nuclear tRNA aminoacylation-dependent pathway may be the principal route used to export tRNA from the nucleus in Saccharomyces cerevisiae. Biochem J 378: 809–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner-Mosonyi M, Leslie DM, Dehghani H, Aitchison JD, Mangroo D (2003) Utp8p is an essential intranuclear component of the nuclear tRNA export machinery of Saccharomyces cerevisiae. J Biol Chem 278: 32236–32245 [DOI] [PubMed] [Google Scholar]
- Strasser K, Masuda S, Mason P, Pfannstiel J, Oppizzi M, Rodriguez-Navarro S, Rondon AG, Aguilera A, Struhl K, Reed R, Hurt E (2002) TREX is a conserved complex coupling transcription with messenger RNA export. Nature 417: 304–308 [DOI] [PubMed] [Google Scholar]
- Takano A, Endo T, Yoshihisa T (2005) tRNA actively shuttles between the nucleus and cytosol in yeast. Science 309: 140–142 [DOI] [PubMed] [Google Scholar]
- Wang CC, Schimmel P (1999) Species barrier to RNA recognition overcome with nonspecific RNA binding domains. J Biol Chem 274: 16508–16512 [DOI] [PubMed] [Google Scholar]
- Wolin SL, Matera AG (1999) The trials and travels of tRNA. Genes Dev 13: 1–10 [DOI] [PubMed] [Google Scholar]
- Zasloff M, Rosenberg M, Santos T (1982) Impaired nuclear transport of a human variant transfer Rnaimet. Nature 300: 81–84 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information


