Abstract
HIV-1 gene expression is the major determinant regulating the rate of virus replication and, consequently, AIDS progression. Following primary infection, most infected cells produce virus. However, a small population becomes latently infected and constitutes the viral reservoir. This stable viral reservoir seriously challenges the hope of complete viral eradication. Viewed in this context, it is critical to define the molecular mechanisms involved in the establishment of transcriptional latency and the reactivation of viral expression. We show that Suv39H1, HP1γ and histone H3Lys9 trimethylation play a major role in chromatin-mediated repression of integrated HIV-1 gene expression. Suv39H1, HP1γ and histone H3Lys9 trimethylation are reversibly associated with HIV-1 in a transcription-dependent manner. Finally, we show in different cellular models, including PBMCs from HIV-1-infected donors, that HIV-1 reactivation could be achieved after HP1γ RNA interference.
Keywords: chromatin, HIV, latency, reactivation, transcription
Introduction
HIV-1 infection is a dynamic process involving continuous rounds of infection, replication and cell death. Continuous viral replication causes the loss of CD4+ T cells and, therefore, determines the rate of progression to immunodeficiency in infected individuals. The use of highly active anti-retroviral therapy (HAART) has recently raised the possibility of a cure for HIV-infected individuals (Finzi and Siliciano, 1998). Despite this success, there have been reports of viral rebound after interruption of HAART in patients in whom HIV plasma viremia was undetectable (Chun et al, 1999). Virus rebound is due to a small population of non-productively infected resting T cells that constitute the viral reservoir (Chun and Fauci, 1999). This latent reservoir of HIV, in the pool of resting CD4+ T cells, is established rapidly in primary HIV infection and can persist for an extended period of time (Chun et al, 1998a). Two forms of viral latency have been described (Bisgrove et al, 2005). Preintegration latency (unstable) is due to infected resting T cells in which HIV remains in the cytoplasm, whereas post-integration latency is due to cells containing integrated, transcriptionally inactive provirus. This latter latent state is stable as its half-life is same as that of infected cells and its progeny (Pomerantz, 2002). Thus, a detailed understanding of viral latency and reactivation is required to design new therapies that can eradicate HIV (Marcello, 2006).
Following integration into the host cell genome, the HIV-1 provirus is organized, similar to cellular genes into chromatin (Marcello et al, 2004). It has been proposed that the chromatin environment near the viral integration site may play a role in the transcriptional silencing of the HIV-1 genome (Jordan et al, 2003). Integrated HIV-1 provirus has positioned nucleosomes in its 5′ LTR and a correlation between HIV-1 gene expression and nucleosome remodeling has been reported. Activation of HIV-1 gene expression by histone deacetylase inhibitors, cytokines and the viral transactivator, Tat, is accompanied by the loss or rearrangement of a positioned nucleosome, nuc-1, near the viral mRNA start site (Verdin et al, 1993; Van Lint et al, 1996; El Kharroubi et al, 1998). Moreover, TSA strongly induces cell-free transcription of a chromatinized HIV-1 promoter, suggesting that HDAC activity may play a role in LTR repression (Sheridan et al, 1997). Collectively, these studies strongly suggest that chromatin is an important regulatory component of HIV-1 transcription. Relevant to the role of chromatin in HIV-1 gene expression, Tat has been shown to associate with histone acetyltransferases (HATs) p300/CBP, p300/CBP-associated factor (PCAF) and hGCN5 (Benkirane et al, 1998; Hottiger et al, 1998; Marzio et al, 1998). Tat-recruited HATs presumably acetylate histones in LTR-proximal nucleosomes to potentiate transcription (Benkirane et al, 1998). Indeed, in the absence of Tat, LTR-associated nucleosomes are hypoacetylated (Lusic et al, 2003). Upon activation by Tat, recruitment of Tat coactivators, PCAF, CBP and GCN5, was observed and correlated to nucleosomal hyperacetylation (Lusic et al, 2003). Knock-down of PCAF and p300 dramatically reduced Tat transactivation of an integrated HIV-1 promoter (Bres et al, 2002). Thus, a limiting step in Tat-mediated activation of the LTR is to relieve the chromatin repression mediated by positioned nucleosomes. Several reports have implicated HDAC activity in chromatin-mediated repression of HIV-1 promoter. First, HDAC inhibitors are potent activators of the HIV-1 LTR (Van Lint et al, 1996). Transcription factors such as YY1, NF-κB p50 homodimer and thyroid hormone receptor have been shown to mediate recruitment of HDAC1 to the LTR and consequently inhibit transcription from the viral promoter (Coull et al, 2000; He and Margolis, 2002; Hsia and Shi, 2002; Williams et al, 2006). However, TSA is also a potent activator of a non-integrated promoter (Kiernan et al, 1999; Quivy et al, 2002; Pagans et al, 2005). TSA cooperates with NF-κB in the activation of a transiently transfected LTR (Quivy et al, 2002). Additionally, several transcription factors involved in the activation of the HIV-1 LTR are regulated by reversible acetylation, including NF-κB and Sp1 (Chen et al, 2001; Kiernan et al, 2003; Huang et al, 2005). Thus, whether activation of the HIV-1 promoter by TSA is due to inhibition of HDAC or FDAC (factor deacetylase) activities or both is not clear.
In addition to histone hypoacetylation, trimethylation of histone H3 lysine 9 (H3 Lys9) is correlated with heterochromatin assembly and gene silencing (Grewal and Moazed, 2003). H3 Lys9 trimethylation is mediated by a conserved methyltransferase Su(var)3-9 in Drosophila, Suv39H1 in humans and Clr4 in fission yeast (Rea et al, 2000; Nakayama et al, 2001). Swi6 in fission yeast and HP1 (heterochromatin protein 1) in Drosophila and humans are conserved proteins found associated with H3Lys9 methyltransferases Suv39H1 and Clr4 (Aagaard et al, 1999). Additionally, HP1 and Swi6 bind specifically to H3 Lys9 trimethylated by Suv39H1 and Clr4, respectively (Bannister et al, 2001; Lachner et al, 2001). Thus, it has been proposed that H3Lys9 methyltransferases are required to initiate formation of heterorochromatin by recruiting HP1 (Cheutin et al, 2003), and that heterochromatin propagation and maintenance involves a ‘self-sustaining' loop, in which methylated H3Lys9 binds to HP1, which, in turn, recruits more Suv39H1 (Grewal and Moazed, 2003; Maison and Almouzni, 2004). In humans, three isoforms of HP1 protein exist that differ by their localization (Maison and Almouzni, 2004). Whereas HP1α and β are concentrated at pericentric heterochromatin, HP1γ also localizes to euchromatic sites (Minc et al, 2000; Nielsen et al, 2001). A consequence of HP1 recruitment is the establishment of a chromatin repressive state that leads to gene silencing (Grewal and Moazed, 2003; Maison and Almouzni, 2004).
Although important progress has been made in our understanding of the molecular mechanism involved in viral promoter activation (Marcello et al, 2001; Ott et al, 2004), the identity of cellular chromatin modifiers and the mechanism involved in the repression of HIV-1 promoter is still unclear. We hypothesized that heterochromatin machinery and repressive histone marks may play a determining role in chromatin-mediated HIV-1 transcriptional silencing and consequently participate in the establishment of the viral reservoir. Here, we present evidence that Suv39H1, HP1γ and histone H3 lysine 9 trimethylation are a major cellular determinant responsible for chromatin-mediated transcriptional silencing of HIV-1.
Results
SUV39H1 and HP1γ mediate repression of the HIV-1 LTR in a chromatin-dependent manner
In order to evaluate the role of Suv39H1 and HP1 in chromatin-mediated repression of the HIV-1 promoter, we employed RNA interference in HeLa cells containing a single copy of an integrated LTR-luciferase reporter gene (HeLa LTR-luc) or HeLa cells transiently transfected with LTR-luc construct. The expression level of endogenous Suv39H1 was reduced using a specific small-interfering (siRNA) both in HeLa and HeLa LTR-luc. Knock-down of Suv39H1 was efficient, as measured by RT–PCR (Figure 1A, top panel). Knock-down expression of Suv39H1 in HeLa LTR-luc cells had no effect on basal expression, whereas it enhanced Tat-mediated activation by up to five-fold (Figure 1A, lower panel). Interestingly, no significant effect was observed when an LTR-luc construct was transiently transfected in HeLa cells (Figure 1A, lower panel). We next investigated the role of HP1 proteins in both basal LTR activity and Tat transactivation. We used specific siRNAs for each isoform of HP1 (Figure 1B and Supplementary Figure S1). Whereas knock-down of HP1α and HP1β had only a minor effect on LTR basal activity (Supplementary Figure S1) and Tat-mediated transactivation (data not shown), knock-down of HP1γ both enhanced LTR basal expression level (up to 10-fold) and facilitated Tat transactivation of the HIV1 LTR (up to 100-fold) (Figure 1B, lower left panel). Knock-down of HP1γ had no effect on a transiently transfected LTR-luc reporter gene (Figure 1B, right panel). Taken together, these results show that Suv39H1 and HP1γ play a significant role in the chromatin-mediated repression of the viral promoter. Our results, showing that HP1γ but not HP1α or β is involved in LTR repression, are consistent with the recent findings that integrated HIV-1 (Schroder et al, 2002) and HP1γ are both localized in euchromatin, whereas HP1α and β are concentrated at pericentric heterochromatin (Minc et al, 2000; Nielsen et al, 2001). It should be noted that cells were transduced with GST-Tat, rather than transfected with a Tat expression plasmid (throughout our study), as we observed that knock-down of HP1 proteins affected the expression of genes from the CMV and TK promoters (Supplementary Figure S2 and data not shown). Enhanced LTR basal activity observed after HP1γ knock-down may result from general transcriptional derepression or from its direct involvement in the regulation of the viral promoter. Thus, we analyzed the consequence of HP1γ knock-down on the expression of cellular genes, such as IκBα, p53 and GAPDH, by RT–PCR. Whereas mRNAs corresponding to luciferase and IκBα were enhanced, no change in mRNA levels of p53 and GAPDH was observed (Figure 1C). Additionally, we found that knock-down of HP1γ did not affect expression of PCAF, p300 or CDK9 as monitored by Western blotting (Figure 4B).
Figure 1.

SUV39H1 and HP1γ mediate repression of the HIV-1 LTR in a chromatin-dependent manner. (A) HeLa LTR-Luc and HeLa cells were transfected twice with siRNA as indicated. For HeLa cells, LTR-Luc construct was cotransfected with siRNA in the second round of transfection. Twenty-four hours after the second round of transfection, cells were transduced with GST or GST-Tat as indicated. Expression of Suv39H1 and GAPDH mRNA was analyzed by RT–PCR using specific oligonucleotides (top panel). Twenty-four hours after transduction, extracts were prepared and luciferase activity was measured (lower panels). Results are presented as fold activation relative to GST-treated cells. The mean relative luciferase activities were obtained from three independent experiments. (B) Experiment was performed as in (A), except that siRNA specific for each isoform of HP1 was used. Expression levels of HP1α, β (Supplementary Figure S1) and γ were analyzed by Western blotting using specific antibodies (top panel). Results are presented as fold activation relative to GST-treated cells. The mean relative luciferase activities were obtained from three independent experiments. (C) Total RNA was prepared from cells transfected with control siRNA (Sc) or HP1γ-specific siRNA (experiment shown in (B)). RT–PCR was performed using oligonucleotides specific for the indicated genes. PCR products were analyzed on agarose gels and visualized by ethidium bromide.
Figure 4.
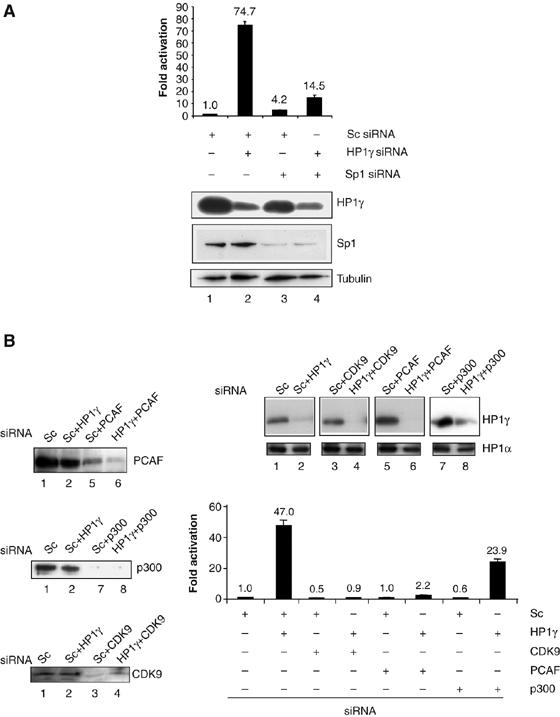
Transcriptional activation of the HIV-1 LTR observed after HP1γ knock-down requires Sp1, P-TEFb and PCAF. HeLa LTR-Luc cells were transfected with siRNAs specific for HP1γ, Sp1, CDK9, PCAF, p300 or control (Sc), as indicated. Luciferase activity was measured 48 h after transfection. Expression levels of HP1γ, Sp1, CDK9, PCAF, p300 and HP1α were determined by immunoblotting. Results are presented as fold activation relative to transfection of control siRNA. The mean relative luciferase activities were obtained from three independent experiments.
Dynamic association of Suv39H1, HP1γ and H3TriMetK9 with the HIV-1 LTR
Next, we investigated the direct involvement of Suv39H1 and HP1γ in Tat-mediated activation of an integrated viral promoter by performing quantitative chromatin immunoprecipitation assay (Q-ChIP) (Figure 2). HeLa LTR-luc cells were treated for 4 h with GST or GST-Tat. We investigated, by RT–PCR using oligonucleotides specific for the luciferase reporter gene that a 4-h treatment with GST-Tat is sufficient to induce transcription from the integrated LTR (Figure 2A). Consistent with transcriptional activation, GST-Tat was recruited to the viral promoter (Figure 2B). The amount of promoter immunoprecipitated using RNA polII, Sp1, p300 and CyclinT1 antibodies in the absence of GST-Tat was higher than that in the control immunoprecipitation sample (mock), suggesting that RNA polII, Sp1, p300 and CyclinT1 are associated with the promoter in the absence of Tat. GST-Tat treatment resulted in enhanced recruitment of p300 and CyclinT1, whereas the amounts of RNA polII and Sp1 were not affected. By contrast, PCAF was specifically recruited to the viral promoter after GST-Tat treatment. Interestingly, in the absence of GST-Tat, the amount of promoter region immunoprecipitated with Suv39H1, HP1γ and H3triMetK9 antibodies was higher than that in the control IP. Treatment of cells with GST-Tat resulted in promoter activation and reduction of promoter-associated Suv39H1 and HP1γ together with a reduced amount of H3Lys9 trimethylation mark. As a control, the amount of H3 was not affected by GST-Tat, whereas H3 acetylation was enhanced after GST-Tat treatment. Taken together, our results show that in the non-transcribing state, Suv39H1 and HP1γ are associated with the promoter and that H3 is trimethylated at K9. Tat-mediated activation of the LTR is accompanied by loss of Suv39H1 and HP1γ and recruitment of transcriptional coactivators such as PCAF, p300 and P-TEFb. At the level of histones, we found that loss of H3TriMetK9 after GST-Tat treatment was accompanied by H3 acetylation. To further analyze the dynamic association of HP1γ with the HIV-1 promoter, we performed in situ hybridization combined with immunofluoresence using anti-HP1γ-specific antibody. For this purpose, we used U2OS cells harboring an integrated HIV-1 vector containing the Rev responsive element (RRE) and Ψ located between the major splice donor and acceptor site 7, and an additional RNA tag inserted within this intron to facilitate its visualization by in situ hybridization. Previous experiments have shown that introns mostly accumulate at their transcription site, and control experiments where HIV-1 DNA and RNA were detected simultaneously showed that the foci detected with the intronic probe indeed corresponded to HIV-1 transcription sites (not shown). In the absence of transcription, integrated HIV-1 associated with HP1γ foci in the nucleus (Figure 2C, upper panels). Upon activation of transcription by Tat, HIV-1 was no longer associated with HP1γ (Figure 2C, lower panels). Taken together, these experiments clearly demonstrate that physical and functional interaction between HIV-1 and HP1γ is dynamic.
Figure 2.

Tat-mediated activation of the HIV-1 LTR is associated with reduction of Suv39H1, HP1γ and H3TriMetK9 associated with the LTR and recruitment of coactivators. HeLa LTR-luc cells were treated for 4 hours with GST or GST-Tat. Tat-mediated LTR activation was measured by RT–PCR using luciferase-specific oligonucleotides and GAPDH oligonucleotide as control (A). Chromatin prepared from GST- and GST-Tat-treated cells was subjected to immunoprecipitation with the indicated antibodies (B). As a control, immunoprecipitation was performed in the absence of antibody (Mock). Input and immunoprecipitated DNAs were subjected to real-time PCR using LTR-specific oligonucleotides. The amount of immunoprecipitated material was normalized to the input DNA. Data are representative of three independent experiments. (C) HIV-1 promoters are associated with HP1γ foci in the absence of Tat. U2OS cells containing an integrated HIV-1 reporter were transfected with a Tat expression vector or a control plasmid. The HIV-1 transcription sites were visualized with a fluorescent oligonucleotide probe against the intronic part of the vector (left panels and red color in the merged image), and HP1γ was labelled with a specific antibody (middle panels and green color in the merged image). In the absence of Tat (upper panels), HIV-1 transcription sites colocalized with HP1γ foci with a high frequency (four out of four in the field shown, and 90% of randomly selected cells; n=60). In contrast, in the presence of Tat (lower panels), HIV-1 transcription sites were located in regions containing low concentrations of HP1γ (two out of two in the field shown, and 95% of randomly selected cells; n=60). Note that the contrasts in the RNA images were scaled differently with and without Tat to allow for their simultaneous visualization. Quantification of the signals in the deconvolved images showed that about 20 times more RNA accumulated at the transcription sites in the presence of Tat. Each field was 69 × 54 μm. Insets: zoom of the boxed region in the merged images (4.4 × 4.4 μm).
Transcriptional activation of the HIV-1 LTR observed after HP1γ knock-down requires Sp1, P-TEFb and PCAF
Next, we investigated the mechanism involved in HP1γ-mediated repression of the LTR by performing ChIP assays to determine promoter occupancy by repressors and activators in cells expressing HP1γ compared to cells where HP1γ level was reduced using specific siRNA (Figure 3). Consistent with experiments shown in Figure 1B, knock-down of HP1γ resulted in a 10-fold increase in LTR basal activity (Figure 3A). By ChIP, the amount of promoter region immunoprecipitated using anti-HP1γ antibody was reduced in cells transfected with HP1γ-specific siRNA compared with control cells (Figure 3B). Interestingly, reduction of Suv39H1 and H3TriMetK9 was observed in HP1γ knock-down cells. Loss of H3TriMetK9 was accompanied by acetylation of H3. While the amount of RNA polII, Sp1 and p300 bound to the promoter was not affected by HP1γ knock-down, we observed a recruitment of PCAF and CyclinT1. These experiments suggest a role for PCAF and P-TEFb in transcriptional activation observed after HP1γ knock-down. As physical and functional interactions between Sp1 and cyclinT1 (Yedavalli et al, 2003), and Sp1 and PCAF have been reported (Chu et al, 2005), we decided to perform a double knock-down assay to address the requirement for Sp1, CyclinT1 and PCAF in LTR derepression (Figure 4). Knock-down of HP1γ resulted in 74-fold activation of the LTR (Figure 4A). This transcriptional activation was reduced by five-fold when both HP1γ and Sp1 levels were reduced in cells suggesting that the observed LTR activation is dependent on Sp1 transcription factor. Similarly, the enhanced expression from the LTR observed after HP1γ knock-down was dependent on the expression of CDK9 and PCAF (Figure 4B). Requirement for CTD kinase activity in the LTR activation observed after HP1γ knock-down was confirmed by using DRB, which is known to inhibit RNAPII CTD phosphorylation by CDK9, or by expressing a CDK9 dominant-negative mutant in HP1γ knock-down HeLa LTR-luc cells (data not shown). Consistent with ChIP assays, double knock-down of HP1γ and p300 had only a minor effect (two-fold reduction of LTR activity). Taken together, these experiments show that enhanced LTR basal activity after knock-down of HP1γ is mediated by Sp1, P-TEFb and PCAF.
Figure 3.
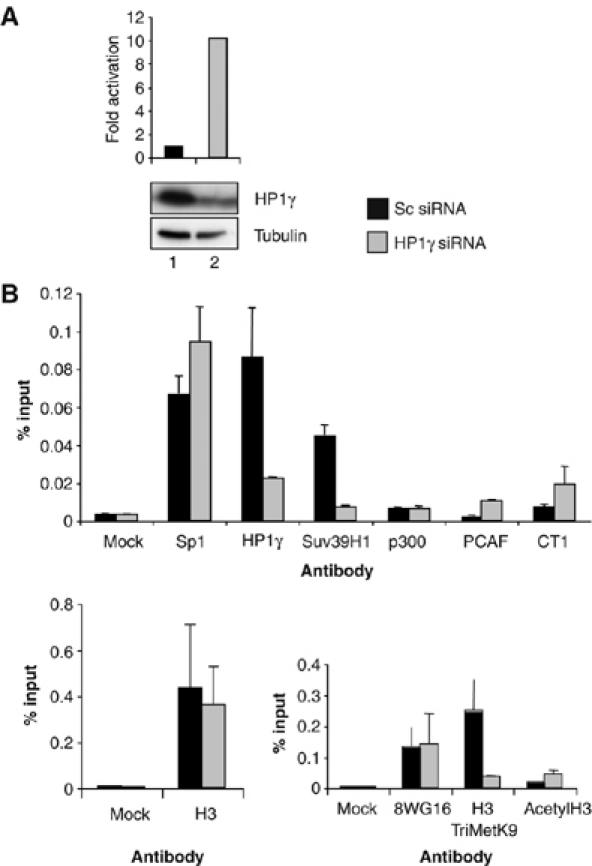
HP1γ knock-down results in reduction of Suv39H1 and H3TriMetK9 associated with the LTR and recruitment of PCAF and P-TEFb. HeLa LTR-Luc cells were transfected with control Sc siRNA (A, lane 1; B) or HP1γ-specific siRNA (A, lane 2; B). Forty-eight hours post-transfection, cells were harvested. An aliquot of cells was used to analyze luciferase activity (presented as fold activation relative to Sc siRNA-transfected cells) and HP1γ and tubulin expression by Western blotting (A). ChIP assay was performed as in Figure 2 using the indicated antibodies (B). Data are representative of three independent experiments.
Knock-down of HP1γ leads to HIV-1 reactivation in latently infected cells
Our data highlight the role of Suv39H1and HP1γ in chromatin-mediated repression of the HIV-1 promoter. We next evaluated the role of HP1γ in HIV-1 replication by employing HP1γ RNA interference. Recent findings have shown that stochastic fluctuations in Tat influence the commitment to viral latency decision (Weinberger et al, 2005). Low Tat expression or low Tat transcriptional activity (due to mutations in the gene or low expression of a coactivator) leads to phenotypic bifurcation and commitment toward latency. In this regard, latently infected U1 cells, which express mutant Tat unable to efficiently activate the LTR and consequently produce low amount of virus (Emiliani et al, 1998), represent an interesting model to study the involvement of HP1γ in HIV-1 transcriptional latency. Thus, we analyzed the consequence of HP1γ knock-down on HIV-1 production in U1 cells. As shown in Figure 5A, knock-down of HP1γ (left panel) results in increased HIV-1 production compared with control, HP1α or HP1β siRNA-transfected cells, suggesting a role of HP1γ in HIV-1 transcriptional latency in U1 cells. This experiment also confirms the lack of function of HP1α and HP1β in the viral promoter activity observed in Supplementary Figure S1. Interestingly, TNFα-mediated activation of HIV-1 was enhanced two-fold in HP1γ knock-down U1 cells and not affected when HP1α and HP1β levels were reduced (Figure 5). Similarly, Knock-down of HP1γ, but not HP1α, induced viral LTR activity in Jurkat cells harboring a latent HIV-1 retroviral vector lacking the tat gene and in which gfp is under the control of the HIV-1 LTR (Jordan et al, 2003) (Supplementary Figure S3). Next, we analyzed the role of HP1γ in HIV-1 infection of Jurkat T-cell line. Jurkat cells that were transfected with control siRNA or HP1γ-specific siRNA (Figure 5B) were infected with HIV-1. Virus replication was monitored by measuring the production of p24 in the supernatant every 3 days post-infection. Comparison of HIV-1 replication reveals faster replication kinetics of HIV-1 in HP1γ knock-down Jurkat cells (Figure 5C). This effect was observed early post-infection (days 3, 7 and 10: Supplementary Figure S4). Given that the knock-down of HP1γ is only transient, the observed effect on virus replication suggests that HP1γ-mediated repression of viral gene expression, and consequently virus replication, takes place early during infection. To verify this hypothesis, we evaluated the effect of HP1γ knock-down in a single-round infection assay, where cells were infected with HIV-1 lacking its envelope gene, pseudotyped with vesicular stomatitis virus (VSV.G) envelope and harboring luciferase gene (HIV-1VSV-Luc). Interestingly, in a single-round infection assay, we observed that knock-down of HP1γ resulted in increased virus production compared with control siRNA-transfected Jurkat cells, as measured by luciferase activity (Figure 5D). As expected, knock-down of p300 resulted in reduction of virus production. Furthermore, we found that HP1γ knock-down did not affect reverse transcription or the number of HIV-1 integration events (Supplementary Figure S5). Taken together, these experiments show that HP1γ-mediated chromatin repression of HIV-1 gene expression is established early during infection at a post-integration step. Enhanced virus production observed after HP1γ knock-down is due to either enhanced transcription from integrated HIV-1 or an enhanced number of transcribing HIV-1-integrated proviruses. To further characterize the involvement of HP1γ in chromatin-mediated HIV-1 repression in more physiological assays, we analyzed the effect of HP1γ knock-down on HIV-1 replication in PBMCs from HIV-1-infected untreated donors (Figure 6A). CD8+ T-cell-depleted PBMCs were transfected with control siRNA or HP1γ siRNA. As shown in Figure 6, knock-down of HP1γ in PBMCs from HIV-1-infected patients resulted in faster HIV-1 replication kinetics compared with control siRNA-transfected PBMCs. Interestingly, when PBMCs were isolated from HAART-treated patients with undetectable viremia, virus could be isolated from HP1γ siRNA-, but not from control siRNA-transfected PBMCs (Figure 6B). These results demonstrate the involvement of HP1γ in HIV-1 silencing in naturally infected cells.
Figure 5.

(A) Knock-down of HP1γ results in increased virus production in U1 cells, a cellular model for HIV-1 latency. U1 cells were transfected with siRNA as indicated in the figure. Efficiency of HP1α, HP1β and HP1γ knock-down was analyzed 48 h after transfection by Western blotting using a specific antibody. Virus production was monitored 48 h post-transfection by measuring p24 antigen in culture supernatant. Cells were treated with TNFα for 12 h when indicated. (B–D) HP1γ-mediated inhibition of HIV-1 replication is established early after virus infection. Jurkat cells were transfected with either control siRNA or HP1γ-, p300- or HP1α-specific siRNA. At 48 h post-transfection, cells were either (B) analyzed for expression levels of HP1γ, HP1α and p300 or (C) infected with HIV-1, and virus replication was monitored every 3 days after infection by measuring p24 viral antigen in culture supernatant or (D) infected with a single-round infectious virus (HIV-1VSV-luc) and virus production was monitored 48 h after infection by measuring luciferase activity in cell extracts.
Figure 6.

Knock-down of HP1γ results in faster HIV-1 replication in PBMCs from infected donors. CD4+ T cells purified from HIV-1-infected untreated (A) or HAART-treated (B) patients were transfected with control siRNA or HP1γ-specific siRNA using nucleofection. siRNA-transfected cells were cocultured with activated PBMCs from a healthy donor. Virus replication was monitored every 3 days after co-culture by measuring p24 viral antigen in culture supernatant (P1: patient 1; P2: patient 2). Knock-down of HP1γ was analyzed by Western blotting 48 h post-transfection in cells that were not subjected to coculture.
Discussion
In this paper, we have shown that Suv39H1, HP1γ and H3Lys9 trimethylation plays a major role in chromatin-mediated repression of HIV-1 gene expression in several systems, including HeLa cells containing a single integrated copy of the HIV-1 LTR-luc reporter gene, an HIV-1-infected T-cell line and PBMCs from infected individuals. These findings further advance our understanding of Tat-mediated activation of viral gene expression and the mechanism involved in transcriptional latency, which leads to the establishment of a viral reservoir observed in HIV-infected patients.
Currently, it is not clear how transcriptional silencing of HIV-1 genes is established. HDAC1 has been implicated in chromatin-mediated LTR silencing. Both YY1 and the NF-κB p50 homodimer have been shown to recruit HDAC1 to the LTR to repress its basal transcription activity (Coull et al, 2000; He and Margolis, 2002; Williams et al, 2006). Knock-down of p50 stimulated basal LTR activity up to three-fold (Williams et al, 2006). We observed that knock-down of HDAC1 had a minor effect on LTR basal activity (two-fold: Supplementary Figure S6), suggesting that inhibition of HDAC1 activity is not sufficient for efficient LTR derepression. In contrast, the deacetylase inhibitor TSA induces high promoter activity despite the modest effect of HDAC1 knock-down. One possibility is that HDAC-mediated repression of the viral promoter involves more than one HDAC and knock-down of one of the HDACs is not sufficient for full promoter de-repression. Alternatively, it has recently been shown that treatment with TSA reduces the amount of HP1 proteins, particularly HP1γ, in cells (Bartova et al, 2005). Thus, TSA, by reducing the expression level of HP1γ, mimics the effect of HP1γ-specific siRNA on LTR activity. In support of this, we found that TSA-mediated activation of an integrated LTR was maintained after HDAC1 knock-down, whereas it decreased five-fold when HP1γ expression level was reduced using specific siRNA (Supplementary Figure S6), suggesting that the effect of TSA is dependent on HP1γ. Further experiments are required to analyze the effect of HDAC1 knock-down and TSA treatment on HP1γ removal from the viral promoter. Recently, it has been shown that activation of HIV-1 from latently infected Jurkat cells by NF-κB requires the recruitment of TFIIH (Kim et al, 2006), suggesting that limitation in TFIIH may lead to transcriptional silencing and latency.
In microglial cells, which are targets of HIV infection within the central nervous system, COUP-TF interacting protein 2, CTIP2, represses LTR activity through its interaction with COUP-TF and Sp1 transcription factors (Marban et al, 2005). Whether CTIP2 plays a repressive role in T cells is not known. A surprising but interesting observation was that knock-down of Sp1 transcription factor enhanced basal LTR activity up to four-fold (Figure 4A). Thus, knock-down of Sp1 may lead to removal of CTIP2 from the viral promoter and consequently its de-repression. Further work is needed to evaluate the role of CTIP2 in LTR activity in T cells.
HP1γ is recruited to the HIV-1 promoter
Our experiments clearly show that HP1γ plays a major role in chromatin-mediated repression of viral gene expression. Up to 100-fold activation of basal LTR activity could be observed depending on the efficiency of HP1γ knock-down. How the heterochromatin complex is recruited to the HIV-1 promoter is unknown. In cells, it is known however that the repetitive nature of satellite repeats and transposons plays a role in the recruitment of heterochromatin machinery leading to silencing of nearby genes (Grewal and Moazed, 2003). Additionally, non-coding RNA molecules, including dsRNA, are known to play a role in heterochromatin formation presumably by a mechanism involving a direct interaction between chromodomain proteins and RNA (Grewal and Moazed, 2003). Whether Suv39H1 and/or HP1γ can bind to the HIV-1 short transcripts produced in the absence of Tat and thus be recruited to the viral promoter to establish a silent chromatin state is not known. It has been proposed that HP1, bound to methylated H3Lys9, mediates recruitment of Suv39H1 for heterochromatin propagation and maintenance in a ‘self-sustaining' loop (Grewal and Moazed, 2003; Maison and Almouzni, 2004). Accordingly, when we analyzed the consequence of HP1γ knock-down on LTR occupancy (Figure 7), we found that Suv39H1 was displaced and the H3TriMetK9 repressive mark was reduced. In contrast, coactivators, such as PCAF and P-TEFb, were recruited. Interestingly, whereas knock-down of Suv39H1 did not affect basal expression from an integrated HIV-1 LTR (Figure 1A) or virus production in U1 cells (data not shown), HIV-1 replicated with faster kinetics when Jurkat cells were transfected with Suv39H1-specific siRNA and subsequently infected with HIV-1 (Supplementary Figure S7). This experiment suggests that Suv39H1 is required for the deposition of the repressive H3Lys9 methylation mark, which consequently leads to recruitment of HP1γ to maintain the repressive state. Derepression of the viral promoter observed after HP1γ knock-down is dependent on the presence of Sp1 transcription factor. Interestingly, it has recently been shown that Sp1 can recruit P-TEFb and PCAF to Sp1 target promoters. Sp1 transcriptional activity is dependent on the CTD-kinase activity of P-TEFb. Indeed, Sp1 interacts directly with the regulatory subunit of P-TEFb, CyclinT1 (Yedavalli et al, 2003). Activation of the TbetaRII promoter involves recruitment of PCAF and p300 into a complex containing Sp1/NF-Y/HDAC1 (Huang et al, 2005). Acetylation of Sp1 by PCAF leads to dissociation of HDAC1 from the Sp1/NF-Y complex (Huang et al, 2005). Consistent with this, we found that P-TEFb and PCAF are required for Sp1-mediated activation of the viral promoter after HP1γ knock-down, suggesting a similar mechanism. Alternatively, NF-κB can also recruit P-TEFb to the viral promoter (Barboric et al, 2001). However, knock-down of p65 did not affect promoter derepression observed after HP1γ knock-down (Supplementary Figure S8).
Figure 7.

A proposed model for the involvement of chromatin in the regulation of HIV-1 promoter activity.
Tat-mediated transactivation of the HIV-1 promoter involves chromatin derepression and recruitment of coactivators
In addition to its effect on basal transcription from the LTR, we observed that Tat-dependent activation of the integrated LTR was also enhanced 100-fold after HP1γ knock-down, suggesting that knock-down of HP1γ also facilitates Tat-mediated transactivation. The increased Tat transcriptional activity was dependent on its binding to TAR RNA, as mutation of TAR RNA, which leads to loss of Tat binding, inhibited Tat transactivation of the LTR in both control and HP1γ knocked-down cells (Supplementary Figure S9). By contrast, activation of basal expression of the LTR observed after HP1γ knock-down was not affected by mutation of TAR (Supplementary Figure S9). Interestingly, Tat recruitment to the TAR element is accompanied by recruitment of coactivators such as P-TEFb, PCAF and p300 and concomitant reduction of Suv39H1, HP1γ and H3TriMetK9 associated with the promoter (Figure 7). Whereas HP1γ knock-down results in recruitment of PCAF and P-TEFb to the promoter, p300 was not recruited, unlike in cells treated with Tat. Thus, for optimal transactivation, Tat needs to relieve chromatin-mediated repression and recruit specific coactivators to the promoter (Figure 7). Whether p300 plays a role in Tat-specific de-repression of the LTR remains to be determined. Indeed, it is not clear how Tat relieves chromatin repression of the LTR. One possibility is that Tat associates with an enzymatic activity responsible for direct modification of H3TriMetK9 (Chen et al, 2006; Cloos et al, 2006; Klose et al, 2006; Yamane et al, 2006) or modification of local histones that destabilizes HP1γ binding (Fischle et al, 2005). Alternatively, a reported interaction between Tat and silent information regulator, SirT1 (Pagans et al, 2005), may contribute to the release of heterochromatin complex from the viral promoter. Indeed, Sir proteins are known to play a central role in the recruitment of heterochromatin complexes by inducing a local hypoacetylation of histones (Grewal and Moazed, 2003). Tat has been shown to be deacetylated by SirT1(Pagans et al, 2005). Thus, Tat may decoy the HDAC activity of SirT1, leading to a local hyperacetylation of histones and consequently the release of HP1γ and Suv39H1 from the LTR. Further work is needed to elucidate the mechanism involved in Tat-mediated chromatin derepression within the viral promoter.
Involvement of HP1γ in chromatin-mediated HIV-1 transcriptional silencing and latency
Transcriptional silencing of HIV-1 provirus in CD4+T-cells is the major mechanism responsible for viral latency and establishment of a stable viral reservoir (Bisgrove et al, 2005). This reservoir is established early during primary infection. Results from several experiments presented in this paper suggest a role of HP1γ in HIV-1 transcriptional latency and the establishment of the viral reservoir. (1) HP1γ-mediated repression of HIV-1 replication was observed in PBMCs from infected donors and in U1 cells, a cellular model for HIV-1 transcriptional latency. (2) HP1γ-mediated repression of the viral promoter can be observed in a single-round infection assay, suggesting that promoter repression is established early during infection. (3) Transient knock-down of HP1γ results in faster HIV-1 replication kinetics in Jurkat cells. (4) HP1γ does not affect the number of integrated proviruses, but rather the rate of transcription from integrated HIV-1.
A challenge in AIDS treatment is the reactivation of the viral reservoir in combination with HAART to completely eradicate the virus (Pomerantz, 2002; Lassen et al, 2004). In this respect, several molecules have been used to reactivate viral reservoir, including prostratin (Kulkosky et al, 2001), valproic acid (Lehrman et al, 2005), IL-2 (Chun et al, 1998b), IL-7 (Brooks et al, 2003) and the viral transactivator Tat (Lin et al, 2003). How conceivable is the use of HP1γ knock-down in viral reservoir reactivation? Although long-term knock-down of HP1γ would certainly be cytotoxic, HIV-1 derepression requires only a transient knock-down of HP1γ, which may be tolerated. Indeed, HP1γ knock-down was not cytotoxic as measured by cell viability assay (data not shown). siRNA-based therapy has been successfully used to protect mice from lethal herpes simplex virus 2 infection (Palliser et al, 2006). Additionally, antagomirs were successfully used in mice to inhibit the function of a cellular microRNA. Thus, it will be interesting to evaluate the feasibility of a combinational therapy involving HP1γ-specific siRNA and HAART in eradicating the HIV-1 reservoir in infected individuals.
Materials and methods
Cell culture, transfection and transduction
HeLa and HeLa cells containing a single copy of integrated HIV-1LTR-luciferase reporter construct (HeLa LTR-Luc) (Treand et al, 2006) were propagated in DMEM with 10% FBS. Jurkat, U1 and PBMCs were propagated in RPMI containing 10% FBS. Oligofectamine was used for siRNA transfection into HeLa cells. Jurkat, U1 and PBMCs were transfected using nucleofection technology according to the manufacturer's protocol (Amaxa). Transduction with GST and GST-Tat was carried out as previously described (Bres et al, 2005). Luciferase activity was measured 48 h after transduction according to the manufacturer's protocol (Promega). Luciferase activity was normalized to protein concentration using Bradford (Biorad).
The HeLa LTRHIV-1wt-Luc and HeLa LTRHIV-1TARm-Luc cells are derived from HeLa cells in which one copy of a luciferase gene under the control of the entire HIV-1 BRU promoter (LTR from −454 to +338) wt or ΔTAR (deletion of the 3-nt bulge of TAR, nt 476–478) has been inserted by recombination using the ‘Flp-InTM system' kit (Invitrogen) according to the manufacturer's protocol (see Supplementary data).
Plasmids
The LTR-Luc reporter gene construct was previously described (Kiernan et al, 1999).
Small-interfering RNAs
RNA oligonucleotides corresponding to SUV39H1 (forward: 5′-ACCUCUUUGACCUGGACUATT-3′; reverse: 5′-UAGUCCAGGUCAAAGAGGUTT-3′), HP1α (forward: 5′-GCUUUGAGAGAGGACUGGAACTT-3′; reverse: 5′-GUUCCAGUCCUCUCUCAAAGCTT-3′), HP1β (forward: 5′-GACUCCAGUGGAGAGCUCAUGTT-3′; reverse: 5′-CAUGAGCUCUCCACUGGAGUCTT-3′), HP1γ (forward: 5′-GAGGCAGAGCCUGAAGAAUTT-3′; reverse: 5′-AUUCUUCAGGCUCUGCCUCTT-3′), Sp1 (forward: 5′-UGCAUGGUGUUGAGAAGAATT-3′; reverse: 5′-UUCUUCUCAACACCAUCCATT-3′), CDK9 (forward: 5′-CCAAAGCUUCCCCCUAUAATT-3′; reverse: 5′-UUAUAGGGGGAAGCUUUGGTT-3′), PCAF (forward: 5′-UCGCCGUGAAGAAAGCGCATT-3′; reverse: 5′-UGCGCUUUCUUCACGGCGATT-3′) and p300 (forward: 5′-CAGAGCAGUCCUGGAUUAGTT-3′; reverse: 5′-CUAAUCCAGGACUGCUCUUGTT-3′) were synthesized (MWG Biotech, Ebersberg, Germany). Control Scramble II siRNAs were purchased from MWG Biotech.
Western blot analysis
Western blot analysis was performed as previously described (Kiernan et al, 1999). Proteins were visualized by chemiluminescence (Pierce). Primary antibodies used were anti-HP1α, anti-HP1β, anti-HP1γ (Euromedex), anti-PCAF (Santa Cruz), anti-p300 (Pharmingen), anti-Sp1 (Santa Cruz) and monoclonal anti-tubulin (Sigma).
Chromatin immunoprecipitation
See Supplementary data.
RT–PCR
Total RNA was extracted from HeLa cells using the NucleoSpin RNA kit (Machery Nagel, Duren, Germany) and reverse-transcribed using SuperScript First-Strand Synthesis System for RT–PCR (Invitrogen). RT products were PCR-amplified using oligonucleotides specific for luciferase (forward: 5′-AAG AGA TAC GCC CTG GTT CCT G-3′; reverse: 5′-CGG TAG GCT GCG AAA TGT TCA-3′), GAPDH (forward: 5′-GTA TTG GGC GCC TGG TCA CC-3′; reverse: 5′-CGCTCC TGG AAG ATG GTG ATG G-3′), p53 (forward: 5′-CAG CCA AGT CTG TGA CTT GCA CGT AC-3′; reverse: 5′-CTA TGT CGA AAA GTG TTT CTG TCA TC-3′), IκBα (forward: 5′-GCA GCA GCT CAC CGA GGA CG-3′; reverse: 5′-CAG GAC TGA GTC AGG ACT CC-3′) and Suv39H1 (forward: 5′-AGGACTTAGAAAGGGAGCTGC-3′; reverse: 5′-TACAGTGATGCGTCCCAGATG-3′). PCR products were resolved on 1% agarose/TAE gels containing ethidium bromide.
In situ hybridization
Human U2OS cells were cotransfected with a hygro marker and a previously described HIV-1 vector containing the LTRs, RRE and Ψ (Zufferey et al, 1998). The RRE and Ψ were located between the major splice donor and acceptor site 7 and an additional RNA tag was inserted within this intron to facilitate its visualization by in situ hybridization, as described previously (Fusco et al, 2003). Stable clones were then selected by hygromycin and individual clones were analyzed for their ability to induce transcription when transfected with a Tat-expressing vector. Clone 2E-11 showed a strong response to Tat and was used in further experiments.
Clone 2E-11 was transfected by Tat or an empty vector and cells were fixed 24 h later. In situ hybridization was performed as previously described, with a Cy3-labeled oligonucleotide probe specific for the tagged intron (Fusco et al, 2003). For HP1γ detection, cells were then re-permeabilized in 0.1% Triton/PBS for 10 min at room temperature and incubated with HP1γ-specific antibody. Cells were imaged on a DMRA microscope (Leica) equipped with a Cool-snap Fx camera (Roper scientific). Image stacks were collected at 0.2 μm intervals and deconvolved using Huygens2 (Bitplane).
PBMC isolation and CD8+ T-cell depletion
Peripheral blood mononuclear cells of HIV-1-infected, untreated patients from University Hospital of Montpellier were isolated by Ficoll–Paque (Eurobio) density centrifugation. PBMCs (106 cells/ml) were then incubated for 1 h at 4°C with anti-CD8 monoclonal antibody B9–11 (1 μg/ml; Beckman-Coulter) in PBS supplemented with 0.1% FCS. Cells were washed twice and incubated for an additional hour at 4°C with 100 μl of magnetic beads coated with anti-mouse Ig (CELLection™ Pan Mouse IgG kit, Dynal Biotech). The mixture was placed in a magnet and cells that were not bound to beads were collected. The collected cells contained less than 5% CD8+ T cells.
Coculture assay for viral production
PBMCs from healthy donors were preactivated using 5 μg/ml PHA (phytohemagglutinin-P; DIFCO)/10 U/ml IL-2 (interleukin-2, Roche) for 72 h. They were then washed once with PBS and once with RPMI medium before coculture assay. The siRNA-electroporated HIV-positive PBMCs (106 cells/ml) were cocultured with preactivated PBMCs (106 cells/ml) from healthy donors in the presence of 10 U/ml IL-2. The culture medium was collected every 3 or 4 days. Fresh preactivated healthy PBMC were added to the culture every 7 days. Viral production was measured by quantifying the amount of p24 in the culture medium using an ELISA kit (Beckman-Coulter).
Infection
Jurkat cells were infected with HIV-1 for 2 h at 37°C, washed and resuspended in RPMI containing 10% FCS. Virus production was monitored in culture supernatant every 3 days by measuring p24 antigen (Beckman Coulter).
Pseudotyped virion production and single-round infection
The plasmid pNL4.3-env−-Luc+, harboring a luciferase gene (obtained from AIDSreagent Program), was cotransfected with the envelope plasmid pMD.2G encoding the G protein of VSV.G into human embryonic kidney cells 293T. The virions, named HIV-1VSV-Luc, were collected and filtered (0.45-μm filters) 48 h post-transfection.
For single-round infection, Jurkat cells were transfected with siRNA and 48 h later were incubated with the VSV-Luc virions or heat-inactivated virions (56°C, 30 min) at an MOI of 0.01 for 18 h. They were then washed twice with PBS and cultured in RPMI-1640 medium containing 10% FCS. Luciferase activity was measured 48 h post-infection using a luminometer. Total DNA was also extracted from a portion of infected cells for real-time PCR assay.
Integration (Brussel and Sonigo 2003)
Supplementary Material
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Supplementary Figure S4
Supplementary Figure S5
Supplementary Figure S6
Supplementary Figure S7
Supplementary Figure S8
Supplementary Figure S9
Supplementary Materials
Acknowledgments
We thank Kuan-Teh Jeang for critical reading of the manuscript. We are grateful to Slimane Ait-Si-Ali for sharing HP1 siRNA sequences and E Verdin for J-Lat cells. This work was supported by grants from the Human Frontier Science program (young investigator program), EU012182, SIDACTION and ANRS to MB. IC was successively funded by the Ecole Normale Supérieur de Lyon and the Ministère de la Recherche. YL was supported by SIDACTION fellowship.
References
- Aagaard L, Laible G, Selenko P, Schmid M, Dorn R, Schotta G, Kuhfittig S, Wolf A, Lebersorger A, Singh PB, Reuter G, Jenuwein T (1999) Functional mammalian homologues of the Drosophila PEV-modifier Su(var)3-9 encode centromere-associated proteins which complex with the heterochromatin component M31. EMBO J 18: 1923–1938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, Allshire RC, Kouzarides T (2001) Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410: 120–124 [DOI] [PubMed] [Google Scholar]
- Barboric M, Nissen RM, Kanazawa S, Jabrane-Ferrat N, Peterlin BM (2001) NF-kappaB binds P-TEFb to stimulate transcriptional elongation by RNA polymerase II. Mol Cell 8: 327–337 [DOI] [PubMed] [Google Scholar]
- Bartova E, Pachernik J, Harnicarova A, Kovarik A, Kovarikova M, Hofmanova J, Skalnikova M, Kozubek M, Kozubek S (2005) Nuclear levels and patterns of histone H3 modification and HP1 proteins after inhibition of histone deacetylases. J Cell Sci 118: 5035–5046 [DOI] [PubMed] [Google Scholar]
- Benkirane M, Chun RF, Xiao H, Ogryzko VV, Howard BH, Nakatani Y, Jeang KT (1998) Activation of integrated provirus requires histone acetyltransferase p300. and P/CAF are coactivators for HIV-1 Tat. J Biol Chem 273: 24898–24905 [DOI] [PubMed] [Google Scholar]
- Bisgrove D, Lewinski M, Bushman F, Verdin E (2005) Molecular mechanisms of HIV-1 proviral latency. Expert Rev Anti Infect Ther 3: 805–814 [DOI] [PubMed] [Google Scholar]
- Bres V, Gomes N, Pickle L, Jones KA (2005) A human splicing factor, SKIP, associates with P-TEFb and enhances transcription elongation by HIV-1 Tat. Genes Dev 19: 1211–1226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bres V, Tagami H, Peloponese JM, Loret E, Jeang KT, Nakatani Y, Emiliani S, Benkirane M, Kiernan RE (2002) Differential acetylation of Tat coordinates its interaction with the co-activators cyclin T1 and PCAF. EMBO J 21: 6811–6819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks DG, Hamer DH, Arlen PA, Gao L, Bristol G, Kitchen CM, Berger EA, Zack JA (2003) Molecular characterization, reactivation, and depletion of latent HIV. Immunity 19: 413–423 [DOI] [PubMed] [Google Scholar]
- Brussel A, Sonigo P (2003) Analysis of early human immunodeficiency virus type 1 DNA synthesis by use of a new sensitive assay for quantifying integrated provirus. J Virol 77: 10119–10124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Fischle W, Verdin E, Greene WC (2001) Duration of nuclear NF-kappaB action regulated by reversible acetylation. Science 293: 1653–1657 [DOI] [PubMed] [Google Scholar]
- Chen Z, Zang J, Whetstine J, Hong X, Davrazou F, Kutateladze TG, Simpson M, Mao Q, Pan CH, Dai S, Hagman J, Hansen K, Shi Y, Zhang G (2006) Structural insights into histone demethylation by JMJD2 family members. Cell 125: 691–702 [DOI] [PubMed] [Google Scholar]
- Cheutin T, McNairn AJ, Jenuwein T, Gilbert DM, Singh PB, Misteli T (2003) Maintenance of stable heterochromatin domains by dynamic HP1 binding. Science 299: 721–725 [DOI] [PubMed] [Google Scholar]
- Chu BY, Tran K, Ku TK, Crowe DL (2005) Regulation of ERK1 gene expression by coactivator proteins. Biochem J 392: 589–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun TW, Davey RT Jr, Engel D, Lane HC, Fauci AS (1999) Re-emergence of HIV after stopping therapy. Nature 401: 874–875 [DOI] [PubMed] [Google Scholar]
- Chun TW, Engel D, Berrey MM, Shea T, Corey L, Fauci AS (1998a) Early establishment of a pool of latently infected, resting CD4(+) T cells during primary HIV-1 infection. Proc Natl Acad Sci USA 95: 8869–8873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun TW, Engel D, Mizell SB, Ehler LA, Fauci AS (1998b) Induction of HIV-1 replication in latently infected CD4+ T cells using a combination of cytokines. J Exp Med 188: 83–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun TW, Fauci AS (1999) Latent reservoirs of HIV: obstacles to the eradication of virus. Proc Natl Acad Sci USA 96: 10958–10961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloos PA, Christensen J, Agger K, Maiolica A, Rappsilber J, Antal T, Hansen KH, Helin K (2006) The putative oncogene GASC1 demethylates tri- and dimethylated lysine 9 on histone H3. Nature 442: 307–311 [DOI] [PubMed] [Google Scholar]
- Coull JJ, Romerio F, Sun JM, Volker JL, Galvin KM, Davie JR, Shi Y, Hansen U, Margolis DM (2000) The human factors YY1 and LSF repress the human immunodeficiency virus type 1 long terminal repeat via recruitment of histone deacetylase 1. J Virol 74: 6790–6799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Kharroubi A, Piras G, Zensen R, Martin MA (1998) Transcriptional activation of the integrated chromatin-associated human immunodeficiency virus type 1 promoter. Mol Cell Biol 18: 2535–2544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emiliani S, Fischle W, Ott M, Van Lint C, Amella CA, Verdin E (1998) Mutations in the tat gene are responsible for human immunodeficiency virus type 1 postintegration latency in the U1 cell line. J Virol 72: 1666–1670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finzi D, Siliciano RF (1998) Viral dynamics in HIV-1 infection. Cell 93: 665–671 [DOI] [PubMed] [Google Scholar]
- Fischle W, Tseng BS, Dormann HL, Ueberheide BM, Garcia BA, Shabanowitz J, Hunt DF, Funabiki H, Allis CD (2005) Regulation of HP1-chromatin binding by histone H3 methylation and phosphorylation. Nature 438: 1116–1122 [DOI] [PubMed] [Google Scholar]
- Fusco D, Accornero N, Lavoie B, Shenoy SM, Blanchard JM, Singer RH, Bertrand E (2003) Single mRNA molecules demonstrate probabilistic movement in living mammalian cells. Curr Biol 13: 161–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grewal SI, Moazed D (2003) Heterochromatin and epigenetic control of gene expression. Science 301: 798–802 [DOI] [PubMed] [Google Scholar]
- He G, Margolis DM (2002) Counterregulation of chromatin deacetylation and histone deacetylase occupancy at the integrated promoter of human immunodeficiency virus type 1 (HIV-1) by the HIV-1 repressor YY1 and HIV-1 activator Tat. Mol Cell Biol 22: 2965–2973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hottiger MO, Felzien LK, Nabel GJ (1998) Modulation of cytokine-induced HIV gene expression by competitive binding of transcription factors to the coactivator p300. EMBO J 17: 3124–3134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsia SC, Shi YB (2002) Chromatin disruption and histone acetylation in regulation of the human immunodeficiency virus type 1 long terminal repeat by thyroid hormone receptor. Mol Cell Biol 22: 4043–4052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Zhao S, Ammanamanchi S, Brattain M, Venkatasubbarao K, Freeman JW (2005) Trichostatin A induces transforming growth factor beta type II receptor promoter activity and acetylation of Sp1 by recruitment of PCAF/p300 to a Sp1. NF-Y complex. J Biol Chem 280: 10047–10054 [DOI] [PubMed] [Google Scholar]
- Jordan A, Bisgrove D, Verdin E (2003) HIV reproducibly establishes a latent infection after acute infection of T cells in vitro. EMBO J 22: 1868–1877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiernan R, Bres V, Ng RW, Coudart MP, El Messaoudi S, Sardet C, Jin DY, Emiliani S, Benkirane M (2003) Post-activation turn-off of NF-kappa B-dependent transcription is regulated by acetylation of p65. J Biol Chem 278: 2758–2766 [DOI] [PubMed] [Google Scholar]
- Kiernan RE, Vanhulle C, Schiltz L, Adam E, Xiao H, Maudoux F, Calomme C, Burny A, Nakatani Y, Jeang KT, Benkirane M, Van Lint C (1999) HIV-1 tat transcriptional activity is regulated by acetylation. EMBO J 18: 6106–6118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YK, Bourgeois CF, Pearson R, Tyagi M, West MJ, Wong J, Wu SY, Chiang CM, Karn J (2006) Recruitment of TFIIH to the HIV LTR is a rate-limiting step in the emergence of HIV from latency. EMBO J 25: 3596–3604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose RJ, Yamane K, Bae Y, Zhang D, Erdjument-Bromage H, Tempst P, Wong J, Zhang Y (2006) The transcriptional repressor JHDM3A demethylates trimethyl histone H3 lysine 9 and lysine 36. Nature 442: 312–316 [DOI] [PubMed] [Google Scholar]
- Kulkosky J, Culnan DM, Roman J, Dornadula G, Schnell M, Boyd MR, Pomerantz RJ (2001) Prostratin: activation of latent HIV-1 expression suggests a potential inductive adjuvant therapy for HAART. Blood 98: 3006–3015 [DOI] [PubMed] [Google Scholar]
- Lachner M, O'Carroll D, Rea S, Mechtler K, Jenuwein T (2001) Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature 410: 116–120 [DOI] [PubMed] [Google Scholar]
- Lassen K, Han Y, Zhou Y, Siliciano J, Siliciano RF (2004) The multifactorial nature of HIV-1 latency. Trends Mol Med 10: 525–531 [DOI] [PubMed] [Google Scholar]
- Lehrman G, Hogue IB, Palmer S, Jennings C, Spina CA, Wiegand A, Landay AL, Coombs RW, Richman DD, Mellors JW, Coffin JM, Bosch RJ, Margolis DM (2005) Depletion of latent HIV-1 infection in vivo: a proof-of-concept study. Lancet 366: 549–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X, Irwin D, Kanazawa S, Huang L, Romeo J, Yen TS, Peterlin BM (2003) Transcriptional profiles of latent human immunodeficiency virus in infected individuals: effects of Tat on the host and reservoir. J Virol 77: 8227–8236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lusic M, Marcello A, Cereseto A, Giacca M (2003) Regulation of HIV-1 gene expression by histone acetylation and factor recruitment at the LTR promoter. EMBO J 22: 6550–6561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maison C, Almouzni G (2004) HP1 and the dynamics of heterochromatin maintenance. Nat Rev Mol Cell Biol 5: 296–304 [DOI] [PubMed] [Google Scholar]
- Marban C, Redel L, Suzanne S, Van Lint C, Lecestre D, Chasserot-Golaz S, Leid M, Aunis D, Schaeffer E, Rohr O (2005) COUP-TF interacting protein 2 represses the initial phase of HIV-1 gene transcription in human microglial cells. Nucleic Acids Res 33: 2318–2331 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Marcello A (2006) Latency: the hidden HIV-1 challenge. Retrovirology 3: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcello A, Lusic M, Pegoraro G, Pellegrini V, Beltram F, Giacca M (2004) Nuclear organization and the control of HIV-1 transcription. Gene 326: 1–11 [DOI] [PubMed] [Google Scholar]
- Marcello A, Zoppe M, Giacca M (2001) Multiple modes of transcriptional regulation by the HIV-1 Tat transactivator. IUBMB Life 51: 175–181 [DOI] [PubMed] [Google Scholar]
- Marzio G, Tyagi M, Gutierrez MI, Giacca M (1998) HIV-1 tat transactivator recruits p300 and CREB-binding protein histone acetyltransferases to the viral promoter. Proc Natl Acad Sci USA 95: 13519–13524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minc E, Courvalin JC, Buendia B (2000) HP1gamma associates with euchromatin and heterochromatin in mammalian nuclei and chromosomes. Cytogenet Cell Genet 90: 279–284 [DOI] [PubMed] [Google Scholar]
- Nakayama J, Rice JC, Strahl BD, Allis CD, Grewal SI (2001) Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science 292: 110–113 [DOI] [PubMed] [Google Scholar]
- Nielsen AL, Oulad-Abdelghani M, Ortiz JA, Remboutsika E, Chambon P, Losson R (2001) Heterochromatin formation in mammalian cells: interaction between histones and HP1 proteins. Mol Cell 7: 729–739 [DOI] [PubMed] [Google Scholar]
- Ott M, Dorr A, Hetzer-Egger C, Kaehlcke K, Schnolzer M, Henklein P, Cole P, Zhou MM, Verdin E (2004) Tat acetylation: a regulatory switch between early and late phases in HIV transcription elongation. Novartis Found Symp 259: 182–193; discussion 193–186, 223–185 [PubMed] [Google Scholar]
- Pagans S, Pedal A, North BJ, Kaehlcke K, Marshall BL, Dorr A, Hetzer-Egger C, Henklein P, Frye R, McBurney MW, Hruby H, Jung M, Verdin E, Ott M (2005) SIRT1 regulates HIV transcription via Tat deacetylation. PLoS Biol 3: e41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palliser D, Chowdhury D, Wang QY, Lee SJ, Bronson RT, Knipe DM, Lieberman J (2006) An siRNA-based microbicide protects mice from lethal herpes simplex virus 2 infection. Nature 439: 89–94 [DOI] [PubMed] [Google Scholar]
- Pomerantz RJ (2002) Eliminating HIV-1 reservoirs. Curr Opin Investig Drugs 3: 1133–1137 [PubMed] [Google Scholar]
- Quivy V, Adam E, Collette Y, Demonte D, Chariot A, Vanhulle C, Berkhout B, Castellano R, de Launoit Y, Burny A, Piette J, Bours V, Van Lint C (2002) Synergistic activation of human immunodeficiency virus type 1 promoter activity by NF-kappaB and inhibitors of deacetylases: potential perspectives for the development of therapeutic strategies. J Virol 76: 11091–11103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rea S, Eisenhaber F, O'Carroll D, Strahl BD, Sun ZW, Schmid M, Opravil S, Mechtler K, Ponting CP, Allis CD, Jenuwein T (2000) Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature 406: 593–599 [DOI] [PubMed] [Google Scholar]
- Schroder AR, Shinn P, Chen H, Berry C, Ecker JR, Bushman F (2002) HIV-1 integration in the human genome favors active genes and local hotspots. Cell 110: 521–529 [DOI] [PubMed] [Google Scholar]
- Sheridan PL, Mayall TP, Verdin E, Jones KA (1997) Histone acetyltransferases regulate HIV-1 enhancer activity in vitro. Genes Dev 11: 3327–3340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treand C, du Chene I, Bres V, Kiernan R, Benarous R, Benkirane M, Emiliani S (2006) Requirement for SWI/SNF chromatin-remodeling complex in Tat-mediated activation of the HIV-1 promoter. EMBO J 25: 1690–1699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Lint C, Emiliani S, Ott M, Verdin E (1996) Transcriptional activation and chromatin remodeling of the HIV-1 promoter in response to histone acetylation. EMBO J 15: 1112–1120 [PMC free article] [PubMed] [Google Scholar]
- Verdin E, Paras P Jr, Van Lint C (1993) Chromatin disruption in the promoter of human immunodeficiency virus type 1 during transcriptional activation. EMBO J 12: 3249–3259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberger LS, Burnett JC, Toettcher JE, Arkin AP, Schaffer DV (2005) Stochastic gene expression in a lentiviral positive-feedback loop: HIV-1 Tat fluctuations drive phenotypic diversity. Cell 122: 169–182 [DOI] [PubMed] [Google Scholar]
- Williams SA, Chen LF, Kwon H, Ruiz-Jarabo CM, Verdin E, Greene WC (2006) NF-kappaB p50 promotes HIV latency through HDAC recruitment and repression of transcriptional initiation. EMBO J 25: 139–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamane K, Toumazou C, Tsukada Y, Erdjument-Bromage H, Tempst P, Wong J, Zhang Y (2006) JHDM2A, a JmjC-containing H3K9 demethylase, facilitates transcription activation by androgen receptor. Cell 125: 483–495 [DOI] [PubMed] [Google Scholar]
- Yedavalli VS, Benkirane M, Jeang KT (2003) Tat and trans-activation-responsive (TAR) RNA-independent induction of HIV-1 long terminal repeat by human and murine cyclin T1 requires Sp1. J Biol Chem 278: 6404–6410 [DOI] [PubMed] [Google Scholar]
- Zufferey R, Dull T, Mandel RJ, Bukovsky A, Quiroz D, Naldini L, Trono D (1998) Self-inactivating lentivirus vector for safe and efficient in vivo gene delivery. J Virol 72: 9873–9880 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Supplementary Figure S4
Supplementary Figure S5
Supplementary Figure S6
Supplementary Figure S7
Supplementary Figure S8
Supplementary Figure S9
Supplementary Materials


