Abstract
In Drosophila, the partition defective (Par) complex containing Par3, Par6 and atypical protein kinase C (aPKC) directs the polarized distribution and unequal segregation of the cell fate determinant Numb during asymmetric cell divisions. Unequal segregation of mammalian Numb has also been observed, but the factors involved are unknown. Here, we identify in vivo phosphorylation sites of mammalian Numb and show that both mammalian and Drosophila Numb interact with, and are substrates for aPKC in vitro. A form of mammalian Numb lacking two protein kinase C (PKC) phosphorylation sites (Numb2A) accumulates at the cell membrane and is refractory to PKC activation. In epithelial cells, mammalian Numb localizes to the basolateral membrane and is excluded from the apical domain, which accumulates aPKC. In contrast, Numb2A is distributed uniformly around the cell cortex. Mutational analysis of conserved aPKC phosphorylation sites in Drosophila Numb suggests that phosphorylation contributes to asymmetric localization of Numb, opposite to aPKC in dividing sensory organ precursor cells. These results suggest a model in which phosphorylation of Numb by aPKC regulates its polarized distribution in epithelial cells as well as during asymmetric cell divisions.
Keywords: asymmetric cell division, epithelial cells, Numb, phosphorylation, PKC
Introduction
Asymmetric cell division is a conserved mechanism used to generate cell diversity (Betschinger and Knoblich, 2004; Roegiers and Jan, 2004). Central to this process is cell polarization and the segregation of cell-fate determinants that influence developmental potential in progeny cells. The establishment of cell polarity depends on a conserved protein complex containing the partition defective (Par) proteins, Par3 and Par6, as well as atypical protein kinase C (aPKC) (Henrique and Schweisguth, 2003; Macara, 2004). This complex is conserved among metazoa and functions in the polarization of epithelial and neuronal cells, in oriented cell migration and in asymmetric cell division (Nelson, 2003).
In Drosophila, the unequal distribution of the cell-fate determinant Numb plays an essential role in binary cell-fate decisions, following asymmetric cell divisions in the developing nervous system (Rhyu et al, 1994; Betschinger and Knoblich, 2004; Roegiers and Jan, 2004). From prophase onwards, Numb becomes localized to a crescent at the anterior lateral cortex of sensory organ precursor (pI) cells that divide asymmetrically in pupal nota, to generate the Numb-inheriting pIIb cell and a pIIa cell. The Bazooka (Par3)-Par6-aPKC complex localizes opposite to Numb at the posterior lateral cortex and is required to direct the anterior localization of cell-fate determinants including Numb (Bellaiche et al, 2001b; Roegiers et al, 2001a; Betschinger et al, 2003; Rolls et al, 2003). The Par complex acts, at least in part, via the aPKC-dependent phosphorylation of Lethal(2) giant larvae (Lgl) (Betschinger et al, 2003). Additional yet unknown mechanisms are predicted to regulate the polarized distribution of Numb as asymmetric localization of Numb in dividing pI cells can be observed in the complete absence of lgl activity (Justice et al, 2003; Langevin et al, 2005).
In mammals, Numb is essential for normal development (Zhong et al, 2000; Zilian et al, 2001). Asymmetric segregation of Numb has been observed in dividing cells of the ventricular zone as well as in retinal progenitors and isolated cortical progenitors supporting a conserved role in asymmetric cell division and cell-fate determination (Zhong et al, 1996; Cayouette et al, 2001; Shen et al, 2002). In polarized epithelial cells Numb localization is restricted to the basolateral membrane suggesting that cell polarity may also influence the distribution of mammalian Numb (Dho et al, 2006). However, the mechanisms which regulate Numb localization in mammalian cells are unknown. Here, we show that both mammalian and Drosophila Numb are direct substrates of aPKC, and demonstrate that Numb phosphorylation serves as a conserved mechanism to regulate its asymmetric localization.
Results
Mammalian Numb is phosphorylated by aPKC
Recently, we demonstrated that cortical membrane localization of mammalian Numb is dynamically regulated in response to activation of G protein-coupled receptors and protein kinase C (PKC)-dependent signaling pathways (Dho et al, 2006). These studies suggested that Numb might be a direct target of serine/threonine protein kinases in mammalian cells. To directly test whether Numb is phosphorylated in vivo, we labeled HEK293T cells with 32P-labeled orthophosphate and immunoprecipitated endogenous Numb. Immunoprecipitates resolved by SDS–PAGE and subjected to autoradiography revealed that Numb is a phosphoprotein (Figure 1A). Treatment of cells with TPA, which activates classical and novel PKC isozymes, caused a two-fold increase in 32P-labeled incorporation. The increase in phosphorylation observed following TPA treatment suggested that Numb might be a substrate for a member of the PKC family. To test whether Numb was a direct substrate of one or more PKC isozymes, we conducted in vitro kinase assays. Numb immunoprecipitates were incubated with γ-32P-labeled ATP and recombinant active PKC isozymes from all three subfamilies (Figure 1B). Ten percent of the total in vitro kinase reaction mixture was resolved by SDS–PAGE and visualized by autoradiography. All of the PKC isoforms tested showed activity in the in vitro reactions. An immunokinase reaction with no exogenously added PKC was also developed to exclude the possibility that the kinase activity detected came from a co-precipitating kinase (Figure 1B upper). To resolve the Numb-specific bands from the reaction mixture, we boiled and denatured 90% of the reaction and re-immunoprecipitated with anti-Numb antibodies (Figure 1B, middle). A doublet, representing the protein isoforms of Numb (p65/p66 and p71/p72) was observed. Numb proteins were most efficiently phosphorylated by PKCα and by the atypical PKCζ isozymes, but several other PKC family members had significant activity against Numb in this assay. The membrane was blotted with anti-Numb to confirm the identity of the phosphorylated bands (Figure 1B, lower).
Figure 1.
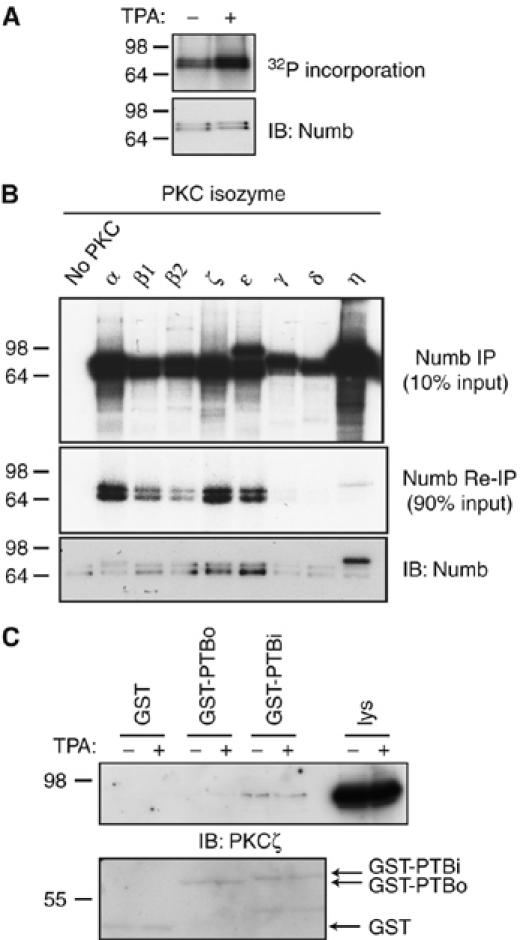
Numb is phosphorylated in vivo. (A) Autoradiograph of Numb immunoprecipitates from 32P-labeled orthophosphate-labeled HEK293T cells treated with the phorbol ester TPA. Equal loading confirmed by Numb immunoblotting (lower panel). (B) Numb immunoprecipitates were incubated with [γ-32P]ATP and either no PKC, or with the indicated recombinant PKC isozymes. Ten percent of the kinase reaction was subjected to autoradiography to confirm activity (upper panel). Ninety percent of each kinase reaction was boiled, diluted, and Numb was re-immunoprecipitated and subjected to autoradiography (middle panel). Immunoblotting with anti-Numb antibody confirmed the presence of Numb protein in each reaction (lower panel). Note that the band migrating just above Numb in the PKCη reaction likely represents 32P-labeled autophosphorylated PKCη that was not removed by the denaturing immunoprecipitation step. (C) GST-Numb PTBi and not GST-Numb PTBo or GST alone bound to endogenous PKCζ from MDCK cells lysates (upper panel). Equivalent input of GST-PTB fusion proteins was confirmed by Coomassie blue staining of membrane.
In Caenorhabditis elegans, the putative Numb ortholog, CKA1, was isolated in a yeast two-hybrid screen as a binding partner of the aPKC, PKC3 (Zhang et al, 2001). Furthermore, the interaction with PKC3 was shown to be mediated by the CKA1 PTB domain. Mammalian Numb protein isoforms contain two variant forms of the PTB domain. The PTBi variant contains a lysine-rich 11 amino-acid insert compared to the PTBo variant, and the protein isoforms that include PTBi (p72 and p66) are predominantly localized to the membrane cortex, whereas forms containing the PTBo domain (p65 and p71) are cytosolic (Dho et al, 1999). To examine whether mammalian Numb interacts with PKC, we performed in vitro binding experiments. GST fusion proteins of the Numb PTB domain were incubated with lysates from MDCK cells, which express both the atypical PKC isoforms λ and ζ as well as PKCα (data not shown). Binding of atypical PKC to GST-Numb-PTBi, but not GST alone or a GST fusion of the splice variant form, PTBo (Figure 1C), was observed by immunoblotting with an antibody that detects both isoforms. No binding of GST-PTBi or PTBo to PKCα was observed (data not shown). This result suggests that the Numb–PKCζ interaction is selective for the membrane-localized forms of Numb. We cannot exclude the possibility that Numb proteins bind to other PKC isoforms in a manner not detectable by this approach. As Numb is phosphorylated in vivo in response to TPA treatment, and is also phosphorylated in vitro, by classical and novel PKC family members, Numb may be coupled in vivo to several distinct PKC isozymes.
Previously, we observed that treatment of HeLa cells with TPA caused a rapid loss of Numb from the cortical membrane, implying that activation of classical or novel PKC alters the subcellular distribution of Numb (Dho et al, 2006). To test whether atypical PKC activation might also cause changes in Numb localization, we transfected HeLa cells with a constitutively active (CA) or dominant negative (DN) form of PKCζ. Expression of a CA PKCζ caused a redistribution of endogenous Numb from cortical membrane puncta to the cytoplasm (Figure 2A). In contrast, expression of a DN PKCζ did not induce relocalization to the cytosol (Figure 2B). In support of these observations, expression of CA PKCζ also resulted in an increase in the amount of Numb recovered from the Triton-X 100 soluble fraction (S), and a decrease in the Triton-insoluble fraction (P) of HeLa cell lysates compared to control cells (Figure 2C). Expression of DN PKCζ had no effect on the amount of Numb in the Triton soluble or insoluble fractions (Supplementary Figure 1). These data indicate that Numb is a substrate for PKC, and that phosphorylation may play a role in regulating its membrane localization.
Figure 2.
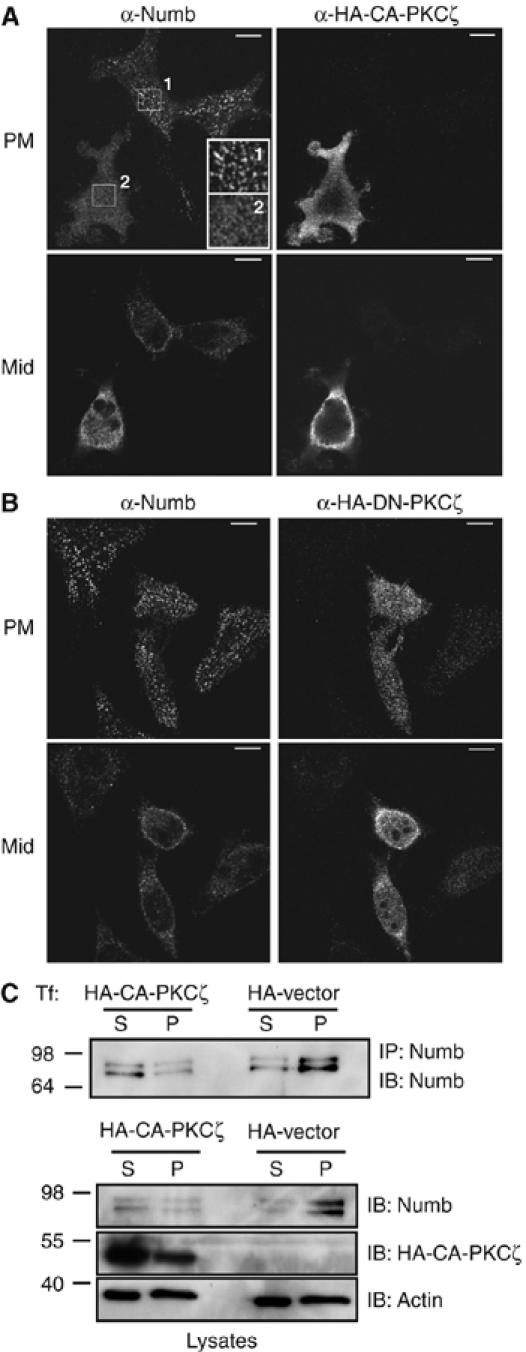
Expression of CA PKCζ causes redistribution of membrane-associated Numb to the cytoplasm. (A) Cortical membrane localization of endogenous Numb is lost in cells expressing CA PKCζ (HA-CA-PKCζ) compared to untransfected cells (upper left panels). Boxed regions have been digitally enlarged to show plasma membrane puncta staining of Numb in an untransfected cell (inset box 1) compared to a HA-CA-PKCζ-transfected cell (inset box 2). Numb cytosolic staining is increased in the presence of HA-CA-PKCζ (lower left panel). (B) Transfection of HeLa cells with DN PKCζ (HA-DN-PKCζ) had no effect on plasma membrane Numb localization. Size bars represent 10 μm. (C) Biochemical fractionation of HeLa cells transfected with HA-CA-PKCζ or HA vector. Numb immunoprecipitations were performed on the triton soluble fraction (S) and triton insoluble pellet (P) and immunoblotted with anti-Numb antibody (upper panel). Twenty μg of protein from each fraction was separated by SDS–PAGE and blotted with anti-Numb, anti-PKCζ to confirm expression levels, and anti-actin to confirm equal protein loading (lower panels).
Phosphorylation of Numb at serine residues 7 and 295 regulates localization to the cortical membrane
Analysis of the mammalian Numb amino-acid sequence using Netphos (http://www.cbs.dtu.dk/services/NetPhos) identified 40 potential phosphorylation sites. Phospho-tryptic peptide mapping of Numb, which was immunoprecipitated from 32P-labeled labeled HEK293T cells revealed five phosphopeptides (Figure 3A). Treatment of cells with TPA did not change the number or pattern of tryptic peptides observed indicating that Numb is basally phosphorylated on multiple sites and that acute stimulation does not induce phosphorylation on new sites. Numb was immunoprecipitated from untreated or TPA-treated cells and subjected to mass spectrometry analysis to map individual sites of phosphorylation. A total of nine phosphorylation sites were identified including serine residues (Ser) 7 and Ser295 (Figure 3B). Both of these sites conform to the consensus site for PKC phosphorylation ((R/K)1−3-X-(S/T)-X-(R/K)1−3) and are conserved across species (Figure 3C). In particular, the site corresponding to Ser7 in C. elegans CKA1 (Ser65) was identified by Zhang et al (2001) as a phosphorylation site for PKC3. As well, Ser295 lies within a serine- and threonine-rich region of Numb that we previously demonstrated was required for Numb mobilization in response to TPA stimulation or GPCR activation (Dho et al, 2006). Within this region, we also identified phosphorylation of Ser276, which was previously shown to be phosphorylated by Ca2+/calmodulin-dependent protein kinase I (CaM-KI) in vitro ((Tokumitsu et al, 2005) and our unpublished data).
Figure 3.
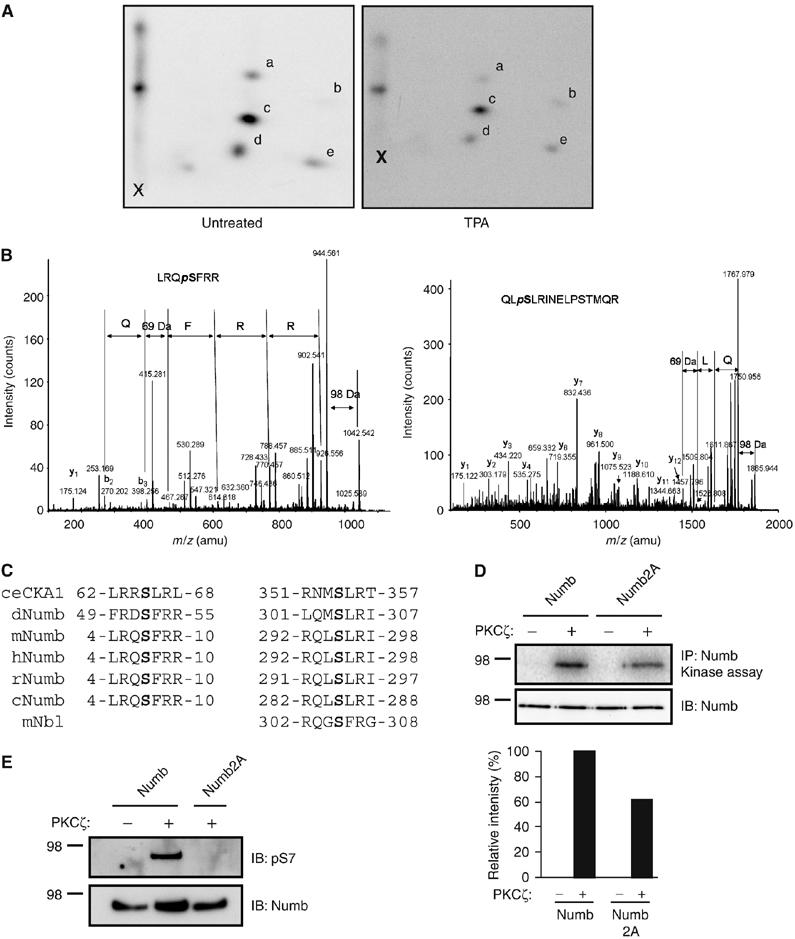
Identification of mammalian Numb phosphorylation sites by phosphopeptide mapping and mass spectrometry. (A) Numb was immunoprecipitated from 32P orthophosphate-labeled, untreated or TPA-treated HEK293T cells and resolved by SDS–PAGE and autoradiography. Bands corresponding to radiolabeled Numb (confirmed by parallel immunoblot) were excised from the membrane and tryptic fragments were purified and resolved by two-dimensional thin layer electrophoresis and ascending chromatography. Five phosphopeptides were recovered in samples from untreated (left panel) or TPA-treated cells (right panel; spots a–e). (B) Mass spectrometry analysis of Numb proteins purified by immunoprecipitation. MS/MS spectra of peptides LRQSFRR, and QLSLRINELPSTMQR indicating phosphorylated residues Ser7 (left) and Ser295 (right) are shown. (C) Sequence conservation surrounding Ser7 and Ser295 in Numb protein sequence alignment from: C. elegans (ceCKA1; Q9XTY6), Drosophilia melanogaster (dNumb; P16554), mouse (mNumb; AAD47836), human (hNumb; NP001005744), rat (rNumb; ABC69734), chicken (cNumb; AAD49434), and related mouse Numblike protein (mNbl; NP035080). (D) HeLa cells were transfected with HA-tagged Numb or Numb2A, and immunoprecipitated Numb subjected to in vitro kinase reactions with PKCζ as described in Figure 1B. Immunoblotting with anti-Numb antibodies revealed equivalent loading in all lanes (lower panel). Storm Phosphoimager analysis was used to detect 32P-labeled incorporation, and analysis performed using ImageQuant 5.0 software. A histogram of 32P-labeled incorporation from a representative experiment of four independent repeats is shown. (E) Immunoprecipitated Numb or Numb2A subjected to kinase assays with PKCζ as described in Figure 1B (in the presence of cold ATP), resolved by SDS–PAGE and immunoblotted with affinity-purified anti-pS7 antibody (upper panel) or anti-Numb (lower panel).
As both serines 7 and 295 matched conserved PKC consensus phosphorylation sites, we chose to further evaluate the function of these two residues by creating mutant forms of Numb in which Ser7 and Ser295 were changed to alanine independently and in combination. We chose the p66 Numb isoform for this analysis because it is abundantly expressed in most mammalian cell types and has previously been characterized with respect to function and subcellular localization (Dho et al, 1999, 2006; McGill and McGlade, 2003; Smith et al, 2004). The phospho-site mutants were expressed in HEK293T cells and subjected to phospho-tryptic peptide mapping. The tryptic peptide profile of the S7A and S295A mutants revealed the loss of a single phosphopeptide spot (Supplementary Figure 2A; spot b and a, respectively) and both spots were lost in the double mutant, Numb2A, independently confirming that Ser7 and Ser295 are phosphorylated in vivo. To assess whether these sites are directly phosphorylated by PKCζ, we performed an in vitro kinase assay on HA-tagged wild type or Numb2A immunoprecipitated from HeLa cells. In a representative experiment, incorporation of 32P into Numb2A was reduced by 39% compared to wild-type Numb, indicating that one or both of these sites is phosphorylated by PKCζ in vitro (Figure 3D). In contrast, phosphorylation of Numb2A by PKCα was reduced by only 11% in a representative experiment (Supplementary Figure 2B). Numb2A retains four additional serine residues that conform to the PKC consensus sequence, and could account for the residual in vitro phosphorylation of Numb2A. However, these data suggest that Ser7 and Ser295 are preferred sites for phosphorylation by PKCζ.
A Numb antibody that recognizes phosphorylated serine 7 was generated and used in Western blots of Numb immunoprecipitates subjected to in vitro kinase reactions with PKCζ in the absence of 32P-labeled ATP. Anti-pS7 recognized wild-type Numb that had been incubated with PKCζ in vitro, but not Numb2A, or wild-type Numb incubated in a control reaction without PKCζ (Figure 3E), indicating that serine 7 is a direct target of PKCζ in vitro.
In HeLa cells, Numb is normally localized to both the cortical membrane and intracellular vesicles (Santolini et al, 2000; Smith et al, 2004; Dho et al, 2006). Numb interacts with and colocalizes at the plasma membrane with the α-adaptin subunit of AP2 (Santolini et al, 2000; Dho et al, 2006). Consistent with this, GFP-Numb expressed in HeLa cells localizes to both the cortical membrane with α-adaptin, and within the cytosol (Figure 4A, left). In contrast, the double phospho-site mutant (GFP-Numb2A) appeared to localize almost exclusively to cortical membrane patches where it also colocalized with AP2 (Figure 4A, right). Mean GFP fluorescence intensity measurements at the membrane and in the cytosol confirmed a significant difference between the membrane to cytosolic ratio (m:c) of wild-type GFP-Numb (m:c=1.5) and GFP-Numb2A (m:c=2.4; n=20, P=8 × 10−7). The single phospho-site mutants exhibited modest changes in membrane localization compared to GFP-Numb (Supplementary Figure 2C). In addition, we also analyzed the extent of colocalization of GFP-Numb or GFP-Numb2A with AP2. There was no significant difference between the mean Pearson's coefficients for GFP-Numb (0.881) and GFP-Numb2A (0.899; n=10, P=0.3), suggesting that both wild type and Numb2A are able to associate with the AP2 complex. Together, these results indicate that phosphorylation of Numb regulates its subcellular localization.
Figure 4.

Numb phosphorylation at Ser7 and Ser295 regulates cortical membrane localization. (A) HeLa cells were transfected with either GFP-Numb (left panels) or GFP-Numb2A (right panels), and immunostained with anti-α-adaptin. GFP-Numb is localized at the basal substratum-associated membrane with α-adaptin (left panels, PM) and to cytoplasmic vesicles in optical sections taken at the midpoint through the cell (left panels, Mid). GFP-Numb2A was predominantly localized at the cortical membrane (right panels, PM) and little cytosolic/vesicle-associated localization is observed (right panels, Mid). The relative association of GFP-Numb and GFP-Numb2A at the plasma membrane and in the cytosol was determined by calculating a ratio of the mean GFP fluorescence intensity using Improvision Volocity 3.7 software program. Values are given in the text. Bottom panels show merged images of the boxed regions that have been digitally enlarged. (B) Coexpression of CA PKCζ (HA-CA-PKCζ) resulted in loss of GFP-Numb at the cortical membrane (left panels, PM), and increased cytosolic staining (left panels, mid). HA-CA-PKCζ expression has no effect on GFP-Numb2A plasma membrane localization (right panels). Boxed regions have been digitally enlarged and represented as panels below. Size bars represent 10 μm.
Expression of CA aPKCζ (CA-PKCζ) relocalizes endogenous Numb from the plasma membrane to the cytosolic compartment (Figure 2A). To determine whether this effect requires Numb phosphorylation at Ser7 and/or Ser295, we examined the consequence of expression of CA-PKCζ on the localization of GFP-Numb2A. Coexpression of CA-PKCζ resulted in the loss of GFP-Numb from the cortical membrane, and in its accumulation in the cytosol (Figure 4B, left). In contrast, CA-PKCζ expression had no effect on the localization of GFP-Numb2A, which accumulated at the plasma membrane (Figure 4B, right). In addition, live cell imaging demonstrated that while GFP-Numb is rapidly lost from the plasma membrane following the addition of TPA, GFP-Numb2A is insensitive to TPA treatment (Supplementary Figure 2D and Supplementary movie 1 and 2). Therefore, the redistribution of Numb from the cortical membrane to the cytosol in response to PKC activation appears to be mediated by phosphorylation of Ser7 and 295.
Mammalian Numb localization is restricted to the basolateral membrane in polarized MDCK cells
aPKC is an integral part of the conserved polarity complex containing Par3 and Par6, which resides at the tight junction of mammalian epithelial cells and plays a conserved role in establishing cell polarity (Macara 2004). Previously, we have shown that Numb is localized to the basolateral membrane of polarized MDCK cells (Dho et al, 2006). To investigate whether the Par/aPKC protein complex might influence the distribution of mammalian Numb in MDCK cells, we examined the subcellular distribution of Numb in relation to that of aPKC and Par3 (Figure 5A) in optical sections (X–Y) of polarized MDCK cells acquired at 1-μm intervals. Figure 5A shows representative images taken in the plane of the cell where aPKC and Par3 are concentrated at the membrane (Figure 5A schematic, X–Y section I). In this region of the cell, cortical membrane localization of Numb is not observed (Figure 5A). In optical sections 2–4 μm below (Figure 5A schematic, X–Y section II), where distinct membrane staining of aPKC and Par3 is not observed, Numb is concentrated at the lateral membrane (Figure 5A, II).
Figure 5.
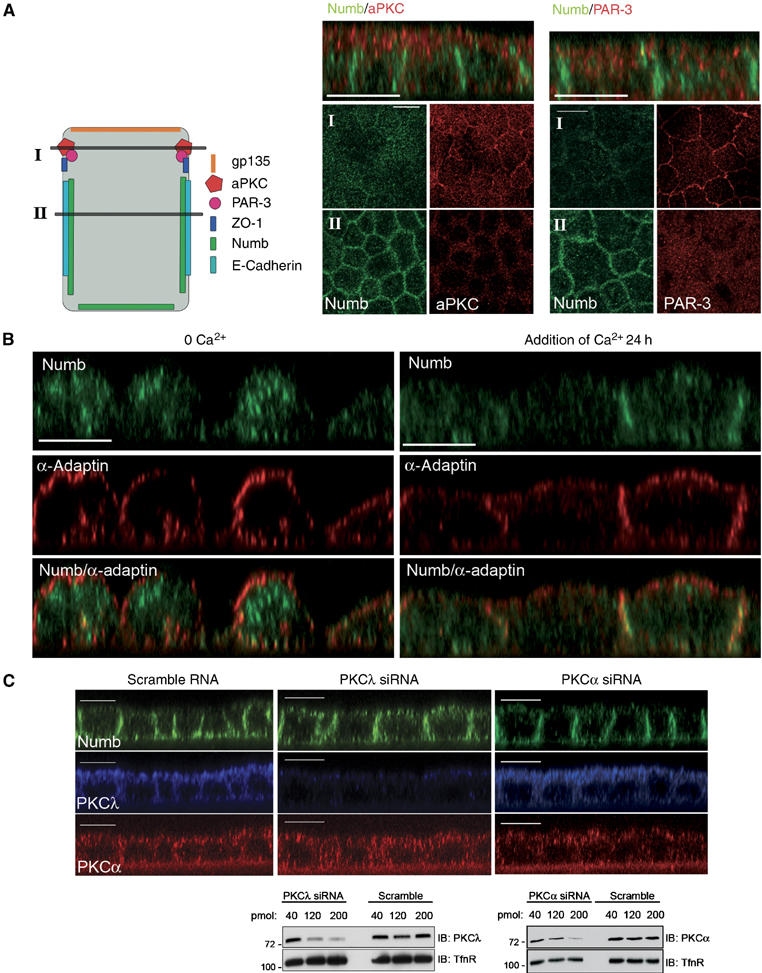
Cell polarity determines Numb localization in MDCK cells. (A) Polarized MDCK cells were fixed and co-stained with anti-NumbC (green) and either anti-aPKC or anti-Par3 (red). Shown are confocal X–Y sections taken at the subapical region of the cells (I) and 2–4 μm below at the basolateral region (II) as depicted in the diagramatic representation of the cells to the left of the images. Accumulation of Numb at the lateral membranes, below the subapical aPKC and Par3 staining, is shown in the Z sections shown above. (B) Polarized MDCK cells were grown in low calcium medium for 20 h (calcium switch) followed by re-addition of normal media for 24 h. Fixed cells were co-stained with anti-NumbC (green) and anti-α-adaptin (red). (C) MDCK cells were treated with 200 pmol aPKCλ or PKCα siRNA oligonucleotides and grown on filters for 72 h. Following fixation and permeabilization, the monolayers were co-stained with anti-aPKC (which also recognizes aPKCλ; blue), anti-PKCα (red), and anti-NumbC (green). Size bars indicate 10 μm. In lower panel MDCK cells were transfected with 40, 120, or 200 pmol of aPKCλ or PKCα siRNA oligonucleotides, or scrambled RNA control as indicated. In total, 20 μg of protein lysate for each condition was separated by SDS–PAGE and blotted with anti-aPKC or anti-PKCα. Membranes were reprobed with anti-Transferrin receptor as a loading control.
To test whether the establishment of cell polarity affects Numb localization, we performed Ca2+ switch experiments. Removal of Ca2+ induces the dissolution of adherens junctions, apical protein complexes, and consequently the depolarization of MDCK cells. In Ca2+-depleted cells, both Numb and the Numb-associated protein α-adaptin are observed in puncta uniformly distributed around the cortical membrane. Numb is also observed on cytoplasmic vesicles (Figure 5B, left panels). Addition of Ca2+ resulted in the redistribution of Numb to the cortical membrane and subsequent concentration at the lateral membrane after 24 h (Figure 5B, right panels), while the Numb-associated protein α-adaptin remains largely distributed around the cortical membrane. These results suggest that the establishment of cell polarity in MDCK cells has a role in directing the polarized distribution of Numb.
aPKC activity is important for the establishment of the apical and basolateral domains in MDCK cells, and for the lateral membrane localization of proteins such as Lgl and Par1 (Yamanaka et al, 2003; Suzuki et al, 2004). Therefore, we investigated the involvement of aPKC in regulating the localization of Numb. We used siRNA to deplete cells of PKCλ, the predominant isoform of aPKC expressed in MDCK cells, as described previously (Suzuki et al, 2004). Knockdown of PKCλ resulted in a disruption of endogenous Numb localization (Figure 5C, middle panels) and accumulation in the apical membrane region when compared to control scrambled RNAs (Figure 5C, left panels). The localization of the lateral membrane marker, E-cadherin, was minimally disrupted by PKCλ depletion (Supplementary Figure 3). Depletion of PKCα by siRNA did not affect Numb or E-cadherin localization in MDCK cells (Figure 5C, right panels, and Supplementary Figure 3). The efficiency of PKC knockdown was confirmed by Western blotting (Figure 5C lower panels). These results further support the notion that the activity of aPKC and the establishment of cell polarity regulates the basolateral membrane localization of Numb.
Polarized distribution of mammalian Numb in epithelial cells is regulated by phosphorylation
To test the effects of phosphorylation of Numb at serine 7 and 295 on basolateral localization in polarized cells, we expressed GFP-Numb or GFP-Numb2A in MDCK cells. Similar to endogenous Numb, GFP-Numb was predominantly localized to the lateral membrane, below the level of the tight junctions (Figure 6A, optical section II). GFP-Numb also appeared to localize to the apical domain in 39% (n=57) of cells, perhaps as a consequence of overexpression (Supplementary Figure 4). In the region where aPKC and ZO-1 were concentrated at the membrane, GFP-Numb was mainly cytoplasmic (Figure 6A, optical section I). A similar pattern of GFP-Numb distribution was also observed relative to Par3 and ZO-1 (Supplementary Figure 5A). In contrast, in 95% of expressing cells (n=40), GFP-Numb2A was concentrated at the membrane in both the lateral and apical domains, coincident with aPKC and ZO-1 membrane localization (Figure 6B, and Supplementary Figures 4 and 5B). The single site mutants, GFP-NumbS7A and GFP-NumbS295A, were also mislocalized, but to a lesser degree (Supplementary Figure 4).
Figure 6.

Phosphorylation at Ser7 and 295 is required for lateral membrane localization of Numb. GFP-Numb2A localization overlaps with aPKC at the apical region of the cell. Polarized MDCK cells transfected with GFP-Numb (A) or GFP-Numb2A (B) were fixed and co-stained with anti-aPKC (red) and anti-ZO-1 (blue). Confocal X–Y sections were taken as indicated in Figure 5 to show the subapical localization of aPKC (I) and the basolateral accumulation of Numb (II). Optical sections taken in the Z-axis are shown above. (C) MDCK cells transfected with GFP-Numb or GFP-Numb2A were stained anti-gp135 to mark the apical membrane. Merged images of GFP-Numb or GFP-Numb2A (green), and gp135 (red) staining in optical sections taken in the Z-axis are shown. Size bars indicate 10 μm.
To further define the apical membrane domain we stained GFP-Numb- and GFP-Numb2A-expressing cells with the apical marker gp135 (Ojakian and Schwimmer, 1988). Both gp135 and GFP-Numb2A were observed in patches or puncta in the apical membrane region that appeared to partially overlap while wild-type GFP-Numb did not colocalize (Figure 6C). These results suggest that phosphorylation of Numb at Ser7 and 295 is required to restrict its localization to the lateral membrane of polarized MDCK cells.
To examine whether phosphorylation mediates the dynamic regulation of Numb localization in response to epithelial cell polarization, we performed a Ca2+ switch experiment in MDCK cells expressing GFP-Numb and GFP-Numb2A. Upon Ca2+ depletion, MDCK cells depolarized and both wild type and Numb2A distributed over the entire membrane and into cytosolic vesicles (Figure 7A and B, panels labeled 0 Ca2+). Within 24 h after addition of Ca2+, GFP-Numb was concentrated at the lateral membrane and at regions of contact between cells (Figure 7A, upper panels), and was mainly cytosolic at the level of aPKC, Par3, and ZO-1 (Figure 7A and Supplementary Figure 5C). In contrast, GFP-Numb2A remained uniformly distributed around the cell cortex (Figure 7B and Supplementary Figure 5D). To further assess the distribution of wild-type GFP-Numb and GFP-Numb2A following Ca2+ addition, we used the apical marker gp135. This analysis revealed minimal colocalization between gp135 and wild-type GFP-Numb (mean Pearson's coefficient=0.138), while colocalization of gp135 with GFP-Numb2A was significantly higher (mean Pearson's coefficient=0.58; n-25, P=1.7 × 10−11; Figure 7C). These data demonstrate that phosphorylation of Numb not only regulates its localization to the cortical membrane, but also that, as a consequence of the activity of the Par polarity complex, aPKC-dependent phosphorylation may restrict Numb localization to the lateral domain during cell polarization.
Figure 7.
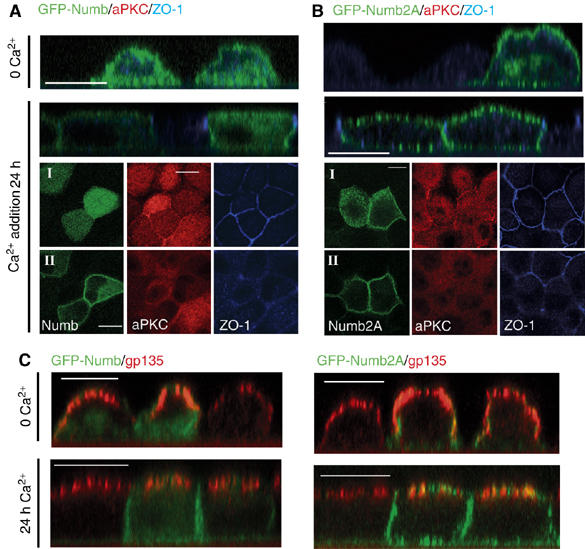
Phosphorylation is required for Numb localization to the basolateral membrane in response to cell polarization. Polarized MDCK cells transfected with GFP-Numb (A) or GFP-Numb2A (B) were switched to low-calcium media (0 Ca2+) for 20 h followed by Ca2+ re-addition for 24 h to allow for repolarization. Changes in the localization of GFP-Numb, aPKC, and ZO-1 were determined by immunostaining as in Figure 6 (A, B). (C) Colocalization of Numb with the apical membrane marker gp135 in a calcium switch experiment was determined as in Figure 6. Merged images of GFP-Numb or GFP-Numb2A (green), and gp135 (red) staining in optical sections taken in the Z-axis are shown. Colocalization of GFP-Numb (left panels) and GFP-Numb2A (right panels) with gp135 after calcium readdition was quantified using the colocalization module of Improvision Volocity 3.7 software program. Values are given in the text. Size bars indicate 10 μm.
Conservation of aPKC phosphorylation sites in Drosophila Numb
To establish whether aPKC-dependent phosphorylation is a conserved mechanism for regulating the cortical membrane localization of Numb, we first examined whether a myc-tagged version of Drosophila Numb forms a complex with PKCζ in HEK293 cells. Co-immunoprecipitation of Drosophila Numb with PKCζ indicates that this interaction is conserved (Figure 8A). We next analyzed the sequence of Drosophila Numb. A total of five evolutionarily conserved aPKC phosphorylation sites were revealed including Ser52 and Ser304, corresponding to Ser7 and Ser295 in murine Numb (isoform p66; Figure 3C). We tested whether PKC could phosphorylate Drosophila Numb in an in vitro kinase assay. Both PKCα and PKCζ, the human orthologue of Drosophila aPKC, phosphorylated Numb in an immune-complex assay (Figure 8B). PKCζ also phosphorylated a GST-Numb fusion protein (Figure 8C). Mutations of all five of the conserved aPKC sites (Numb5A) reduced the in vitro phosphorylation by PKCζ, indicating that some of these sites are the targets of PKCζ (Figure 8C). A form of Numb in which Ser52 is left intact, while the other four serines were mutated to alanine (Numb4A), was still efficiently phosphorylated by PKCζ indicating that Ser52 is one of the acceptor sites in vitro. However, mutation of Ser52 into alanine did not significantly reduce the in vitro phosphorylation of GST-Numb, suggesting that PKCζ phosphorylates additional sites (Figure 8C). NanoLC-MS–MS analyses of the in vitro phosphorylated GST-Numb identified a total of eight aPKC sites (in red in Figure 8E). As an example, Figure 8D shows the ion chromatogram of the doubly protonated peptide ions m/z 457.7 and m/z 497.7 for control and PKCζ-treated GST-Numb. Confirmation of the phosphorylated Ser52 residue was obtained from the MS–MS spectrum of m/z 497.7 (Figure 8D). Five PKCζ phosphorylation sites that do not appear conserved were identified in this analysis (Ser31, Ser35, Ser48, Ser161, and Ser297) (Figure 8E). These sites likely account for the residual phosphorylation of GST-Numb5A (Figure 8C). Although these analyses provide direct identification of aPKC phosphorylated residues, other potential phosphorylation sites remained elusive. For example, the early eluting tryptic peptide QMS304LR was observed only in the control sample. Its absence in the aPKC-treated sample strongly suggests that Ser304 (in blue in Figure 8E) is in fact phosphorylated and could not be detected owing to nonretention of this hydrophilic peptide during reverse phase LC. We conclude that aPKC phosphorylates Numb at several sites in both Drosophila and mouse, including at the conserved Ser7 and Ser295 sites (Ser52 and Ser304 in Drosophila Numb).
Figure 8.

Phosphorylation regulates asymmetric localization of Drosophila Numb. (A) Interaction of Drosophila Numb with PKCζ. Cell lysates obtained from HEK293 cells transfected with myc-tagged Numb or an empty control vector were immunoprecipitated with an anti-myc antibody (IP) and immunoprecipitates were immunoblotted (IB) with PKCζ antibody. Whole-cell lysates were analyzed by Western blotting (IB) to control for expression. (B) Autoradiography demonstrating that Drosophila Numb is an in vitro substrate for PKC isozymes alpha (α) and zeta (ζ) (upper panel). Immunoblot with anti-Numb antibodies confirms presence of Numb protein in each reaction (lower panel). (C) PKCζ phosphorylates Drosophila Numb on conserved serine residues in vitro. Right panel is a Coomassie-stained SDS–PAGE gel showing the purified protein samples used in the kinase assay. Autoradiography (left panel) of the same gel showing phosphorylation of GST-Numb but not GST alone. PKCζ also efficiently phosphorylates GST-Numb S52A and GST-Numb 4A, but only poorly phosphorylates GST-Numb 5A. The compared phosphorylation of GST-Numb 5A and GST-Numb 4A suggests that Ser52 is one of the site phosphorylated in vitro and that Ser52 is not the sole acceptor site. The asterisk marks autophosphorylated PKCζ. (D) Mass spectrometry analysis. Ion chromatograms for the doubly charged peptide SFRDSFR (m/z 457.7) and its corresponding phosphopeptide at Ser52 (m/z 497.7) obtained from the aPKC-treated (top left) and control (top right) GST-Numb tryptic digests. Middle panel shows the MS–MS spectrum of m/z 497.7 for the phosphopeptide SFRDpSFR with characteristic b- and y-type fragment ions, indicating the position of the modified residue at Ser52. (E) The full sequence of Drosophila Numb. The residues that are underlined correspond to the sequenced peptides (65% coverage). The phosphorylated sites are indicated in red. Indirect evidence suggest that Ser304 (in blue) is also phosphorylated (see text). (F) Localization of aPKC (blue) and myc-tagged Numb (green) in dividing pI cells (senseless in red). Numb localized at the anterior cortex of pI cells, whereas aPKC localized at the posterior cortex opposite to Numb at prometaphase and metaphase. Numb4A and NumbS52A also localized opposite to aPKC at the anterior cortex. In contrast, the distribution of Numb5A was not restricted to the anterior cortex.
Phosphorylation of Numb regulates asymmetric localization in dividing SOP cells
We next investigated the localization of Drosophila Numb in dividing sensory organ precursor pI cells. The pI cells divide asymmetrically within the plane of the notum epithelium and along the body axis. In these cells, Numb localizes at the anterior cortex, opposite to aPKC, which relocalizes from the apical cortex to the lateral posterior cortex upon mitosis (Bellaiche et al, 2001a; Roegiers et al, 2001b). We examined the possible role of phosphorylation in the regulation of Drosophila Numb localization by studying the distribution of Myc-tagged versions of Numb, Numb4A, NumbS52A, and Numb5A that were expressed in pI cells using the neurPGAL4 driver. Importantly, overexpression of Numb4A, NumbS52A, or Numb5A in pI cells led to cell-fate transformation in the bristle lineage indicative of gain of Numb function (data not shown). This indicates that these Numb mutant proteins are functional. Similar to endogenous Numb (Bellaiche et al, 2001b; Schaefer and Knoblich, 2001), Myc-Numb localized at the anterior cortex, opposite to aPKC, in all cells at prometaphase and metaphase (n=44; Figure 8F). Consistently, myc-Numb colocalized with Pins (Supplementary Figure 6). In contrast, the crescent formed by Numb5A appeared to extend posteriorly in 91% of the dividing pI cells at prometaphase (n=32; Figure 8F). Interestingly, a recent study has shown that a mutant Numb protein, NumbS52F, fails to localize properly in dividing pI cells (Bhalerao et al, 2005). Thus, one possible interpretation of our data is that the mislocalization of Numb5A is due to the S52A mutation. We therefore studied the localization of NumbS52A and found that it localized asymmetrically in 84% of the pI cells at prometaphase (n=50; Figure 8F). Thus, the S52A mutation alone cannot be responsible for the defective localization of Numb5A. Additionally, mutations of the four other serine residues in Numb4A did not significantly change the asymmetric distribution of Numb (n=27; Figure 8F). Therefore, we conclude that the defects in Numb5A distribution in dividing pI cells depends on the combination of at least two mutations, S52A and a mutation in one of the four conserved aPKC consensus sites, possibly Ser304. Thus, our data support the notion that aPKC-mediated phosphorylation of Drosophila Numb contributes to the asymmetric distribution of Numb in dividing pI cells.
Discussion
This study provides evidence for a conserved mechanism regulating the asymmetric distribution of the cell-fate determinant Numb. We demonstrate that mammalian and Drosophila Numb proteins are substrates for aPKC and that phosphorylation regulates Numb localization at the cortical membrane. Our data also indicate that aPKC-dependent phosphorylation regulates the polarized distribution of Numb in mammalian epithelial cells and Drosophila sensory organ precursor cells.
The aPKC/Par3/Par6 complex plays a conserved role in establishing polarity in a variety of cellular contexts, including during asymmetric cell divisions in C. elegans and Drosophila, and in apical–basal polarity of epithelial tissues. In mammalian epithelial cells, aPKC is required for the establishment and maintenance of apical–basal polarity (Suzuki et al, 2001; Suzuki and Ohno, 2006). In this context, several targets of aPKC have been identified, including the conserved proteins, Lgl and Par1, whose activities also contribute to cell polarity (Plant et al, 2003; Yamanaka et al, 2003; Hurov et al, 2004; Suzuki et al, 2004). In mammalian cells, Lgl plays a role in adherens junction disassembly and phosphorylation of Lgl by aPKC restricts its localization to the lateral cell membrane (Yamanaka et al, 2003, 2006). Similarily, aPKC-dependent phosphorylation of Par1 restricts its localization to the basolateral membrane of polarized MDCK cells (Suzuki et al, 2004). Our data indicate that Numb is also a downstream target of aPKC in polarized cells, and that phosphorylation at Ser7 and 295 mediates exclusion from the apical domain and accumulation at the lateral domain.
Previously, we and others have demonstrated a role for mammalian Numb in receptor endocytosis and recycling (Santolini et al, 2000; Nishimura et al, 2003; Smith et al, 2004; Hutterer and Knoblich, 2005). Our findings suggest that in polarized epithelial cells the trafficking function of Numb may be restricted to the basolateral membrane by aPKC-dependent phosphorylation. Thus, Numb may serve as a link between the Par/aPKC polarity complex and the endocytic machinery, and function to regulate the trafficking of membrane proteins at the basolateral membrane. In agreement with such a model, Numb has previously been implicated in the polarized endocytosis of the neuronal cell adhesion molecule L1 (Nishimura et al, 2003). Although the relevant membrane targets of Numb in epithelial cells are currently unknown, components of the Notch pathway are attractive candidates as Numb antagonizes Notch receptor signaling pathway in both Drosophila and in mammalian cells (Frise et al, 1996; Guo et al, 1996; Berezovska et al, 1999; French et al, 2002; McGill and McGlade, 2003).
In Drosophila, the Par complex has previously been shown to direct the asymmetric localization of Numb, Pon, and Miranda via the aPKC-mediated inhibitory phosphorylation of Lgl (Betschinger et al, 2003). However, Numb asymmetric localization could still be observed in 30% of lgl mutant pI cells (Justice et al, 2003; Langevin et al, 2005), suggesting that additional mechanisms may exist to regulate the asymmetric localization of Numb. Thus, we propose that the aPKC-dependent phosphorylation of Numb may account for the observed Lgl-independent asymmetric localization of Numb. This proposal implicitly assumes that this Lgl-independent process is aPKC-dependent. To verify this assumption, we have generated clones of apkc mutant cells. Unfortunately, large apkc mutant clones could not easily be recovered in the pupal notum, preventing us from studying the distribution of Numb in apkc mutant pI cells. A mutation in one of the Numb sites shown here to be phosphorylated by aPKC, Ser52, has previously been characterized (Bhalerao et al, 2005). The mutant protein, NumbS52F, fails to localize asymmetrically in pI at mitosis. The defective localization of NumbS52F contrasts with the asymmetric localization of NumbS52A. One possible interpretation is that the S52F, but not the S52A, mutation alters the conformation of Numb such that it prevents the phosphorylation of other essential aPKC sites or inhibits the actin-dependent cortical localization of Numb that is mediated by the N-terminal region of Numb (Knoblich et al, 1997).
In addition to the aPKC-dependent regulation of Numb localization, our results raise the possibility that a hierarchy of phosphorylation sites may be responsible for controlling additional aspects of Numb localization and function. In addition to serines 7 and 295, we have identified seven additional in vivo phosphorylation sites on mammalian Numb. Several of these do not conform to PKC consensus sites (unpublished data) yet are conserved in Drosophila. Recently, Tokumitsu et al (2005) described Ser276 as a target of CaMK, and we also identified this site in our mass spectral analysis (data not shown). Although the functional consequences of phosphorylation at this site were not addressed, the authors demonstrate that phosphorylation confers binding to 14-3-3 proteins suggesting this site has a regulatory role. In addition, the Drosophila Numb-associated kinase (NAK), which was isolated in a yeast two-hybrid screen as a Numb interactor (Chien et al, 1998), is highly related to mammalian adaptin-associated kinase (AAK) (Conner and Schmid, 2002), raising the possibility that members of this family of protein kinases might also phosphorylate Numb in a manner that regulates its association with α-adaptin or other endocytic proteins. Further functional analysis of Numb phosphorylation site mutants and identification of upstream kinases will yield insight into the conserved signaling pathways that regulate the localization and function of Numb and also will reveal areas of divergence.
Materials and methods
Immunoprecipitation and Western blotting
Cell lysates were prepared from transiently transfected cultured HeLa, HEK293T or MDCK cells, and immunoprecipitations were performed as described previously (Dho et al, 1999, Smith et al, 2004) The immune complexes were eluted in boiling SDS–Laemmli sample buffer. The proteins were separated by SDS–PAGE, transferred to PVDF membrane (Pall Corp) and incubated with primary antibodies for 1 h at room temperature. Immunoblots were developed using HRP-conjugated Protein A (BioRad) or goat anti-mouse HRP and ECL (Amersham). Preparation of Triton-X 100 soluble and insoluble cell extracts from confluent 10 cm2 culture plates of HeLa cells transfected with HA-CA-PKCζ, HA-DN- PKCζ, or empty vector was performed as by Dho et al (2006). Numb was immunoprecipitated from each fraction using anti-NumbC antibodies, separated by SDS–PAGE, and visualized by Western blotting and ECL.
In vitro kinase assays
Numb immunoprecipitates or purified GST-Numb fusion proteins were incubated for 15 min (30°C) in kinase buffer (25 mM Tris–HCl pH 7.5, 25 mM NaCl, 5 mM MgCl2, 1 mM DTT, 0.5 mM EGTA) containing 1 μCi γ-32P-labeled ATP and 15 ng of active PKC isozymes (Calbiochem). Reactions were stopped by addition of an equal volume of 2 × SDS–Laemmli sample buffer, boiled, and 10% resolved by SDS–PAGE followed by autoradiography. The remaining 90% was diluted with NP40 lysis buffer and re-immunoprecipitated with anti-NumbC antibodies, resolved by SDS–PAGE, transferred to PVDF and detected by autoradiography. To assess the in vitro phosphorylation of Drosophila Numb, HeLa-SPR cells were transfected with FLAG-Numb, lysed, and immunoprecipitated with anti-FLAG (Sigma), and kinase reactions performed as described above. Following autoradiography, anti-Numb Western blots were performed to confirm equal loading.
In vivo 32P-labeling and tryptic peptide analysis
HEK293T cells were labeled in 3 ml phosphate-free DMEM with 10% dialyzed FBS, and 0.5 mCi/ml of 32P-labeled orthophosphate for 4 h. Numb was immunoprecipitated with anti-NumbC antibodies and phospho-tryptic peptide analysis carried out as described by Boyle et al (1991) and in Supplementary Experimental Procedures.
Mass spectrometry
Details of the isolation of tryptic peptides and mass spectral analyses are given in Supplementary Experimental Procedures. Mass spectrometry analyses were performed in positive ion mode on a QStar XL mass spectrometer (Applied Biosystems/Sciex, Concord, Ontario, Canada) via a nanoelectrospray (nanoLC-MS for in vitro aPKC experiments) or a MALDI (gel band analyses). The data generated by MS and MS/MS experiments were processed by Bioanalyst software 1.1.5, and database searches were performed against the NCBI nonredundant protein database using Prospector MS-Fit (http://prospector.ucsf.edu) and Mascot (http://www.matrixscience.com). For data validation, identity of the peptide sequences and phosphorylation sites was manually interpreted on each MS/MS spectrum.
Immunocytochemistry
HeLa cell transfection has been described previously (Dho et al, 2006). MDCK cells were transfected at 75% confluency in six-well dishes using Lipofectamine 2000. Cells were trypsinized and seeded at high density on Costar cell culture inserts (0.4 μm pore) 4 h post transfection. For the Ca2+-switch experiments, MDCK cells were grown in Ca2+-free media (GIBCO S-MEM plus 2% dialyzed FBS) for 20 h. Normal Ca2+ was re-established by addition of DMEM plus 2% dialyzed FBS. Cells were fixed and stained as described by Dho et al (2006), and antibodies used are detailed in Supplementary Experimental Procedures. Confocal images were acquired using a Zeiss Axiovert 100 microscope (Carl Zeiss MicroImaging Inc.) with a 100 × oil-immersion objective (NA 1.5). Images were collected at 8-bit depth, with a resolution of 1024 × 1024 pixels. Live-imaging was performed as by Dho et al (2006). Additional details of image analysis can be found in Supplementary Experimental Procedures.
Pupal nota were dissected from staged pupae and fixed and stained as described by Bellaiche et al (2001a). Primary antibodies were rabbit anti-phospho-histone 3 (Upstate Biotechnology;1:1000), mouse anti-myc (9E10, obtained from DSHB; 1:500), rat anti-Pins (gift from C Doe; 1:500), and rabbit anti-aPKC (Santa Cruz Biotechnology: 1:500). The Cy3- and Cy5-coupled secondary antibodies were from Jackson's Laboratories and Alexa-488-coupled secondary antibodies were from Molecular Probes. Images were acquired on a Leica SP2 confocal microscope. All images were processed and assembled using LSM or ImageJ and Adobe Photoshop.
siRNA experiments
An siRNA oligonucleotide targeted against canine PKCλ (5′-AGTTCTGTTGGTGCGATTA-3′) has been described previously (Suzuki et al, 2004). The siRNA oligonucleotide used to target PKCα (5′-CTATGGCGTCCTATTATAT-3′) was designed using siDESIGN Center (Dharmacon). MDCK cells were seeded on six-well plates and transfected with 200 pmol siRNA duplex or scrambled RNA using Lipofectamine 2000 (Invitrogen). Cells were re-seeded on Falcon cell culture inserts (six-well format) 5 h post-transfection and cells were fixed and stained 72 h post-transfection.
Fly stocks
DNA encoding the Drosophila myc-tagged Numb4A, NumbS52A, and Numb5A were cloned into the pUAST vector. Transgenic flies were generated by DNA injection in w strains using the Δ2.3 helper plasmid. The UAS-Numb-myc line has been described previously (Yaich et al, 1998). Expression of UAS-Numb-myc, UAS-Numb4A-myc, UAS-NumbS52A-myc, and UAS-Numb5A-myc under the control of neurPGal4 and tub-Gal80ts was achieved by shifting pupae at 0 h APF to 28°C.
Supplementary Material
Supplementary Movie 1
Supplementary Movie 2
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3
Supplementary Figure 4
Supplementary Figure 5
Supplementary Figure 6
Acknowledgments
We thank V Stambolic for use of his Hunter Apparatus, J Trejo for the HeLa-SPR cells, Michael Quon for PKCζ expression constructs, J Fawcett and T Pawson for anti-Par3 antibodies, G Ojakian for anti-gp135, and Mike Woodside and Paul Paroutis for assistance with image analysis. We also thank R Bodmer, C Doe, M Gonzalez-Gaitan, and the Developmental Studies Hybridoma Bank (DSHB, Iowa University) for flies and antibodies. We thank members of the McGlade, Thibault, and Schweisguth Laboratories for helpful discussions. This work was supported by the Association pour la Recherche sur le Cancer (grant #3458 to FS) and the National Cancer Institute of Canada with funds from the Canadian Cancer Society (to CJM).
References
- Bellaiche Y, Gho M, Kaltschmidt JA, Brand AH, Schweisguth F (2001a) Frizzled regulates localization of cell-fate determinants and mitotic spindle rotation during asymmetric cell division. Nat Cell Biol 3: 50–57 [DOI] [PubMed] [Google Scholar]
- Bellaiche Y, Radovic A, Woods DF, Hough CD, Parmentier ML, O'Kane CJ, Bryant PJ, Schweisguth F (2001b) The partner of inscuteable/discs-large complex is required to establish planar polarity during asymmetric cell division in Drosophila. Cell 106: 355–366 [DOI] [PubMed] [Google Scholar]
- Berezovska O, McLean P, Knowles R, Frosh M, Lu FM, Lux SE, Hyman BT (1999) Notch1 inhibits neurite outgrowth in postmitotic primary neurons. Neuroscience 93: 433–439 [DOI] [PubMed] [Google Scholar]
- Betschinger J, Knoblich JA (2004) Dare to be different: asymmetric cell division in Drosophila, C. elegans and vertebrates. Curr Biol 14: R674–R685 [DOI] [PubMed] [Google Scholar]
- Betschinger J, Mechtler K, Knoblich JA (2003) The Par complex directs asymmetric cell division by phosphorylating the cytoskeletal protein Lgl. Nature 422: 326–330 [DOI] [PubMed] [Google Scholar]
- Bhalerao S, Berdnik D, Torok T, Knoblich JA (2005) Localization-dependent and -independent roles of numb contribute to cell-fate specification in Drosophila. Curr Biol 15: 1583–1590 [DOI] [PubMed] [Google Scholar]
- Boyle WJ, van der Geer P, Hunter T (1991) Phosphopeptide mapping and phosphoamino acid analysis by two-dimensional separation on thin-layer cellulose plates. Methods Enzymol 201: 110–149 [DOI] [PubMed] [Google Scholar]
- Cayouette M, Whitmore AV, Jeffery G, Raff M (2001) Asymmetric segregation of Numb in retinal development and the influence of the pigmented epithelium. J Neurosci 21: 5643–5651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien CT, Wang S, Rothenberg M, Jan LY, Jan YN (1998) Numb-associated kinase interacts with the phosphotyrosine binding domain of Numb and antagonizes the function of Numb in vivo. Mol Cell Biol 18: 598–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conner SD, Schmid SL (2002) Identification of an adaptor-associated kinase, AAK1, as a regulator of clathrin-mediated endocytosis. J Cell Biol 156: 921–929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dho SE, French MB, Woods SA, McGlade CJ (1999) Characterization of four mammalian numb protein isoforms. Identification of cytoplasmic and membrane-associated variants of the phosphotyrosine binding domain. J Biol Chem 274: 33097–33104 [DOI] [PubMed] [Google Scholar]
- Dho SE, Siderovski DP, Trejo J, McGlade CJ (2006) Dynamic regulation of Mammalian Numb by GPCR and PKC activation: structural determinants of Numb association with the cortical membrane. Mol Biol Cell 17: 4142–4155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- French MB, Koch U, Shaye RE, McGill MA, Dho SE, Guidos CJ, McGlade CJ (2002) Transgenic expression of numb inhibits notch signaling in immature thymocytes but does not alter T cell fate specification. J Immunol 168: 3173–3180 [DOI] [PubMed] [Google Scholar]
- Frise E, Knoblich JA, Younger-Shepherd S, Jan LY, Jan YN (1996) The Drosophila Numb protein inhibits signaling of the Notch receptor during cell–cell interaction in sensory organ lineage. Proc Natl Acad Sci USA 93: 11925–11932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo M, Jan LY, Jan YN (1996) Control of daughter cell fates during asymmetric division: interaction of Numb and Notch. Neuron 17: 27–41 [DOI] [PubMed] [Google Scholar]
- Henrique D, Schweisguth F (2003) Cell polarity: the ups and downs of the Par6/aPKC complex. Curr Opin Genet Dev 13: 341–350 [DOI] [PubMed] [Google Scholar]
- Hurov JB, Watkins JL, Piwnica-Worms H (2004) Atypical PKC phosphorylates PAR-1 kinases to regulate localization and activity. Curr Biol 14: 736–741 [DOI] [PubMed] [Google Scholar]
- Hutterer A, Knoblich JA (2005) Numb and alpha-adaptin regulate Sanpodo endocytosis to specify cell fate in Drosophila external sensory organs. EMBO Rep 6: 836–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Justice N, Roegiers F, Jan LY, Jan YN (2003) Lethal giant larvae acts together with numb in notch inhibition and cell fate specification in the Drosophila adult sensory organ precursor lineage. Curr Biol 13: 778–783 [DOI] [PubMed] [Google Scholar]
- Knoblich JA, Jan LY, Jan YN (1997) The N terminus of the Drosophila Numb protein directs membrane association and actin-dependent asymmetric localization. Proc Natl Acad Sci USA 94: 13005–13010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langevin J, Le Borgne R, Rosenfeld F, Gho M, Schweisguth F, Bellaiche Y (2005) Lethal giant larvae controls the localization of notch-signaling regulators numb, neuralized, and Sanpodo in Drosophila sensory-organ precursor cells. Curr Biol 15: 955–962 [DOI] [PubMed] [Google Scholar]
- Macara IG (2004) Parsing the polarity code. Nat Rev Mol Cell Biol 5: 220–231 [DOI] [PubMed] [Google Scholar]
- McGill MA, McGlade CJ (2003) Mammalian numb proteins promote Notch1 receptor ubiquitination and degradation of the Notch1 intracellular domain. J Biol Chem 278: 23196–23203 [DOI] [PubMed] [Google Scholar]
- Nelson WJ (2003) Adaptation of core mechanisms to generate cell polarity. Nature 422: 766–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura T, Fukata Y, Kato K, Yamaguchi T, Matsuura Y, Kamiguchi H, Kaibuchi K (2003) CRMP-2 regulates polarized Numb-mediated endocytosis for axon growth. Nat Cell Biol 5: 819–826 [DOI] [PubMed] [Google Scholar]
- Ojakian GK, Schwimmer R (1988) The polarized distribution of an apical cell surface glycoprotein is maintained by interactions with the cytoskeleton of Madin–Darby canine kidney cells. J Cell Biol 107: 2377–2387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plant PJ, Fawcett JP, Lin DC, Holdorf AD, Binns K, Kulkarni S, Pawson T (2003) A polarity complex of mPar-6 and atypical PKC binds, phosphorylates and regulates mammalian Lgl. Nat Cell Biol 5: 301–308 [DOI] [PubMed] [Google Scholar]
- Rhyu MS, Jan LY, Jan YN (1994) Asymmetric distribution of numb protein during division of the sensory organ precursor cell confers distinct fates to daughter cells. Cell 76: 477–491 [DOI] [PubMed] [Google Scholar]
- Roegiers F, Jan YN (2004) Asymmetric cell division. Curr Opin Cell Biol 16: 195–205 [DOI] [PubMed] [Google Scholar]
- Roegiers F, Younger-Shepherd S, Jan LY, Jan YN (2001a) Bazooka is required for localization of determinants and controlling proliferation in the sensory organ precursor cell lineage in Drosophila. Proc Natl Acad Sci USA 98: 14469–14474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roegiers F, Younger-Shepherd S, Jan LY, Jan YN (2001b) Two types of asymmetric divisions in the Drosophila sensory organ precursor cell lineage. Nat Cell Biol 3: 58–67 [DOI] [PubMed] [Google Scholar]
- Rolls MM, Albertson R, Shih HP, Lee CY, Doe CQ (2003) Drosophila aPKC regulates cell polarity and cell proliferation in neuroblasts and epithelia. J Cell Biol 163: 1089–1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santolini E, Puri C, Salcini AE, Gagliani MC, Pelicci PG, Tacchetti C, Di Fiore PP (2000) Numb is an endocytic protein. J Cell Biol 151: 1345–1352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer M, Knoblich JA (2001) Protein localization during asymmetric cell division. Exp Cell Res 271: 66–74 [DOI] [PubMed] [Google Scholar]
- Shen Q, Zhong W, Jan YN, Temple S (2002) Asymmetric Numb distribution is critical for asymmetric cell division of mouse cerebral cortical stem cells and neuroblasts. Development 129: 4843–4853 [DOI] [PubMed] [Google Scholar]
- Smith CA, Dho SE, Donaldson J, Tepass U, McGlade CJ (2004) The cell fate determinant numb interacts with EHD/Rme-1 family proteins and has a role in endocytic recycling. Mol Biol Cell 15: 3698–3708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki A, Hirata M, Kamimura K, Maniwa R, Yamanaka T, Mizuno K, Kishikawa M, Hirose H, Amano Y, Izumi N, Miwa Y, Ohno S (2004) aPKC acts upstream of PAR-1b in both the establishment and maintenance of mammalian epithelial polarity. Curr Biol 14: 1425–1435 [DOI] [PubMed] [Google Scholar]
- Suzuki A, Ohno S (2006) The PAR-aPKC system: lessons in polarity. J Cell Sci 119: 979–987 [DOI] [PubMed] [Google Scholar]
- Suzuki A, Yamanaka T, Hirose T, Manabe N, Mizuno K, Shimizu M, Akimoto K, Izumi Y, Ohnishi T, Ohno S (2001) Atypical protein kinase C is involved in the evolutionarily conserved par protein complex and plays a critical role in establishing epithelia-specific junctional structures. J Cell Biol 152: 1183–1196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokumitsu H, Hatano N, Inuzuka H, Sueyoshi Y, Yokokura S, Ichimura T, Nozaki N, Kobayashi R (2005) Phosphorylation of Numb family proteins. Possible involvement of Ca2+/calmodulin-dependent protein kinases. J Biol Chem 280: 35108–35118 [DOI] [PubMed] [Google Scholar]
- Yaich L, Ooi J, Park M, Borg JP, Landry C, Bodmer R, Margolis B (1998) Functional analysis of the Numb phosphotyrosine-binding domain using site-directed mutagenesis. J Biol Chem 273: 10381–10388 [DOI] [PubMed] [Google Scholar]
- Yamanaka T, Horikoshi Y, Izumi N, Suzuki A, Mizuno K, Ohno S (2006) Lgl mediates apical domain disassembly by suppressing the PAR–3–aPKC-PAR-6 complex to orient apical membrane polarity. J Cell Sci 119: 2107–2118 [DOI] [PubMed] [Google Scholar]
- Yamanaka T, Horikoshi Y, Sugiyama Y, Ishiyama C, Suzuki A, Hirose T, Iwamatsu A, Shinohara A, Ohno S (2003) Mammalian Lgl forms a protein complex with PAR-6 and aPKC independently of PAR-3 to regulate epithelial cell polarity. Curr Biol 13: 734–743 [DOI] [PubMed] [Google Scholar]
- Zhang L, Wu SL, Rubin CS (2001) A novel adapter protein employs a phosphotyrosine binding domain and exceptionally basic N-terminal domains to capture and localize an atypical protein kinase C: characterization of Caenorhabditis elegans C kinase adapter 1, a protein that avidly binds protein kinase C3. J Biol Chem 276: 10463–10475 [DOI] [PubMed] [Google Scholar]
- Zhong W, Feder JN, Jiang MM, Jan LY, Jan YN (1996) Asymmetric localization of a mammalian numb homolog during mouse cortical neurogenesis. Neuron 17: 43–53 [DOI] [PubMed] [Google Scholar]
- Zhong W, Jiang MM, Schonemann MD, Meneses JJ, Pedersen RA, Jan LY, Jan YN (2000) Mouse numb is an essential gene involved in cortical neurogenesis. Proc Natl Acad Sci USA 97: 6844–6849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilian O, Saner C, Hagedorn L, Lee HY, Sauberli E, Suter U, Sommer L, Aguet M (2001) Multiple roles of mouse Numb in tuning developmental cell fates. Curr Biol 11: 494–501 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Movie 1
Supplementary Movie 2
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3
Supplementary Figure 4
Supplementary Figure 5
Supplementary Figure 6


