Abstract
K+-selective ion channels (K+ channels) have been found in bacteria, archaea, eucarya, and viruses. In Paramecium and other ciliates, K+ currents play an essential role in cilia-based motility. We have retrieved and sequenced seven closely related Paramecium K+-channel gene (PAK) sequences by using previously reported fragments. An additional eight unique K+-channel sequences were retrieved from an indexed library recently used in a pilot genome sequencing project. Alignments of these protein translations indicate that while these 15 genes have diverged at different times, they all maintain many characteristics associated with just one subclass of metazoan K+ channels (CNG/ERG type). Our results indicate that most of the genes are expressed, because all predicted frameshifts and several gaps in the homolog alignments contain Paramecium intron sequences deleted from reverse transcription-PCR products. Some of the variations in the 15 genomic nucleotide sequences involve an absence of introns, even between very closely related sequences, suggesting a potential occurrence of reverse transcription in the past. Extrapolation from the available genome sequence indicates that Paramecium harbors as many as several hundred of this one type of K+-channel gene. This quantity is far more numerous than those of K+-channel genes of all types known in any metazoan (e.g., ∼80 in humans, ∼30 in flies, and ∼15 in Arabidopsis). In an effort to understand this plurality, we discuss several possible reasons for their maintenance, including variations in expression levels in response to changes in the freshwater environment, like that seen with other major plasma membrane proteins in Paramecium.
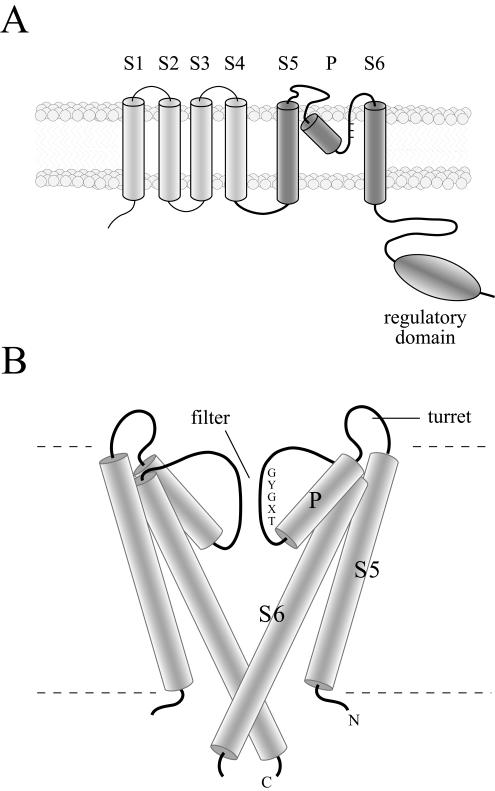
The potassium ion (K+) is a major cytoplasmic solute maintained at high concentrations by the transportation of the ion from outside the cell against a concentration gradient. As cells expend substantial amounts of energy to establish this gradient, there is a controlled release of K+ passively through K+-selective ion channels (K+ channels). This passive permeation of K+ governs the resting potential of the membrane and therefore the excitability and behavior of both animals (15) and protists (20). The rapidly growing number of sequenced genomes has revealed that all of the major taxa (bacteria, archea, eucarya, and viruses), with the exception of a few parasitic forms, contain genes that encode K+-channel proteins (15). These proteins can have between two and eight transmembrane domains or segments and are likely to function as tetrameric structures (3, 37). All of the known Paramecium K+-channel proteins are most similar to a large number of proteins predicted to have six transmembrane segments (Fig. 1A) (22). The primary sequence of K+-channel proteins is recognized by a highly conserved core element that includes a canonical filter sequence (TXGYG) connecting two conserved α-helical transmembrane segments (S5 and S6; Fig. 1B). Recently crystallized K+-channel proteins from bacteria revealed that the two transmembrane segments donated by each of the four subunits form an inverted tepee enclosing a central aqueous pathway lined by carbonyl oxygens extending from the TXGYG amino acid residues (Fig. 1B) (5, 38). The pore helix, between S5 and the filter sequence, holds this filter in a precise position that mimics oxygen donated by water molecules in solution and allows ions to permeate at near-diffusion-limiting rates (>107 per s) through a highly selective filter. The analysis also revealed that an extended portion of S6 encloses a water-filled vestibule, restricted from the cytoplasm, where small changes can prevent K+ from flowing through the channel (Fig. 1B).
FIG. 1.
(A) Diagrams representing the topology of K+-channels most closely related to the PaK proteins. (A) The predicted transmembrane topology of a single K+-channel subunit related to the Paramecium sequences examined in this report. The core segments and regulatory domain are highlighted (dark gray). (B) Core structure of two of the four subunits of a representative K+ channel. Shown are the relative positions of the core elements, including the fifth and sixth transmembrane segments (S5 and S6, respectively), the connecting pore helix (P), and the K+ filter seen in crystals of bacterial K+ channels.
K+ channels have been extensively investigated by electrophysiological means (mostly in metazoan animals) by examining the opening and closing of the channel molecules upon exposure to various stimuli (“gating principles”), such as voltage, external ligands, Ca2+, and mechanical force (15). Among protists, Paramecium tetraurelia is the most electrophysiologically studied species. Extensive analyses of this unicell revealed several different K+ currents activated by depolarization, hyperpolarization, and Ca2+-dependent depolarization and hyperpolarization, as well as by mechanical stimulation (20, 31; 32 and citations therein). Since the resting membrane potential is largely governed by the resting permeation of K+ in protists, as in metazoans, there are also K+ channels that open at rest (20; 32 and citations therein). This resting permeation has been shown to be sensitive to cyclic AMP (cAMP) (14).
K+-channel sequences from P. tetraurelia were first discovered by PCR amplification with degenerate oligonucleotides designed on the basis of the Shaker family of K+ channels (16). Five of the cloned PCR products, referred to as PAK1 to PAK5, contained sequences that translated into amino acid residues expected from the filter sequence to near the end of S6 of K+ channels (Fig. 1) (16). PAK1 and PAK2 were used to find longer fragments by Southern hybridization of a Paramecium genomic lambda phage library (16) (GenBank accession numbers U19907 and U19908, respectively). Additional probing resulted in PAK11, a homolog so similar to PAK1 that micronuclear copies were cloned to establish that these were in fact not the same gene (23). Several cDNA sequences modified the originally deduced open reading frame (ORF) of PAK1 (22). The overexpression of either the PAK1 or the PAK11 ORF produced a behavioral phenotype and the suppression of two Ca2+-activated K+ currents (22).
Here we report that additional probing has led to more versions of both PAK1- and PAK2-type genes, and a pilot project to sequence the genome of P. tetraurelia has revealed a startling number of short sequences that appear to be additional K+-channel genes (4, 35). We have retrieved these cloned fragments, and extended sequencing has revealed that these are indeed K+-channel genes. Sequences of reverse transcription (RT)-PCR products from cDNAs have indicated that many of them are apparently expressed. The variability in primary sequence over the most conserved portion of the proteins is restricted to two particular regions. In addition, intron positions in the genes show some variations; in some locations, a precise absence of introns in very closely related nucleotide sequences suggests that RT may have occurred. This report summarizes knowledge of this family of K+ channels to date and discusses the puzzle of P. tetraurelia having more members of this family than are currently known in any metazoan.
MATERIALS AND METHODS
Stocks and cultures.
P. tetraurelia stocks 51s (+/+) (34) and nd6 (nd6/nd6) (21) were cultured at 20 to 25°C in a growth medium containing buffered wheatgrass extract inoculated with Enterobacter aerogenes (34). nd6 mutants are wild type except for their inability to discharge trichocysts.
PAK nomenclature.
With the discovery of so many related sequences and the likelihood of so many more additional sequences to be found, a systematic nomenclature is used. Each gene is given a number representing a subtype, followed by a decimal point, and an additional number representing that particular unique family member. Sequences that translate into proteins with amino acids that are >65% identical are assigned to the same subtype. Previously cloned PAK1, PAK11, and PAK2 are now PAK1.1, PAK1.2, and PAK2.1, respectively (16, 22).
Cloning of PAK1.3, PAK1.4, PAK2.2, and PAK2.3.
A λ library of P. tetraurelia genomic DNA (gift from J. Forney) was probed with a 32P-labeled 0.85-kb fragment of PAK1 DNA (5′-ATGCTCTCAATAAAG…ACACTCACACTTGGC-3′; GenBank accession number AF424539) or 0.8-kb fragment of PAK2 DNA (5′-GGTGGAATATATACA…CAACCTTAAAAAATT-3′; GenBank accession number AF432226) (22). Samples positive for 10-kb particles were identified through several rounds of purification by Southern hybridization plaque lifts, the DNA was purified, and various restriction enzymes were used to produce smaller fragments. Southern hybridization-positive bands of these fragments were purified and cloned into pBluescript II KS(+) (Stratagene, La Jolla, Calif.).
Cloning of PAK2.4, PAK3.1, PAK3.2, PAK4.1, PAK5.1, PAK6.1, PAK 7.1, and PAK8.1.
An indexed macronuclear DNA library (courtesy of J. Cohen) originally constructed (18) for cloning by complementation (8, 9) was used in the pilot Paramecium genome sequencing project (4). Universal T3 and T7 primers were used to sequence from both ends of 1,800 cloned fragments, resulting in 2,990 sequences that were an average of 500 bases long (4). BlastX comparisons with previously characterized K+-channel proteins identified eight sequences as potential fragments of K+-channel genes (4) (GenBank accession numbers AL446894, AL448367, AL448113, AL447143, AL447926, AL446195, AL447013, and AL448883). The library was replicated, corresponding clones were isolated, and plasmids were purified.
Sequencing.
Unique oligonucleotides (sequences available upon request) were designed and used to extend both strands of the target sequence from the plasmids, PCR products from genomic DNA, and RT-PCR products from cDNA (see below). Automatic sequencing reactions with an ABI PRISM Big Dye Terminator cycle sequencing kit (Applied Biosystems, Foster City, Calif.) were done with a programmable thermal controller (model 100; MJ Research, Watertown, Mass.). The reaction products were purified by using CleanSEQ (Agencourt, Beverly, Mass.) and sequenced at the University of Wisconsin Biotechnology Center, Madison. Sequencing results were analyzed by using Edit View (Applied Biosystems) and SeqMan II (DNAStar, Madison, Wis.).
Preparation of total DNA and total RNA from Paramecium.
Standard molecular biology techniques were used for the preparation of total DNA and total RNA (2, 33). Total DNA was extracted from 51s and nd6 cultures as previously described (11). Briefly, cells were harvested, washed in 10 mM Tris (pH 7.4), and dissolved in a lysis buffer containing 50 mM NaOH, 35 mM EDTA, 0.5% sodium dodecyl sulfate, and 1.0 mg of proteinase K/ml. Samples were incubated for 4 h, followed by standard phenol-chloroform extraction and ethanol precipitation, and were resuspended in 10 mM Tris-1 mM EDTA (pH 8.0). To extract and purify total RNA, nd6 cells were harvested and washed in 10 mM Tris-buffered solution (pH 7.4). Approximately 250-μl aliquots of cells (2 × 105 cells) were lysed in 750 μl of Tri-Reagent (Molecular Research Center, Cincinnati, Ohio) and processed according to the protocol recommended by the manufacturer. Minipreparations of plasmid DNA from bacterial clones were purified by using spin columns and a commercial protocol (Qiagen, Chatsworth, Calif.)
PCR and subcloning of the seven K+-channel fragments identified in the pilot genomic sequencing project from genomic DNA.
To verify the cloned K+-channel sequences found in the library, fragments were PCR amplified from purified total Paramecium DNA by using the MJ Research model 100 programmable thermal controller, unique primer pairs (sequences available upon request), and Advantage cDNA polymerase mix (Clontech Laboratories Inc., Palo Alto, Calif.). The PCR products were either directly sequenced after resin purification (Qiagen) or cloned into pBluescript II KS(+) by use of additional unique restriction sites on the 5′ ends of the primers (sequences available upon request). At least two independent PCR products were sequenced either directly or as subcloned fragments.
Synthesis of the first-strand cDNA and RT-PCR products of K+-channel sequences from total RNA.
The first-strand cDNA was synthesized from purified nd6 total RNA by using either fragment-specific or poly(dT) oligonucleotides and a SuperScript first-strand synthesis system (Invitrogen Corporation, Carlsbad, Calif.), followed by RT-PCR (oligonucleotide sequences available upon request). Two or three clones or samples of RT-PCR products were sequenced as stated above.
Sequence comparison and secondary structure prediction.
BlastX and BlastP were used to identify sequences homologous to both previously published Paramecium K+-channel sequences and homologous sequences in other organisms found in the databases (GenBank/EMBL/DDBJ at the National Center for Biotechnology Information [NCBI]). The RPS-BLAST algorithm was used to identify domain sequences in the Conserved Domain Database, NCBI. Secondary structures were examined by using PROTEAN (DNAStar); protein and nucleotide sequences were aligned with MEGALIGN (DNAStar) or Pileup (Genetics Computer Group). The dendrogram was generated from distances calculated by using Paup 4.0 (Genetics Computer Group) to generate a heuristic bootstrap 50% majority-rule consensus tree (100 replicates) from a ClustalW alignment (MEGALIGN; Gonnet 250).
Nucleotide sequence accession numbers.
The GenBank accession numbers for the extended genomic sequences are as follows: AY302085 (PAK1.4), AY302087 (PAK2.2), AY302088 (PAK2.3), AY302089 (PAK2.4), AY302089 (PAK3.1), AY302091 (PAK3.2), AY302092 (PAK4.1), AY302093 (PAK5.1), AY302094 (PAK6.1), AY302095 (PAK7.1), and AY302096 (PAK8.1).
RESULTS
Targeted searching of K+-channel core sequences.
We have expanded the work of Ling et al. on PAK1.1 (previously PAK1), PAK1.2 (previously PAK11), and PAK2.1 (previously PAK2) (22, 23). We report here the cloning and sequencing of two PAK1 types (PAK1.3 and PAK1.4) and two PAK2.1-type isoforms (PAK2.2 and PAK2.3). These sequences were discovered by using the respective cloned family members as probes and were retrieved from a lambda phage genomic library (Y. Saimi et al., unpublished data). We have extended and verified these new sequences by obtaining multiple PCR products with specific oligonucleotides. Currently, we have determined at least 1,100 nucleotides of genomic sequence for each of the new isoform ORFs.
PAK-like sequences revealed in the pilot Paramecium genome sequencing project.
The pilot Paramecium genome sequencing project randomly surveyed 1 to 2% of the somatic genome of P. tetraurelia (4, 35). Analyses showed that 8 of the 841 protein-coding genes apparently encode K+-channel-like fragments. Surprisingly, none of these matched the PAK sequences previously retrieved on the basis of homology. These are M22B03r (now referred to as PAK7.1), M05G09r (PAK3.1), R14A01r (PAK3.2), M20F01r (PAK6.1), M19F02u (PAK2.4), R25B11r (PAK5.1), M11E01r (PAK4.1), and R16B08u (PAK8.1) (see Material and Methods for accession numbers). The corresponding library clones were retrieved, and the sequences were extended toward the one end of the gene available in each fragment. Currently, the sequences of the clones include a minimum of 1,500 nucleotides and, as determined on the basis of homology, are beyond the end of the published PAK1-type isoforms. The available library sequences also include a fragment, R15EO3u, recognized as the far 5′ portion of a PAK-like gene (35). This sequence has not been extended to the core and was excluded from the analyses.
PAK translations have key features of K+ channels.
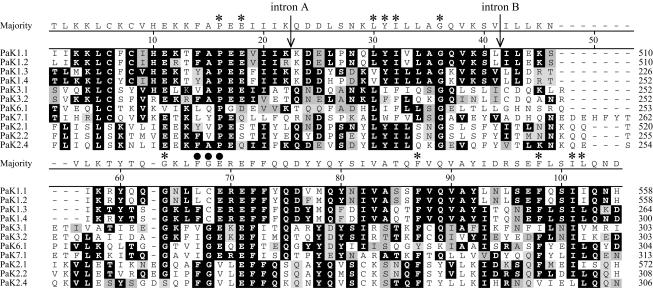
The translations of all of the PAK sequences are homologous to known potassium channels in the S5-S6 core segment (BlastP Expect values of <1e-08 and bit scores of >62). Although the homology extends beyond this domain, we focus on it, since it is the crux of K+ filtration, gating, and regulation (5). The hypothetical translations of the S5-pore helix-S6 region require the removal of intron sequences in PAK4.1, PAK5.1, PAK6.1, and PAK7.1. These sequences were removed from cDNAs (see below). By use of secondary structure predictions, biochemical attributes of residue side chains, and multiple alignments, the S5-S6 region can be structurally modeled on the basis of the many other channel sequences and the available crystal structure (Fig. 2). The residues TXGYG(D/E) (amino acids [aa] 82 to 87; Fig. 2) that constitute the K+ filter in the tetrameric channel protein are conserved in all 15 translations. Y69, Y74, and aromatic residue 75 match the key residues in the pore helix known to hold the filter in position. The S6 residues of the PaK proteins begin the characteristic distance filter with side chains at the start (negative, positive, and then hydrophobic residues; aa 95 to 99) and midsegment (glycines and aromatic residues; aa 108 to 112) of other K+ channels.
FIG. 2.
ClustalW (Gonnet 250) alignment of PaK channel sequences. The alignment shows high similarity among the PAK gene products. Two transmembrane segments (S5 and S6), the turret, pore helix, and filter are designated based on the apparent homology between the PaK translations and the K+ channels of other organisms. The topology is inferred from the bacterial crystal structure (Fig. 1) (5) (see the text for more details). Light gray, dark gray, and black highlighting indicates residues that are within 2, 1, and 0 distance units, respectively. The majority sequence is numbered across the alignment and includes gaps. The residue numbers on the right are relative to either the beginning of this alignment or that of previously published translations.
The residues that correspond to the S5 region of all of the PAK translations include a highly conserved H28 and one or two cysteines (C30 and/or C32) that are generally conserved in many CNG/ERG/EAG channels from other organisms (Fig. 2). Distinctive substitutions are found in residues directly flanking the S5 region, where the numbers of charged and polar versus hydrophobic residues are distinctly different. For example, the amino-terminal end of the PaK1-type S5 region contains more hydrophobic residues than that of the PaK2 type. This variation is also seen in K+ channels from other organisms.
The region labeled the “turret” is a variable linkage between the S5 region and the pore helix. This sequence (∼30 residues) is about twice as long as that found in voltage-dependent K+ channels (∼15 residues) but shorter than that found in some EAG/ERG channels, which can be as long as 50 residues. The length of the turret and especially the conserved tryptophan and immediate flanking residues (positions 51 to 53) are common in many related metazoan channels.
All PaK proteins are closely related.
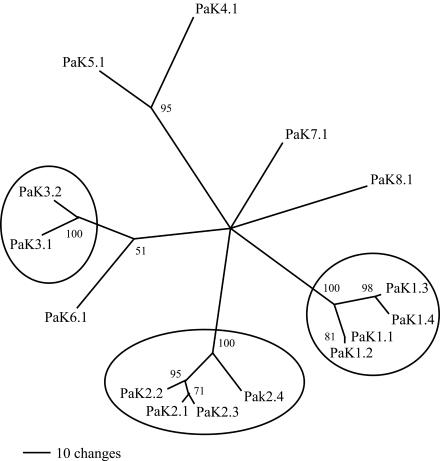
The amino acid residues of the S5-pore helix-S6 region of the 15 PaK proteins are clearly related. In fact, for all of the translations that extend toward a putative regulatory domain (see below), only a single amino acid residue gap is required to align the residues, with the exception of the turret and the region of the regulatory domain (Fig. 2; see Fig. 4). In alignments with a large number of metazoan K+ channels, PaK proteins appear closer to certain plant channels but nonetheless form a distinct group (data not shown). A comparison of the 15 channel core sequences (Fig. 2) resulted in a simple distance tree with three distinct groups (Fig. 3, circles). PaK1.1, PaK1.2, PaK1.3, and PaK1.4 are so very closely related, even when their nucleotide sequences are compared (see below), that they can be considered isoforms (>60% identity; Fig. 3, right circle). PaK2.1, PaK2.2, and PaK2.3 are also very closely related (Fig. 3, bottom circle). PaK2.4, while a little more distant at the nucleotide level, is consistently grouped with the related PaK2-type proteins. In addition, PaK3.1 and PaK3.2 are grouped (Fig. 3, left circle), and while PaK4.1 and PaK5.1 do not meet our criteria for isoforms, they are often paired with several different alignment parameters (Fig. 3, upper branch point). The remaining three PaK proteins (PaK6.1, PaK7.1, and PaK8.1), while still more closely related to each other than to any other metazoan channels, could not be reliably grouped.
FIG. 4.
ClustalW (Gonnet 250) alignment of putative regulatory domain sequences. All PaK sequences that could be extended ∼90 amino acids downstream of S6 share a conserved stretch with homology to cyclic nucleotide binding domain sequences (reference numbers 14778, 9173, 9037, and 3826 in the Conserved Domain Database, NCBI). The most highly conserved amino acids in this domain are marked with asterisks. Black circles mark the three residues that have a consensus sequence identical to that of residues known to interact with cyclic nucleotides in cyclic nucleotide-gated (CNG) channels, but none of the individual Paramecium translations are precisely conserved. For an explanation of the highlighting, see the legend to Fig. 2. The residue numbers on the right are relative to those in Fig. 2.
FIG. 3.
Simple distance tree for PaK proteins. The alignment in Fig. 2 was used to calculate a distance tree showing the clustering of some of the more similar sequences. Sequences that have >65% identity of amino acid residues and highly repeatable branch points include the PaK1 cluster, the PaK2 cluster, and the PaK3 pair (circled).
Protein sequence comparison of a possible regulatory domain.
As with many similar metazoan K+ channels, at ∼90 amino acids downstream of the S6 region in all of the PaK channels for which data are available, there is a cytoplasmic region similar to a putative cyclic nucleotide binding domain. These primary sequences are similar to those found in cyclic nucleotide gated channels, several kinases, and the Escherichia coli catabolite gene activator (Fig. 4; see also Fig. 1A and the Conserved Domain Database at NCBI). While the PaK proteins align with other known domains and have a number of highly conserved residues in common with both channels and cyclic nucleotide binding kinases (Fig. 4, asterisks), there are several critical substitutions. In particular, positions 67 to 69 (Fig. 4, black circles) align with known cyclic nucleotide binding domains which have a conserved FGE sequence at these positions, believed to be a part of the binding site for cAMP or cGMP, particularly in cyclic nucleotide gated channels (17). While the conservation allowing for the alignment is clearly suggestive of a distant relationship, it is not clear whether this region binds cyclic nucleotides or some other, related ligand.
Missing introns in DNA sequences of PAK genes.
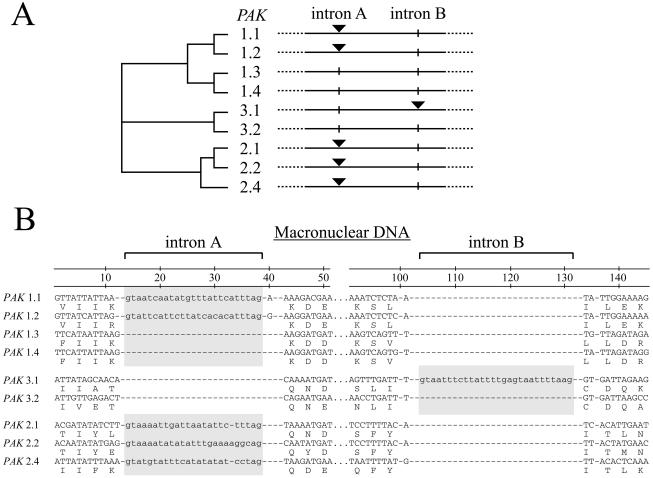
The PAK1-type isoforms are closely related at the nucleotide level (>70% identity in the ORF sequence available). The PAK2-type nucleotide sequences are just as similar to each other (>70% identity), except for PAK2.4, which is slightly more distant (>60% identity). In addition, PAK3.1 and PAK3.2 resemble each other (>60% identity), whereas most of the remaining sequences are less similar (33 to 50% identity). Despite the sequence conservation, a surprising result is that the entire intron sequences in very closely related members (>65% sequence identity) of this multigene family are occasionally missing (Fig. 5). While variations in intron length and sequence have been repeatedly observed between highly homologous sequences (e.g., reference 26) and distantly related proteins do not always show conserved intron positions, we are unaware of any published intron deletions within a duplicated sequence family this closely related (Fig. 5). Intron A, found at a particular position within the regulatory domain (Fig. 4, between positions 23 and 24), is conserved in several PAK1.x-type and PAK2.x-type isoforms (Fig. 5A, black triangles) but is absent from the PAK1.3 and PAK1.4 genes (Fig. 5A, vertical lines). While the complete ORFs of PAK1.3 and PAK1.4 have yet to be analyzed (only 1,179 and 1,804 nucleotides are known), at least one additional intron, upstream of the region shown, is conserved in PAK1.1-, PAK1.2-, and PAK1.4-type isoforms (data not shown). A second intron found in the regulatory domain of only PAK3.1 is also missing from the closely related PAK3.2 sequence (Fig. 5B, intron B; see also Fig. 4, between positions 41 and 42). The potential removal of the intron sequence during macronuclear development is unlikely because the sequence removed does not have the canonical 5′ and 3′ TA repeats universally found at the ends of micronucleus-specific internal eliminated sequences (IESs) (Fig. 5B).
FIG. 5.
Intron positions do not correlate with sequence similarity. (A) A distance tree highlights the relatedness of several closely related groups of PaK translations. While intron A is conserved in two distantly related groups (PaK1 type and PaK2 type, black triangles), two of the closely related PaK1-type sequences (PaK1.3 and PaK1.4) do not have this intron (vertical lines). A second example of two closely related sequences not sharing an intron was found for PaK3.1 (intron B [black triangle]) and PaK3.2 (vertical line). (B) Sequence of introns A and B and their flanks. Note the canonical 5′-GT…AG-3′ intron boundaries and the similarity of amino acid residues in the previously defined groups (PaK1 type, PaK2 type, and PaK3 type).
Although gaps in the protein alignments are common in the turret loop and in the putative regulatory domain, the positions of introns A and B are unambiguous and do not introduce gaps in the alignment (Fig. 4). Across the other regions of the PAK genes, we have discovered additional conserved intron positions in more distantly related sequences, but without more members of the gene family, it is difficult to determine how these introns have changed (data not shown).
PAK gene expression.
Expression was assessed by using RT-PCR. Putative introns in different areas of the ORFs were predicted from the frameshifts that they cause and/or by sequence comparisons with published PAK or related K+-channel genes in databases. The products of RT-PCR indeed confirmed that putative introns were spliced from the RNA. Ling et al. previously reported on the intron splicing of PAK1.1, PAK1.2, and PAK2.1 (22, 23). Here, we report the splicing of a single intron from various regions of PAK4.1, PAK5.1, PAK6.1, PAK7.1, and PAK8.1 (Fig. 6). The cDNAs from closely related isoforms, such as PAK1.1 and PAK1.2 or PAK2.1, PAK2.2, and PAK2.3, have nucleotide substitutions in the corresponding exons to warrant distinct identification. Thus, for every one of the 12 PAK genes predicted to have at least one intron, the expected spliced RNA has been found. It should be noted that PAK3.2 is not currently predicted to have any introns. Currently, only PAK1.1 and PAK1.2 have been extended to their farthest 5′ and 3′ RT-PCR-amplifiable ends. Although the RT-PCR products do not cover the entire ORFs of any of the 12 genes with verified introns or any part of isoforms PAK1.3 and PAK1.4, the results indicate that most, if not all, of the PAK genes are expressed in routine laboratory cultures.
FIG. 6.
Examples of intron sequences removed from various positions in the RT-PCR products of the PAK genes tested. All of these introns were predicted by using BlastX and ClustalW alignments to find gaps in conserved sequences or frameshifts in the putative translations prior to the production of cDNA and the amplified RT-PCR products.
DISCUSSION
An astonishing number of K+-channel genes exist in P. tetraurelia.
The core sequences, especially the TXGYGD filter sequence, are so well conserved in Paramecium that they allow unambiguous identification of K+ channels. We have so far encountered 15 very similar K+-channel genes in the somatic genome of P. tetraurelia (16, if R15E03u is included). Eight of these were recognized in a set of 841 identified protein-coding ORFs from a random genome sequencing project (4, 35). This frequency of occurrence suggests that there may be more K+-channel genes in the Paramecium genome than are currently known in any other organism.
The number of K+ channels in completely sequenced genomes ranges from 1 in budding yeasts to about 75 in humans and worms (15, 36). This means that the fraction of eukaryotic genomes dedicated to K+-channel genes can vary from very low to as high as 0.25% (in humans, 75 out of an estimated 30,000 protein-coding ORFs) (36). If the actual fraction of K+-channel genes in Paramecium were ≈0.2%, the probability of finding eight or more such gene fragments in 841 randomly recognized ORFs would be very low (P = 0.0003). On the other hand, an estimation that 1% (∼300 out of 30,000 ORFs) of the genome is dedicated to K+-channel genes is surprising, and there are several caveats. Since the library was constructed from fragments of DNA from polyploid (somatic) macronuclei and estimations of the genome size are imprecise, the absolute number of members of any multigene family is difficult to estimate (29, 35). With only a small fraction of the genome sequenced and reports of under- and overamplification of particular loci, it is plausible that the sample is some how biased (6). In addition, the fact that Paramecium uses only one stop codon (TGA), combined with the discovery of several very unusual genes (8-10, 12), makes the number of valid ORFs in the available library sequences difficult to precisely determine. Regardless of these caveats, the number of unique, expressed, and structurally related K+-channel genes represents one of the larger gene families in Paramecium.
Functional K+-channel genes or pseudogenes?
Variations among the 15 sequences are not experimental artifacts. Even the two most similar sequences, PAK1.1 and PAK1.2 (previously PAK1 and PAK11) (Fig. 3), upon independent and repeated amplifications, differed by the same substitutions. The two genes contained different 5′ and 3′ untranslated regions, and while the intron positions were conserved, the intron sequences were different. The cloning of their micronuclear (germ line) copies verifies that the PAK1.1 and PAK1.2 sequences are separate genes, each with unique IESs removed during the development of the polyploid macronucleus (22). While there is precedence for describing other duplicated genes found in Paramecium as potential psuedogenes (6), we have been unable to find any indication that the 15 ORFs described here harbor missense or nonsense mutations. All of our results collected thus far support the idea that these sequences are functional. While BlastX comparisons of sequences revealed frameshifts and gapping compared with other known sequences, closer examination of the sequences revealed probable intron sequences (35). Sequencing results for RT-PCR-amplified cDNAs made by use of gene-specific or poly(dT) oligonucleotides revealed that intron sequences were indeed removed from the RNA template in 14 out of the 15 ORFs examined (the 15th is a complete ORF without introns).
Mechanisms of diversification.
The usual evolutionary explanation for multiple paralogs is gene duplication through retention of duplicated DNA copies in the same nucleus. However, the PAK genes appear to have multiplied in the Paramecium genome by retaining duplicated DNA paralogs, some of which contain a particular intron that is missing in other closely related nucleotide sequences. Furthermore, this variation in specific introns is not coupled to the number of nucleotide substitutions in the exon sequence. For example, PAK1.1 and PAK1.4 are closely related by exon sequences (and the presence of other introns), but PAK1.1 has intron A and PAK1.4 does not (Fig. 5) (22). On the other hand, PAK1.1 and PAK2.4 are distantly related by exon sequences, but both have intron A in the same positions (Fig. 5). A second example of this phenomenon has been found in that intron B in PAK3.1 is missing in PAK3.2, although again the two genes are judged to be very similar on the basis of exon sequences and other characteristics (Fig. 5). The simplest hypothesis seems to be that reverse-transcribed copies of the genes were somehow retained, resulting in PAK1.4 and PAK3.2. The frequency of this event is surprising, especially since we are aware of only one other report of an intron insertion or deletion (indel) event resulting in a polymorphism in a single species (7, 25).
As reported in the Paramecium literature and in the two PAK genes previously analyzed (PAK1.1 and PAK1.2), IESs and introns have distinct features, and introns are retained during the development of the somatic nucleus (macronucleus) (28, 30). Paramecium intron sequences that are spliced during mRNA maturation have a narrow size range (typically between 19 and 30 bp) and require the typical splicesome-type 5′-GT and AG-3′ on the ends of the eliminated sequences (35). IES fragments can range in size from 28 bases up to hundreds of bases long and are always removed at 5′- and 3′-TA dinucleotide repeats (19, 27). The intron sequences that are absent from the closely related K+-channel genes in this study cannot be easily explained by the removal of IESs. We are therefore inclined to consider that these variations exist in the germ line, appearing during evolution of the genome and not during macronuclear development. Currently, there is no evidence that the sequences with recently deleted introns are expressed (PAK1.3, PAK1.4, and PAK3.2). PAK1.4 can be used to pursue this question because additional introns (on the basis of homology) appear to be present in the sequence (data not shown).
Paramecium sequences are high in AT content (35), and Paramecium introns are even more AT rich and short. A point mutation resulting in an in-frame intron that cannot be spliced could in theory add a short peptide (6 to 10 aa) that might not disrupt function. This scenario could provide a mechanism that results in selectable but functional differences among K+ channels. While there is no direct evidence for this scenario, the diversity of additional intron positions in the PAK genes and the existence of very AT-rich sequences that show gaps in alignments are intriguing (data not shown).
Physiological function(s)?
Functional paralogs in other eukaryotic genomes are usually few in numbers and have diverged enough to serve different functions in different tissues or at different developmental stages. PaK channels are puzzling because there are so many of them, they are so very similar, and they all appear to be expressed. Assuming that all PAK genes are functional, as the RT-PCR results seem to indicate, the selective advantage that they offer to allow for their maintenance in the genome is currently unknown.
The technique of microinjecting DNA fragments of an ORF, specifically excluding 3′ untranslated regions, into the macronucleus has successfully mimicked null mutations and silenced the effects of a particular protein involved in ion transfer (9). Although injecting plasmids with cloned PAK gene fragments not containing 3′ untranslated regions did not reduce K+ currents (unpublished data), full-length in-frame PAK1.1 or PAK1.2 (but not PAK2.1) genes resulted in the reduction of the depolarization and hyperpolarization of Ca2+-activated K+ currents in Paramecium under a voltage clamp, without significantly affecting other K+ currents (22). The simplest interpretation is that these two gene products are involved in the two affected currents. It is possible that a combination of injection experiments with functionally tagged as well as intentionally nonfunctional sequences from the other PAK genes may help to clarify their biological role(s).
Assuming that all PaK proteins are functional channel subunits, the plurality is indeed puzzling. Their structural differences, although often minute, may reflect their functional differences. Alignment of the proteins reveals that the greatest variations are in the turret region facing the outside and the cytoplasmic ligand binding domain facing the cytoplasm. The longer turret of the related ERG/EAG channels appears to influence the opening and closing of the channels and could influence the external binding of ions (24). Complex cooperativity within the ligand binding domain of CNG heteromultimers has been shown to result in changes in both the selection and the binding of ligands (13, 17). It is therefore plausible that these two regions function as sensors of the external and internal milieus to regulate the probability of gate opening. One could imagine a freshwater cell needing to adjust its resting membrane potential through frequent regulation of the probability of the opening of its resting K+ channels, perhaps by regulating the orientation of ciliary beats, but the use of a very large panel of potential channel subunits remains counterintuitive.
The surface antigen genes (IAgs) constitute a large family of proteins that are mutually exclusively expressed in response to changes in environmental factors (ionic strength, temperature, pH, and so forth). A direct survey of Paramecium membrane proteins indicates that several other membrane proteins also change coordinately with changes in IAgs (1), although the genes that encode these other proteins are unknown. This finding indicates that the kinds of environmental factors that might influence the functions of these K+ channels are already known to alter the expression of other highly regulated surface proteins. Changes in certain residues that form the outer portion of a channel near the outer opening of the channel might affect how they respond to signals from the environment.
These notions can be tested. The possible alternative expression or even changes in the amounts of expression of different PAK genes at different ion concentrations, osmolarities, temperatures, and so forth can be examined. Alternative expression of groups of channel genes may also be found to correlate with that of the IAg genes and can be verified. Quantitative changes in expression at the RNA level (or perhaps even at the DNA level) in terms of under- or overamplification of certain loci may also be detected. We hope to be able to discover how this potentially very large family of channel genes is involved in regulating the finely tuned responses that paramecia make in response to their ever-changing environmental conditions.
Acknowledgments
We especially acknowledge Laura Oesterle for technical assistance. In addition, we thank Matt Pink, Shah Dodwad, and other undergraduate students in our laboratory who have contributed to our efforts.
This work was supported by NIH grant GM22714.
Footnotes
Dedicated to the memory of Andre Adoutte, who first recognized the coordinated protein remodeling of the Paramecium surface membrane.
REFERENCES
- 1.Adoutte, A., K. Y. Ling, S. Chang, F. Huang, and C. Kung. 1983. Physiological and mutational protein variations in the ciliary membrane of Paramecium. Exp. Cell Res. 148:387-404. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 2003. Current protocols in molecular biology. Greene Publishing Associates & Wiley Interscience, New York, N.Y.
- 3.Choe, S. 2002. Potassium channel structures. Nat. Rev. Neurosci. 3:115-121. [DOI] [PubMed] [Google Scholar]
- 4.Dessen, P., M. Zagulski, R. Gromadka, H. Plattner, R. Kissmehl, E. Meyer, M. Betermier, J. E. Schultz, J. U. Linder, R. E. Pearlman, C. Kung, J. Forney, B. H. Satir, J. L. Van Houten, A. M. Keller, M. Froissard, L. Sperling, and J. Cohen. 2001. Paramecium genome survey: a pilot project. Trends Genet. 17:306-308. [DOI] [PubMed] [Google Scholar]
- 5.Doyle, D. A., J. Morais Cabral, R. A. Pfuetzner, A. Kuo, J. M. Gulbis, S. L. Cohen, B. T. Chait, and R. MacKinnon. 1998. The structure of the potassium channel: molecular basis of K+ conduction and selectivity. Science 280:69-77. [DOI] [PubMed] [Google Scholar]
- 6.Dubrana, K., and L. Amar. 2000. Programmed DNA under-amplification in Paramecium primaurelia. Chromosoma 109:460-466. [DOI] [PubMed] [Google Scholar]
- 7.Feiber, A. L., J. Rangarajan, and J. C. Vaughn. 2002. The evolution of single-copy Drosophila nuclear 4f-rnp genes: spliceosomal intron losses create polymorphic alleles. J. Mol. Evol 55:401-413. [DOI] [PubMed] [Google Scholar]
- 8.Froissard, M., A. M. Keller, and J. Cohen. 2001. ND9P, a novel protein with armadillo-like repeats involved in exocytosis: physiological studies using allelic mutants in Paramecium. Genetics 157:611-620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haynes, W. J., C. Kung, Y. Saimi, and R. R. Preston. 2002. An exchanger-like protein underlies the large Mg2+ current in Paramecium. Proc. Natl. Acad. Sci. USA 99:15717-15722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haynes, W. J., K. Y. Ling, R. R. Preston, Y. Saimi, and C. Kung. 2000. The cloning and molecular analysis of pawn-B in Paramecium tetraurelia. Genetics 155:1105-1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haynes, W. J., K. Y. Ling, Y. Saimi, and C. Kung. 1995. Induction of antibiotic resistance in Paramecium tetraurelia by the bacterial gene APH-3′-II. J. Eukaryot. Microbiol. 42:83-91. [DOI] [PubMed] [Google Scholar]
- 12.Haynes, W. J., B. Vaillant, R. R. Preston, Y. Saimi, and C. Kung. 1998. The cloning by complementation of the pawn-A gene in Paramecium. Genetics 149:947-957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.He, Y., and J. W. Karpen. 2001. Probing the interactions between cAMP and cGMP in cyclic nucleotide-gated channels using covalently tethered ligands. Biochemistry 40:286-295. [DOI] [PubMed] [Google Scholar]
- 14.Hennessey, T., H. Machemer, and D. L. Nelson. 1985. Injected cyclic AMP increases ciliary beat frequency in conjunction with membrane hyperpolarization. Eur. J. Cell Biol. 36:153-156. [PubMed] [Google Scholar]
- 15.Hille, B. 2001. Ion channels of excitable membranes, 3rd ed. Sinauer, Sunderland, Mass.
- 16.Jegla, T., and L. Salkoff. 1995. A multigene family of novel K+ channels from Paramecium tetraurelia. Receptors Channels 3:51-60. [PubMed] [Google Scholar]
- 17.Kaupp, U. B., and R. Seifert. 2002. Cyclic nucleotide-gated ion channels. Physiol. Rev. 82:769-824. [DOI] [PubMed] [Google Scholar]
- 18.Keller, A. M., and J. Cohen. 2000. An indexed genomic library for Paramecium complementation cloning. J. Eukaryot. Microbiol. 47:1-6. [DOI] [PubMed] [Google Scholar]
- 19.Ku, M., K. Mayer, and J. D. Forney. 2000. Developmentally regulated excision of a 28-base-pair sequence from the Paramecium genome requires flanking DNA. Mol. Cell. Biol. 20:8390-8396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kung, C., and Y. Saimi. 1982. The physiological basis of taxes in Paramecium. Annu. Rev. Physiol. 44:519-534. [DOI] [PubMed] [Google Scholar]
- 21.Lefort-Tran, M., K. Aufderheide, M. Pouphile, M. Rossignol, and J. Beisson. 1981. Control of exocytotic processes: cytological and physiological studies of trichocyst mutants in Paramecium tetraurelia. J. Cell Biol. 88:301-311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ling, K. Y., W. J. Haynes, L. Oesterle, C. Kung, R. R. Preston, and Y. Saimi. 2001. K(+)-channel transgenes reduce K(+) currents in Paramecium, probably by a post-translational mechanism. Genetics 159:987-995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ling, K. Y., B. Vaillant, W. J. Haynes, Y. Saimi, and C. Kung. 1998. A comparison of internal eliminated sequences in the genes that encode two K(+)-channel isoforms in Paramecium tetraurelia. J. Eukaryot. Microbiol. 45:459-465. [DOI] [PubMed] [Google Scholar]
- 24.Liu, J., M. Zhang, M. Jiang, and G. N. Tseng. 2002. Structural and functional role of the extracellular s5-p linker in the HERG potassium channel. J. Gen. Physiol. 120:723-737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lynch, M., and A. O. Richardson. 2002. The evolution of spliceosomal introns. Curr. Opin. Genet. Dev. 12:701-710. [DOI] [PubMed] [Google Scholar]
- 26.Madeddu, L., M. C. Gautier, L. Vayssie, A. Houari, and L. Sperling. 1995. A large multigene family codes for the polypeptides of the crystalline trichocyst matrix in Paramecium. Mol. Biol. Cell 6:649-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meyer, E., and S. Duharcourt. 1996. Epigenetic programming of developmental genome rearrangements in ciliates. Cell 87:9-12. [DOI] [PubMed] [Google Scholar]
- 28.Preer, J. R., Jr. 2000. Epigenetic mechanisms affecting macronuclear development in Paramecium and Tetrahymena. J. Eukaryot. Microbiol. 47:515-524. [DOI] [PubMed] [Google Scholar]
- 29.Prescott, D. M. 1994. The DNA of ciliated protozoa. Microbiol. Rev. 58:233-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Prescott, D. M. 1997. Origin, evolution, and excision of internal eliminated segments in germline genes of ciliates. Curr. Opin. Genet. Dev. 7:807-813. [DOI] [PubMed] [Google Scholar]
- 31.Preston, R. R., Y. Saimi, and C. Kung. 1990. Evidence for two K+ currents activated upon hyperpolarization of Paramecium tetraurelia. J. Membr. Biol. 115:41-50. [DOI] [PubMed] [Google Scholar]
- 32.Saimi, Y., S. H. Loukin, X. L. Zhou, B. Martinac, and C. Kung. 1999. Ion channels in microbes. Methods Enzymol. 294:507-524. [DOI] [PubMed] [Google Scholar]
- 33.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 34.Sonneborn, T. M. 1970. Methods in Paramecium research, p. 241-339. In D. M. Prescott (ed.), Methods in cell physiology, vol. 4. Academic Press, Inc., New York, N.Y.
- 35.Sperling, L., P. Dessen, M. Zagulski, R. E. Pearlman, A. Migdalski, R. Gromadka, M. Froissard, A. M. Keller, and J. Cohen. 2002. Random sequencing of Paramecium somatic DNA. Eukaryot. Cell 1:341-352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Venter, J. C., M. D. Adams, E. W. Myers, P. W. Li, R. J. Mural, G. G. Sutton, H. O. Smith, M. Yandell, C. A. Evans, R. A. Holt, J. D. Gocayne, P. Amanatides, R. M. Ballew, D. H. Huson, J. R. Wortman, Q. Zhang, C. D. Kodira, X. H. Zheng, L. Chen, M. Skupski, G. Subramanian, P. D. Thomas, J. Zhang, G. L. Gabor Miklos, C. Nelson, S. Broder, A. G. Clark, J. Nadeau, V. A. McKusick, N. Zinder, A. J. Levine, R. J. Roberts, M. Simon, C. Slayman, M. Hunkapiller, R. Bolanos, A. Delcher, I. Dew, D. Fasulo, M. Flanigan, L. Florea, A. Halpern, S. Hannenhalli, S. Kravitz, S. Levy, C. Mobarry, K. Reinert, K. Remington, J. Abu-Threideh, E. Beasley, K. Biddick, V. Bonazzi, R. Brandon, M. Cargill, I. Chandramouliswaran, R. Charlab, K. Chaturvedi, Z. Deng, V. Di Francesco, P. Dunn, K. Eilbeck, C. Evangelista, A. E. Gabrielian, W. Gan, W. Ge, F. Gong, Z. Gu, P. Guan, T. J. Heiman, M. E. Higgins, R. R. Ji, Z. Ke, K. A. Ketchum, Z. Lai, Y. Lei, Z. Li, J. Li, Y. Liang, X. Lin, F. Lu, G. V. Merkulov, N. Milshina, H. M. Moore, A. K. Naik, V. A. Narayan, B. Neelam, D. Nusskern, D. B. Rusch, S. Salzberg, W. Shao, B. Shue, J. Sun, Z. Wang, A. Wang, X. Wang, J. Wang, M. Wei, R. Wides, C. Xiao, C. Yan, et al. 2001. The sequence of the human genome. Science 291:1304-1351. [DOI] [PubMed] [Google Scholar]
- 37.Yellen, G. 2002. The voltage-gated potassium channels and their relatives. Nature 419:35-42. [DOI] [PubMed] [Google Scholar]
- 38.Zhou, Y., J. H. Morais-Cabral, A. Kaufman, and R. MacKinnon. 2001. Chemistry of ion coordination and hydration revealed by a K+ channel-Fab complex at 2.0 A resolution. Nature 414:43-48. [DOI] [PubMed] [Google Scholar]