Abstract
The expression of individual chemoreceptor (CR) genes in Caenorhabditis elegans is regulated by multiple environmental and developmental cues, possibly enabling C. elegans to modulate its sensory responses. We had previously shown that KIN-29, a member of the salt-inducible kinase family, acts in a subset of chemosensory neurons to regulate the expression of CR genes, body size and entry into the alternate dauer developmental stage. Here, we show that KIN-29 regulates these processes by phosphorylating the HDA-4 class II histone deacetylase (HDAC) and inhibiting the gene repression functions of HDA-4 and an MEF-2 MADS domain transcription factor. MEF-2 binds directly to the CR gene regulatory sequences, and is required only to repress but not activate CR gene expression. A calcineurin phosphatase antagonizes the KIN-29/MEF-2-regulated pathway to modulate levels of CR gene expression. Our results identify KIN-29 as a new regulator of MEF2/HDAC functions in the nervous system, reveal cell-specific mechanisms of action of this pathway in vivo and demonstrate remarkable complexity in the regulation of CR gene expression in C. elegans.
Keywords: C. elegans, chemoreceptor, HDAC, MEF2, SIK
Introduction
Animals modify their behavior and development in response to changing external and internal conditions. These adaptive changes are mediated by changes in the functions of individual organs and cells, and ultimately, by changes in the expression of individual genes. Gene regulatory regions must, therefore, integrate information from multiple signaling pathways so as to fine-tune the spatiotemporal levels of gene expression (Wyrick and Young, 2002; Cases and de Lorenzo, 2005).
The Caenorhabditis elegans sensory nervous system provides an excellent system in which to explore the mechanisms by which developmental and environmental signals converge to regulate gene expression. Unlike vertebrate and Drosophila olfactory neurons, each chemosensory neuron in C. elegans expresses multiple G protein-coupled chemoreceptor (CR) genes, each of which may respond to distinct subsets of chemicals (Lanjuin and Sengupta, 2004). We and others have shown that regulation of CR gene expression is complex, such that in addition to developmental pathways, distinct subsets of CR genes are regulated by levels of a constitutively produced pheromone, food and neuronal activity (Troemel et al, 1999; Peckol et al, 2001; Lanjuin and Sengupta, 2002; Nolan et al, 2002; Tobin et al, 2002). How information from these pathways is integrated to modulate levels of CR gene expression is not known.
A family of proteins implicated in the regulation of gene expression in response to external signals is the AMPK/SNF1 family of Ser/Thr kinases. AMPK/SNF1 kinases play major roles in regulating cellular and whole-body energy homeostasis via activation and repression of key metabolic genes in response to cellular and pathological stress (Kahn et al, 2005; Kim and Lee, 2005). The salt-inducible kinases (SIKs) are members of this kinase family and, as implied by their name, were originally identified as kinases upregulated in adrenocortical cells in rats fed on high-salt diets (Wang et al, 1999). SIKs are also upregulated in response to ACTH signaling, and depolarization and kainate-induced seizures in adrenocortical tumor cells and in the nervous system, respectively (Feldman et al, 2000; Okamoto et al, 2004). Recently, SIK1 has also been shown to be upregulated in mouse primary hepatocytes upon fasting, and SIK and AMPK play critical roles in the regulation of hepatic gluconeogenic gene expression in response to fasting and feeding (Koo et al, 2005). SIK activity is further promoted by phosphorylation by upstream kinases, such as LKB1 (Lizcano et al, 2004). However, although SIKs are expressed in multiple cell types including neurons, their roles and targets in most tissue types have not been identified.
The MEF2 class of MADS domain transcription factors also plays an essential role in the regulation of gene expression in response to environmental signals in multiple cell types. In muscles, MEF2 regulates the expression of fiber type-specific genes in response to altered patterns of electrical stimulation (Wu et al, 2000; Liu et al, 2005), whereas MEF2 is critical for activity-dependent survival of cultured post-mitotic neurons (Mao et al, 1999; Li et al, 2001; Gaudilliere et al, 2002). Recently, MEF2 has also been shown to regulate synapse number in response to activity in cultured hippocampal and cerebellar granule neurons (Flavell et al, 2006; Shalizi et al, 2006). The gene regulatory functions of MEF2 are extensively modulated via interaction with different proteins in different cell types and via cell-specific post-translational mechanisms (McKinsey et al, 2002; Heidenreich and Linseman, 2004; Flavell et al, 2006; Gregoire et al, 2006; Riquelme et al, 2006; Shalizi et al, 2006). In particular, MEF2 interacts directly with class II histone deacetylases (HDACs) to repress gene expression (Lu et al, 2000b). Although multiple kinases have been implicated in the regulation of MEF2/HDAC functions, the role of AMPK family members in MEF2/HDAC-mediated gene expression has been suggested only in skeletal muscle in response to exercise via as yet uncharacterized mechanisms (Al-Khalili et al, 2004; Holmes et al, 2005; McGee and Hargreaves, 2006).
We previously showed that the C. elegans mutant for the SIK homolog kin-29 exhibits phenotypes consistent with defects in the acquisition and transduction of sensory information (Lanjuin and Sengupta, 2002). In kin-29 mutants, the expression of a subset of CR genes is altered, and animals exhibit reduced body size and deregulated entry into the alternate dauer developmental stage (Lanjuin and Sengupta, 2002; Maduzia et al, 2005). All examined kin-29 phenotypes could be rescued by expression of kin-29 in chemosensory neurons alone, leading us to suggest that misregulation of CR gene expression in kin-29 mutants was causal to the observed physiological abnormalities. Here, we show that KIN-29 acts by antagonizing the gene repressive functions of the C. elegans MEF2 ortholog MEF-2 and the class II HDAC protein HDA-4 in chemosensory neurons. We show that phosphorylation of HDA-4 by KIN-29 is essential for alleviation of repression, and that constitutive Ca2+ signaling partly bypasses the requirement for KIN-29 in this pathway. We demonstrate that MEF-2 interacts directly with a cis-regulatory element in a KIN-29-regulated CR gene promoter, but is not required to promote CR gene expression. Finally, we show that the calcineurin Ser/Thr phosphatase antagonizes the KIN-29 pathway. Taken together, our results identify an SIK family member as a new regulator of MEF2/HDAC functions in the nervous system, and provide insights into the complexity of signaling pathways that regulate CR gene expression in C. elegans.
Results
Mutations in mef-2 and hda-4 suppress the kin-29 phenotype of downregulated CR gene expression
To identify the mechanisms by which KIN-29 regulates CR gene expression, we carried out a suppressor screen. Expression of the str-1 and sra-6 CR genes is reduced, although not abolished, in kin-29 mutants in the AWB and ASH chemosensory neurons, respectively (Lanjuin and Sengupta, 2002). kin-29(oy39) missense mutants carrying stably integrated copies of a str-1∷gfp fusion gene were mutagenized and mutants isolated in which expression of str-1∷gfp in the AWB neurons was restored to wild-type levels (Figure 1A–E). Subsequent mapping and complementation experiments indicated that a subset of the identified alleles defined two complementation groups representing the mef-2 transcription factor and the hda-4 class II histone deactelyase (HDAC) genes (Dichoso et al, 2000; Choi et al, 2002) (Figure 1F). oy63 and oy65 are predicted to result in missense mutations in highly conserved amino acids in the conserved DNA-binding MADS box domain of MEF-2 (Figure 1F). The mef-2(gv1) allele (Dichoso et al, 2000) removes most of the MADS and MEF domains that are required for MEF-2 function, and is likely a null allele (Figure 1F). oy59 results in a premature stop codon in the first exon of hda-4, whereas the spontaneously arising oy57 suppressor allele is a point mutation in the splice acceptor site of the sixth exon, which is predicted to result in a truncated HDA-4 protein lacking the HDAC domain (Figure 1F). The hda-4(ok518) deletion allele was obtained from the C. elegans Gene Knockout Consortium. Sequences encoding part of the HDAC domain are deleted in hda-4(ok518) (Figure 1F).
Figure 1.
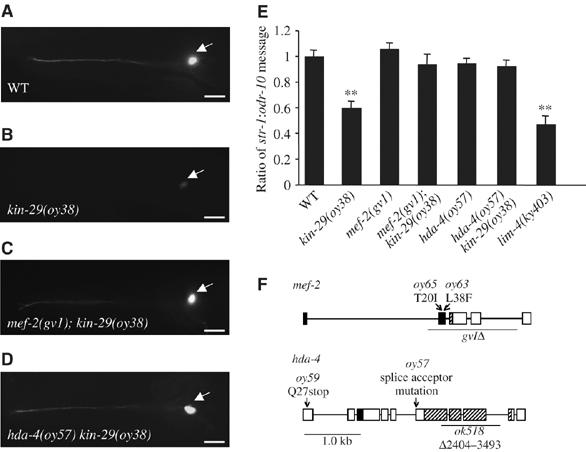
Mutations in the mef-2 MEF-2 transcription factor and hda-4 histone deacetylase genes suppress the downregulated str-1 CR gene expression phenotype of kin-29 mutants. (A–D) Expression of str-1∷gfp in an AWB neuron in animals of the indicated genotypes. Arrow points to the AWB cell body. Images were acquired at the same exposure time. Lateral view: anterior is on the left. Scale, 15 μm. (E) Reduced levels of endogenous str-1 message in kin-29 mutants are restored to wild-type levels in mef-2; kin-29 and kin-29 hda-4 double mutant animals. The ratio of endogenous str-1 message to endogenous odr-10 message in animals of the indicated genotypes is shown. The mean of the ratios from 8 to 12 independent RT–PCR experiments performed on multiple days is shown. Error bars denote the s.e.m. Double asterisks mark levels that are different from wild type at P<0.001 using a two-sample t-test. (F) Genomic structures of mef-2 and hda-4. The molecular identities of mutations in mef-2 (top) and hda-4 (bottom) are shown. The extent of the deletions in mef-2(gv1) and hda-4(ok518) are indicated by underlines. Filled and hatched boxes indicate the MADS and MEF2 domains, respectively, in MEF-2, and the MEF2-binding and HDAC domains, respectively, in HDA-4. The mutation in oy57 results in an hda-4 cDNA that uses an alternate splice acceptor site in intron 6, resulting in a truncated HDA-4 protein.
We further characterized the CR gene expression phenotypes of mef-2 and hda-4 mutants. Mutations in hda-4 and mef-2 suppressed the downregulation of str-1∷gfp expression in both the kin-29(oy39) missense and the kin-29(oy38) null background at all developmental stages, although no gross effects were observed on str-1∷gfp expression in a wild-type background (Table I). However, hda-4(ok518) failed to suppress the str-1∷gfp expression phenotype in kin-29 mutant animals. It is likely that hda-4(ok518) is a hypomorph, as the str-1∷gfp expression phenotype of kin-29(oy38) mutants was fully suppressed in hda-4(oy57)/hda-4(ok518) transheterozygotes (Table I). Mutations in mef-2 and hda-4 also fully suppressed the downregulation of expression of sra-6∷gfp in the ASH neurons of kin-29 mutants (Table I), indicating that MEF-2 and HDA-4 regulate the expression of multiple CR genes in different sensory neuron subtypes. Suppression of the str-1∷gfp expression phenotype was abolished upon introduction of hda-4 or mef-2 genomic sequences into kin-29 hda-4 or mef-2; kin-29 double mutants, respectively (Table I). The observed effects on transgene expression were confirmed by semiquantitative RT–PCR, which showed that levels of endogenous str-1 message were comparable among mef-2; kin-29 and kin-29 hda-4 double mutants and wild-type animals, but significantly reduced, although not abolished, in kin-29 mutants or in animals mutant for the LIM homeobox gene lim-4 required for specification of AWB fate (Sagasti et al, 1999) (Figure 1E).
Table 1.
Mutations in mef-2 and hda-4 suppress the reduced CR gene expression phenotype of kin-29 mutants
| Straina | % expressing str-1∷gfp at WT levels | n | P-values |
|---|---|---|---|
| Wild type | 100 | 280 | |
| kin-29(oy39) | 0 | 218 | <0.001 |
| kin-29(oy38) | 0 | 267 | <0.001 |
| mef-2(gv1) | 100 | 155 | |
| mef-2(oy65) | 100 | 223 | |
| hda-4(oy57) | 100 | 189 | |
| mef-2(oy63); kin-29(oy39) | 100 | 204 | |
| mef-2(oy65); kin-29(oy39) | 100 | 158 | |
| mef-2(oy65); kin-29(oy38) | 100 | 188 | |
| mef-2(gv1); kin-29(oy39) | 100 | 209 | |
| mef-2(gv1); kin-29(oy38) | 100 | 194 | |
| mef-2(gv1); kin-29(oy39); Ex[gfp-tagged mef-2 genomic] | 5 | 121 | <0.001 |
| mef-2(oy65); kin-29(oy39); Ex[srd-23∷mef-2] | 25 | 153 | <0.001 |
| mef-2(oy65); kin-29(oy39); Ex[lim-4∷mef-2] | 27 | 174 | <0.001 |
| kin-29(oy38) hda-4(oy57) | 100 | 201 | |
| kin-29(oy39) hda-4(oy59) | 100 | 211 | |
| kin-29(oy38) hda-4(ok518) | 0 | 224 | <0.001 |
| kin-29(oy39) hda-4(ok518) | 0 | 228 | <0.001 |
| kin-29(oy38) hda-4(oy57)/ hda-4(ok518) | 96 | 144 | |
| rrf-3(pk1426); kin-29(oy38) hda-4(RNAi)b | 21 | 65 | <0.001c |
| kin-29(oy38) hda-4(oy57); Ex[gfp-tagged hda-4 genomic] | 11 | 155 | <0.001 |
| kin-29(oy38) hda-4(oy57); Ex[odr-4∷hda-4∷gfp] | 8 | 146 | <0.001 |
| kin-29(oy38) hda-4(oy57); Ex[lim-4∷hda-4] | 10 | 188 | <0.001 |
| Straina | % expressing sra-6∷gfp at WT levels | n | P-values |
| Wild type | 100 | 234 | |
| kin-29(oy38) | 0 | 245 | <0.001 |
| mef-2(gv1) | 100 | 153 | |
| hda-4(oy57) | 100 | 101 | |
| mef-2(gv1); kin-29(oy38) | 100 | 167 | |
| kin-29(oy38) hda-4(oy57) | 100 | 189 | |
| Adult animals grown at 20°C were examined. Shown are the percentages of animals expressing str-1∷gfp or sra-6∷gfp in at least one AWB or ASH neuron, respectively, at wild-type levels (× 50 magnification). P-values were determined using a two-sample t-test between proportions. Only significant differences (P<0.001), compared to wild type unless indicated otherwise, are shown. | |||
| aFor strains carrying extrachromosomal arrays, data shown are the averages of two or more independent transgenic lines. All strains carry integrated copies of str-1∷gfp or sra-6∷gfp. | |||
| bAnimals were fed with bacteria expressing hda-4 dsRNA, starting at the L4 larval stage, and str-1∷gfp expression was examined 24 h later. | |||
| cData compared to kin-29(oy38). | |||
To determine whether loss of HDA-4 function at any developmental stage is sufficient to upregulate str-1∷gfp expression in kin-29 mutants, we fed late larval stage kin-29 mutant animals with bacteria expressing hda-4 dsRNA. Downregulation of hda-4 by RNAi at late larval stages led to partial suppression of the str-1∷gfp expression phenotype in adults (Table I), indicating that HDA-4 is required through adulthood to modulate CR gene expression.
Additional phenotypes of kin-29 mutants are also suppressed by mutations in mef-2 and hda-4
In addition to altered CR gene expression, kin-29 mutants exhibit a small body size and hypersensitivity to the constitutively produced dauer pheromone, such that they inappropriately enter the dauer stage at low pheromone concentrations (Lanjuin and Sengupta, 2002; Maduzia et al, 2005). However, kin-29 hda-4 and mef-2; kin-29 double mutants exhibited body lengths that were not significantly different from those of wild-type animals (Figure 2A), indicating that mutations in hda-4 and mef-2 suppress the decreased body length phenotype of kin-29 mutants. Interestingly, similar to kin-29 mutants, mef-2 single mutants were significantly smaller in body length than wild-type animals (Dichoso et al, 2000), whereas hda-4 single mutants were slightly thinner than wild-type animals, but exhibited normal body length (Figure 2A). Mutations in hda-4 and mef-2 also fully suppressed the pheromone hypersensitivity phenotype of kin-29 mutants, but did not result in pheromone hypersensitivity on their own (Figure 2B). hda-4(ok518) only partly suppressed the body length and pheromone hypersensitivity phenotypes of kin-29(oy39) mutants, and failed to suppress the phenotypes of kin-29(oy38) animals (data not shown). These results suggest that KIN-29 acts by antagonizing the functions of MEF-2 and HDA-4 in the regulation of CR gene expression, body size and dauer pheromone hypersensitivity.
Figure 2.
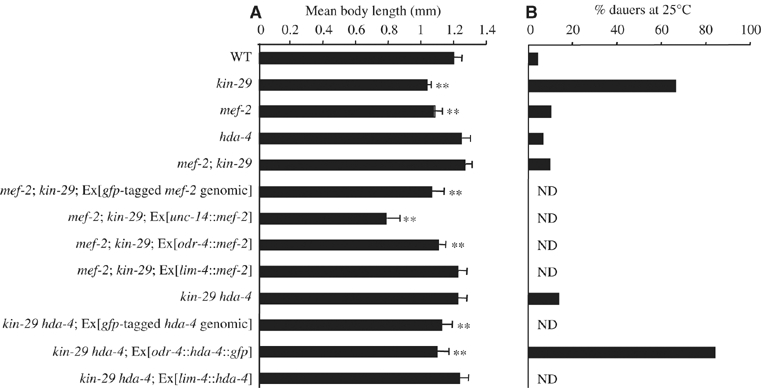
lf mutations in mef-2 and hda-4 suppress the decreased body size and pheromone hypersensitivity phenotypes of kin-29 mutants. (A, B) Body lengths (A) and dauer pheromone hypersensitivity (B) of animals of the indicated genotypes. Alleles used were kin-29(oy38), kin-29(oy39), mef-2(gv1), mef-2(oy65) and hda-4(oy57). No significant differences were observed between animals carrying different alleles of a gene. For strains carrying extrachromosomal arrays, data shown are the average of two or more independent transgenic lines, which were not significantly different from each other using a two-sample t-test. For dauer assays, data shown are from a single experiment with all strains assayed in parallel, with the exception of strains carrying transgenic arrays. These were tested independently together with animals that have lost the array as an internal control. All strains contain stably integrated str-1∷gfp fusion genes. Error bars denote the s.d.; n=35–48 (A) and 200–400 (B). Double asterisks denote values that are different from wild type at P<0.001. n.d., not done.
MEF-2 and HDA-4 act in the chemosensory neurons to suppress all kin-29 phenotypes
Previous reports indicated that MEF-2 and HDA-4 proteins were broadly expressed in multiple cell types (Dichoso et al, 2000; Choi et al, 2002). To determine whether MEF-2 and HDA-4 were expressed in the AWB and ASH chemosensory neurons, we generated gfp-tagged mef-2 and hda-4 genomic constructs. These constructs fully rescued the mef-2- and hda-4-mediated suppression, respectively, of CR∷gfp gene expression and body size phenotypes of kin-29 mutants (Table I and Figure 2A). GFP was localized to the nuclei of multiple sensory (including the AWB and ASH chemosensory) and non-sensory neurons, as well as in non-neuronal cells (data not shown). Nuclear localization was observed at all developmental stages, including the dauer stage and under different environmental conditions, including exposure to high concentrations of pheromone, absence of food and heat stress.
We previously showed that KIN-29 acts cell-autonomously to regulate CR gene expression and non-cell-autonomously in chemosensory neurons to regulate body size and entry into the dauer stage (Lanjuin and Sengupta, 2002). We explored the site(s) of action of MEF-2 and HDA-4 by expressing mef-2 and hda-4 cDNAs specifically in different cell types using cell- and tissue-specific promoters and assaying for loss of suppression of kin-29 mutant phenotypes. Expression of mef-2 and hda-4 in the AWB neurons under the lim-4 (Sagasti et al, 1999) or srd-23 (Colosimo et al, 2004) promoters resulted in reduced str-1∷gfp expression levels similar to those observed in kin-29 single mutants, but did not lead to decreased body length (Table I and Figure 2A). However, expression of both genes under the pan-neural unc-14 promoter (Ogura et al, 1997) or the odr-4 promoter, which drive expression in most chemosensory neurons (Dwyer et al, 1998), was sufficient to result in loss of suppression of other kin-29 phenotypes (Table I and Figure 2). These results indicate that similar to KIN-29, expression of MEF-2 and HDA-4 in the AWB neurons is sufficient to regulate CR gene expression, whereas expression in the chemosensory neurons is sufficient to regulate body size and entry into the dauer stage.
Phosphorylation of HDA-4 by KIN-29 is necessary for str-1 expression
In mammals, class II HDACs are phosphorylated at two conserved residues by upstream kinases, including CaMKII. Phosphorylated HDACs translocate from the nucleus to the cytoplasm, thereby alleviating HDAC-mediated repression of gene expression (McKinsey et al, 2000a, 2000b; Kao et al, 2001; Chawla et al, 2003; Linseman et al, 2003). Mutations of two CaMK phosphorylation sites in HDAC5 have been shown to result in increased nuclear localization and constitutive repression of gene expression (Lu et al, 2000a; McKinsey et al, 2000a, 2000b). Alignment of HDA-4 sequences with those of class II HDACs identified S198 in HDA-4 as analogously located to the CaMK target S259 residue in mammalian HDAC5 (Figure 3A), although no residue analogous to the S498 target amino acid could be definitively identified (data not shown). We generated full-length genomic hda-4 genes encoding either wild-type HDA-4 or HDA-4(S198A) driven under the hda-4 promoter. Introduction of either wild-type HDA-4 or HDA-4(S198A) fully abolished the suppression of str-1∷gfp expression phenotype in kin-29 hda-4 mutants (Table II), indicating that the HDA-4(S198A) mutant protein was capable of rescuing the hda-4 mutant phenotype. However, we observed significant downregulation of str-1∷gfp expression upon introduction of HDA-4(S198A), but not wild-type HDA-4, into wild-type or hda-4(oy57) animals (Table II). Both wild-type and HDA-4(S198A) retained nuclear localization in wild-type and kin-29 mutant backgrounds (data not shown). These results suggest that phosphorylation of HDA-4 at S198 may be required to alleviate HDA-4-mediated repression of str-1∷gfp gene expression, and that HDA-4(S198A) acts dominantly to repress str-1∷gfp expression.
Figure 3.
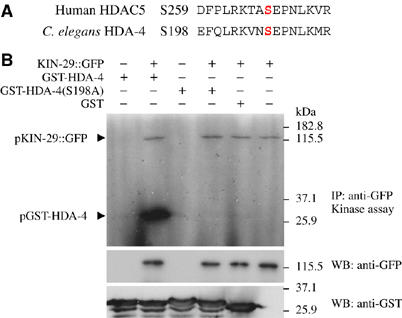
KIN-29 phosphorylates HDA-4 at residue S198 in vitro. (A) Alignment of the CaMK/SIK phosphorylation target sequences in HDAC5 and HDA-4. The targeted S259 and S198 residues in HDAC5 and HDA-4, respectively, are in red. (B) In vitro kinase assay. Phosphorylated proteins (pKIN-29∷GFP and pGST-HDA-4) were detected by autoradiography. Substrates used were bacterially purified wild-type or S198A mutated HDA-4 (residues 191–204) fused to GST or GST alone. Western blotting was performed with anti-GFP and anti-GST antibodies to detect KIN-29∷GFP and GST fusion proteins, respectively.
Table 2.
The HDA-4(S198A) mutation constitutively represses str-1∷gfp expression
| Straina,b | % expressing str-1∷gfp at WT levels | n | P-values |
|---|---|---|---|
| Wild type | 100 | 120 | |
| kin-29(oy38) | 0 | 120 | <0.001 |
| hda-4(oy57) | 100 | 109 | |
| Ex[hda-4∷hda-4] | 100 | 110 | |
| Ex[hda-4∷hda-4 (S198A)] | 18 | 112 | <0.001 |
| kin-29(oy38); Ex[hda-4∷hda-4] | 0 | 102 | <0.001 |
| kin-29(oy38); Ex[hda-4∷hda-4 (S198A)] | 0 | 105 | <0.001 |
| hda-4(oy57); Ex[hda-4∷hda-4] | 100 | 117 | |
| hda-4(oy57); Ex[hda-4∷hda-4 (S198A)] | 17 | 115 | <0.001 |
| kin-29(oy38) hda-4(oy57) | 100 | 119 | |
| kin-29(oy38) hda-4(oy57); Ex[hda-4∷hda-4] | 10 | 108 | <0.001 |
| kin-29(oy38) hda-4(oy57); Ex[hda-4∷hda-4 (S198A)] | 11 | 110 | <0.001 |
| Adult animals grown at 20°C were examined. Shown are the percentages of animals expressing str-1∷gfp in at least one AWB neuron at wild-type levels (× 50 magnification). P-values were determined using a two-sample t-test between proportions. Only significant differences (P<0.001), compared to wild type, are shown. | |||
| aAll strains contain stably integrated str-1∷gfp fusion genes. | |||
| bFor strains carrying extrachromosomal arrays, data shown are the averages of two or more independent transgenic lines. All hda-4 genomic sequences are tagged with gfp coding sequences. | |||
The consensus motif for SIK phosphorylation has been determined (Horike et al, 2003; Screaton et al, 2004). In HDAC5, this motif is identical to the motif targeted by CaMKs (McKinsey et al, 2000a; Screaton et al, 2004), and SIK2 can phosphorylate HDAC5 at this site in vitro (Screaton et al, 2004). We explored whether KIN-29 can also directly phosphorylate HDA-4. Functional GFP-tagged KIN-29 was immunoprecipitated from worm extracts with anti-GFP antibodies and incubated with bacterially purified HDA-4 peptides in the presence of [γ-32P]ATP. As shown previously, KIN-29 undergoes autophosphorylation at an unidentified site(s) (Maduzia et al, 2005) (Figure 3B). We also observed phosphorylation of HDA-4 by KIN-29. Consistent with the genetic data presented above, phosphorylation of HDA-4 was abrogated by the S198A mutation (Figure 3B). We did not detect phosphorylation of bacterially produced MEF-2 under similar conditions (data not shown). These results suggest that KIN-29 may directly target HDA-4 at the S198 residue.
Constitutive Ca2+ signaling can partly bypass the requirement for KIN-29 in the regulation of CR gene expression
As Ca2+ signaling has been shown to regulate MEF2/HDAC functions, we explored the possibility that Ca2+ signaling also plays a role in regulating str-1 expression via the KIN-29/MEF-2/HDA-4-mediated pathway. Loss-of-function mutations in the voltage-gated Ca2+ channel genes unc-36 and egl-19 (Schafer et al, 1996; Lee et al, 1997), and the unc-43 CaMKII gene (Reiner et al, 1999) did not result in altered str-1∷gfp expression in either wild-type or kin-29 mutant backgrounds (Table 3). However, a unc-43(gf) and egl-19(gf) allele partially suppressed the str-1∷gfp gene expression phenotype in kin-29 mutants (Table III). No gross alterations were observed in str-1∷gfp gene expression in a wild-type background. The gf mutation in unc-43 is predicted to result in a Ca2+-independent, constitutively activated CaMKII enzyme (Reiner et al, 1999), whereas the gf mutation in egl-19 is predicted to prolong depolarization and result in sustained Ca2+ influx (Lee et al, 1997). Interestingly, unlike in mef-2; kin-29 or kin-29 hda-4 double mutants, in which upregulation of str-1∷gfp expression was evident at all developmental stages, upregulation of gene expression was observed primarily at later larval stages and in adults (1% of L1, 21% of L2, 67% of L3 and 82% of unc-43(gf); kin-29(oy39) L4 larvae and adults exhibited stronger str-1∷gfp expression; n=100). Moreover, unc-43(gf) failed to suppress the dauer pheromone hypersensitivity phenotype and the sra-6∷gfp expression downregulation phenotype of kin-29 mutants (Supplementary Table 2). These results indicate that increased or constitutive Ca2+ signaling is able to partially bypass the requirement for a subset of KIN-29 functions in a subset of cells at later developmental stages. HDA-4(S198A) reduced the unc-43(gf)-mediated upregulation of str-1∷gfp expression in kin-29 mutants (Table III), suggesting, but not proving, that constitutively activated UNC-43 may bypass KIN-29 by phosphorylating HDA-4 at the S198 residue.
Table 3.
Constitutive Ca2+ signaling partially bypasses the requirement for KIN-29 in regulating str-1∷gfp expression
| Straina | % expressing str-1∷gfp at WT levels | n | P-values |
|---|---|---|---|
| Wild type | 100 | 200 | |
| kin-29(oy39) | 0 | 200 | <0.001 |
| kin-29(oy38) | 0 | 200 | <0.001 |
| CaMKII | |||
| unc-43(e408)lf | 100 | 134 | |
| unc-43(n498)gf | 100 | 153 | |
| unc-43(e408)lf; kin-29(oy39) | 0 | 112 | b |
| unc-43(e408)lf; kin-29(oy38) | 0 | 140 | b |
| unc-43(n498)gf; kin-29(oy39) | 66 | 167 | <0.001b |
| unc-43(n498)gf; kin-29(oy38) | 72 | 122 | <0.001b |
| unc-43(n498)gf; kin-29(oy38); Ex[593 bp str-1∷gfp] | 78 | 115 | <0.001b |
| unc-43(n498)gf; kin-29(oy38); Ex[hda-4∷hda-4 (S198A)] | 34 | 135 | <0.001b |
| Ca2+ channels | |||
| unc-36(e251) | 100 | 120 | |
| egl-19(n582)lf | 100 | 100 | |
| egl-19(n2368)gf | 100 | 130 | |
| unc-36(e251); kin-29(oy39) | 0 | 125 | b |
| egl-19(n582)lf; kin-29(oy39) | 0 | 150 | b |
| egl-19(n2368)gf; kin-29(oy39) | 56 | 34 | <0.001b |
| Data shown are from adult animals grown at 20°C. Shown are the percentages of animals expressing str-1∷gfp in at least one AWB neuron at wild-type levels (× 50 magnification). P-values were determined using a two-sample t-test between proportions. Only significant differences (P<0.001), compared to wild type or kin-29 as indicated, are shown. | |||
| aAll strains contain stably integrated copies of str-1∷gfp with the exception of unc-43(n498)gf; kin-29(oy38), which carries an extrachromosomal array of a construct containing 593-bp str-1 promoter sequences fused to gfp. The hda-4 genomic sequence is tagged with a gfp-coding sequence. | |||
| bData compared to kin-29(oy38) and kin-29(oy39). | |||
Calcineurin acts in parallel to the KIN-29-regulated pathway to modulate CR gene expression
The Ca2+-dependent Ser/Thr phosphatase calcineurin dephosphorylates and antagonizes SIK-mediated phosphorylation of the CREB coactivator TORC2 in the regulation of gene expression in cultured cells (Screaton et al, 2004). Calcineurin also activates MEF2 via desphosphorylation of a specific residue, coupled with suppression of sumoylation at a neighboring residue (Mao and Wiedmann, 1999; Wu et al, 2001; Gregoire et al, 2006; Kang et al, 2006; Shalizi et al, 2006). To determine the role of calcineurin in KIN-29/MEF-2-regulated CR gene expression, we examined str-1∷gfp expression in animals mutant for the cnb-1 calcineurin regulatory and the tax-6 calcineurin catalytic subunit genes (Bandyopadhyay et al, 2002; Kuhara et al, 2002). However, lf mutations in either gene did not affect str-1∷gfp expression (Table IV). We next determined whether mutations in tax-6/cnb-1 could bypass the requirement for KIN-29 in the regulation of str-1∷gfp expression. lf mutations in both tax-6 and cnb-1 partially suppressed the str-1∷gfp expression phenotypes of both kin-29(oy38) null and kin-29(oy39) missense mutants (Table IV), suggesting that calcineurin antagonizes KIN-29 to regulate CR gene expression. Calcineurin targets the S408/S444 residue in MEF2A/D for dephosphorylation, which, in turn, has been shown to be targeted for phosphorylation by the CDK5 Ser/Thr kinase (Gong et al, 2003; Flavell et al, 2006; Shalizi et al, 2006). Mutation of the corresponding conserved residue (S321) in MEF-2, or lf mutations in the cdk-5 ortholog, did not affect MEF-2 functions (Table IV), suggesting that calcineurin may target another site in MEF-2 or target alternate regulatory proteins.
Table 4.
lf mutations in calcineurin suppress kin-29(lf) mutations
| Straina,b | % expressing str-1∷gfp at WT levels | n | P-values |
|---|---|---|---|
| Wild type | 100 | 250 | |
| kin-29(oy39) | 0 | 250 | <0.001 |
| kin-29(oy38) | 0 | 250 | <0.001 |
| Calcineurin | |||
| cnb-1(jh103) | 100 | 206 | |
| tax-6(p675) | 100 | 233 | |
| cnb-1(jh103); kin-29(oy39) | 27 | 180 | <0.001c |
| cnb-1(jh103); kin-29(oy38) | 36 | 145 | <0.001c |
| tax-6(p675); kin-29(oy39) | 32 | 155 | <0.001c |
| SUMO/CDK5 | |||
| smo-1(ok359) | 100 | 102 | |
| cdk-5(ok626) | 100 | 155 | |
| smo-1(ok359); kin-29(oy39) | 0 | 108 | c |
| cdk-5(ok626); kin-29(oy39) | 0 | 167 | c |
| mef-2(oy65); kin-29(oy39) | 100 | 240 | |
| mef-2(oy65); kin-29(oy39); Ex[mef-2∷mef-2 genomic] | 9 | 195 | <0.001 |
| mef-2(oy65); kin-29(oy39); Ex[mef-2∷mef-2 (K316R)] | 6 | 164 | <0.001 |
| mef-2(oy65); kin-29(oy39); Ex[mef-2∷mef-2 (S321A)] | 6 | 200 | <0.001 |
| Ex[mef-2∷mef-2 genomic] | 100 | 145 | |
| Ex[mef-2∷mef-2 (K316R)] | 100 | 134 | |
| Adult animals grown at 20°C were examined. P-values were determined using a two-sample t-test between proportions. Only significant differences (P<0.001), compared to wild type or kin-29 as indicated, are shown. | |||
| aAll strains contain stably integrated str-1∷gfp fusion genes. | |||
| bFor strains carrying extrachromosomal arrays, data shown are the average of three or more independent transgenic lines. | |||
| cData compared to kin-29(oy38) and kin-29(oy39). | |||
As sumoylation has also been implicated in the regulation of MEF2 function, we next determined whether sumoylation plays a role in modulating MEF-2 activity. The residue targeted for sumoylation in MEF2 proteins is conserved in MEF-2. However, an MEF-2(K316R) mutation did not affect str-1∷gfp expression in wild-type animals, and fully rescued the mef-2-mediated suppression of the str-1∷gfp expression phenotype in kin-29 mutants (Table IV). Consistent with this result, lf mutations in smo-1, the sole SUMO ortholog encoded by the C. elegans genome (Broday et al, 2004), also did not alter str-1∷gfp expression levels (Table IV). This result suggests that sumoylation does not play a major role in this gene regulatory process.
MEF-2 directly binds CR upstream regulatory sequences to regulate gene expression
We determined whether MEF-2 regulates CR gene expression directly or indirectly, by carrying out a deletion analysis of the str-1 upstream regulatory sequences to define KIN-29- and MEF-2-regulated sites (Figure 4A). Previously, 1.8 kb of str-1 upstream sequences were shown to drive KIN-29-regulated gfp expression in the AWB neurons (Troemel et al, 1997; Lanjuin and Sengupta, 2002). We narrowed the required minimal sequences to 593 bp upstream of the start codon. Further deletion analyses within these minimal sequences defined two regions required for correct regulation of str-1∷gfp expression. Although ∼200 bp sequences upstream of the start codon drove gfp expression in the AWB neurons, expression driven by these sequences was no longer regulated by KIN-29 (Figure 4A). Deletion of a 29 bp sequence located −237 to −208 relative to the start codon also fully abolished KIN-29-mediated regulation of expression in the context of either the 1.8 kb or the 593 bp minimal str-1 regulatory sequences (Figure 4A). These results suggest that the element required for activation of str-1∷gfp expression in the AWB neurons is distinct from the element required for KIN-29-mediated modulation of gene expression.
Figure 4.
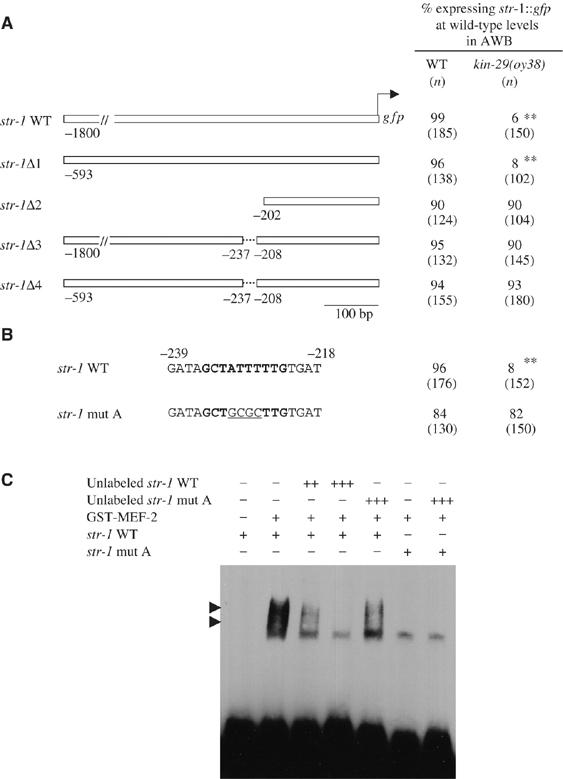
MEF-2 binds directly to str-1 regulatory sequences to regulate gene expression. (A) Expression of gfp driven by the indicated str-1 regulatory sequences in wild-type and kin-29(oy38) mutant animals. Numbers shown are the average of at least two independent transgenic lines carrying each construct. Animals were examined at × 50 magnification. Double asterisks denote values different from wild type at P<0.001 using a two-sample t-test between proportions. (B) A predicted MEF-2-binding site is in bold. Mutations generated in this site are underlined. Numbers shown are the average of two independent transgenic lines carrying the mutated MEF-2 site introduced into the 593 bp str-1 promoter. Animals were examined at × 50 magnification. Double asterisks denote values different from wild type at P<0.001 using a two-sample t-test between proportions. (C) The interaction of bacterially expressed GST-MEF-2 fusion protein with a predicted MEF-2-binding site from the str-1 promoter was examined using EMSA. Two strongly shifted bands are shown (arrowheads), presumably representing homodimeric or oligomeric complexes, as described previously (Dichoso et al, 2000). Concentrations of sequences used are as follows: +=10 fmol; ++=5 pmol; +++=50 pmol.
Target sequences recognized by MEF2 family members, including C. elegans MEF-2, have been well characterized (Gossett et al, 1989; Pollock and Treisman, 1991; Dichoso et al, 2000). We identified a site related to the MEF-2-binding element within the 29 bp sequence required for KIN-29-mediated regulation of str-1∷gfp expression. Mutating the core of the MEF-2-binding site alone resulted in upregulation of str-1∷gfp expression levels in kin-29 mutants (Figure 4B). Sequence analysis of the ∼3.5 kb sra-6 upstream regulatory sequences also identified a single predicted MEF-2-binding element, which, when deleted, resulted in upregulation of sra-6∷gfp expression in kin-29 mutants (Supplementary Figure 1). No effect was observed on str-1∷gfp or sra-6∷gfp expression upon mutation of the predicted MEF-2 site in a wild-type background. To confirm that MEF-2 directly regulates str-1 expression, we carried out an electrophoretic mobility shift assay (EMSA) using epitope-tagged MEF-2 protein and the MEF-2 recognition site derived from str-1 upstream sequences. MEF-2 specifically bound these sequences in vitro, as evidenced by the retarded mobility of the DNA–protein complex (Figure 4C). No binding was observed to a mutated MEF-2-binding site. These results suggest that MEF-2 directly binds CR gene regulatory sequences to modulate gene expression.
Insertion of MEF-2-binding site sequences is sufficient to confer KIN-29-mediated regulation on CR genes
We had previously shown that expression of only a subset of CR genes was altered in kin-29 mutants (Lanjuin and Sengupta, 2002). In recent work, we identified additional CR genes expressed in the AWB and other chemosensory neurons (Colosimo et al, 2004). However, expression of srsx-3, srd-23 and sru-38 CR∷gfp fusion genes was unaltered in kin-29(oy38) animals, and we did not identify predicted MEF-2-binding elements in their upstream regulatory sequences (Figure 5A and C, Supplementary Table 1). We wondered whether insertion of an MEF-2-binding site into CR gene regulatory sequences would be sufficient to confer KIN-29-mediated regulation of gene expression. As the element necessary to drive expression in the AWB neurons and the MEF-2 recognition element were located in close proximity in the str-1 regulatory sequences, we inserted a 26 bp sequence containing the predicted MEF-2-binding site derived from str-1 into the regulatory sequences of the srsx-3 gene in close proximity to an element identified as necessary to drive expression in the AWB and AWC neurons (Nokes et al, in preparation) (Figure 5B). Insertion of these sequences resulted in significant downregulation of gfp expression in both the AWB and AWC neurons in a kin-29 mutant background (Figure 5B and C). This downregulation was suppressed in mef-2; kin-29 double mutants (Figure 5C). However, insertion of the MEF-2 sequences further upstream failed to confer KIN-29-mediated regulation of expression (Figure 5C). These results indicate that the identified MEF-2-binding site is both necessary and sufficient to confer KIN-29-mediated regulation onto CR genes, but that the relative location of this site with respect to additional regulatory sites is likely to be critical for this regulation.
Figure 5.
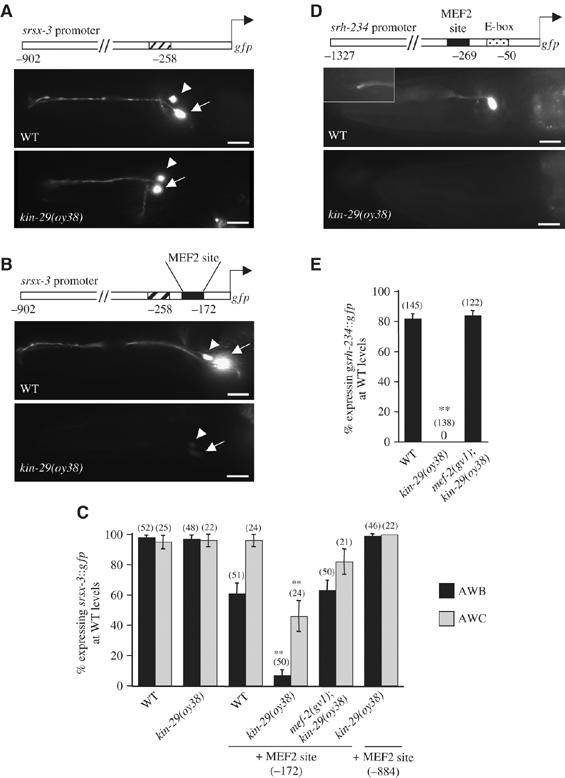
Location of an MEF-2 element in close proximity to cell-specific regulatory elements confers KIN-29-mediated regulation onto CR genes. (A, B) Expression of gfp driven by 902 bp srsx-3 regulatory sequences without (A) or with (B) an inserted mef-2 element (filled box) in AWB (arrowhead) and AWC (arrow) neurons of wild-type (top) and kin-29(oy38) mutant (bottom) animals. Hatched box indicates sequences required to drive gfp expression in the AWB and AWC neurons. Numbers indicate the position of each element in the srsx-3 regulatory sequences with respect to the start codon of srsx-3. Lateral view: anterior is on the left. Scale, 15 μm. (C) The percentage of AWB or AWC neurons of the indicated genotypes expressing srsx- 3∷gfp at wild-type levels is indicated. Numbers indicate the position of the inserted MEF-2 site in the srsx-3 upstream regulatory sequences with respect to the start codon. Adult animals grown at 20°C were examined at × 400 magnification. Numbers in parentheses indicate numbers of animals examined. Error bars denote the SEP. Double asterisks indicate values different from the matched wild-type controls at P<0.001 using a two-sample t-test between proportions. (D) Expression of srh-234∷gfp in an ADL neuron of wild-type (top) or kin-29(oy38) mutant animals. The inset shows the characteristic doublet sensory cilia of an ADL neuron. Filled box indicates the mef-2 element; stippled box indicates the E-box shown to be required to drive CR gene expression in the ADL neurons (McCarroll et al, 2005). Anterior is on the left. Scale, 15 μm. (E) Percentage of animals of the indicated genotypes expressing srh-234∷gfp in at least one ADL neuron. Adult animals grown at 20°C were examined at × 50 magnification. Error bars denote the SEP. Double asterisks indicate values different from wild type at P<0.001 using a two-sample t-test between proportions.
Identification of additional KIN-29/MEF-2-regulated CR genes
To identify additional KIN-29/MEF-2-regulated CR genes, we searched the genome for candidate CRs that contain a predicted MEF-2-binding site, as well as a cell-specific regulatory element in close proximity. Although elements driving gene expression in specific chemosensory neuron types are largely unknown, recently, an E-box motif was identified as being necessary to drive CR gene expression in the ADL chemosensory neurons (McCarroll et al, 2005). However, none of the identified ADL-expressed CR genes contained the predicted MEF-2-binding sites in their upstream regulatory sequences, and were not regulated by KIN-29 (Supplementary Table 1). We identified three additional CR genes (srh-60, srz-24, srh-234) that contained the E-box motif as well as a single predicted MEF-2-binding site in their upstream regulatory sequences. The E-box and the predicted MEF-2 site were located in close proximity upstream of the srz-24 and srh-234, but not of the srh-60 genes (Supplementary Table 1). All three gene regulatory sequences drove gfp expression in the ADL neurons (Figure 5D and Supplementary Table 1). Expression driven by the srh-234 but not the srz-24 or srh-60 regulatory sequences was strongly downregulated in kin-29 mutants (Figure 5D and E and Supplementary Table 1). This downregulation of expression was fully suppressed by mutations in mef-2 (Figure 5E). These results further suggest that the location of the predicted MEF-2 site relative to other regulatory elements is one important parameter conferring KIN-29/MEF-2-mediated regulation of gene expression onto CR genes.
Discussion
The KIN-29 SIK regulates gene expression via MEF-2/HDAC
Results presented in this work indicate that the KIN-29 SIK targets the HDA-4 class II HDAC for phosphorylation, and thereby alleviates MEF-2/HDA-4-mediated repression of CR gene expression in chemosensory neurons. Although AMPK has been suggested to regulate skeletal muscle gene expression via MEF2 and associated proteins in response to exercise (Al-Khalili et al, 2004; Holmes et al, 2005), to our knowledge, this work is the first to demonstrate that an SIK regulates MEF2/HDAC functions in the nervous system, and that this regulation is biologically relevant in vivo.
KIN-29 may target HDA-4 at the S198 residue, and an HDA-4(S198A) mutation results in constitutive repression of CR gene expression. Thus, we suggest that in kin-29 mutants, HDA-4 is constitutively dephosphorylated, resulting in localized chromatin condensation (reviewed in Yang and Gregoire, 2005) (Figure 6A and B). This alteration may prevent activation of gene expression by protein(s) binding to sites located in close proximity, such as activators required to drive str-1 expression in the AWB neurons. Upon phosphorylation by KIN-29 and/or CaMKs such as CaMKII, this repression is alleviated. In mammalian cells, phosphorylation of HDACs results in translocation of HDACs to the cytoplasm (McKinsey et al, 2000a; Kao et al, 2001; Chawla et al, 2003). Moreover, phosphorylation of TORC2 by SIK also results in cytoplasmic sequestering (Screaton et al, 2004; Koo et al, 2005). Unlike in other cultured cell types, we did not observe phosphorylation state-dependent nucleoplasmic shuttling of either HDA-4 or MEF-2. Similarly, we also observed cytoplasmic localization of KIN-29 under all conditions examined (Lanjuin and Sengupta, 2002). Thus, although in both cases we are unable to exclude transient shuttling events or shuttling of a subset of molecules, it remains formally possible that KIN-29 acts via a downstream kinase. This kinase is unlikely to be solely UNC-43 CaMKII, as lf mutations in unc-43 have no effects on CR gene expression.
Figure 6.
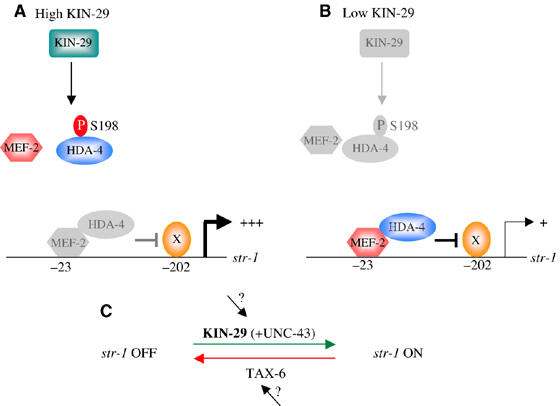
A model for regulation of str-1 CR expression by KIN-29/MEF-2/HDA-4. (A) Under conditions of high KIN-29 activity, HDA-4 is phosphorylated by KIN-29 at S198, inhibiting MEF-2/HDA-4-mediated repression of gene expression. X represents a transcriptional activator that binds to sequences within −200 bp of the start codon to drive str-1 expression specifically in the AWB neurons. The gene activation functions of this protein may be inhibited by DNA-bound MEF-2/HDA-4. (B) Under conditions of low KIN-29 activity, the majority of HDA-4 molecules are unphosphorylated and interact with MEF-2 to repress str-1 expression. (C) str-1 CR gene expression is regulated by the antagonistic actions of the KIN-29 kinase and the TAX-6 calcineurin phosphatase. Phosphorylation of HDA-4 by KIN-29 and other kinases such as UNC-43 CaMKII results in activation of gene expression, whereas dephosphorylation by TAX-6 results in repression of gene expression. The inputs into KIN-29 and TAX-6 functions are not yet known.
Our results also reveal a new role of MEF2 proteins in the regulation of gene expression. In neurons and other cell types, phosphorylation of class II HDACs has been shown to result in the dissociation of HDACs from MEF2, thereby allowing MEF2 to promote gene expression in association with transcriptional coactivators (Lu et al, 2000a). However, we did not detect a requirement for MEF-2 in the positive regulation of CR gene expression, such that levels of str-1 expression are not appreciably altered in an mef-2 mutant background. MEF-2 and HDA-4 have been shown to physically interact (Choi et al, 2002), and we find that mutations in these genes act similarly to suppress kin-29 phenotypes. Thus, MEF-2 and HDA-4 likely act in a complex to regulate str-1 gene expression. These observations suggest that in the absence of KIN-29 function, direct DNA binding by MEF-2 may be required to recruit HDA-4 to the str-1 regulatory sequences, but that unlike in other systems, MEF-2 is not itself required to drive str-1 expression.
What is the role of calcineurin in the regulation of CR gene expression via KIN-29/MEF-2? In cultured cells, calcineurin antagonizes SIK to regulate TORC2 function by regulating its subcellular localization (Screaton et al, 2004). Thus, the balance between phosphorylation and dephosphorylation of TORC2 by SIK and calcineurin, respectively, regulates the gene activation properties of TORC2. A similar mechanism may operate in the regulation of CR gene expression by the KIN-29 pathway (Figure 6C). In this case, phosphorylation of the MEF-2/HDA-4 complex results in gene expression, whereas dephosphorylation results in repression. In the simplest model, we suggest that the MEF-2/HDA-4 complex is targeted by KIN-29 as well as by an additional kinase(s) such as UNC-43 CaMKII, leading to inhibition of its repressive functions. In the absence of KIN-29-mediated phosphorylation, but in the presence of calcineurin, the MEF-2/HDA-4 complex represses gene expression. Loss of calcineurin function may partly compensate for the decreased levels of phosphorylation in kin-29 mutants to alleviate this repression, thereby leading to partial suppression of the kin-29 mutant phenotype. The site(s) targeted by calcineurin in the MEF-2/HDA-4 complex in the AWB neurons is not yet known.
Multiple cis-regulatory elements drive correct levels of CR gene expression
In this analysis, we identified an element in the str-1 regulatory sequences that is required to drive str-1∷gfp expression in the AWB neurons developmentally, as well as a distinct element required to interact with MEF-2. The MEF-2-interacting site is both necessary and sufficient to modulate expression levels, but does not itself dictate spatial expression patterns. Introduction of a single MEF-2-binding site into a KIN-29-independent CR gene regulatory sequence was sufficient to confer KIN-29-mediated regulation, although the relative position of the MEF-2 site is important. These results suggest that CR gene expression is directed by multiple regulatory modules, where a module consists of a cis-regulatory site and the trans-acting factor(s) interacting with the site (Yuh and Davidson, 1996; Markstein et al, 2004). Each of these modules may be regulated by different developmental or environmental conditions, thereby conferring appropriate regulation of CR gene expression. However, it remains possible that in some CR gene regulatory sequences, multiple signaling pathways converge onto a single module.
Physiological consequences of CR gene regulation by the KIN-29/MEF-2/HDA-4 pathway
In contrast to more complex nervous systems, the C. elegans nervous system is relatively shallow, consisting of only one or a few layers of interneurons between peripheral sensory neurons and motor neurons that drive locomotion (White et al, 1986). We and others have previously suggested that perhaps to compensate for the relative simplicity of its nervous system structure, peripheral sensory neurons carry out complex functions in C. elegans, thereby contributing extensively to behavioral and developmental plasticity (Peckol et al, 2001; Lanjuin and Sengupta, 2002; Clark et al, 2006). Thus, modulation of CR gene expression via integration of external and internal cues may provide a simple mechanism for C. elegans to rapidly alter its physiological responses to changing conditions.
We show that expression of kin-29, hda-4 and mef-2 in the chemosensory neurons is sufficient not only to rescue the CR gene expression defects, but also to rescue the body size and dauer developmental defects (Lanjuin and Sengupta, 2002; this work). Correct acquisition and transduction of both external and internal sensory cues have been shown to be critical for the regulation of these physiological processes (Albert et al, 1981; Golden and Riddle, 1984; Kimura et al, 1997; Sze et al, 2000; Fujiwara et al, 2002; Lanjuin and Sengupta, 2002). The SIK family of kinases mediates feedback-mediated regulation of gluconeogenic gene expression in response to fasting or feeding, and has also been implicated in the regulation of insulin signaling in adipocytes (Horike et al, 2003; Koo et al, 2005). Moreover, the activity of AMPK in the hypothalamus in response to internal metabolic state regulates food intake and body weight (Minokoshi et al, 2004). It is thus tempting to speculate that the KIN-29 pathway may also respond to internal metabolic state, so as to appropriately modulate sensory neuron gene expression. The subset of sensory genes regulated by KIN-29 may play particularly critical roles in recognizing and responding to biologically relevant chemicals, and in regulating sensory neuron function and food intake. We note that kin-29, mef-2 and hda-4 are also expressed in multiple additional neuronal and non-neuronal cell types (Dichoso et al, 2000; Choi et al, 2002; Lanjuin and Sengupta, 2002), and are likely to act in other pathways. It will be interesting to determine whether SIKs also act via regulation of MEF2/HDAC protein functions to regulate gene expression in the nervous system of other organisms. Unlike SIK, KIN-29 may not be activated via phosphorylation by upstream kinases such as LKB1 (Lizcano et al, 2004), as mutations in the predicted LKB1 target sites did not affect KIN-29 functions (A Lanjuin and P Sengupta, unpublished observations). Thus, we expect that these molecules regulate gene expression in response to external signals via a plethora of distinct mechanisms in different cell types in vivo, thereby allowing a conserved signaling module to be utilized in multiple contexts for different gene regulatory functions.
Materials and methods
Strains
Worms were grown as previously described (Brenner, 1974). Double and triple mutant strains were constructed using standard methods and confirmed via PCR, sequencing or complementation tests.
Isolation, mapping and cloning of mef-2 and hda-4
A kin-29(oy39) mutant strain carrying integrated copies of str-1∷gfp (kyIs104) was mutagenized with EMS. mef-2(oy65), mef-2(oy63) and hda-4(oy59) were isolated as suppressors of the reduced str-1∷gfp expression phenotype of kin-29(oy39) mutants in a screen of ∼25 000 haploid genomes. hda-4(oy57) was isolated as a spontaneous suppressor of kin-29(oy38). Mutations were mapped using standard methods, and mutant phenotypes rescued by wild-type genomic sequences. The molecular identities of the mutations were determined by sequencing. Suppressor strains were outcrossed 2–4 times before characterization.
Expression constructs and generation of transgenic animals
Promoter∷gfp constructs were generated by amplifying upstream genomic sequences, which were then cloned into the pPD95.77 expression vector (gift of A Fire) or fused to gfp-coding sequences by PCR (Hobert, 2002). Site-directed mutageneses were carried out with the QuikChange site-directed mutagenesis kit (Stratagene) and confirmed by sequencing. For each promoter∷gfp construct, expression from the same extrachromosomal array was examined in wild-type and kin-29 mutant animals. mef-2 and hda-4 cDNAs were kind gifts from J Ahnn. Details of constructs are available upon request. Transgenic animals were generated using the unc-122∷dsRed or pRF4 co-injection markers.
Detection of endogenous str-1 transcripts by RT–PCR
Total RNA isolated from mixed-stage populations of animals was reverse-transcribed using an oligo-d(T) primer. Primers specific for str-1- and odr-10-coding sequences were then used to amplify str-1 and odr-10 sequences, respectively, in the same amplification reaction. The amplified PCR products were electrophoresed and quantitated using Molecular Analyst (BioRad) software. Levels of odr-10 expression are not affected by mutations in kin-29 (Lanjuin and Sengupta, 2002).
Identification of MEF-2-binding sites upstream of additional CR genes
Deletion and site-directed mutagenesis of str-1 and sra-6 upstream sequences identified a DNA motif highly similar to the mammalian consensus of MEF2 (CTA(A/T)4TA(G/A)) (Gossett et al, 1989; Pollock and Treisman, 1991; Wasserman and Fickett, 1998) that was both necessary and sufficient to confer MEF2-mediated gene regulation. We searched for MEF-2-binding sites in 1.5 kb upstream sequences of CRs containing the E-box motif (McCarroll et al, 2005) using a simple matrix-based motif detection program, Possum (http://zlab.bu.edu/~mfrith/possum/). The matrix used for the search was generated using the mammalian consensus MEF2 site and the MEF-2 sites experimentally identified in the str-1 and sra-6 promoters, together with the E-box motif. Score threshold was set at 5.
RNA interference of hda-4
RNAi was performed in the rrf-3(pk1426) background to enhance neuronal RNAi (Simmer et al, 2003). L4 larval stage rrf-3; kin-29 double mutant animals carrying stably integrated str-1∷gfp transgenes were placed on an NGM agar plate containing 1 mM IPTG and seeded with bacterial food, producing dsRNA directed against the hda-4 gene. str-1∷gfp expression was quantitated after 24 h. The L4440 empty vector was used as a control.
Electrophoretic mobility shift assay
To obtain purified MEF-2 protein, a full-length mef-2 cDNA was cloned into pGEX4T-1 (a kind gift from J Ahnn) and expressed in JM109 Escherichia coli cells. The GST-MEF-2 fusion protein was purified on a glutathione–Sepharose column following manufacturer's recommended protocols (Amersham Biosciences). EMSA was performed by incubating 1–2 μg GST-MEF-2 fusion protein with biotin-labeled and/or unlabeled wild-type or mutant sequences. Samples were electrophoresed on a 6% native polyacrylamide gel and visualized by chemiluminescence (Pierce). Sequences used were the following: str-1 wild type: 5′-GATAGCTATTTTTGTGAT: str-1 mut A: 5′-GATAGCTGCGCTTGTGAT. Mutated bases are underlined.
Kinase assay
Worm lysates from a mixed-stage population of wild-type animals carrying a stably integrated functional gfp-tagged kin-29 transgene (oyIs41) were incubated with rabbit polyclonal anti-GFP antibody (Clontech) and immunoprecipitated with protein A Sepharose 4B beads (Zymed). Immune complexes were mixed with or without substrates and divided into two aliquots. One aliquot was analyzed by Western blotting using anti-GFP and anti-GST antibodies (Amersham). The second aliquot was analyzed for KIN-29 kinase activity by incubation with 10 μCi of [γ-32P]ATP (3000 μCi/mmol). Samples were electrophoresed on an 11% denaturing polyacrylamide gel and visualized by autoradiography. Substrates used were GST-HDA-4 (aa 191–204), GST-HDA-4(S198A) (aa 191–204) or GST alone. GST fusion proteins were purified from bacteria as described above.
Examination of CR gene expression, body length and pheromone hypersensitivity phenotypes
All animals were grown at 20°C for one or two generations in well-fed conditions, using HB101 as a food source before analyses. Levels of CR promoter∷gfp expression were examined in animals under × 50 or × 400 magnification using a dissection or compound microscope equipped with epifluorescence. For body-length measurements, adult animals were measured 24 h after the final molt under × 100 magnification using Nomarski optics and an eyepiece micrometer. For pheromone hypersensitivity assays, animals were allowed to lay 50–100 eggs at room temperature on 6-cm plates with or without pheromone. Parents were then removed and the plates were incubated at 25°C. Dauer and non-dauer animals were counted 48 h later.
Supplementary Material
Supplementary Table 1
Acknowledgments
We thank Anne Lanjuin for identification of hda-4(oy57), Caron Gauthier and Mayumi Shibuya for technical assistance, Michael Greenberg, Joohong Ahnn and Man-Wah Tan for communication of results before publication, Praseeda Mullasseril for assistance with kinase assays, the Caenorhabditis Genetics Center and the C. elegans Gene Knockout Consortium for strains, Andy Fire for expression plasmids, Joohong Ahnn and Cori Bargmann for reagents and Cori Bargmann and Anne Lanjuin for comments on the manuscript. This work was supported by the NIH (F32 DC05310 (KN) and NINDS P30 NS45713) and the NSF (IBN 0129370 (PS)).
References
- Al-Khalili L, Chibalin AV, Yu M, Sjodin B, Nylen C, Zierath JR, Krook A (2004) MEF2 activation in differentiated primary human skeletal muscle cultures requires coordinated involvement of parallel pathways. Am J Physiol Cell Physiol 286: C1410–C1416 [DOI] [PubMed] [Google Scholar]
- Albert PS, Brown SJ, Riddle DL (1981) Sensory control of dauer larva formation in Caenorhabditis elegans. J Comp Neurol 198: 435–451 [DOI] [PubMed] [Google Scholar]
- Bandyopadhyay J, Lee J, Lee J, Lee JI, Yu JR, Jee C, Cho JH, Jung S, Lee MH, Zannoni S, Singson A, Kim do H, Koo HS, Ahnn J (2002) Calcineurin, a calcium/calmodulin-dependent protein phosphatase, is involved in movement, fertility, egg laying, and growth in Caenorhabditis elegans. Mol Biol Cell 13: 3281–3293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S (1974) The genetics of Caenorhabditis elegans. Genetics 77: 71–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broday L, Kolotuev I, Didier C, Bhoumik A, Gupta BP, Sternberg PW, Podbilewicz B, Ronai Z (2004) The small ubiquitlin-like modifier (SUMO) is required for gonadal and uterine-vulval morphogenesis in Caenorhabditis elegans. Genes Dev 18: 2380–2391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cases I, de Lorenzo V (2005) Promoters in the environment: transcriptional regulation in its natural context. Nat Rev Microbiol 3: 105–118 [DOI] [PubMed] [Google Scholar]
- Chawla S, Vanhoutte P, Arnold FJ, Huang CL, Bading H (2003) Neuronal activity-dependent nucleocytoplasmic shuttling of HDAC4 and HDAC5. J Neurochem 85: 151–159 [DOI] [PubMed] [Google Scholar]
- Choi KY, Ji YJ, Jee C, Kim do H, Ahnn J (2002) Characterization of CeHDA-7, a class II histone deacetylase interacting with MEF-2 in Caenorhabditis elegans. Biochem Biophys Res Commun 293: 1295–1300 [DOI] [PubMed] [Google Scholar]
- Clark DA, Biron D, Sengupta P, Samuel ADT (2006) The AFD sensory neurons encode multiple functions underlying thermotactic behavior in C. elegans. J Neurosci 26: 7444–7451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colosimo ME, Brown A, Mukhopadhyay S, Gabel C, Lanjuin AE, Samuel AD, Sengupta P (2004) Identification of thermosensory and olfactory neuron-specific genes via expression profiling of single neuron types. Curr Biol 14: 2245–2251 [DOI] [PubMed] [Google Scholar]
- Dichoso D, Brodigan T, Chwoe KY, Lee JS, Llacer R, Park M, Corsi AK, Kostas SA, Fire A, Ahnn J, Krause M (2000) The MADS-Box factor CeMEF2 is not essential for Caenorhabditis elegans myogenesis and development. Dev Biol 223: 431–440 [DOI] [PubMed] [Google Scholar]
- Dwyer ND, Troemel ER, Sengupta P, Bargmann CI (1998) Odorant receptor localization to olfactory cilia is mediated by ODR-4, a novel membrane-associated protein. Cell 93: 455–466 [DOI] [PubMed] [Google Scholar]
- Feldman JD, Vician L, Crispino M, Hoe W, Baudry M, Herschman HR (2000) The salt-inducible kinase, SIK, is induced by depolarization in brain. J Neurochem 74: 2227–2238 [DOI] [PubMed] [Google Scholar]
- Flavell SW, Cowan CW, Kim TK, Greer PL, Lin Y, Paradis S, Griffith EC, Hu LS, Chen C, Greenberg ME (2006) Activity-dependent regulation of MEF2 transcription factors suppresses excitatory synapse number. Science 311: 1008–1012 [DOI] [PubMed] [Google Scholar]
- Fujiwara M, Sengupta P, McIntire S (2002) Regulation of body size and behavioral state of C. elegans by sensory perception and the EGL-4 cGMP-dependent protein kinase. Neuron 36: 1091–1102 [DOI] [PubMed] [Google Scholar]
- Gaudilliere B, Shi Y, Bonni A (2002) RNA interference reveals a requirement for myocyte enhancer factor 2A in activity-dependent neuronal survival. J Biol Chem 277: 46442–46446 [DOI] [PubMed] [Google Scholar]
- Golden JW, Riddle DL (1984) The Caenorhabditis elegans dauer larva: developmental effects of pheromone, food, and temperature. Dev Biol 102: 368–378 [DOI] [PubMed] [Google Scholar]
- Gong X, Tang X, Wiedmann M, Wang X, Peng J, Zheng D, Blair LA, Marshall J, Mao Z (2003) Cdk5-mediated inhibition of the protective effects of transcription factor MEF2 in neurotoxicity-induced apoptosis. Neuron 38: 33–46 [DOI] [PubMed] [Google Scholar]
- Gossett LA, Kelvin DJ, Sternberg EA, Olson EN (1989) A new myocyte-specific enhancer-binding factor that recognizes a conserved element associated with multiple muscle-specific genes. Mol Cell Biol 9: 5022–5033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregoire S, Tremblay AM, Xiao L, Yang Q, Ma K, Nie J, Mao Z, Wu Z, Giguere V, Yang XJ (2006) Control of MEF2 transcriptional activity by coordinated phosphorylation and sumoylation. J Biol Chem 281: 4423–4433 [DOI] [PubMed] [Google Scholar]
- Heidenreich KA, Linseman DA (2004) Myocyte enhancer factor-2 transcription factors in neuronal differentiation and survival. Mol Neurobiol 29: 155–166 [DOI] [PubMed] [Google Scholar]
- Hobert O (2002) PCR fusion-based approach to create reporter gene constructs for expression analysis in transgenic C. elegans. Biotechniques 32: 728–730 [DOI] [PubMed] [Google Scholar]
- Holmes BF, Sparling DP, Olson AL, Winder WW, Dohm GL (2005) Regulation of muscle GLUT4 enhancer factor and myocyte enhancer factor 2 by AMP-activated protein kinase. Am J Physiol Endocrinol Metab 289: E1071–E1076 [DOI] [PubMed] [Google Scholar]
- Horike N, Takemori H, Katoh Y, Doi J, Min L, Asano T, Sun XJ, Yamamoto H, Kasayama S, Muraoka M, Nonaka Y, Okamoto M (2003) Adipose-specific expression, phosphorylation of Ser794 in insulin receptor substrate-1, and activation in diabetic animals of salt-inducible kinase-2. J Biol Chem 278: 18440–18447 [DOI] [PubMed] [Google Scholar]
- Kahn BB, Alquier T, Carling D, Hardie DG (2005) AMP-activated protein kinase: ancient energy gauge provides clues to modern understanding of metabolism. Cell Metab 1: 15–25 [DOI] [PubMed] [Google Scholar]
- Kang J, Gocke CB, Yu H (2006) Phosphorylation-facilitated sumoylation of MEF2C negatively regulates its transcriptional activity. BMC Biochem 7: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao HY, Verdel A, Tsai CC, Simon C, Juguilon H, Khochbin S (2001) Mechanism for nucleocytoplasmic shuttling of histone deacetylase 7. J Biol Chem 276: 47496–47507 [DOI] [PubMed] [Google Scholar]
- Kim MS, Lee KU (2005) Role of hypothalamic 5′-AMP-activated protein kinase in the regulation of food intake and energy homeostasis. J Mol Med 83: 514–520 [DOI] [PubMed] [Google Scholar]
- Kimura KD, Tissenbaum HA, Liu Y, Ruvkun G (1997) daf-2, an insulin receptor-like gene that regulates longevity and diapause in Caenorhabditis elegans. Science 277: 942–946 [DOI] [PubMed] [Google Scholar]
- Koo SH, Flechner L, Qi L, Zhang X, Screaton RA, Jeffries S, Hedrick S, Xu W, Boussouar F, Brindle P, Takemori H, Montminy M (2005) The CREB coactivator TORC2 is a key regulator of fasting glucose metabolism. Nature 437: 1109–1111 [DOI] [PubMed] [Google Scholar]
- Kuhara A, Inada H, Katsura I, Mori I (2002) Negative regulation and gain control of sensory neurons by the C. elegans calcineurin TAX-6. Neuron 33: 751–763 [DOI] [PubMed] [Google Scholar]
- Lanjuin A, Sengupta P (2002) Regulation of chemosensory receptor expression and sensory signaling by the KIN-29 Ser/Thr kinase. Neuron 33: 369–381 [DOI] [PubMed] [Google Scholar]
- Lanjuin A, Sengupta P (2004) Specification of chemosensory neuron subtype identities in Caenorhabditis elegans. Curr Opin Neurobiol 14: 22–30 [DOI] [PubMed] [Google Scholar]
- Lee RY, Lobel L, Hengartner M, Horvitz HR, Avery L (1997) Mutations in the alpha1 subunit of an L-type voltage-activated Ca2+ channel cause myotonia in Caenorhabditis elegans. EMBO J 16: 6066–6076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Linseman DA, Allen MP, Meintzer MK, Wang X, Laessig T, Wierman ME, Heidenreich KA (2001) Myocyte enhancer factor 2A and 2D undergo phosphorylation and caspase-mediated degradation during apoptosis of rat cerebellar granule neurons. J Neurosci 21: 6544–6552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linseman DA, Bartley CM, Le SS, Laessig TA, Bouchard RJ, Meintzer MK, Li M, Heidenreich KA (2003) Inactivation of the myocyte enhancer factor-2 repressor histone deacetylase-5 by endogenous Ca(2+)//calmodulin-dependent kinase II promotes depolarization-mediated cerebellar granule neuron survival. J Biol Chem 278: 41472–41481 [DOI] [PubMed] [Google Scholar]
- Liu Y, Randall WR, Schneider MF (2005) Activity-dependent and -independent nuclear fluxes of HDAC4 mediated by different kinases in adult skeletal muscle. J Cell Biol 168: 887–897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lizcano JM, Goransson O, Toth R, Deak M, Morrice NA, Boudeau J, Hawley SA, Udd L, Makela TP, Hardie DG, Alessi DR (2004) LKB1 is a master kinase that activates 13 kinases of the AMPK subfamiy, including MARK/PAR-1. EMBO J 23: 833–843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, McKinsey TA, Nicol RL, Olson EN (2000a) Signal-dependent activation of the MEF2 transcription factor by dissociation from histone deacetylases. Proc Natl Acad Sci USA 97: 4070–4075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, McKinsey TA, Zhang CL, Olson EN (2000b) Regulation of skeletal myogenesis by association of the MEF2 transcription factor with class II histone deacetylases. Mol Cell 6: 233–244 [DOI] [PubMed] [Google Scholar]
- Maduzia LL, Roberts AF, Wang H, Lin X, Chin LJ, Zimmerman CM, Cohen S, Feng XH, Padgett R (2005) C. elegans serine-threonine kinase KIN-29 modulates TGFβ signaling and regulates body size formation. BMC Dev Biol 5: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Z, Bonni A, Xia F, Nadal-Vicens M, Greenberg ME (1999) Neuronal activity-dependent cell survival mediated by transcription factor MEF2. Science 286: 785–790 [DOI] [PubMed] [Google Scholar]
- Mao Z, Wiedmann M (1999) Calcineurin enhances MEF2 DNA binding activity in calcium-dependent survival of cerebellar granule neurons. J Biol Chem 274: 31102–31107 [DOI] [PubMed] [Google Scholar]
- Markstein M, Zinzen R, Markstein P, Yee KP, Erives A, Stathopoulos A, Levine M (2004) A regulatory code for neurogenic gene expression in the Drosophila embryo. Development 131: 2387–2394 [DOI] [PubMed] [Google Scholar]
- McCarroll SA, Li H, Bargmann CI (2005) Identification of transcriptional regulatory elements in chemosensory receptor genes by probabilistic segmentation. Curr Biol 15: 347–352 [DOI] [PubMed] [Google Scholar]
- McGee SL, Hargreaves M (2006) Exercise and skeletal muscle glucose transporter 4 expression: molecular mechanisms. Clin Exp Pharm Phys 33: 395–399 [DOI] [PubMed] [Google Scholar]
- McKinsey TA, Zhang CL, Lu J, Olson EN (2000a) Signal-dependent nuclear export of a histone deacetylase regulates muscle differentiation. Nature 408: 106–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinsey TA, Zhang CL, Olson EN (2000b) Activation of the myocyte enhancer factor-2 transcription factor by calcium/calmodulin-dependent protein kinase-stimulated binding of 14-3-3 to histone deacetylase 5. Proc Natl Acad Sci USA 97: 14400–14405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinsey TA, Zhang CL, Olson EN (2002) MEF2: a calcium-dependent regulator of cell division, differentiation and death. Trends Biochem Sci 27: 40–47 [DOI] [PubMed] [Google Scholar]
- Minokoshi Y, Alquier T, Furukawa N, Kim YB, Lee A, Xue B, Mu J, Foufelle F, Ferre P, Birnbaum MJ, Stuck BJ, Kahn BB (2004) AMP-kinase regulates food intake by responding to hormonal and nutrient signals in the hypothalamus. Nature 428: 569–574 [DOI] [PubMed] [Google Scholar]
- Nolan KM, Sarafi-Reinach TR, Horne JG, Saffer AM, Sengupta P (2002) The DAF-7 TGF-beta signaling pathway regulates chemosensory receptor gene expression in C. elegans. Genes Dev 16: 3061–3073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogura K, Shirakawa M, Barnes TM, Hekimi S, Ohshima Y (1997) The UNC-14 protein required for axonal elongation and guidance in Caenorhabditis elegans interacts with the serine/threonine kinase UNC-51. Genes Dev 11: 1801–1811 [DOI] [PubMed] [Google Scholar]
- Okamoto M, Takemori H, Katoh Y (2004) Salt-inducible kinase in steroidogenesis and adipogenesis. Trends Endocrinol Metab 15: 21–26 [DOI] [PubMed] [Google Scholar]
- Peckol EL, Troemel ER, Bargmann CI (2001) Sensory experience and sensory activity regulate chemosensory receptor gene expression in C. elegans. Proc Natl Acad Sci USA 98: 11032–11038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollock R, Treisman R (1991) Human SRF-related proteins: DNA-binding properties and potential regulatory targets. Genes Dev 5: 2327–2341 [DOI] [PubMed] [Google Scholar]
- Reiner DJ, Newton EM, Tian H, Thomas JH (1999) Diverse behavioural defects caused by mutations in Caenorhabditis elegans unc-43 CaM kinase II. Nature 402: 199–203 [DOI] [PubMed] [Google Scholar]
- Riquelme C, Barthel KK, Liu X (2006) SUMO-1 modification of MEF2A regulates its transcriptional activity. J Cell Mol Med 10: 132–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagasti A, Hobert O, Troemel ER, Ruvkun G, Bargmann CI (1999) Alternative olfactory neuron fates are specified by the LIM homeobox gene lim-4. Genes Dev 13: 1794–1806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer WR, Sanchez BM, Kenyon CJ (1996) Genes affecting sensitivity to serotonin in Caenorhabditis elegans. Genetics 143: 1219–1230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Screaton RA, Conkright MD, Katoh Y, Best JL, Canettieri G, Jeffries S, Guzman E, Niessen S, Yates JR III, Takemori H, Okamoto M, Montminy M (2004) The CREB coactivator TORC2 functions as a calcium- and cAMP-sensitive coincidence detector. Cell 119: 61–74 [DOI] [PubMed] [Google Scholar]
- Shalizi A, Gaudilliere B, Yuan Z, Stegmuller J, Shirogane T, Ge Q, Tan Y, Schulman B, Harper JW, Bonni A (2006) A calcium-regulated MEF2 sumoylation switch controls postsynaptic differentiation. Science 311: 1012–1017 [DOI] [PubMed] [Google Scholar]
- Simmer F, Moorman C, Van Der Linden AM, Kuijk E, Van Den Berghe PV, Kamath R, Fraser AG, Ahringer J, Plasterk RH (2003) Genome-wide RNAi of C. elegans using the hypersensitive rrf-3 strain reveals novel gene functions. PLoS Biol 1: 77–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sze JY, Victor M, Loer C, Shi Y, Ruvkun G (2000) Food and metabolic signalling defects in a Caenorhabditis elegans serotonin-synthesis mutant. Nature 403: 560–564 [DOI] [PubMed] [Google Scholar]
- Tobin D, Madsen D, Kahn-Kirby A, Peckol E, Moulder G, Barstead R, Maricq A, Bargmann CI (2002) Combinatorial expression of TRPV channel proteins defines their sensory functions and subcellular localization in C. elegans neurons. Neuron 35: 307–318 [DOI] [PubMed] [Google Scholar]
- Troemel ER, Kimmel BE, Bargmann CI (1997) Reprogramming chemotaxis responses: sensory neurons define olfactory preferences in C. elegans. Cell 91: 161–169 [DOI] [PubMed] [Google Scholar]
- Troemel ER, Sagasti A, Bargmann CI (1999) Lateral signaling mediated by axon contact and calcium entry regulates asymmetric odorant receptor expression in C. elegans. Cell 99: 387–398 [DOI] [PubMed] [Google Scholar]
- Wang Z, Takemori H, Halder SK, Nonaka Y, Okamoto M (1999) Cloning of a novel kinase (SIK) of the SNF1/AMPK family from high salt diet-treated rat adrenal. FEBS Lett 453: 135–139 [DOI] [PubMed] [Google Scholar]
- Wasserman WW, Fickett JW (1998) Identification of regulatory regions which confer muscle-specific gene expression. J Mol Biol 278: 167–181 [DOI] [PubMed] [Google Scholar]
- White JG, Southgate E, Thomson JN, Brenner S (1986) The structure of the nervous system of the nematode Caenorhabditis elegans. Phil Trans R Soc London B 314: 1–340 [DOI] [PubMed] [Google Scholar]
- Wu H, Naya FJ, McKinsey TA, Mercer B, Shelton JM, Chin ER, Simard AR, Michel RN, Bassel-Duby R, Olson EN, Williams RS (2000) MEF2 responds to multiple calcium-regulated signals in the control of skeletal muscle fiber type. EMBO J 19: 1963–1973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Rothermel B, Kanatous S, Rosenberg P, Naya FJ, Shelton JM, Hutcheson KA, DiMaio JM, Olson EN, Bassel-Duby R, Williams RS (2001) Activation of MEF2 by muscle activity is mediated through a calcineurin-dependent pathway. EMBO J 20: 6414–6423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyrick JJ, Young RA (2002) Deciphering gene expression regulatory networks. Curr Opin Genet Dev 12: 130–136 [DOI] [PubMed] [Google Scholar]
- Yang XJ, Gregoire S (2005) Class II histone deacetylases: from sequence to function, regulation, and clinical implication. Mol Cell Biol 25: 2873–2884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuh CH, Davidson EH (1996) Modular cis-regulatory organization of Endo16, a gut-specific gene of the sea urchin embryo. Development 122: 1069–1082 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1


