Abstract
Nutrient starvation or rapamycin treatment, through inhibition of target of rapamycin, causes condensation of ribosomal DNA (rDNA) array and nucleolar contraction in budding yeast. Here we report that under such conditions, condensin is rapidly relocated into the nucleolus and loaded to rDNA tandem repeats, which is required for rDNA condensation. Rpd3-dependent histone deacetylation is necessary and sufficient for condensin's relocalization and loading to rDNA array, suggesting that histone modification plays a regulatory role for condensin targeting. Rapamycin independently, yet coordinately, inhibits rDNA transcription and promotes condensin loading to rDNA array. Unexpectedly, we found that inhibition of rDNA transcription in the absence of condensin loading leads to rDNA instability. Our data suggest that enrichment of condensin prevents rDNA instability during nutrient starvation. Together, these observations unravel a novel role for condensin in the maintenance of regional genomic stability.
Keywords: condensin, extrachromosomal rDNA circle, ribosomal DNA, target of rapamycin, the nucleolus
Introduction
Control of cell growth requires that cells rapidly change their protein biosynthetic capacity by altering ribosome concentration in response to nutrient availability. In budding yeast, this is accomplished by regulating the transcription of ribosomal protein (RP) and ribosomal RNA (rRNA) genes, which represent over 90% of total transcription (Warner, 1999; Moss, 2004; Rudra and Warner, 2004). In budding yeast, there are on average 150 tandem copies of rRNA genes located on chromosome XII. In addition to encoding for 35S and 5S rRNAs, ribosomal DNA (rDNA) array serves as an organizer of the nucleolus. rDNA array is unique in two ways: first, it is the most highly transcribed region of the genome; second, it is intrinsically unstable because of the highly repetitive nature that makes it an easy target for homologous recombination. Extrachromosomal rDNA circle (ERC) is a hallmark for the instability of rDNA repeats. When there is a double-strand break at rDNA, the free DNA end will invade a complementary sequence within rDNA array. After recombination, this may lead to the formation of an ERC containing varying number of rDNA repeats, depending on which repeat is being paired, and a loss of the same number of repeats from rDNA array. Under normal conditions, nevertheless, rDNA array remains stable, as indicated by its relatively stable size and lack of detectable ERCs. Under certain pathological situations such as aging, however, the number of rDNA repeats can fluctuate, which is accompanied by ERC accumulation as the recombination by-product. Mutations in genes such as SIR2, TOP1/2 and RPA135 are known to cause formation of ERCs and/or shrinkage of rDNA array (Brewer et al, 1992; Kaeberlein et al, 1999).
Condensation of chromosomal DNA is pivotal for chromosome segregation and accurate transmission of genetic information during cell divisions. Chromosome condensation is mediated by a highly conserved protein machinery called condensin, an abundant non-histone component of the metaphase chromosomes, which belongs to the structural maintenance of chromosomes (SMC) protein family (Milutinovich and Koshland, 2003; Legagneux et al, 2004; Hirano, 2005). Condensin is a large five-subunit complex with two core components of Smc2 and Smc4, and three non-SMC subunits called Brn1, Ycs4 and Ycs5/Ycg1 in yeast (Legagneux et al, 2004; Hirano, 2005). Both genetic and biochemical evidences indicate that condensin is critical for mitotic chromosome compaction and segregation. For example, mutations in different condensin subunits block chromosome condensation and subsequent chromosome segregation (Strunnikov et al, 1995; Lavoie et al, 2000; Ouspenski et al, 2000; Bhalla et al, 2002). In a Xenopus in vitro chromosome assembly assay, antibody blocking or immunodepletion of condensin subunits abrogates mitotic DNA condensation (Hirano and Mitchison, 1994). Condensin compacts DNA in an ATP-dependent manner: the binding of condensins to DNA in vitro in the presence of ATP introduces global positive writhe and produces positive supercoils (Kimura and Hirano, 1997; Kimura et al, 1999). A recent study with a single DNA molecule nanomanipulation technique shows in real time that condensin is able to physically compact DNA in the presence of ATP (Strick et al, 2004). In budding yeast, condensin is especially important for promoting the condensation and resolution of rDNA sister chromatids at anaphase (D'Amours et al, 2004; Sullivan et al, 2004; Wang et al, 2004). This process requires the Cdc14 phosphatase, but the mechanism is still unclear. More recently, condensin mutations have been found to affect non-mitotic/meiotic events linked to chromatin dynamics such as transcriptional repression and recombination (Hirano, 2005). However, more direct evidence is needed to rule out the possibility that global defects in chromatin structure are responsible for these diverse phenotypes observed in the condensin mutants.
Target of rapamycin (TOR) is a key regulator of ribosome biogenesis in yeast and mammals in response to nutrient conditions (Schneper et al, 2004; Tsang and Zheng, 2004). TOR regulation of ribosome biogenesis involves all three RNA polymerases, and inhibition of TOR by rapamycin or starvation leads to a rapid decrease in the expression of ribosomal genes, including 35S rRNA (by Pol I), RP (by Pol II) and 5S rRNA (by Pol III) (Zaragoza et al, 1998; Powers and Walter, 1999). We have recently observed that nutrient starvation or rapamycin treatment rapidly causes rDNA condensation and nucleolar contraction (Tsang et al, 2003). Here we present new results on the mechanism and physiological role of rDNA condensation. Our data suggest that starvation-induced rDNA condensation is a condensin-mediated and histone deacetylation-regulated event, which is important for preventing rDNA instability after rDNA transcription is turned off.
Results
Condensin is required for initiation and maintenance of rDNA condensation and nucleolar contraction
The role of condensin in mitotic chromosomal compaction suggests that condensin or a condensin-like factor is involved in starvation-induced rDNA condensation. To explore this possibility, we examined the ability of rDNA to undergo condensation when condensin is inactivated using the temperature-sensitive alleles. In the rich medium, rDNA staining of the wild-type (WT) cells typically shows large, punctate structure organized in a crescent shape in the periphery of the nucleus, occupying approximately 1/3 of the nucleus. Shifting to the starvation medium (−N) leads to rapid condensation of rDNA into a single granular or fibrillar structure (Figure 1A) (Tsang et al, 2003). The same result is obtained with ycs4-2 mutant at the permissive temperature (23°C). When Ycs4-2 is inactivated, however, rDNA structure remains in the large, crescent shape during starvation (−N). Inhibition of TOR by rapamycin also causes rDNA condensation (Tsang et al, 2003), which is also blocked by the condensin mutations (Figure 1B). Identical results were obtained for two other non-SMC condensin mutants (brn1-9 and ycs5-1) (Supplementary Figure 1A and C). These observations indicate that a functional condensin is required for starvation-induced rDNA condensation.
Figure 1.
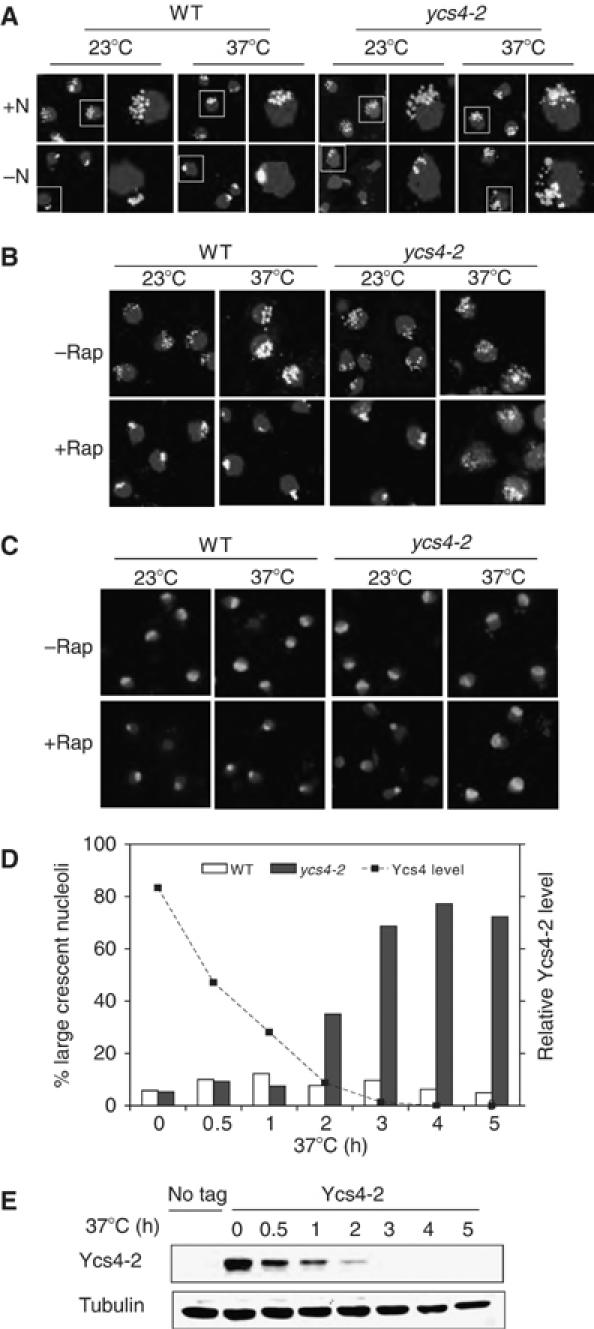
Condensin is required for condensation of rDNA array during starvation. (A) Loss of condensin blocks rDNA condensation during starvation. Early log-phase WT and ycs4-2 cells in synthetic complete (SC) medium (+N) were shifted from 23 to 37°C or were allowed to remain at 23°C for 2 h and then changed to the nutrient starvation (−N) medium or fresh SC medium (+N) and incubated for 30 min. rDNA structure was analyzed by FISH with a 25S rDNA probe (bright white). The boxed cell is enlarged on the right. Nuclear DNA was stained with DAPI (light gray). A color version of this figure is available at The EMBO Journal Online. (B) Condensin is required for rDNA condensation during rapamycin treatment. Early log-phase cultures of WT, brn1-9, ycs4-2 and ycs5-1 cells in YPD were shifted from 23 to 37°C or were allowed to remain at 23°C for 2 h, which was followed by incubation without or with 200 nM rapamycin for 30 min. rDNA and nuclei were analyzed by FISH (bright white) and DAPI (light gray), respectively. A color version of this figure is available at The EMBO Journal Online. (C) Condensin is required for rapamycin-induced nucleolar contraction. The same as Figure 1B except nucleolar structure was analyzed by IF with a Nop1 antibody (bright white). Nuclear DNA was stained with DAPI (light gray). A color version of this figure is available at The EMBO Journal Online. (D) Condensin is required for maintaining the condensed nucleolar structure in the presence of rapamycin. Early log-phase WT and ycs4-2 cells were treated with 200 nM rapamycin for 1 h at 23°C to induce nucleolar contraction and then shifted to 37°C. The condensed nucleolus is quantitated (N=100 for each data point) and plotted against Ycs4-2 level. (E) Rapid loss of Ycs4-2 at the restrictive temperature. Ycs4-2 with chromosomally tagged Myc12 in the ycs4-2 strain is analyzed by Western blot at different conditions.
As condensin mutations may globally affect the overall genomic structure, we analyzed rDNA structural change using the restriction ratio (the area of rDNA fluorescence in situ hybridization (FISH) signal divided by the area of DAPI signal) (Freeman et al, 2000). This measurement more accurately reflects the intrinsic change of rDNA by taking the entire nuclear DNA into consideration. Cells with a restriction ratio ⩾0.3 are scored as having a normal or open rDNA (O), whereas a restriction ratio ⩽0.15 indicates a condensed rDNA (C). At both temperatures, rDNA structure in the majority of WT cells is in the open conformation (O) in the rich medium, but changes to the closed conformation upon starvation (Supplementary Figure 1B and D). Essentially, the same results were obtained with condensin mutants at the permissive temperature. After condensin is inactivated in ycs4-2 cells at 37°C, however, rDNA structure remains largely in the open conformation during starvation. The nucleolus undergoes contraction during starvation or rapamycin treatment, which is closely correlated with rDNA condensation (Tsang et al, 2003). By indirect immunofluorescence (IF) staining, we found that nucleolar contraction is similarly blocked by the condensin mutations (Figure 1C). Because rDNA condensation is always accompanied by nucleolar contraction, for convenience, we typically use IF to follow both nucleolar contraction and rDNA condensation in our subsequent experiments.
We next asked whether condensin is necessary for maintaining the contracted state of the nucleolus. To address this question, we first contracted the nucleolus of ycs4-2 cells by treating them with rapamycin at the permissive temperature (23°C). Cells were then shifted to 37°C to inactivate condensin and the nucleolar structure was followed by Nop1 staining. The nucleolus begins to resume the large, crescent shape within 2 h of shifting to 37°C and the nucleolar structure in nearly all of the cells has de-contracted by 3 h (Figure 1D and Supplementary Figure 1F). Nucleolar de-contraction is closely correlated with the disappearance of Ycs4-2 (Figure 1E). Taken together, the above results demonstrate that condensin is essential for both initiation and maintenance of rDNA condensation and nucleolar contraction during nutrient starvation.
Rapamycin causes enrichment of condensin to the nucleolus and rDNA array
To better understand how condensin is involved in rDNA condensation, we examined Ycs4-Myc12 localization by IF. Ycs4 is distributed throughout the entire nucleus during normal growth in rich medium (Figure 2A). The same whole-nucleus localization pattern of Ycs4 during normal growth is also observed by Murray and co-workers (Bhalla et al, 2002) and Amon and co-workers (D'Amours et al, 2004). Upon rapamycin treatment, however, Ycs4 is found almost exclusively in the nucleolus. Rapamycin does not affect Ycs4 protein level (Supplementary Figure 2B). Similarly, Smc2-HA6 and Smc4-HA6 are distributed throughout the nucleus in early log-phase cells in rich medium, but become highly concentrated in the nucleolus upon rapamycin treatment (Supplementary Figure 2C). Thus, rapamycin treatment appears to relocate the entire canonical condensin complex into the nucleolus.
Figure 2.
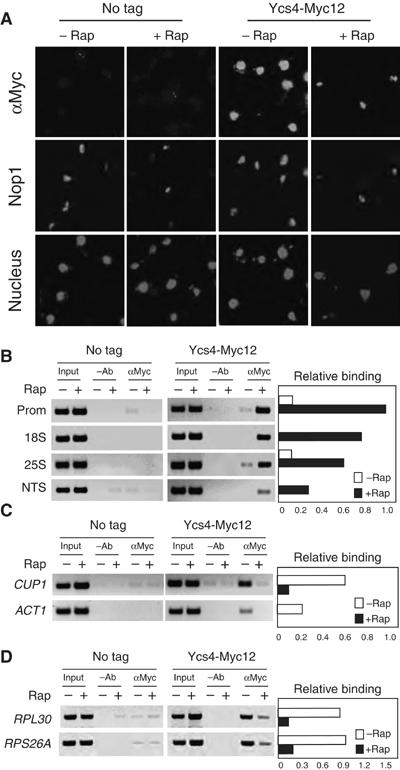
Rapamycin causes rapid relocation of condensin into the nucleolus and loading of condensin to rDNA array. (A) Rapamycin causes Ycs4 to rapidly relocate into the nucleolus. Early log-phase cultures of untagged (BLY03) and Ycs4-Myc12 chromosomally tagged (ZW206) cells in YPD were treated with or without 200 nM rapamycin for 1 h at 23°C. Nucleolar structure and distribution of Ycs4-Myc12 were examined by IF with a monoclonal Nop1 antibody (bright white) and the A14 rabbit polyclonal anti-Myc antibody (bright white), respectively. The nuclei were stained with DAPI (light gray). A color version of this figure is available at The EMBO Journal Online. (B) Rapamycin causes enrichment of Ycs4 on rDNA array. Early log-phase untagged (BLY03) and Ycs4-Myc12 (ZW206) cells were treated with 200 nM rapamycin for 30 min. Ycs4-Myc12 protein associated with rDNA chromatin was determined by ChIP with an anti-Myc antibody (9E10). Polymerase chain reactions (PCR) were performed with rDNA primer pairs as indicated in Supplementary Figure 2D. –Ab, no antibody as a negative control. The right-hand panel shows quantification of the level of Ycs4-Myc12 bound to rDNA chromatin. (C) Rapamycin causes dissociation of Ycs4 from other chromosomal regions. Samples from (B) were analyzed for Ycs4-Myc12 associated with CUP1 and ACT1. The right-hand panel shows quantification of the level of Ycs4-Myc12 bound to the CUP1 and ACT1 loci. (D) Rapamycin causes dissociation of Ycs4 from ribosomal protein (RP) genes. Samples from (B) were analyzed for Ycs4-Myc12 associated with RPL30 and RPS26A. The right-hand panel shows quantification of the level of Ycs4-Myc12 bound to the RPL30 and RPS26A loci.
To determine whether condensin is loaded to the rDNA, we performed chromatin immunoprecipitation (ChIP). In the absence of the Myc tag or Myc antibody, little or no detectable DNA was precipitated, demonstrating the specificity of the ChIP assay (Figure 2B–D). Ycs4 normally associates with rDNA at a low level (Figure 2B). Upon rapamycin treatment, however, rDNA-bound Ycs4 is significantly increased. Interestingly, the highest Ycs4 level is found at the promoter, which is followed by 18S, 25S and NTS (non-transcribed spacer) (Figure 2B). This condensin gradient may be generated by loading condensin first at the promoter and then spreading unidirectionally toward the transcribed regions. Alternatively, it may reflect a functional need for loading variable amount of condensins to different rDNA sequences. Further study in this area is needed to clarify these possibilities. In contrast, Ycs4 association with CUP1, ACT1 and RP genes is significantly decreased by rapamycin treatment (Figure 2C and D). These observations suggest that rapamycin causes a redistribution of condensins to rDNA array from other genomic regions, and possibly from an unbound pool of condensins as well.
Histone deacetylation promotes condensin enrichment to the nucleolus and the rDNA
Rpd3 is a yeast histone deacetylase (HDAC) that becomes associated with rDNA array upon nutrient starvation, which is responsible for histone H4 deacetylation at K5,12 in the 35S rDNA chromatin region (Tsang et al, 2003). To ask whether histone deacetylation plays a role in condensin recruitment, we analyzed Ycs4-Myc9 localization in the absence of Rpd3. Upon rapamycin treatment, Ycs4 becomes highly enriched in the nucleolus in the WT but not rpd3Δ strain (Figure 3A). In addition, rpd3Δ mutation abrogates rapamycin-induced Ycs4 loading to the rDNA (Figure 3B and Supplementary Figure 3B). As the rpd3Δ mutation does not affect Ycs4-Myc9 protein level (Supplementary Figure 3C), these results indicate that Rpd3 is required for condensin enrichment to the nucleolus and the rDNA. To further study the role of H4 deacetylation, we analyzed Ycs4 localization in histone H4 K5,12G and K5,12R mutant strains. H4 K5,12G mutant mimics the acetylated state of histone H4, whereas H4 K5,12R resembles the deacetylated state (Megee et al, 1990; Ma et al, 1998). We found that Ycs4 is distributed throughout the entire nucleus of the H4 K5,12G mutant, even after the cells are treated with rapamycin (Figure 4A). In contrast, Ycs4 is enriched in the nucleolus of H4 K5,12R cells even in the rich medium in the absence of rapamycin. Ycs4 is enriched to the rDNA in H4 K5,12R cells, but not in H4 K5,12G cells, regardless of the presence of rapamycin (Figure 4B and Supplementary Figure 4B). These H4 mutations do not affect Ycs4 protein level (Supplementary Figure 4C). Together, the above data suggest that Rpd3-dependent histone H4 deacetylation is necessary and sufficient to cause condensin enrichment to the nucleolus and the rDNA.
Figure 3.
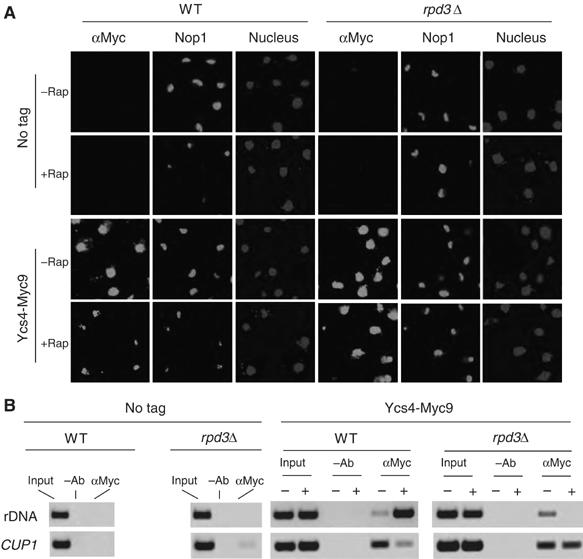
Rpd3 is required for rapamycin-induced nucleolar contraction and condensin loading to rDNA. (A) Rpd3 is required for rapamycin induction of condensin relocation to the nucleolus and nucleolar contraction. WT and rpd3Δ cells expressing Ycs4-Myc9 were treated with 200 nM rapamycin for 1 h. Nucleolar structure and Ycs4-Myc9 localization were analyzed by IF staining with Nop1 (bright white) and Myc antibodies (bright white), respectively. Nuclear DNA was stained with DAPI (light gray). A color version of this figure is available at The EMBO Journal Online. (B) The samples in (A) were analyzed for Ycs4 loading to the promoter of 35S rDNA by ChIP. CUP1 was used as a control.
Figure 4.
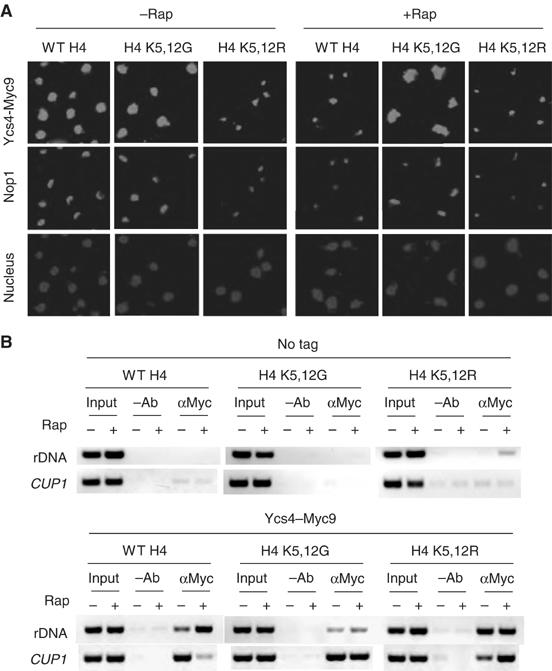
Histone deacetylation at H4 K5,12 is necessary and sufficient to cause condensin enrichment to the nucleolus and rDNA array. (A) Deacetylation at H4 K5,12 is necessary and sufficient to relocalize Ycs4-Myc9 to the nucleolus. WT, H4 K5,12G and H4 K5,12R strains carrying Ycs4-Myc9 were treated without or with rapamycin for 1 h and analyzed for Ycs4-Myc9 localization by IF with a Myc antibody. The nucleolus and nucleus are marked by IF with a Nop1 antibody and DAPI staining, respectively. A color version of this figure is available at The EMBO Journal Online. (B) Deacetylation at H4 K5,12 is necessary and sufficient to cause condensin loading to rDNA. The samples in Figure 4A were analyzed for Ycs4 loading to 35S rDNA promoter by ChIP.
Nucleolar contraction and condensin loading to the rDNA are independent of rDNA transcription
It is generally anticipated that transcriptionally repressed chromosomal domains have a more compacted chromatin structure. Hence, condensin loading and condensin-mediated DNA condensation during starvation may simply be an indirect effect of rDNA transcriptional repression. To test this idea, we used the rrn3-1/syc1-8 mutation to repress rDNA transcription. Rrn3 is a crucial transcription factor for Pol I, which is required for 35S rRNA synthesis and cell growth (Cadwell et al, 1997). After extended incubation at the restrictive temperature (38°C), however, the nucleolus remains in the large crescent shape (Figure 5A). Under such condition, condensin is loaded to the rDNA only in the presence but not in the absence of rapamycin (Figure 5B). Essentially, the same results were obtained with rpa190-1 and rpa190-3 mutants. Rpa190 is the largest subunit of Pol I. Incubation of these mutant cells at the restrictive temperature inhibits cell growth and 35S rRNA synthesis (Wittenkind et al, 1989). However, it does not induce nucleolar contraction (Figure 5C). In addition, histone H4 acetylation is not affected (Figure 5D). Together, these results show that repression of rDNA transcription per se is insufficient to cause histone deacetylation, condensin loading or rDNA condensation. Therefore, histone deacetylation and condensin loading appear to operate independent of rDNA transcription. Rapamycin also inhibits the transcription of RP genes (Powers and Walter, 1999), but causes condensin dissociation from these genomic loci (Figure 2D). Therefore, condensin association also is not related to the transcription of Pol II-dependent genes.
Figure 5.
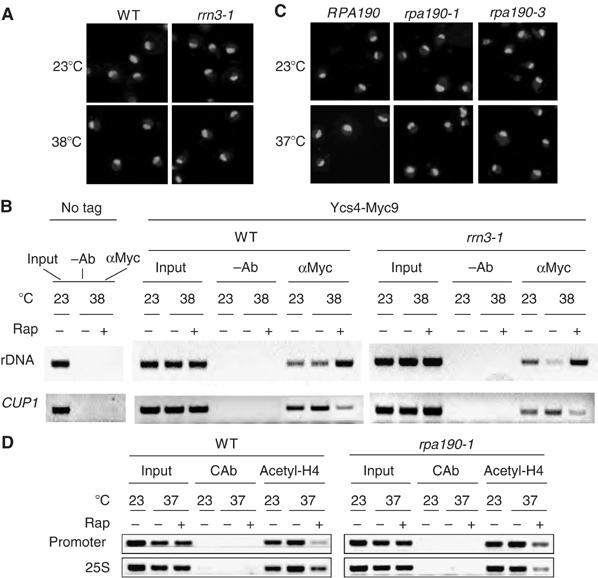
Transcriptional repression does not cause nucleolar contraction and condensin loading. (A) Loss of Rrn3 is insufficient to cause nucleolar contraction. Early log-phase WT and rrn3-1 cells carrying chromosomally tagged YCS4-Myc9 in YPD were shifted from 23 to 38°C and incubated for 3 h. Merged pictures of nuclear (bright white) and nucleolar (light gray) staining are shown. (B) Loss of Rrn3 is insufficient to cause condensin loading to rDNA array. Samples in (A) were examined for Ycs4-Myc9 association with 35S rDNA promoter and CUP1 by ChIP with Myc antibody (9E10). –Ab, no antibody was used as a negative control. (C) Loss of Pol I is insufficient to cause nucleolar contraction. Early log-phase WT (RPA190), rpa190-1 and rpa190-3 cells in YPD were shifted from 23 to 37°C and incubated for 3 h. Merged staining of Nop1 and DAPI is shown. (D) Loss of Pol I is insufficient to cause H4 deacetylation. Samples in (C) were examined for H4 acetylation level at 35S rDNA promoter and 25S transcribing regions by ChIP with anti-hyperacetylated histone H4 antibody. CAb, rabbit pre-serum was used as a negative control.
Condensin is required for maintaining the integrity of the nucleolus and the rDNA when rDNA transcription is inhibited by rapamycin
We have noticed that prolonged rapamycin treatment (⩾3 h) of the ycs4-2 mutant at the restrictive temperature leads to many cells with a fragmented nucleolus (Figure 6A and B and Supplementary Figure 5A). This phenotype has been previously shown as a result of accumulation of ERCs (Sinclair and Guarente, 1997). The top1Δtop2-4 mutant also contains elevated ERCs (Kim and Wang, 1989). We found indeed that top1Δtop2-4 cells have the same fragmented nucleolar phenotype (Figure 6C), suggesting that the fragmented nucleolar phenotype is a common feature for ERCs. Because ERCs are extrachromosomal, we hypothesized that cells with a high ERC content would show rDNA distributed throughout the entire nucleus. Indeed, FISH staining indicates that rDNA is dispersed throughout the entire nucleus in top1Δtop2-4 cells (Figure 6C). Similarly, rDNA is distributed throughout the entire nucleus in a same percentage of ycs4-2 cells as that with fragmented nucleolus (Figure 6D). These results show that FISH staining of rDNA is a good method for detecting cells with a high ERC content.
Figure 6.
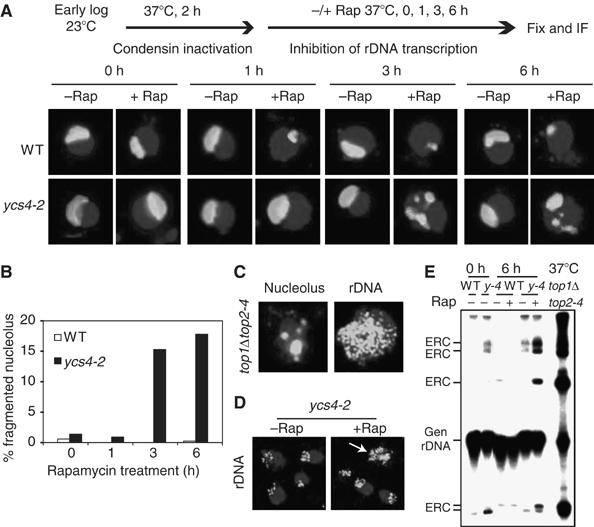
Rapamycin treatment in the absence of condensin causes the fragmented nucleolus phenotype as a result of ERC formation. (A) Rapamycin treatment of condensin mutant causes fragmented nucleolus phenotype. Early log-phase WT and ycs4-2 cells in YPD were shifted from 23 to 37°C for 2 h, followed by incubation with 200 nM rapamycin (+Rap) or a drug carrier (−Rap) for 0, 1, 3 and 6 h. Nucleolar structure was visualized by IF with a Nop1 antibody (bright white). Merged images of representative cells with DAPI staining (light gray) are shown. A color version of this figure is available at The EMBO Journal Online. (B) Quantification of results from (A). N=100. (C) top1Δtop2-4 cells show similar fragmented nucleolus phenotype and distribution of rDNA throughout the entire nucleus. top1Δtop2-4 cells cultured at 23°C were analyzed for nucleolar and rDNA structures by IF and FISH, respectively. Merged images with DAPI staining are shown. A color version of this figure is available at The EMBO Journal Online. (D) Rapamycin treatment in the absence of condensin causes rDNA to distribute throughout the entire nucleus. Early log-phase ycs4-2 cells in YPD were shifted from 23 to 37°C for 2 h, followed by incubation with 200 nM rapamycin (+Rap) or drug carrier (−Rap) for 6 h. rDNA structure was visualized by FISH with a 25S rDNA probe (bright white). Merged images with DAPI staining are shown. The arrowhead points to a cell containing rDNA distributed throughout the entire nucleus. A color version of this figure is available at The EMBO Journal Online. (E) Loss of condensin causes accumulation of extrachromosomal rDNA circles (ERCs). Early log-phase WT and ycs4-2 (y-4) cells in YPD were shifted from 23 to 37°C for 0 and 6 h. Total DNA was isolated, separated by agarose DNA gel electrophoresis and analyzed by Southern blot with a 32P-labeled 25S rDNA probe. top1Δtop2-4 cells cultured at 37°C for 2 h were used as a control. Gen rDNA, genomic rDNA.
To confirm that rapamycin-treated condensin mutants accumulate ERCs, we isolated total DNA from WT and ycs4-2 cells cultured under different conditions and performed Southern blot analysis with a 32P-labeled 25S rDNA probe. Different ERCs migrate slower or faster than chromosome XII DNA during gel electrophoresis in the absence of ethidium bromide, which has been used to detect ERCs (Kim and Wang, 1989; Sinclair and Guarente, 1997). As expected, WT and ycs4-2 cells contain little or no ERCs (Figure 6E). Rapamycin treatment causes a significant increase of ERCs in ycs4-2 but not in WT strain. top1Δtop2-4 cells were used as a positive control. They contain a significantly higher ERC level that is likely to be due to the continuous propagation of ERCs during many passages. We have found that rapamycin still significantly inhibits rDNA transcription in condensin mutants (data not shown), suggesting that condensin may be used to protect rDNA integrity when rDNA transcription is repressed by starvation.
rpa190 mutations repress rDNA transcription, but do not induce condensin loading (Figure 5). We reasoned that if condensin is important to protect the rDNA in the absence of active transcription, elevated ERCs should occur in the Pol I mutants. To test this hypothesis, we incubated rpa190-1 and rpa190-3 cells at 37°C for different times in the rich medium. Indeed, a large population of Pol I mutants show a fragmented nucleolus phenotype and the rDNA is distributed in the entire nucleus after incubation for 4–5 h (Figure 7A and B and Supplementary Figure 6A). The cells with a high ERC content do not show uniform morphology. For example, rpa190-1 cells with a high ERC content after incubation at 37°C for 5 h have the following cell cycle distribution: 44% unbudded with one nucleus (early G1); 51% small and big budded with one nucleus (late G1–early M); 0% big budded with nucleus at the bud neck (metaphase); and 4% big budded with two segregated nuclei (anaphase). This observation suggests that rDNA instability is not caused by a cell cycle-specific event. As rapamycin still induces condensin loading when Pol I is inactive (Figure 5B), we hypothesized that rapamycin treatment would stabilize rDNA in a Pol I mutant if condensin loading is indeed important as suggested from the above experiments. To test this, we first inactivated rpa190-1 and rpa190-3 at 37°C for 2.5 h to inhibit Pol I-dependent transcription. At this point, significant ERCs have not yet appeared (Supplementary Figure 6A). We then used rapamycin to induce condensin loading. We found that rapamycin treatment not only causes rDNA condensation, but also efficiently suppresses ERC formation (Figure 7C and Supplementary Figure 6C and D). To confirm that the protective effect of rapamycin is due to condensin, we generated an rpa190-1 ycs4-2 double mutant and repeated the experiment. In rpa190-1 ycs4-2 cells, rapamycin fails to suppress ERC formation (Figure 7D and Supplementary Figure 6F), confirming that condensin is required to prevent ERC formation. Condensin loading to the rDNA and rDNA transcription are regulated separately but in a coordinated manner, likely through parallel pathways controlled by TOR in response to nutrient availability. This ensures rapid repression of rDNA transcription while maintaining stability in this region.
Figure 7.
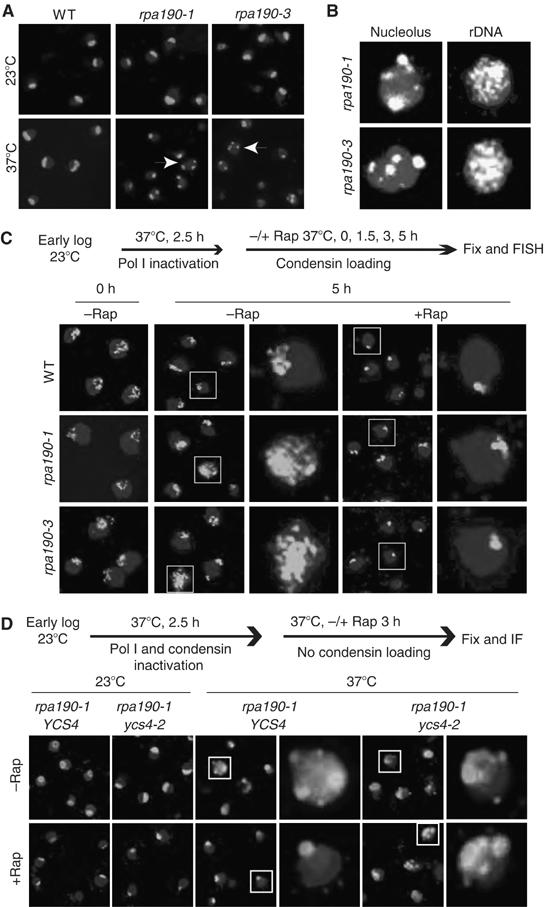
Condensin maintains rDNA stability during transcriptional repression. (A) rpa190-1/3 mutations cause fragmented nucleolus phenotype. Early log-phase WT and rpa190-1/3 cells were shifted from 23 to 37°C in YPD for different times. Merged images of the nucleolus with the nucleus at 5 h are shown. Images of other time points and a color version of this figure are available at The EMBO Journal Online. (B) rpa190-1/3 mutations cause accumulation of cells with rDNA distributed throughout the entire nucleus. rpa190-1/3 cells were incubated at 37°C for 5 h and then analyzed for the nucleolar and rDNA structures by IF and FISH, respectively. Typical images of cells containing fragmented nucleolus or high levels of ERCs are shown. A color version of this figure is available at The EMBO Journal Online. (C) Rapamycin treatment suppresses the ERC formation in Pol I mutants. Early log-phase WT and rpa190-1/3 cells were shifted from 23 to 37°C for 2.5 h to inactivate Pol I, followed by incubation with or without rapamycin for different times. rDNA was analyzed by FISH. The upper panel shows the experimental strategy. Boxed cells were enlarged for better visualization. Images of other time points and a color version of this figure are available at The EMBO Journal Online. (D) Condensin is required for suppression of ERC formation in Pol I mutant in the presence of rapamycin. Single mutant (rpa190-1 YCS4) and double mutant (rpa190-1 ycs4-2) were shifted from 23 to 37°C for 2.5 h to inactivate Pol I and condensin, followed by incubation with or without rapamycin for 3 h. Merged images of the nucleolus with the nucleus are shown. A color version of this figure is available at The EMBO Journal Online.
Discussion
In contrast to global chromosomal DNA compaction during mitosis, DNA condensation during nutrient starvation/rapamycin treatment is restricted to the rDNA, a subchromosomal domain of chromosome XII. In fact, DAPI staining indicates that the size of nuclear DNA increases slightly during starvation or rapamycin treatment (data not shown), possibly reflecting reduced condensin occupancy outside rDNA array. The difference in the scale of the two DNA condensations underscores their specific physiological roles. The global condensation of mitotic chromosomes ensures faithful segregation and transmission of the entire duplicated genome into daughter cells. In contrast, as shown by this study, condensation of rDNA appears to maintain rDNA stability when rDNA transcription is repressed during starvation. Nonetheless, both processes use condensin as the common DNA compacting machinery. Therefore, the rDNA is likely to be compacted in a similar manner (Kimura and Hirano, 1997; Kimura et al, 1999). However, the degree of DNA condensation may differ, which is likely to be influenced by other specific factors involved in each process.
Although mitotic DNA condensation generally correlates with transcriptional repression in higher eukaryotes, this is not the case in budding yeast. In fact, rDNA transcription appears to continue during mitosis because Cdc14-induced condensation does not reduce the levels of nascent rRNA transcripts (Sullivan et al, 2004). In addition, we found that shutting down Pol I-dependent rDNA transcription per se is not sufficient to cause condensin loading to rDNA array (Figure 5). These observations strongly suggest that the TOR-mediated condensin loading on rDNA is independent of rDNA transcription activity, and that TOR regulates these two processes by parallel pathways but in a coordinated manner.
Condensin is controlled in a cell cycle-dependent manner for its mitotic/meiotic functions. Phosphorylation has been shown to play an important role in condensin regulation. In fission yeast, most condensin molecules are cytoplasmic during interphase, and are transported into the nucleus during mitosis in a Cdk1-dependent manner (Sutani et al, 1999). In vertebrates, condensin II is predominantly nuclear, whereas condensin I is sequestered in the cytoplasm during interphase and becomes nuclear during mitosis (Ono et al, 2004). In addition, Cdk1-dependent phosphorylation has been found to stimulate condensin's chromosomal compacting activity in Xenopus egg extracts (Kimura et al, 1998). In budding yeast, condensin is constitutively nuclear throughout the cell cycle (Bhalla et al, 2002), but becomes enriched in the nucleolus during anaphase in a Cdc14-dependent manner, which is important for the resolution of cohesin-independent chromosome linkages at repeated DNA (D'Amours et al, 2004; Sullivan et al, 2004; Wang et al, 2004). In addition, Ycg1/Ycs5 is phosphorylated during mitosis, which is dependent on the mitotic kinase Ipl1/aurora (Lavoie et al, 2004). These observations demonstrate that post-translational modification of condensin itself plays an important role in the global regulation of condensin during mitosis. In this study, we show that histone H4 deacetylation at K5,12 by Rpd3 is necessary and sufficient to control condensin loading to the rDNA during starvation, suggesting that histone modifications have a role in targeting condensins to a specific chromosomal domain. In contrast to the global change of condensin activity by condensin post-translational modifications, chromatin modifications may be advantageous for localizing condensin to a subchromosomal region.
As an expansive tandem repeat, the rDNA is inherently susceptible to homologous recombination. Certain mutations such as those in DNA topoisomerases, or physiological conditions such as aging are known to cause rDNA instability. In WT cells, however, the rDNA is relatively stable, suggesting that there is a mechanism that protects its integrity. Sir2 is an NAD+-dependent HDAC essential for the formation of several silent chromatin domains in yeast, including the rDNA (Smith et al, 1998). In sir2Δ mutant, there is an increase in interchromosomal rDNA recombination during normal growth (Kobayashi et al, 2004) and a higher ERC content, especially in aged sir2Δ cells (Sinclair and Guarente, 1997). Unexpectedly, we found that sir2Δ mutation does not affect nucleolar contraction, rDNA condensation or rDNA stability during rapamycin treatment (unpublished data). These observations suggest that Sir2 is mainly involved in maintaining rDNA stability during normal growth, which is consistent with a model proposed recently (Kobayashi and Ganley, 2005). Sir2 may even cooperate with a low concentration of condensin on rDNA under such condition (Machin et al, 2004), although the precise role of condensin in this particular situation is not clear. Our results show that transcriptional repression without condensin loading is a catastrophic event for rDNA integrity. Sir2 physically interacts with the RNA Pol I transcription machinery (Huang and Moazed, 2003) and mutations in Pol I subunits disrupt Sir2-dependent rDNA silencing (Cioci et al, 2003). One possible scenario is that Sir2 is no longer properly localized to the rDNA when transcription is inhibited. In the absence of condensin loading in Pol I mutants, this could lead to increased rDNA instability and elevated ERC level.
Our results show that rDNA transcriptional repression in the absence of condensin leads to rDNA instability and accumulation of ERCs (Figures 6 and 7), indicating a role of condensin in maintaining rDNA integrity. Our data are also consistent with a recent report that rDNA array is severely shortened in condensin mutants (Johzuka et al, 2006). The elevated rDNA recombination in Pol I mutants we have observed is further in agreement with a much shortened rDNA repeat in these cells (Brewer et al, 1992; Kobayashi et al, 1998). A prevailing model proposes that collision between DNA replication fork and the Pol I transcription machinery causes elevated rDNA recombination and changes in rDNA repeat numbers (Takeuchi et al, 2003). However, a collision is observed only in a fob1 mutant whose rDNA is engineered to ∼20 repeats that are mostly actively transcribed (Takeuchi et al, 2003). It remains to be determined whether such a collision occurs in a fob1 mutant with a normal rDNA array (∼150 repeats). RFB is thought to arrest replication fork in the presence of Fob1 to avoid collision between Pol I machinery and replication fork moving in an opposite direction (Brewer et al, 1992; Kobayashi et al, 1992). It is unclear whether such collisions occur in cells with normal Fob1-mediated RFB activity. A recent study showed that the rDNA can undergo intrachromosomal recombination in the absence of DNA replication (Burkhalter and Sogo, 2004). Together with our data, these new studies indicate that intrachromosomal recombination is not absolutely dependent on DNA replication or active transcription, and that there are potentially multiple mechanisms for homologous recombination within the rDNA.
Materials and methods
Yeast strains and antibodies
The genotypes of yeast strains are shown in Supplementary Table 1. They were obtained from the following sources: BLY03 (WT), BLY04 (ycg1-1) and ZW206 (ycs4-2) (B Lavoie) (Lavoie et al, 2002); MYA-1404 (brn1-9) (ATCC); NBY514 (top1Δtop2-4) (A Murray) (Bhalla et al, 2002); rrn3-1 (J Carbon) (Cadwell et al, 1997); rpa190-1 and rpa190-3 (M Nomura) (Wittenkind et al, 1989). The rpa190-1 ycs4-2 double mutant was constructed by mating of rpa190-1 (NOY259) and ycs4-2 (ZW206) strains and sporulation of the diploid progeny as described (Burke et al, 2000). Yeast strains containing the chromosomal 9xMyc epitope-tagged allele of YCS4 or 6xHA epitope-tagged allele of SMC2 and SMC4 at the C-terminus were generated by the PCR integration approach, as described earlier (Bertram et al, 2000). The antibodies used are as follows: mouse anti-Nop1 (EnCor Biotechnology); rabbit anti-hyperacetylated histone H4 (Upstate); Alexa Fluor 594-conjugated goat anti-mouse, Alexa Fluor 594-conjugated rabbit anti-goat and Alexa Fluor 488-conjugated goat anti-rabbit (Molecular Probes); rabbit anti-Myc (A14) (Santa Cruz Biotechnology); mouse anti-digoxigenin (DIG) (Roche Molecular Biochemicals Diagnostics); mAb anti-Myc (9E10) and anti-HA (12CA5) (Harlan Laboratories); and rat anti-tubulin (Sigma).
Indirect immunofluorescence and fluorescence in situ hybridization
Yeast IF studies were performed as described (Burke et al, 2000). Primary antibody dilution used are as follows: 1:500 anti-Nop1, 1:100 anti-Myc (A14), 1:100 monoclonal anti-HA (12CA5) and 1:500 rabbit polyclonal anti-HA. Alexa Fluor 594- or Alexa Fluor 488-conjugated secondary antibodies were used at a dilution of 1:200. Nuclear DNA was stained with 50 ng/ml DAPI in antifade mounting medium for 15 min. Fluorescent signals were analyzed using an Olympus fluorescence microscope equipped with a digital camera. FISH was preformed as described with minor modifications (Guacci et al, 1994). A DNA probe for 25S rDNA was generated by PCR as described (Kennedy et al, 1997). It was labeled with digoxigenin (DIG) using the BIONICK Labeling System (Invitrogen) and visualized by Alexa Fluor 594-conjugated secondary and tertiary antibodies. rDNA FISH signal was quantified by Quantity One software (Bio-Rad). The restriction ratio was calculated by dividing the area of rDNA FISH signal by the area of DAPI signal. A restriction ratio ⩽0.15 is scored as condensed, whereas a restriction ratio ⩾0.3 is scored as open or puff.
Western blot and chromatin immunoprecipitation assays
Western blot was performed as described before (Ai et al, 2002). ChIP assays were performed as described before (Lieb et al, 2001). For IP, 1 mg of total protein was incubated with 10 μl of anti-Myc (9E10) or 8 μl of anti-hyperacetylated histone H4 overnight at 4°C. Protein G–Sepharose beads were used to recover the antibody–antigen–DNA complexes in all experiments. The total input DNA (input) was prepared in the same way except that the IP steps were omitted. As an internal control, no antibody (−Ab) or rabbit per-serum (CAb) was added to the lysate in parallel to the IP samples. The isogenic WT strains without tagged protein were used as a control to access the background PCR signals. The primer pairs used for PCR detection are as follows: the 35S rDNA promoter, 5′-GTTTTGGTTTCGGTTGTGAA-3′ and 5′-GAAGTACCTCCCAACTACTT-3′; 18S rDNA-encoding region, 5′-TGGCTAACCTTGAGTCCTTG-3′ and 5′-GGCAAATGCTTTCGCAGTAG-3′; 25S rDNA-encoding region, 5′-AGGACGTCATAGAGGGTGAGAATC-3′ and 5′-TTGACTTACGTCGCAGTCCTCAGT-3′; rDNA NTS, 5′-GAGGCAGCGTAAAAGGATGA-3′ and 5′-CCTCCATTTCCCTCTCTTCT-3′; CUP1, 5′-TCTTTTCCGCTGAACCGTTCCAGCA-3′ and 5′-GGCATTGGCACTCATGACCTTCAT-3′; and ACT1, 5′-CGTGATAAGTGATAGTGATATTC-3′ and 5′-CATGATACCTTGGTGTCTTG-3′, RPS26A, 5′-CCAGTTAATCACTAGAAGTGTG-3′ and 5′-CTTTCTTGTTTCTACCGTTGG-3′; RPL30, 5′-GGCGGGAAATAAGAGGTTCA-3′ and 5′-GTTATACTGACCATCTCTGCG-3′. Quantification of ChIP results was performed using Quantity One software (Bio-Rad).
Analysis of extrachromosomal rDNA circles
ERC was detected by Southern blot (D Sinclair, personal communication) with minor modifications. Briefly, cells were spheroplasted by incubating in 500 μl sorbitol solution (0.9 M sorbitol, 0.1 M Tris–HCl, pH 8, 0.1 M EDTA) containing 150 μg/ml zymolyase and 1% (v/v) 2-mercaptoethanol at 30°C with gentle shaking for 15–30 min. An 80 μl volume of 10% SDS was then added and incubated at 65°C for 20 min, followed by incubation with 200 μl 5 M potassium acetate on ice for 30 min. After centrifugation for 3 min at top speed, the supernatant was removed and the DNA was precipitated with 1 ml 100% ethanol. The DNA pellet was resuspended in 300 μl TE containing 0.1 mg/ml RNase and incubated at 37°C for 30 min. The purified DNA was heated with a loading buffer at 55°C for 10 min before loaded onto a 0.6% TBE-buffered agarose gel. Electrophoresis was preformed without ethidium bromide at 2 V/cm for 30 h. Southern blot hybridization was carried out with a 32P-labeled 25S rDNA probe.
Supplementary Material
Supplementary Figures and Table
Acknowledgments
We thank Drs Leroy Liu, Jonathan Warner and David Sinclair for technical advice, Marc Gartenberg and Vasily Studitsky for reading the manuscript, and Jeff Boeke, Brigitte Lavoie, Andrew Murray, Masayasu Nomura and John Carbon for yeast strains. This work was supported by a grant from the National Institutes of Health.
References
- Ai W, Bertram PG, Tsang CK, Chan TF, Zheng XF (2002) Regulation of subtelomeric silencing during stress response. Mol Cell 10: 1295–1305 [DOI] [PubMed] [Google Scholar]
- Bertram PG, Choi J, Carvalho J, Ai WD, Zeng CB, Chan TF, Zheng XFS (2000) Tripartite regulation of Gln3p by TOR, Ure2p and phosphatases. J Biol Chem 275: 35727–35733 [DOI] [PubMed] [Google Scholar]
- Bhalla N, Biggins S, Murray AW (2002) Mutation of YCS4, a budding yeast condensin subunit, affects mitotic and nonmitotic chromosome behavior. Mol Biol Cell 13: 632–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer B, Lockshon D, Fangman W (1992) The arrest of replication forks in the rDNA of yeast occurs independently of transcription. Cell 71: 267–276 [DOI] [PubMed] [Google Scholar]
- Burke D, Dawson D, Stearns T (2000) Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY [Google Scholar]
- Burkhalter M, Sogo J (2004) rDNA enhancer affects replication initiation and mitotic recombination: Fob1 mediates nucleolytic processing independently of replication. Mol Cell 15: 409–421 [DOI] [PubMed] [Google Scholar]
- Cadwell C, Yoon HJ, Zebarjadian Y, Carbon J (1997) The yeast nucleolar protein Cbf5p is involved in rRNA biosynthesis and interacts genetically with the RNA polymerase I transcription factor RRN3. Mol Cell Biol 17: 6175–6183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cioci F, Vu L, Eliason K, Oakes M, Siddiqi I, Nomura M (2003) Silencing in yeast rDNA chromatin: reciprocal relationship in gene expression between RNA polymerase I and II. Mol Cell 12: 135–145 [DOI] [PubMed] [Google Scholar]
- D'Amours D, Stegmeier F, Amon A (2004) Cdc14 and condensin control the dissolution of cohesin-independent chromosome linkages at repeated DNA. Cell 117: 455–469 [DOI] [PubMed] [Google Scholar]
- Freeman L, Aragon-Alcaide L, Strunnikov A (2000) The condensin complex governs chromosome condensation and mitotic transmission of rDNA. J Cell Biol 149: 811–824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guacci V, Hogan E, Koshland D (1994) Chromosome condensation and sister chromatid pairing in budding yeast. J Cell Biol 125: 517–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano T (2005) Condensins: organizing and segregating the genome. Curr Biol 15: R265–R275 [DOI] [PubMed] [Google Scholar]
- Hirano T, Mitchison T (1994) A heterodimeric coiled-coil protein required for mitotic chromosome condensation in vitro. Cell 79: 449–458 [DOI] [PubMed] [Google Scholar]
- Huang J, Moazed D (2003) Association of the RENT complex with nontranscribed and coding regions of rDNA and a regional requirement for the replication fork block protein Fob1 in rDNA silencing. Genes Dev 17: 2162–2176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johzuka K, Terasawa M, Ogawa H, Ogawa T, Horiuchi T (2006) Condensin loaded onto the replication Fork barrier site in the rRNA gene repeats during S phase in a FOB1-dependent fashion to prevent contraction of a long repetitive array in Saccharomyces cerevisiae. Mol Cell Biol 26: 2226–2236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeberlein M, McVey M, Guarente L (1999) The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms 10.1101/gad.13.19.2570. Genes Dev 13: 2570–2580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy B, Gotta M, Sinclair D, Mills K, McNabb D, Murthy M, Pak S, Laroche T, Gasser S, Guarente L (1997) Redistribution of silencing proteins from telomeres to the nucleolus is associated with extension of life span in S. cerevisiae. Cell 89: 381–391 [DOI] [PubMed] [Google Scholar]
- Kim R, Wang J (1989) A subthreshold level of DNA topoisomerases leads to the excision of yeast rDNA as extrachromosomal rings. Cell 57: 975–985 [DOI] [PubMed] [Google Scholar]
- Kimura K, Hirano M, Kobayashi R, Hirano T (1998) Phosphorylation and activation of 13S condensin by Cdc2 in vitro. Science 282: 487–490 [DOI] [PubMed] [Google Scholar]
- Kimura K, Hirano T (1997) ATP-dependent positive supercoiling of DNA by 13S condensin: a biochemical implication for chromosome condensation. Cell 90: 625–634 [DOI] [PubMed] [Google Scholar]
- Kimura K, Rybenkov V, Crisona N, Hirano T, Cozzarelli N (1999) 13S condensin actively reconfigures DNA by introducing global positive writhe: implications for chromosome condensation. Cell 98: 239–248 [DOI] [PubMed] [Google Scholar]
- Kobayashi T, Ganley ARD (2005) Recombination regulation by transcription-induced cohesin dissociation in rDNA repeats. Science 309: 1581–1584 [DOI] [PubMed] [Google Scholar]
- Kobayashi T, Heck DJ, Nomura M, Horiuchi T (1998) Expansion and contraction of ribosomal DNA repeats in Saccharomyces cerevisiae: requirement of replication fork blocking (Fob1) protein and the role of RNA polymerase I. Genes Dev 12: 3821–3830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T, Hidaka M, Nishizawa M, Horiuchi T (1992) Identification of a site required for DNA replication fork blocking activity in the rRNA gene cluster in Saccharomyces cerevisiae. Mol Gen Genet 233: 355–362 [DOI] [PubMed] [Google Scholar]
- Kobayashi T, Horiuchi T, Tongaonkar P, Vu L, Nomura M (2004) SIR2 regulates recombination between different rDNA repeats, but not recombination within individual rRNA genes in yeast. Cell 117: 441–453 [DOI] [PubMed] [Google Scholar]
- Lavoie BD, Hogan E, Koshland D (2002) In vivo dissection of the chromosome condensation machinery: reversibility of condensation distinguishes contributions of condensin and cohesin. J Cell Biol 156: 805–815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavoie BD, Hogan E, Koshland D (2004) In vivo requirements for rDNA chromosome condensation reveal two cell-cycle-regulated pathways for mitotic chromosome folding. Genes Dev 18: 76–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavoie BD, Tuffo KM, Oh S, Koshland D, Holm C (2000) Mitotic chromosome condensation requires Brn1p, the yeast homologue of Barren. Mol Biol Cell 11: 1293–1304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legagneux V, Cubizolles F, Watrin E (2004) Multiple roles of condensins: a complex story. Biol Cell 96: 201–213 [DOI] [PubMed] [Google Scholar]
- Lieb JD, Liu X, Botstein D, Brown PO (2001) Promoter-specific binding of Rap1 revealed by genome-wide maps of protein–DNA association. Nat Genet 28: 327–334 [DOI] [PubMed] [Google Scholar]
- Ma XJ, Wu J, Altheim BA, Schultz MC, Grunstein M (1998) Deposition-related sites K5/K12 in histone H4 are not required for nucleosome deposition in yeast. Proc Natl Acad Sci USA 95: 6693–6698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machin F, Paschos K, Jarmuz A, Torres-Rosell J, Pade C, Aragon L (2004) Condensin regulates rDNA silencing by modulating nucleolar Sir2p. Curr Biol 14: 125–130 [PubMed] [Google Scholar]
- Megee PC, Morgan BA, Mittman BA, Smith MM (1990) Genetic analysis of histone H4: essential role of lysines subject to reversible acetylation. Science 247: 841–845 [DOI] [PubMed] [Google Scholar]
- Milutinovich M, Koshland DE (2003) Molecular biology: SMC complexes—wrapped up in controversy. Science 300: 1101–1102 [DOI] [PubMed] [Google Scholar]
- Moss T (2004) At the crossroads of growth control; making ribosomal RNA. Curr Opin Genet Dev 14: 210–217 [DOI] [PubMed] [Google Scholar]
- Ono T, Fang Y, Spector DL, Hirano T (2004) Spatial and temporal regulation of condensins I and II in mitotic chromosome assembly in human cells. Mol Biol Cell 15: 3296–3308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouspenski II, Cabello OA, Brinkley BR (2000) Chromosome condensation factor Brn1p is required for chromatid separation in mitosis. Mol Biol Cell 11: 1305–1313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers T, Walter P (1999) Regulation of ribosome biogenesis by the rapamycin-sensitive TOR-signaling pathway in Saccharomyces cerevisiae. Mol Biol Cell 10: 987–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudra D, Warner JR (2004) What better measure than ribosome synthesis? Genes Dev 18: 2431–2436 [DOI] [PubMed] [Google Scholar]
- Schneper L, Duvel K, Broach J (2004) Sense and sensibility: nutritional response and signal integration in yeast. Curr Opin Microbiol 7: 624–630 [DOI] [PubMed] [Google Scholar]
- Sinclair DA, Guarente L (1997) Extrachromosomal rDNA circles—a cause of aging in yeast. Cell 91: 1033–1042 [DOI] [PubMed] [Google Scholar]
- Smith JS, Brachmann CB, Pillus L, Boeke JD (1998) Distribution of a limited Sir2 protein pool regulates the strength of yeast rDNA silencing and is modulated by Sir4p. Genetics 149: 1205–1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strick T, Kawaguchi T, Hirano T (2004) Real-time detection of single-molecule DNA compaction by condensin I. Curr Biol 14: 874–880 [DOI] [PubMed] [Google Scholar]
- Strunnikov A, Hogan E, Koshland D (1995) SMC2, a Saccharomyces cerevisiae gene essential for chromosome segregation and condensation, defines a subgroup within the SMC family. Genes Dev 9: 587–599 [DOI] [PubMed] [Google Scholar]
- Sullivan M, Higuchi T, Katis V, Uhlmann F (2004) Cdc14 phosphatase induces rDNA condensation and resolves cohesin-independent cohesion during budding yeast anaphase. Cell 117: 471–482 [DOI] [PubMed] [Google Scholar]
- Sutani T, Yuasa T, Tomonaga T, Dohmae N, Takio K, Yanagida M (1999) Fission yeast condensin complex: essential roles of non-SMC subunits for condensation and Cdc2 phosphorylation of Cut3/SMC4. Genes Dev 13: 2271–2283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi Y, Horiuchi T, Kobayashi T (2003) Transcription-dependent recombination and the role of fork collision in yeast rDNA. Genes Dev 17: 1497–1506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang CK, Bertram PG, Ai W, Drenan R, Zheng XFS (2003) Chromatin-mediated regulation of nucleolar structure and RNA Pol I localization by TOR. EMBO J 22: 6045–6056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang CK, Zheng XFS (2004) Control of ribosome biogenesis by target of rapamycin (TOR). Recent Res Dev Mol Cell Biol 5: 135–147 [Google Scholar]
- Wang B, Yong-Gonzalez V, Strunnikov A (2004) Cdc14p/FEAR pathway controls segregation of nucleolus in S. cerevisiae by facilitating condensin targeting to rDNA chromatin in anaphase. Cell Cycle 3: 71–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner JR (1999) The economics of ribosome biosynthesis in yeast. Trends Biochem Sci 24: 437–440 [DOI] [PubMed] [Google Scholar]
- Wittenkind M, Dodd J, Vu L, Kolb JM, Buhler J-M, Sentenac A, Normura M (1989) Isolation and characterization of temperature-sensitive mutations in RPA190, the gene encoding the largest subunit of RNA polymerase I from Saccharomyces cerevisiae. Mol Cell Biol 8: 3997–4008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaragoza D, Ghavidel A, Heitman J, Schultz MC (1998) Rapamycin induces the G0 program of transcriptional repression in yeast by interfering with the TOR signaling pathway. Mol Cell Biol 18: 4463–4470 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures and Table


