Abstract
In budding yeast, chs5 mutants are defective in chitin synthesis and cell fusion during mating. Chs5p is a late-Golgi protein required for the polarized transport of the chitin synthase Chs3p to the membrane. Here we show that Chs5p is also essential for the polarized targeting of Fus1p, but not of other cell fusion proteins, to the membrane during mating.
Upon cell differentiation, eukaryotic cells undergo dramatic rearrangements on their cell surfaces that are crucial for cell-cell interactions and cellular responses to environmental signals.
The yeast mating response is an excellent model system for study of cell differentiation, cell secretion, and cell fusion. Saccharomyces cerevisiae haploid cells, a or α, exposed to pheromones stop their progression through the cell cycle, undergo polarized cell growth. and form a mating projection, acquiring a pear-shaped morphology called “shmoo.” Polarized mating cells make contact and attach firmly, forming a prezygote. The cell wall separating the partners must then be degraded, and haploid nuclei must merge into a diploid nucleus (8). A number of genetic screens have identified mutants defective in different steps of mating (19). Most proteins required for cell fusion localize to the mating projection, but little is known about the mechanisms responsible for their polarization.
Chs5p is involved in chitin synthesis during vegetative growth and is required for the polarization of the chitin synthase Chs3p to the plasma membrane (9, 10). Additionally, chs5Δ mutants are defective in cell fusion during mating; however, other mutants defective in chitin synthesis mate efficiently, suggesting an independent role of Chs5p in cell fusion (14, 17). By analogy with its role in chitin synthesis, we decided to investigate whether Chs5p is required for the localization of proteins involved in cell fusion.
Strains.
Yeast strains used are presented on Table 1. The strain containing FUS1-3xmyc (YB267) was constructed by a PCR-based method (12). The epitope was inserted at the C terminus of the protein, and the fusion protein is fully functional.
TABLE 1.
List of strains
| Straina | Genotype |
|---|---|
| Y604 | MATaura3-52 lys2-801 ade2-101 trp1-901 his3-Δ200 |
| Y1950 | MATaura3-52 lys2-801 ade2-101 trp1-901 his3-Δ200 chs5::ADE2 |
| Y609 | MATaura3-52 lys2-801 ade2-101 trp1-901 his3-Δ200 spa2::TRP1 |
| YB438 | MATaura3-52 lys2-801 ade2-101 trp1-901 his3-Δ200 chs6::KanMX4 |
| Y603 | MATα ura3-52 lys2-801 ade2-101 trp1-901 his3-Δ200 |
| Y1935 | MATα ura3-52 lys2-801 ade2-101 trp1-901 his3-Δ200 chs5::ADE2 |
| Y601 | MATα ura3-52 lys2-801 ade2-101 trp1-901 his3-Δ200 spa2::URA3 |
| YB442 | MATα ura3-52 lys2-801 ade2-101 trp1-901 his3-Δ200 chs6::KanMX4 |
| Y1936 | MATα ura3-52 lys2-801 ade2-101 trp1-901 his3-Δ200 chs5::ADE2 spa2::URA3 |
| YB446 | MATα ura3-52 lys2-801 ade2-101 trp1-901 his3-Δ200 fus1::URA3 |
| YB450 | MATα ura3-52 lys2-801 ade2-101 trp1-901 his3-Δ200 fus2::URA3 |
| YB447 | MATα ura3-52 lys2-801 ade2-101 trp1-901 his3-Δ200 chs5::ADE2 fus1::URA3 |
| YB451 | MATα ura3-52 lys2-801 ade2-101 trp1-901 his3-Δ200 chs5::ADE2 fus2::URA3 |
| Y431 | MATaura3-52 lys2-801 ade2-101 trp1-901 leu2-Δ98 |
| YB41 | MATaura3-52 lys2-801 ade2-101 trp1-901 leu2-Δ98 chs5::ADE2 |
| YK5 | MATaura3-52 lys2-801 ade2-101 trp1-901 leu2-Δ98 spa2::URA3 |
| YB42 | MATaura3-52 lys2-801 ade2-101 trp1-901 leu2-Δ98 chs5::ADE2 spa2::URA3 |
| YB440 | MATaura3-52 lys2-801 ade2-101 trp1-901 leu2-Δ98 chs6::KanMX4 |
| YB444 | MATaura3-52 lys2-801 ade2-101 trp1-901 leu2-Δ98 fus1::URA3 |
| YB448 | MATaura3-52 lys2-801 ade2-101 trp1-901 leu2-Δ98 fus2::URA3 |
| YB445 | MATaura3-52 lys2-801 ade2-101 trp1-901 leu2-Δ98 chs5::ADE2 fus1::URA3 |
| YB449 | MATaura3-52 lys2-801 ade2-101 trp1-901 leu2-Δ98 chs5::ADE2 fus2::URA3 |
| YB267 | MATaura3-52 lys2-801 ade2-101 trp1-901 his3-Δ200 FUS1::3xmyc |
| YB296 | MATaura3-52 lys2-801 ade2-101 trp1-901 his3-Δ200 FUS1::3xmyc chs5::ADE2 |
| YB271 | MATaura3-52 lys2-801 ade2-101 trp1-901 his3-Δ200 FUS2::3xmyc |
All strains are from this laboratory or from this study.
CHS6, FUS1, and FUS2 were disrupted by a PCR-based method (18). In all cases, the complete open reading frame was replaced by the KanMX4 cassette or the URA3 marker, and disruptions were confirmed by PCR.
Plasmids.
To construct pFUS1::GFP, the coding region of FUS1 plus 300 bp of the 5′ and 3′ flanking regions was amplified by PCR from strain YB267 and cloned as a SacII-SalI fragment into the low-copy-number vector pRS315. The 3xmyc epitope, flanked by NotI sites, was then replaced with GFPS65T in the same reading frame. The Fus1p-green fluorescent protein (GFP) fusion protein is functional because it rescues the mating defect of the fus1 fus2 mutant (data not shown). pFUS2::GFP was constructed by following the same approach described for pFUS1::GFP but using strain YB271. In this case, the high-copy-number vector pRS426 was used. The Fus2p-GFP fusion protein is only partially functional (data not shown), but it localizes as previously described in wild-type cells (4). pSPA2::GFP contains the SPA2 open reading frame plus 700 bp at both the 5′ and 3′ flanking regions in the pRS315 vector. A NotI site was created at the 5′ end by PCR, and GFPS65T was introduced into this site. Spa2p-GFP is fully functional. pFIG1::GFP was a gift of S. Erdman (Syracuse University) and contains the FIG1 gene fused with GFP in the pRS316 centromeric vector. Fig1p-GFP is not functional, but its localization is not affected.
Other methods.
α-Factor treatment, mating assays, and immunoblot analysis were performed as described previously (9, 10) except that cells were mixed in YPDA (1% yeast extract, 2% Bacto Peptone, 2% glucose, 40 mg of adenine/liter) containing 1 M sorbitol.
Fus1p polarization to the membrane requires Chs5p.
Fus1p is a membrane protein required for cell fusion that localizes to the mating projection in pheromone-treated cells (16). As reported previously (15), Fus1p-GFP is found at the membrane of the shmoo projection in wild-type cells (85% of the shmoos). In addition, it is also present in cytoplasmic patches, a phenomenon not described previously (Fig. 1A). By contrast, in chs5 cells, Fus1p-GFP is localized mainly to this internal compartment and is no longer polarized to the cell periphery (Fig. 1A). Only 27% of the chs5 shmoos display a cytoplasmic patch close to the membrane in the projection area (n = 215); this patch rarely reaches the tip (Fig. 1A). These observations suggest that in the absence of Chs5p, Fus1p-GFP is retained in a cytoplasmic compartment and cannot be delivered to the membrane. This defect is specific for chs5 mutants; spa2 mutants, also defective in cell fusion (7), polarize Fus1p properly (Fig. 1A).
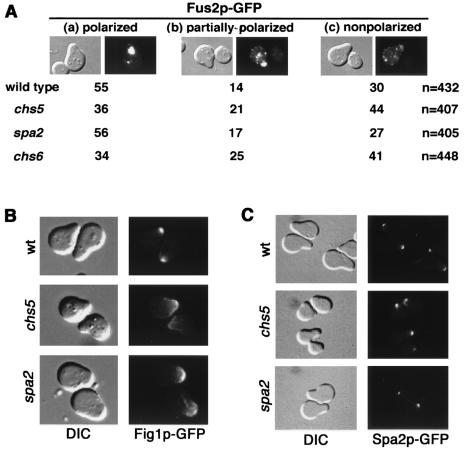
FIG. 1.
Chs5p is required for proper Fus1p localization. (A) Localization of Fus1p-GFP in pheromone-treated cells. MATa strains Y604 (wild type), Y1950 (chs5), Y609 (spa2), and YB438 (chs6), transformed with pFUS1:: GFP, were incubated with α-factor mating pheromone. Fus1p-GFP localization (right panels) and corresponding differential interference contrast (DIC) images (left panels) are shown. Fus1p polarization is lost in the chs5 mutant but not in spa2 or chs6 mutants. (B) Localization of Fus1p-GFP in mating mixtures. (Left) The MATa strain Y604 (wild type) transformed with pFUS1::GFP was mated with the MATα strain Y603 (wild type). (Right) The MATa strain Y1950 (chs5) transformed with pFUS1::GFP was mated with the MATα strain Y1935 (chs5). Localization of Fus1p-GFP in representative examples of prezygotes from the wild-type (left) and chs5 (right) crosses is shown. In wild-type prezygotes, Fus1p-GFP is located in the cell fusion zone, whereas in chs5 prezygotes, Fus1p-GFP is localized in cytoplasmic patches and is not polarized. (C) Strain YB267 (MATa FUS1::3xmyc CHS5) was treated with pheromone, and samples were taken every 30 min. Ten micrograms of protein extracts was separated in SDS-10% polyacrylamide gels, which were blotted and probed with the anti-Myc monoclonal antibody 9E10. Both unglycosylated (asterisk) and O-glycosylated (arrow) forms of Fus1p were detected in wild-type and chs5 strains.
We also examined Fus1p localization in mating mixtures. When a MATα wild-type strain was mated with a MATa wild-type strain containing FUS1-GFP in a centromeric plasmid, Fus1p-GFP was concentrated in the cell fusion zone at the prezygote stage (Fig. 1B). After fusion, the Fus1 protein disappeared (data not shown). In contrast, Fus1p-GFP was present in patches and was not polarized in the prezygotes accumulated in a cross between a MATa chs5 mutant and a MATα chs5 mutant strain expressing FUS1-GFP (Fig. 1B). This suggests that mating cells that are unable to polarize Fus1p to the cell fusion zone are blocked at the prezygote stage.
Fus1p follows the secretory pathway, undergoing O glycosylation during its transport. The unglycosylated and O-glycosylated forms of Fus1p can be distinguished by Western blotting as bands of approximately 56 and 80 kDa, respectively (16) (Fig. 1C). As shown in Fig. 1C, Fus1p-3xmyc is still pheromone induced in the chs5 mutant, although the protein levels are slightly lower than those in wild-type cells. Importantly, the O-glycosylated form of Fus1p can be detected in chs5 cells (Fig. 1C), indicating that Fus1p should be retained in a late-Golgi or post-Golgi compartment in the absence of Chs5p.
Analysis of cell fusion proteins in chs5 cells.
To determine whether the Chs5p-dependent pathway of polarization during mating is specific for Fus1p, we tested the localization of other membrane proteins involved in cell fusion, such as Fus2p and Fig1p, in pheromone-treated chs5 cells. Fus2p is a protein tightly associated with membranes or insoluble particles and localizes to punctate structures under the surface of the shmoo projection (1, 4) (Fig. 2A). Fig1p is a protein with four transmembrane domains localized at the membrane of the tip of the shmoo (5) (Fig. 2B).
FIG. 2.
Chs5p is not required for polarization of Fus2p, Fig1p, or Spa2p. (A) MATa strains Y604 (wild type), Y1950 (chs5), Y609 (spa2), and YB438 (chs6) transformed with pFUS2::GFP were incubated with α-factor. Three patterns of Fus2p-GFP localization were observed; representative differential interference contrast (DIC) and fluorescence images are shown. (a) Polarized, Fus2p-GFP is localized as a large patch at the tip of the shmoo. (b) Partially polarized, Fus2p-GFP is found in a large patch and cytoplasmic dots. (c) Nonpolarized, cytoplasmic-only Fus2p-GFP patches are observed. Quantitation of the localization pattern of Fus2p-GFP in wild-type, chs5, spa2, and chs6 strains is presented as the percentage of the cells in each category. The total number of shmoos analyzed for each strain (n) is given on the right. Pheromone-treated chs5 and chs6 cells show a slight defect in polarization of Fus2p-GFP. (B) Localization of Fig1p. MATa strains Y604 (wild type), Y1950 (chs5), and Y609 (spa2), transformed with pFIG1::GFP, were incubated with α-factor, and Fig1p-GFP was visualized (right panels). Corresponding DIC images are shown (left panels). Fig1p is polarized in all strains. (C) Localization of Spa2p. MATa strains Y431 (wild type), YB41 (chs5), and YK5 (spa2), transformed with pSPA2::GFP, were incubated with α-factor. Spa2p-GFP localization (right panels) and corresponding DIC images (left panels) are shown. Spa2p is polarized in all strains.
We observed only a slight defect in Fus2p polarization in chs5 cells (Fig. 2A). Fig1p-GFP displayed similar localization in wild-type and chs5 cells (Fig. 2B). Thus, these observations indicate that Chs5p is essential for polarization of Fus1p but not for that of all membrane-associated proteins implicated in cell fusion during mating.
We also studied another cortical protein required for mating, Spa2p, which is not associated with the membrane. The Spa2 protein localizes at sites of polarized growth (13); it is involved in polarity processes and is required for cell fusion (7). Spa2p-GFP was polarized normally in pheromone-treated chs5 cells (Fig. 2C), indicating that the chs5 mutant did not have a general defect in polarization.
Chs6p is not required for cell fusion.
Similarly to Chs5p, the Chs6 protein is also required for polarized localization of the chitin synthase Chs3p during vegetative growth (2, 20). In chs6 mutant cells, Chs3p accumulates in an internal endosome-like compartment, resulting in chitin synthesis defects (20). Therefore, it is possible that Chs6p is also required for transport of Fus1p during mating. However, chs6 cells showed no mating defect (Fig. 3A), suggesting that Chs6p does not affect the localization of proteins involved in mating. As expected, Fus1p-GFP was properly localized in pheromone-induced chs6 cells (Fig. 1A). Nevertheless, as in chs5 cells, a slight alteration of Fus2p polarization could be observed in the absence of Chs6p (Fig. 2A). One possibility is that the CHS6 homologues could play a role during mating.
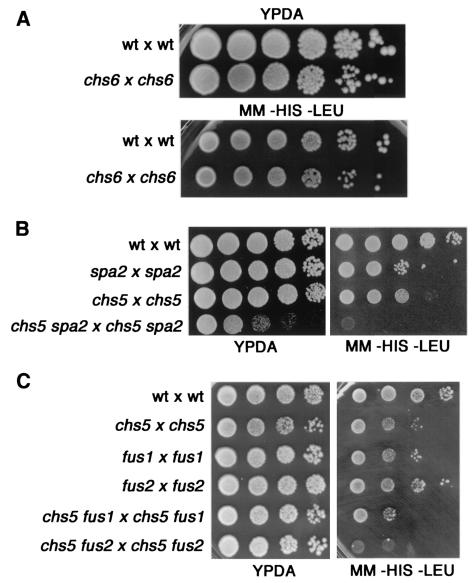
FIG. 3.
Mating efficiency assays. (A) chs6 cells are not defective in mating. The wild-type (wt) strains Y431 (MATa) and Y603 (MATα) and the chs6 mutant strains YB440 (MATa) and YB442 (MATα) were mated quantitatively by the procedure described in Materials and Methods. Serial 1/10 dilutions of mating mixtures were spotted onto YPDA and diploid-selective (−HIS −LEU) plates. (B) The chs5 spa2 double mutant displays a strong mating defect. The mating efficiencies of the following crosses were analyzed as described above: wt × wt (Y431 × Y603), spa2 × spa2 (YK5 × Y601), chs5 × chs5 (YB41 × Y1935), and chs5 spa2 × chs5 spa2 (YB42 × Y1936). (C) Mating efficiencies of chs5 fus1 and chs5 fus2 mutants were analyzed as described above. Results for the following crosses are shown: wt × wt (Y431 × Y603), chs5 × chs5 (YB41 × Y1935), fus1 × fus1 (YB444 × YB446), fus2 × fus2 (YB448 × YB450), chs5 fus1 × chs5 fus1 (YB445 × YB447), and chs5 fus2 × chs5 fus2 (YB449 × YB451). chs5 fus1 double mutants show mating efficiencies similar to those of chs5 or fus1 single mutants; however, chs5 fus2 mutants are clearly more defective than any single mutant.
Genetic interactions of chs5 mutants.
The analysis of Fus1p localization suggests that a lack of Chs5p and a lack of Spa2p alter the cell fusion process for different reasons. In agreement with this hypothesis, the chs5 spa2 double mutant has a more severe mating defect than the chs5 and spa2 single mutants (Fig. 3B). The chs5 spa2 growth defect can be rescued by sorbitol (11); however, the mating efficiency of chs5 spa2 cells is not affected by the presence of sorbitol (data not shown).
We have also analyzed mating efficiency in the chs5 fus1 and chs5 fus2 double mutants. The chs5 fus2 double mutant displays a much more severe mating defect than the chs5 or fus2 single mutant. However, the mating defect of the chs5 fus1 double mutant is only slightly more severe than that of the chs5 single mutant (Fig. 3C). This observation is consistent with the findings of our localization studies, indicating that Chs5p and Fus1p act in the same pathway for cell fusion, whereas Chs5p and Fus2p function in different pathways.
Taken together, our results indicate that chs5 cells are defective in cell fusion during mating because they cannot polarize Fus1p to the membrane. This defect is specific for Fus1p, as evidenced by the fact that chs5 cells can properly localize other polarized proteins such as Fig1p, Spa2p, and Fus2p.
These studies suggest that specific exocytic pathways exist for the polarized secretion of components during cell differentiation and that Chs5p is critical for one of those pathways. It has been shown by electron microscopy that there are secretory vesicles implicated in the cell fusion process during mating in yeast (3, 6). We propose that Chs5p could be specifically required for the incorporation of Fus1p into these vesicles for its transport to the cell surface. Other proteins must use different routes for delivery to the polarized zone in the cell.
Acknowledgments
We thank Shirleen Roeder for providing her microscope for GFP analysis. Scott Erdman provided the pFIG1::GFP plasmid.
This research was supported by NIH grant GM36494.
REFERENCES
- 1.Brizzio, V., A. E. Gammie, and M. D. Rose. 1998. Rvs161p interacts with Fus2p to promote cell fusion in Saccharomyces cerevisiae. J. Cell Biol. 141:567-584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bulawa, C. E. 1993. Genetics and molecular biology of chitin synthesis in fungi. Annu. Rev. Microbiol. 47:505-534. [DOI] [PubMed] [Google Scholar]
- 3.Elia, L., and L. Marsh. 1998. A role for a protease in morphogenetic response during yeast cell fusion. J. Cell Biol. 142:1473-1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Elion, E. A., J. Trueheart, and G. R. Fink. 1995. Fus2 localizes near the site of cell fusion and is required for both cell fusion and nuclear alignment during zygote formation. J. Cell Biol. 130:1283-1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Erdman, S., L. Lin, M. Malczynski, and M. Snyder. 1998. Pheromone-regulated genes required for yeast mating differentiation. J. Cell Biol. 140:461-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gammie, A. E., V. Brizzio, and M. D. Rose. 1998. Distinct morphological phenotypes of cell fusion mutants. Mol. Biol. Cell 9:1395-1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gehrung, S., and M. Snyder. 1990. The SPA2 gene of Saccharomyces cerevisiae is important for pheromone-induced morphogenesis and efficient mating. J. Cell Biol. 111:1451-1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marsh, L., and M. D. Rose. 1997. The pathway of cell and nuclear fusion during mating in S. cerevisiae, p. 827-888. In J. R. Pringle, J. R. Broach, and E. W. Jones (ed.), The molecular and cellular biology of the yeast Saccharomyces cerevisiae: cell cycle and cell biology. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 9.Santos, B., A. Durán, and M. H. Valdivieso. 1997. CHS5, a gene involved in chitin synthesis and mating in Saccharomyces cerevisiae. Mol. Cell. Biol. 17:2485-2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Santos, B., and M. Snyder. 1997. Targeting of chitin synthase 3 to polarized growth sites in yeast requires Chs5p and Myo2p. J. Cell Biol. 136:95-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Santos, B., and M. Snyder. 2000. Sbe2p and Sbe22p, two homologous Golgi proteins involved in yeast cell wall formation. Mol. Biol. Cell 11:435-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schneider, B. L., W. Seufert, B. Steiner, Q. H. Yang, and A. B. Futcher. 1995. Use of polymerase chain reaction epitope tagging for protein tagging in Saccharomyces cerevisiae. Yeast 11:1265-1274. [DOI] [PubMed] [Google Scholar]
- 13.Snyder, M. 1989. The SPA2 protein of yeast localizes to sites of cell growth. J. Cell Biol. 108:1419-1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Trilla, J. A., A. Durán, and C. Roncero. 1999. Chs7, a new protein involved in the control of protein export from the endoplasmic reticulum that is specifically engaged in the regulation of chitin synthesis in Saccharomyces cerevisiae. J. Cell Biol. 145:1153-1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Trueheart, J., J. D. Boeke, and G. R. Fink. 1987. Two genes required for cell fusion during yeast conjugation: evidence for a pheromone-induced surface protein. Mol. Cell. Biol. 7:2316-2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Trueheart, J., and G. Fink. 1989. The yeast cell fusion protein FUS1 is O-glycosylated and spans the plasma membrane. Proc. Natl. Acad. Sci. USA 86:9916-9920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Valdivieso, M. H., P. C. Mol, J. A. Shaw, E. Cabib, and A. Durán. 1991. CAL1, a gene required for activity of chitin synthase 3 in Saccharomyces cerevisiae. J. Cell Biol. 114:101-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wach, A., A. Brachat, R. Pöhlmann, and P. Philipssen. 1994. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast 10:1793-1808. [DOI] [PubMed] [Google Scholar]
- 19.White, J. M., and M. D. Rose. 2001. Yeast mating: getting close to membrane merger. Curr. Biol. 11:R16-R20. [DOI] [PubMed] [Google Scholar]
- 20.Ziman, M., J. S. Chuang, M. Tsung, S. Hamamoto, and R. Schekman. 1998. Chs6p-dependent anterograde transport of Chs3p from the chitosome to the plasma membrane in Saccharomyces cerevisiae. Mol. Biol. Cell 9:1565-1576. [DOI] [PMC free article] [PubMed] [Google Scholar]