Abstract
Recent advances in avian transgenesis have led to the possibility of utilizing the laying hen as a production platform for the large-scale synthesis of pharmaceutical proteins. Ovalbumin constitutes more than half of the protein in the white of a laid egg, and expression of the ovalbumin gene is restricted to the tubular gland cells of the oviduct. Here we describe the use of lentiviral vectors to deliver transgene constructs comprising regulatory sequences from the ovalbumin gene designed to direct synthesis of associated therapeutic proteins to the oviduct. We report the generation of transgenic hens that synthesize functional recombinant pharmaceutical protein in a tightly regulated tissue-specific manner, without any evidence of transgene silencing after germ-line transmission.
Keywords: chicken, genetic modification, lentivirus, pharmaceutical
Many human therapeutic proteins, such as monoclonal antibodies, are produced in industrial bioreactors, but setting up such systems is both time-consuming and expensive. Increasing global demand for recombinant pharmaceutical proteins has resulted in significant research being focused on development of alternative production platforms, including the use of transgenic animals as bioreactors. The basic strategy used has been to target expression of a therapeutic protein to a protein secretory tissue, often the mammary gland, using the regulatory sequences of one of the native proteins synthesized in that tissue. The production of therapeutic proteins has now been demonstrated in transgenic mammals, including sheep, goats, cattle, rabbits (1, 2), and chickens (3–5). The use of transgenic chickens as bioreactors to synthesize therapeutic proteins, as a component of egg yolk or white, may have several advantages over mammalian expression systems, including a shorter timescale for setup and ease of scaleup to a transgenic flock, lower costs associated with husbandry, the potential to produce proteins that are toxic to mammalian cells, beneficial glycosylation profile, and reduced immunogenicity of the purified product (reviewed in ref. 6).
The first reported attempt to generate transgenic hens expressing a therapeutic protein used a vector derived from the avian retrovirus avian leukosis virus, but injection of a low-titer vector preparation into the subgerminal cavity of embryos from new-laid eggs proved to be very inefficient (3). Injection of a high-titer retroviral vector derived from mouse stem cell virus into the heart of embryos that had been incubated for 55 h resulted in both efficient transgenesis and recombinant protein synthesis, but suppression of transgene expression was observed after germ-line transmission (4). This phenomenon would be problematic for effective scaleup. Recent reports have described the culturing and genetic modification of chicken ES cells (ref. 5) or primordial germ cells (ref. 7), which were used to generate chickens that were chimeras between the modified cells and wild-type recipients. High levels of somatic chimerism with chicken ES cells and germ-line contribution by primordial germ cells were demonstrated, but the fidelity of oviduct-specific expression of a human monoclonal antibody in chicken ES cells chimeras was variable. We have shown that lentiviral vectors can be used to generate germ-line transgenic chickens with high efficiency (8), and tissue-specific expression of a transgene delivered using a lentiviral vector has been demonstrated in quail (9).
In this work, we utilize lentiviral vectors derived from equine infectious anemia virus (EIAV) to generate transgenic chickens. This vector has been rendered multiply defective by deletion of all viral coding sequences from the transfer genome. Vectors are produced from separate expression plasmids that are designed to minimize the potential for homologous recombination. The transgenes investigated consist of codon-optimized sequences for either a humanized ScFv-Fc miniantibody, known as miR24, derived from a mouse monoclonal antibody shown to have potential for the treatment of malignant melanoma (10), or human IFN-β-1a (hIFNβ1a), expression of which is directed by regulatory sequences from the chicken ovalbumin gene. Ovalbumin constitutes ≈54% of the protein in the white of hens' eggs, and the steroid-responsive tissue-specific regulation of the ovalbumin gene is well documented (11). We describe the generation of transgenic chickens that synthesize functional recombinant therapeutic protein specifically in the oviduct of laying hens as a component of egg white.
Results
Construct Design.
The regulatory sequences of the ovalbumin gene have been investigated in detail, although the minimum sequence required for oviduct-restricted expression in vivo has not been defined. The EIAV vectors we used to generate transgenic chickens with high efficiency have a capacity of ≈9 kb. Regulatory sequences and parts of the exon/intron structure of the gene were used to generate the following constructs: the construct designated OVA contained the steroid-dependent regulatory element, the negative regulatory element, exon 1, intron 1, and the beginning of exon 2 (OVA, 2.8 kb). The estrogen-responsive enhancer element (ERE) was cloned immediately 5′ to the steroid-dependent regulatory element to generate construct EREOVA (3.5 kb). The endogenous Kozak and start codon were deleted during PCR amplification and replaced with a unique restriction endonuclease site to facilitate frame-independent insertion. A Kozak consensus sequence and start codon were included in the synthetic constructs for the two target proteins. The target proteins were a chimeric ScFv-Fc miniantibody (miR24) and hIFNβ1a. The coding sequence for miR24 was cloned adjacent to the OVA sequence, and the coding sequence for hIFNβ1a was cloned adjacent to both OVA and EREOVA sequences (Fig. 1). The vectors were packaged and injected into embryos from new-laid eggs to generate G0 transgenic birds.
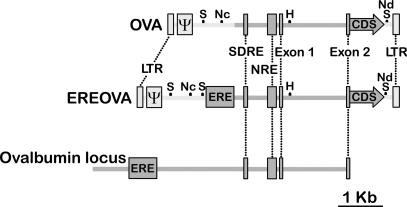
Fig. 1.
Schematic representation of the structure of the vectors described after chromosomal integration. Relative positions of lentiviral elements (light gray) and ovalbumin regulatory sequences (dark gray) are indicated, including the EIAV LTRs and packaging signal (Ψ) and the ovalbumin gene ERE; steroid-dependent regulatory element (SDRE), negative regulatory element (NRE), exon 1, intron 1, and the 5′ segment of exon 2 of the ovalbumin transcribed sequence; and the Kozak sequence and coding sequence of either miR24 or hIFNβ1a (CDS). The positions of restriction sites SpeI (S), NcoI (Nc), HindIII (H), and NdeI (Nd) used in restriction analysis are marked. The U3 region of the 3′ LTR is modified such that the integrated provirus lacks an active promoter within the LTR. Relative positions of regulatory elements in the chicken ovalbumin locus are shown.
Production and Analysis of Transgenic Birds.
A single G0 cockerel (bird OR2-4), hatched after injection of OVA-miR24 vector, was estimated to have vector sequence present in the germ line at a frequency of 5%, by PCR analysis of genomic DNA from semen samples. OR2-4 was crossed to stock hens and 463 chicks screened, among which 19 (4%) transgenic G1 offspring were identified. Genomic DNA from these G1 birds was analyzed by restriction digestion and Southern blot analysis to determine the number of vector insertions in each bird and the number of different insertion events represented in the G1 generation. Genomic DNA digested with HindIII generated junction fragments that revealed that two independent vector insertions were present in these G1 birds (Fig. 2Ai). No G1 birds were found that carried both proviral insertions. SpeI digestion to release a fragment spanning the ovalbumin regulatory sequence and transgene generated the predicted fragment of 5.2 kb in all cases, indicating that the ovalbumin intron present in the vector was retained in the transgenic chickens (data from the first 10 chicks are shown; Fig. 2Aii).
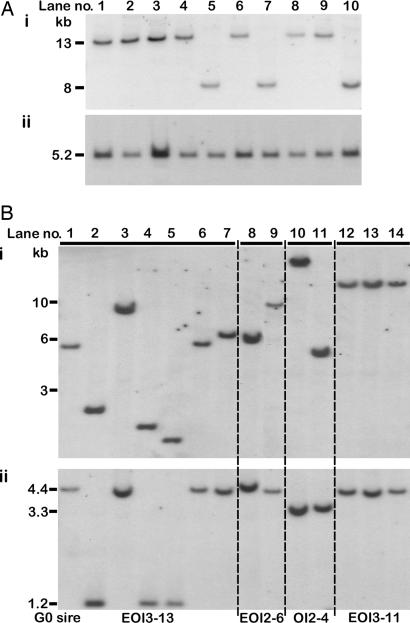
Fig. 2.
Southern transfer analysis of genomic DNA from individual G1 birds. (A and B) Samples from 10 birds carrying the OVA-miR24 provirus (A) were digested with HindIII (Ai) or SpeI (Aii) to generate either a junction fragment or a fragment spanning the ovalbumin regulatory sequence, respectively. Samples from two birds carrying the OVA-IFN provirus and 12 birds carrying the EREOVA-IFN provirus (B) were digested with NcoI (Bi) or NcoI/NdeI (Bii) to generate either a junction fragment or a fragment spanning the ovalbumin regulatory sequence, respectively. The G0 sires of the G1-IFN birds analyzed are indicated below.
A single G0 cockerel (bird OI2-4) transduced with OVA-IFN vector and three G0 cockerels (birds EOI2-6, EOI3-11, and EOI3-13) transduced with EREOVA-IFN vector were crossed to stock hens to generate G1 chicks. Genomic DNA was isolated from two G1 birds carrying the OVA-IFN vector and 12 G1 birds carrying the EREOVA-IFN vector. For Southern transfer analysis, genomic DNA was digested with NcoI to generate junction fragments (Fig. 2Bi) or NcoI/NdeI to release a fragment spanning the ovalbumin regulatory sequence and transgene (Fig. 2Bii). The two OVA-IFN birds represented unique vector insertion events (Fig. 2Bi, lanes 10 and 11), and each contained an intact transgene as evidenced by a 3.3-kb internal fragment (Fig. 2Bii). The 12 EREOVA-IFN birds represented a total of nine independent vector-insertion events. An identical insertion was shared by two offspring of EOI3-13 (Fig. 2Bi, lanes 1 and 6) and another by all three offspring of EOI3-11 (Fig. 2Bi, lanes 12–14). After double digestion, DNA from 9 of the 12 birds produced a fragment of 4.4 kb (Fig. 2Bii), indicating an intact transgene. A truncated fragment of 1.2 kb was detected in the three remaining birds, corresponding to a splicing event during production of the vector between a strong cryptic splice donor in the ERE sequence and the splice acceptor site of exon 2. This splicing event was confirmed by PCR, cloning, and sequence analysis of these samples (data not shown).
Transgene Expression Is Restricted to the Oviduct.
We aimed to direct transgene expression to the oviduct of laying hens by utilizing regulatory sequences of the ovalbumin gene to control expression of the two therapeutic proteins. Tissue samples were collected from oviduct, pancreas, brain, intestine, liver, heart, and breast muscle of adult G1 and G2 hemizygous transgenic hens that were in lay to assess whether transgene expression was restricted to the oviduct of adult hens. Northern blot analysis of total RNA extracted from these tissues was carried out on samples from two G1 and two G2 hens carrying the OVA-miR24 transgene, two G2 hens carrying the EREOVA-IFN transgene, and a nontransgenic control hen. Transcripts for both miR24 and hIFNβ1a were limited to the magnum region of the oviduct (Fig. 3), as was the endogenous ovalbumin transcript. RT-PCR on samples from one of the G2 hens carrying the OVA-miR24 transgene confirmed that the miR24 transcript was restricted to the magnum [supporting information (SI) Fig. 7]. To investigate the site of synthesis of miR24 in more detail, immunohistochemistry on magnum sections with a FITC-conjugated anti-human IgG (Fc-specific) was performed (Fig. 4). These data demonstrate that miR24 protein was localized to tubular gland cells and absent in adjoining epithelial cells. Taken together, these data show that the 2.8- and 3.5-kb 5′ regulatory sequences from the ovalbumin gene utilized in the current study are sufficient to confer tissue-specific expression of transgenes delivered using a lentiviral vector.
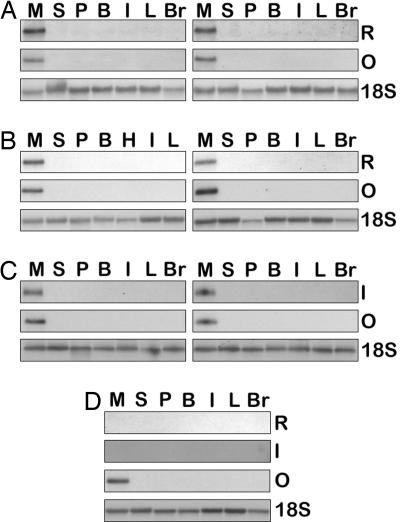
Fig. 3.
Northern blot analysis of total RNA from individual G1 and G2 hens. (A–D) Total RNA was extracted from the magnum portion of the oviduct (M), spleen (S), pancreas (P), brain (B), intestine (I), liver (L), and breast muscle (Br) from: two G1 hens (OR2-4:447 and OR2-4:476, A) and two G2 hens (OR2-4:25:59 and OR2-4:25:171, B) carrying the OVA-miR24 vector; two G2 hens carrying the EREOVA-IFN vector (EOI3-13:148:14 and EOI3-13:148:40, C), and a nontransgenic control hen (D). Blots were sequentially hybridized with probes for either miR24 (R) or hIFNβ1a (I), followed by an ovalbumin probe (O) and then 18S ribosomal RNA (18S), with the exception of the control, which was hybridized sequentially with probes for miR24, hIFNβ1a, ovalbumin, and 18S ribosomal RNA.
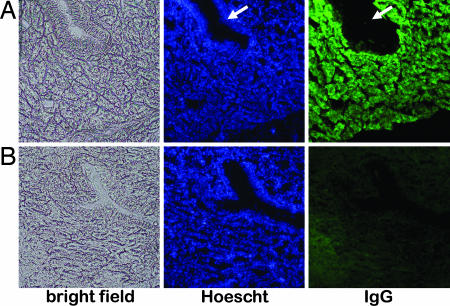
Fig. 4.
Immunohistochemical detection of miR24 in oviduct sections. (A and B) Sections of the magnum portion of the oviduct of hen OR2-4:25:59 (A) and a nontransgenic control (B) were stained with the nuclear stain Hoescht to visualize individual cells or FITC-anti-human IgG to visualize location of miR24. Tubular gland cells expressing the transgene appear green, whereas epithelial cells lining the tubular glands (indicated by arrows) do not produce miR24.
Recombinant Proteins Are Secreted into Egg White.
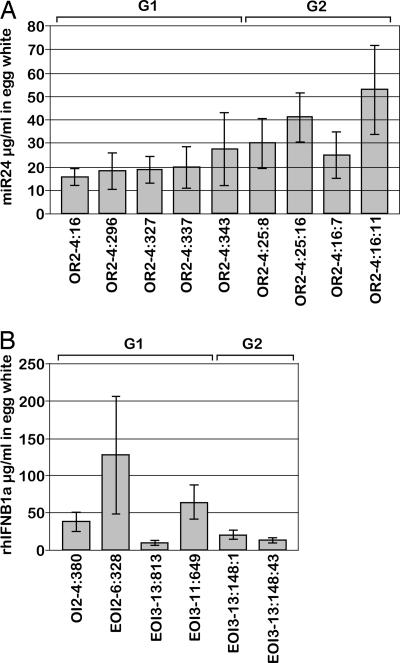
The amount of recombinant protein produced per egg was analyzed by measuring the proteins in a series of eggs laid by individual transgenic birds. Eggs laid by transgenic G1 and G2 hens were collected and the egg white assayed by ELISA for the presence of recombinant protein. Each of the first 20 eggs and then every 10th subsequent egg from each hen was analyzed (up to a maximum of the 150th egg). Recombinant protein was detected in the egg white of all eggs assayed (Fig. 5), and levels remained consistent in eggs from each bird over time. Eggs from nine hens containing the OVA-miR24 vector (5 × G1 and 4 × G2) and representing both independent vector insertions were assayed. The mean values for recombinant miR24 were between 15 and 50 μg/ml (Fig. 5A). Eggs from a single G1 hen containing the OVA-IFN vector, together with three G1 and two G2 hens containing EREOVA-IFN vector, were also assayed. The mean values for recombinant hIFNβ1a protein in eggs from these six hens were more variable than observed for the miR24, ranging from 3.5 to 426 μg/ml (Fig. 5B). Eggs from hen OI2–4:380 (OVA-IFN) contained hIFNβ1a at a mean value of 38 μg/ml, falling within the range measured in eggs from hens with the EREOVA-IFN vector.
Fig. 5.
Concentration of recombinant protein in egg white from transgenic hens. (A and B) Egg-white solution was assayed by ELISA for the presence of recombinant miR24 (A) or hIFNβ1a (B). Columns represent the mean concentration of recombinant protein (±SD) in egg white from each of the first 20 eggs and then each subsequent 10th egg up to a maximum of the 150th egg from the hens indicated. G1 and G2 hens are indicated above, with individual bird numbers below.
Recombinant hIFNβ1a in Egg White Is Functional.
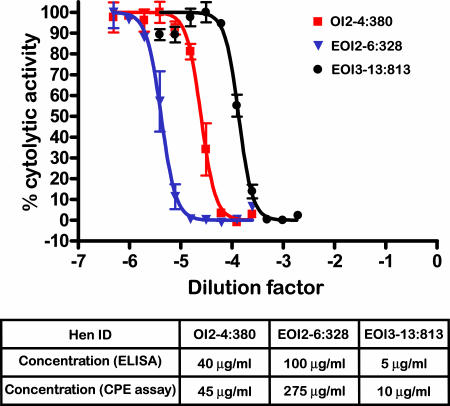
Egg white from hens carrying the hIFNβ1a transgenes was analyzed to determine whether the recombinant protein synthesized was functional. Eggs from a hen carrying the OVA-IFN vector (OI2-4:380) and from two hens carrying the EREOVA-IFN vector (EOI2-6:328 and EOI3-13:813) were collected and egg white assayed for the presence of active hIFNβ1a by using a standard antiviral/cytopathic effect assay. VeroE6 cells were incubated with dilutions of egg white collected from eggs from the transgenic hens, then infected with Semliki Forest Virus. After incubation, the degree to which the egg white and positive control recombinant hIFNβ1a protected cells from infection and subsequent cytolysis was assessed. Egg white from all eggs from transgenic hens demonstrated antiviral activity in this cytopathic effect assay (Fig. 6), but no protection was seen by using control egg white from stock hen eggs. The concentrations of hIFNβ1a present in eggs, as determined by ELISA, correlated with protective activity, as determined by this assay, with the egg from hen EOI2-6:328 containing the highest amount of active hIFNβ1a and the egg from hen EOI3-13:813 containing the lowest amount. These data demonstrate the functional equivalence of recombinant hIFNβ1a between different transgenic birds.
Fig. 6.
Biological activity of recombinant hIFNβ1a in egg white from transgenic hens. Egg white from hens OI2-4:380, EOI2-6:328, and EOI3-13:813 was analyzed in a cytopathic effect assay for antiviral activity. By applying a nonlinear regression (curve fit), the dilution of each sample, which resulted in 50% of cells being protected from Semliki forest virus (EC50), was calculated and compared with the EC50 for reference IFNβ1a to determine hIFNβ1a concentration (activity) in the egg white samples. Estimates of hIFNβ1a measured by ELISA in the same egg white samples are provided for comparison.
Discussion
We previously reported that lentiviral vectors can be utilized for the efficient production of transgenic chickens (8). Multiple lines of transgenic birds were generated, and stable transmission of integrated vector through the germ line was demonstrated with a conserved transgene expression profile in G1 and G2 generations. In the present work, we have progressed this application of EIAV-derived lentiviral vectors to demonstrate tissue-specific expression of heterologous proteins in the oviduct of laying hens, using transgenes derived from regulatory sequences from the chicken ovalbumin gene locus.
The chicken ovalbumin gene encodes more than half of egg white protein, ≈2.2 g of protein per egg (12), with up to 105 copies of the ovalbumin mRNA per cell in the oviduct of laying hens (13). A previous report (5) utilized either 7.5 or 15 kb of the ovalbumin 5′ regulatory sequence coupled with 15 kb of 3′ sequence in an attempt to direct transgene expression specifically to the tubular gland cells of the magnum of laying hens. These constructs were transfected into chicken ES cells and the modified cells used to generate chimeric hens whose oviduct tissue was in part derived from the transgenic chicken ES cells. RT-PCR analysis of RNA from these chimeric hens revealed expression of the ovalbumin-derived transgene in the magnum of the oviduct, but expression was also detected in the intestine, leading the authors (5) to hypothesize that the regulatory regions utilized were not sufficient to obtain transgene expression restricted to the oviduct in vivo.
The constructs investigated in the current study were significantly shorter than those discussed above (5), with the OVA construct spanning only 2.8 kb 5′ to the translation start site. In the EREOVA construct, a 675-bp fragment spanning the ERE, normally located ≈3.3 kb 5′ to the transcription start site (20), was cloned directly 5′ to the end of the OVA construct. Northern blot analysis of mRNA extracted from a range of tissues of transgenic hens in lay showed that transcripts from either transgene were restricted to the magnum portion of the oviduct, coincident with ovalbumin expression. Ectopic expression was not detected in any of the tissues analyzed. Additionally, immunohistochemical staining of sections of the magnum revealed that recombinant protein synthesis was restricted to the region containing the tubular gland cells, the site of ovalbumin synthesis in the magnum, and was absent from adjacent epithelial cells. This observation of tissue-specific transgene expression, using a lentiviral vector to deliver the transgene, is consistent with previous reports from mouse (14), pig (15), quail (9), and rat (16).
The ERE region, included in the EREOVA construct, contains four half-palindromic binding sites for the estrogen receptor that have been shown to act synergistically in vitro, and it was anticipated that inclusion of this element might enhance the quantity of recombinant protein synthesized by transgenic hens. Surprisingly, comparison of eggs from a hen (OI2-4:380) carrying the OVA-IFN transgene with those from hens carrying the EREOVA-IFN transgene (EOI3-13:813 and EOI3-11:649) revealed that the inclusion of this element did not result in an increase in the mean level of hIFNβ1a protein in egg white in the birds analyzed. The exception to this observation was hen EOI2-6:328, where the mean hIFNβ1a protein level in eggs was 2-fold higher than in eggs from other birds carrying the same construct. Because the number of transgenic hens analyzed is limited, it is not possible to draw any firm conclusions about influence of the different ovalbumin regulatory elements on transgene expression levels from these data. Additional factors, such as the vector integration site, the genetic background of the hens, and epigenetic factors, are likely to influence the expression level of any specific transgene integrant. For example, variation in recombinant protein expression level in different hens containing the same vector integration site has been reported (3). It is unlikely that the observed variations in expression level would be an impediment to commercialization when averaged over a production flock.
In recent years, several incremental advances have been made in the use of retroviral vectors for avian transgenesis. Replication defective avian leukosis virus was used to generate hens expressing the reporter β-lactamase from a ubiquitous promoter. While β-lactamase was detected in egg white of transgenic hens, the quantity of protein, efficiency of transgenesis, and germ-line transmission rate were all low (17, 18). Arian leukosis virus was also used to generate chickens expressing human IFN-α-2b at higher levels than had been reported for β-lactamase (3), but the efficiency with which transgenic birds were generated remained very low (a single G1 cockerel from 1,597 chicks hatched), presumably a consequence of the low-titer vector used. Blastodermal injection of high-titer Moloney murine leukemia virus-derived vectors was used to generate transgenic quail with very high efficiency, but transgene silencing prevented detectable expression of the reporter GFP (19). A modified mouse stem cell virus injected into the heart of developing chick embryos generated transgenic hens that expressed an antiprion protein ScFv-Fc from a ubiquitous promoter, but analysis of G0, G1, and G2 generations revealed ≈2-fold transgene suppression in successive generations (4). Although we observed no evidence of transgene silencing, continued assessment of transgene expression levels will be made for several generations to confirm the long-term stability of expression required for commercial protein production. A lentiviral vector was previously used to generate transgenic quail that showed tissue-specific expression without any evidence of ectopic expression or transgene silencing between generations (9). The current report demonstrates the combination of efficient generation of transgenic hens with the synthesis of functional therapeutic proteins at high levels in the egg white.
Methods
Plasmid Construction.
Chimeric mouse–human ScFv-Fc miniantibody miR24 and hIFNβ1a coding sequences were modified to reflect the codon bias in the chicken and to include the signal peptide from chicken lysozyme at the 5′ end. The miR24 coding sequence, synthesized by Entelechon (Regensburg, Germany), was PCR-amplified to introduce SmaI and PmlI flanking restriction endonuclease sites and then cloned into pGEMTeasy (Promega, Madison, WI) to generate pLE11. hIFNβ1a sequence including SmaI flanking restriction endonuclease sites was synthesized by Geneart (Regensburg, Germany) and supplied in PCR-Script (Stratagene, La Jolla, CA).
Primers 5DHR2 (5′-CTTAAGTCCTCAGACTTGGC-3′) and 3Sma (5′-GCCCCGGGTGAACTCTGAGTTGTCTAG-3′) were designed to amplify a 2.8-kb fragment 5′ to the coding sequence of chicken ovalbumin gene. The PCR product was cloned into pGEMTeasy to generate plasmid pLE10 (OVA promoter). Primers LD675for (5′-CTGCAGAAAAATGCCAGGTGG-3′) and LD675rev (5′-TCTAGAGAGAGTAAGCAACAATCTTCT-3′) were designed to amplify a 675-bp region spanning the ERE, a DNaseI hypersensitivity site ≈3.3 kb 5′ to the ovalbumin transcription start site (20). The PCR product was cloned into pGEMTeasy to generate plasmid pLE20a. pLE10 was linearized at the extreme 5′ end of its ovalbumin regulatory sequence with NcoI, and blunt ends were generated with T4 DNA polymerase. The ERE was excised from pLE20a with EcoRI, and blunt ends were generated with T4 DNA polymerase and then ligated into pLE10 to give plasmid pLE21 (EREOVA promoter).
miR24 was excised from pLE11 by SmaI/PmlI digestion and subcloned into the SmaI site of pLE10 to give pRI38. The promoter/transgene was excised from pRI38 by digestion with SacII/MluI and subcloned into the SacII/AscI sites of the minimal EIAV lentiviral vector pONY8.45Nmcs to give pLE38 (OVA-miR24).
hIFNβ1a was subcloned into the SmaI site of pLE21 to give pLE43. The promoter/transgene was excised from pLE43 by digestion with AatII/MluI, and blunt ends were generated with T4 DNA polymerase. pONY8.45Nmcs was digested with SacII/AscI, blunt ends were generated with T4 DNA polymerase, and the promoter/transgene above ligated in to give pLE46 (EREOVA-IFN).
The ERE sequence was excised from pLE43 by digestion with AflII/AatI to generate plasmid pLE42. The promoter/transgene from pLE42 was excised with ApaI/MluI, blunt ends were generated with T4 DNA polymerase, and the fragment was ligated into pONY8.45Nmcs to give pLE45 (OVA-IFN). Schematic representation of integrated constructs is presented in Fig. 1.
Lentivirus Production.
Vector stocks were generated by FuGene6 (Roche, Lewes, U.K.) transfection of HEK293T cells plated on 144-mm dishes with 10.2 μg of vector plasmid/5.1 μg of gag/pol plasmid (pESYNGP)/2.5 μg of rev plasmid (pESYNRev) (21)/0.2 μg of VSV-G plasmid (pRV67) (22). Forty-eight hours after transfection, tissue culture medium was harvested and filtered (0.22 μm), then centrifuged at 6,000 × g for 20 h at 4°C, followed by ultracentrifugation at 50,500 × g for 120 min at 4°C. The pellet was resuspended in formulation buffer (20 mM Tris/100 mM NaCl/10 mg/ml sucrose/10 mg/ml mannitol, pH 7.4). Aliquots of vector were stored at −80°C.
Production and Analysis of Transgenic Birds.
Founder transgenic birds were generated by injection of viral vectors into chick embryos at the new-laid egg stage, followed by culture of the embryos to hatch as described (8). The hatched chicks were raised to sexual maturity, and DNA extracted from semen from adult males was screened by PCR as described (8, 23) to identify cockerels that carried vector sequences in the germ line. Cockerels identified by this method were crossed to wild-type hens, and their offspring were screened by PCR to identify G1 hemizygous transgenic birds. All experiments, animal breeding, and care procedures were carried out under license from the U.K. Home Office.
The vector insertions in individual G1 birds were analyzed by Southern transfer. Genomic DNA (10 μg), extracted from whole blood, was digested with appropriate restriction endonucleases, resolved on a 0.7% wt/vol agarose gel, and transferred to Hybond-N membrane (Amersham Biosciences, Chalfont St. Giles, U.K.). These blots were analyzed by using probes labeled with [32P]dCTP using a RediPrime II kit (Amersham Biosciences). Hybridization was in 500 mM sodium phosphate (pH 7.2)/7% SDS at 65°C and was detected by autoradiography.
RNA and Protein Analysis.
Tissue samples (≈50 mg) were extracted, snap-frozen, and stored at −80°C until required. Total RNA was prepared by using RNA-Bee (AMS Biotech, Abingdon, U.K.), and 10 μg was resolved on a denaturing 1% wt/vol agarose gel. RNA was transferred to Hybond-N membrane (Amersham Biosciences) by using 10× SSC (1× SSC = 0.15 M sodium chloride/0.015 M sodium citrate, pH 7). Hybridization was as for Southern blots.
To assay recombinant protein expressed in egg white, eggs were carefully cracked open and the egg white was recovered free of yolk. The egg white was mixed for at least 60 min at 4°C with 4 vol of ice-cold 50 mM Na acetate buffer (pH 5.0) to remove the bulk of the ovomucin fraction.
For detection of miR24 protein, Immulon 2B plates (Thermo Labsystems, Basingstoke, U.K.) were coated with sheep anti-human IgG (γ-chain specific, AU004; The Binding Site, Birmingham, U.K.) antibody at a concentration of 4 μg/ml 50 mM carbonate/bicarbonate buffer (pH 9.6) and incubated for 1 h at room temperature. The standard used was human IgG1 antibody (BP078; The Binding Site). Plates were washed with PBS/0.05% Tween 20 (PBST) before adsorbing sample egg white solution (pH 5.5). Egg white samples were tested in duplicate and incubated for 1 h at room temperature. Plates were washed with PBST, and the presence of miR24 was revealed by horseradish peroxidase-conjugated anti-sheep/goat IgG antibody at a dilution of 1:1,000 in PBST for 1 h at room temperature (AP004; The Binding Site). The plates were washed with PBST and incubated for 5 min at room temperature with 0.4 mg/ml o-phenylenediamine/0.006% hydrogen peroxide in 50 mM phosphate-citrate buffer (pH 5.0). The reaction was stopped and developed by the addition of 0.25 vol of 12.5% sulfuric acid. Plates were read at 490 nm, and data were analyzed by using a log:logit plot on Microsoft Excel (Microsoft, Redmond, WA). The sensitivity of the ELISA was 8 ng/ml.
hIFNβ1a protein concentration in egg white was determined by using a Human IFN-β ELISA kit (PBL Biomedical Laboratories, Piscataway, NJ; product no. 41400-2A). Data were analyzed by using GraphPad (San Diego, CA) Prism software. The sensitivity of this assay is 250 pg/ml.
Immunohistochemistry.
Adult tissues were isolated and fixed for 30 min in 4% paraformaldehyde in PBS, and tissues were cryo-embedded and sectioned at 14 μm on Superfrost Plus microscope slides (VWR, Lutterworth, U.K.). Slides were blocked in PBST containing 10% heat-inactivated chicken serum for 1 h and incubated for 1 h in secondary antibody solution (1:100 dilution of FITC-conjugated anti-human IgG; Sigma, St. Louis, MO) PBST containing 10% chicken serum. Slides were washed for 1 additional hour in PBST, counterstained with Hoechst, and mounted.
Cytopathic Effect Assay.
VeroE6 cells were seeded at a density of 1 × 104 into each well of a 96-well plate (200 μl per well of DMEM/10% FBS/1% penicillin/streptomycin/glutamine). Cells were incubated at 37°C in 5% CO2 for 24 h. Medium was then replaced with 100 μl of DMEM/2% FBS/1% penicillin/streptomycin/glutamine, except column 2, to which 150 μl of DMEM plus 2.66% FBS/1% penicillin/streptomycin/glutamine was added. Fifty microliters of test egg white samples, undiluted or prediluted 1:1,000 with serum-free medium, was added to column 2.
As controls, Escherichia coli-derived IFNβ1a (PBL Biomedical Laboratories) was spiked into egg white solution from a control egg (undiluted or prediluted 1:1,000 serum-free medium) or into SFM at 4,000 units/ml.
Samples and controls were serially diluted (2-fold) across the 96-well plates from columns 2 to 11. Column 1 was retained as the untreated control, and column 12 was retained as the uninfected control. Plates of cells were then incubated at 37°C 5% CO2 for 24 h before infection of each well in columns 1–11 with 150 plaque-forming units of Semliki Forest virus in 100 μl. Plates of cells were further incubated at 37°C 5% CO2 for 48 h, and virus-induced cytolysis was assessed by using the lactate dehydrogenase cytotoxicity kit (Roche), according to the manufacturer's instructions. Results were analyzed by using GraphPad Prism to compare the titer EC50 for egg white samples with the EC50 determined from a known concentration of E. coli-produced hIFNβ1a.
Plasmids pLE21, pLE45, and pLE46 are available upon request from Viragen (Scotland) Ltd. Plasmids pESYNGP, pESYNRev, and pRV67 are available upon request from Oxford Biomedica Ltd.
Supplementary Material
Acknowledgments
We thank H. Gilhooley, F. Thomson, R. Mitchell, M. Hutchison, and L. Taylor for technical assistance. Financial support was from the Biotechnology and Biological Sciences Research Council and from the Scottish Executive SPUR scheme.
Abbreviations
- EIAV
equine infectious anemia virus
- hIFNβ1a
human IFN-β-1a
- ERE
estrogen-responsive enhancer element.
Footnotes
Conflict of interest statement: I.W. was head of division of this group while he was at Roslin Institute, but he will gain no benefit from publication of this paper. The authors declare no conflict of interest.
See Commentary on page 1739.
This article contains supporting information online at www.pnas.org/cgi/content/full/0610401104/DC1.
References
- 1.Dyck MK, Lacroix D, Pothier F, Sirard MA. Trends Biotechnol. 2003;21:394–399. doi: 10.1016/S0167-7799(03)00190-2. [DOI] [PubMed] [Google Scholar]
- 2.Hunter CV, Tiley LS, Sang HM. Trends Mol Med. 2005;11:293–298. doi: 10.1016/j.molmed.2005.04.001. [DOI] [PubMed] [Google Scholar]
- 3.Rapp JC, Harvey AJ, Speksnijder GL, Hu W, Ivarie R. Transgenic Res. 2003;12:569–575. doi: 10.1023/a:1025854217349. [DOI] [PubMed] [Google Scholar]
- 4.Kamihira M, Ono K, Esaka K, Nishijima K, Kigaku R, Komatsu H, Yamashita T, Kyogoku K, Iijima S. J Virol. 2005;79:10864–10874. doi: 10.1128/JVI.79.17.10864-10874.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhu L, van de Lavoir M, Albanese J, Beenhouwer DO, Cardarelli PM, Cuison S, Deng DF, Deshpande S, Diamond JH, et al. Nat Biotechnol. 2005;23:1159–1169. doi: 10.1038/nbt1132. [DOI] [PubMed] [Google Scholar]
- 6.Lillico SG, McGrew MJ, Sherman A, Sang HM. Drug Discov Today. 2005;10:191–196. doi: 10.1016/S1359-6446(04)03317-3. [DOI] [PubMed] [Google Scholar]
- 7.van de Lavoir M, Diamond JH, Leighton PA, Mather-Love C, Heyer BS, Bradshaw R, Kerchner A, Hooi LT, Gessaro TM, et al. Nature. 2006;441:766–769. doi: 10.1038/nature04831. [DOI] [PubMed] [Google Scholar]
- 8.McGrew MJ, Sherman A, Ellard FM, Lillico SG, Gilhooley HJ, Kingsman AJ, Mitrophanous KA, Sang H. EMBO Rep. 2004;5:728–733. doi: 10.1038/sj.embor.7400171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scott BB, Lois C. Proc Natl Acad Sci USA. 2005;102:16443–16447. doi: 10.1073/pnas.0508437102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nasi ML, Meyers M, Livingston PO, Houghton AN, Chapman PB. Melanoma Res. 1997;7:S155–S162. [PubMed] [Google Scholar]
- 11.Dougherty DC, Sanders MM. Trends Endocrinol Metab. 2005;16:414–419. doi: 10.1016/j.tem.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 12.Burley RW, Vadehra DV. The Avian Egg. New York: Wiley; 1989. p. 72. [Google Scholar]
- 13.Palmiter RD. Cell. 1975;4:189–197. doi: 10.1016/0092-8674(75)90167-1. [DOI] [PubMed] [Google Scholar]
- 14.Lois C, Hong EJ, Pease S, Brown EJ, Baltimore D. Science. 2002;295:868–872. doi: 10.1126/science.1067081. [DOI] [PubMed] [Google Scholar]
- 15.Hofmann A, Kessler B, Ewerling S, Weppert M, Vogg B, Ludwig H, Stojkovic M, Bolehauve M, Brem G, et al. EMBO Rep. 2003;4:1054–1060. doi: 10.1038/sj.embor.7400007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nakagawa T, Feliu-Mojer MI, Wulf P, Lois C, Sheng M, Hoogenraad CC. J Neurosci Methods. 2006;152:1–9. doi: 10.1016/j.jneumeth.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 17.Harvey AJ, Speksnijder G, Baugh LR, Morris JA, Ivarie R. Nat Biotechnol. 2002;19:396–399. doi: 10.1038/nbt0402-396. [DOI] [PubMed] [Google Scholar]
- 18.Harvey AJ, Ivarie R. Poultry Sci. 2003;82:927–930. doi: 10.1093/ps/82.6.927. [DOI] [PubMed] [Google Scholar]
- 19.Mizuarai S, Ono K, Yamaguchi K, Nishijima K, Kamihira M, Iijima S. Biochem Biophys Res Commun. 2001;286:456–463. doi: 10.1006/bbrc.2001.5422. [DOI] [PubMed] [Google Scholar]
- 20.Kato S, Tora L, Yamauchi J, Masushige S, Bellard M, Chambon P. Cell. 2002;68:731–742. doi: 10.1016/0092-8674(92)90148-6. [DOI] [PubMed] [Google Scholar]
- 21.Rohll JB, Mitrophanous KA, Martin-Rendon E, Radcliffe PA, Mazarakis MD, Kingsman SM. Methods Enzymol. 2002;346:466–500. doi: 10.1016/s0076-6879(02)46072-7. [DOI] [PubMed] [Google Scholar]
- 22.Soneoka Y, Cannon PM, Ramsdale EE, Griffiths JC, Romano G, Kingsman SM, Kingsman AJ. Nucleic Acids Res. 1995;23:628–633. doi: 10.1093/nar/23.4.628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sherman A, Dawson A, Mather C, Gilhooley H, Li Y, Mitchell R, Finnegan D, Sang H. Nat Biotechnol. 1998;16:1050–1053. doi: 10.1038/3497. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.