Abstract
An important component of triple-drug anti-AIDS therapy is 2′,3′-dideoxy-3′-thiacytidine (3TC, lamivudine). Single mutations at residue 184 of the reverse transcriptase (RT) in HIV cause high-level resistance to 3TC and contribute to the failure of anti-AIDS combination therapy. We have determined crystal structures of the 3TC-resistant mutant HIV-1 RT (M184I) in both the presence and absence of a DNA/DNA template-primer. In the absence of a DNA substrate, the wild-type and mutant structures are very similar. However, comparison of crystal structures of M184I mutant and wild-type HIV-1 RT with and without DNA reveals repositioning of the template-primer in the M184I/DNA binary complex and other smaller changes in residues in the dNTP-binding site. On the basis of these structural results, we developed a model that explains the ability of the 3TC-resistant mutant M184I to incorporate dNTPs but not the nucleotide analog 3TCTP. In this model, steric hindrance is expected for NRTIs with β- or l- ring configurations, as with the enantiomer of 3TC that is used in therapy. Steric conflict between the oxathiolane ring of 3TCTP and the side chain of β-branched amino acids (Val, Ile, Thr) at position 184 perturbs inhibitor binding, leading to a reduction in incorporation of the analog. The model can also explain the 3TC resistance of analogous hepatitis B polymerase mutants. Repositioning of the template-primer as observed in the binary complex (M184I/DNA) may also occur in the catalytic ternary complex (M184I/DNA/3TCTP) and contribute to 3TC resistance by interfering with the formation of a catalytically competent closed complex.
Introduction
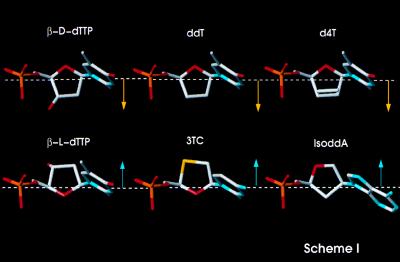
The introduction of combination drug therapies for the treatment of AIDS has resulted in a dramatic decrease in AIDS-related deaths in the United States. One of the standard components in these treatments is 2′,3′-dideoxy-3′-thiacytidine (3TC), a second-generation nucleoside analog reverse transcriptase inhibitor (NRTI) that has a substantially improved pharmacological profile compared with AZT and the dideoxynucleotide inhibitors (1, 2). Like other NRTIs, the triphosphate of 3TC (3TCTP) inhibits RT by causing termination of the growing DNA chain. In lieu of the ribose ring of canonical nucleosides, 3TC has a β-l-oxathiolane ring system (Fig. 1) (3, 4). Although not as efficient an inhibitor of HIV-1 RT as some of the other NRTIs (μM vs. nM Ki), 3TC has much lower toxicity, apparently because it is not a good substrate for mitochondrial DNA polymerases (2, 3).
Figure 1.
Chemical structures of deoxythymidine and various NRTIs, with aligned C1′ and C4′ atoms of the nucleoside rings. Such alignment highlights the unique direction (arrows pointing down) of the β-l-nucleosides ring, projecting in the opposite direction from the other nucleoside and nucleoside analog inhibitors (arrows pointing up).
3TC is also a potent inhibitor of hepadnavirus polymerases (1, 5). Despite the availability of an effective vaccine, about 5% of the world’s population is chronically infected with hepatitis B virus (HBV) (6). In Phase II clinical trials (7), 3TC treatment was shown to reduce HBV viral loads dramatically. As with HIV-1, however, viral resistance to 3TC develops in cell cultures and in clinical studies (8, 9). Clinical treatment of HBV infection by using 3TC has recently been approved.
Structural Background.
A number of crystal structures of HIV-1 RT have been determined, including structures of unliganded HIV-1 RT (10–12), HIV-1 RT complexed with a 19:18 double-stranded DNA template-primer and the Fab fragment of a monoclonal antibody (13, 14), and HIV-1 RT complexed with a 25:21 double-stranded DNA template-primer and dTTP (15). HIV-1 RT is a heterodimer, composed of a larger 560-residue chain (p66) and a second smaller subunit (p51) that contains the N-terminal 440 residues of the p66 subunit. Although both subunits contain the fingers, palm, thumb, and connection subdomains, only p66 has a polymerase active site and a DNA-binding cleft formed by the p66 fingers, palm, and thumb subdomains (16). In HIV-1 RT, M184 is the second residue of the highly conserved YXDD (Tyr-X-Asp-Asp) motif, which contains two of the three essential aspartic acids that define the polymerase active site (17, 18). In all HIV-1 RT structures, the YXDD motif makes an unusual type II′ turn that positions the side chains of the four consecutive amino acids (YXDD) on the surface of the p66 palm subdomain, allowing for interactions with template-primer, dNTP, and metal cofactors. Most retroviral RTs (including HIV-1 RT) and the HBV polymerase contain a methionine residue as amino acid X; in Moloney murine leukemia virus (MuLV) RT the motif is YVDD (19). In HBV polymerase, residue X is M552. Nonretroviral RTs contain the alternative motifs YIDD, YLDD, and YADD (19–21). The dNTP-binding site in HIV-1 RT is composed of both protein and nucleic acid. Among the residues that contribute, either directly or indirectly, to the formation of the dNTP-binding site are Y115, F116, F160, Q151, and L74 of the p66 subunit (15, 22–27).
Resistance to 3TC.
Initially, resistance to 3TC is associated with the substitution of isoleucine for methionine at position 184 of HIV-1 RT, which results from a single base change (ATG → ATA). The M184I variant is rapidly replaced by the variant M184V during 3TC therapy (28–35). Similar mutations at the equivalent position in HBV polymerase and in simian immunodeficiency virus and feline immunodeficiency virus RTs confer resistance to 3TC, suggesting a common mechanism of resistance to β-l-nucleoside inhibitors (36–39). In HBV polymerase, 3TC resistance is associated either with a single mutation of methionine to isoleucine at position 552 in the YMDD motif or with two mutations that occur concomitantly (M552V/L528M) (8).
Despite the plethora of biochemical and clinical data on NRTI-resistant mutants of HIV-1 RT, the structural details that underlie NRTI resistance are not well understood. On the basis of a combination of biochemical, crystallographic, and molecular modeling experiments, we have proposed that alteration of positioning of the nucleic acid template-primer can play an important role in NRTI resistance (22, 23, 40, 41). We now know from the structure of RT/DNA complexed with dTTP (RT/DNA/dTTP) (15) that the ternary complex is a closed structure with the fingers folded down toward the dNTP-binding site and the extended template overhang bent away from the DNA duplex. Many of the NRTI-resistance mutations are closer to the bound dNTP than were previously predicted. Nevertheless, repositioning of template-primer is still likely to affect dNTP and NRTI binding by distorting the dNTP-binding pocket.
In wild-type RT, M184 contacts both the 3′ end of the primer and the dNTP. Presteady and steady-state kinetic studies with the 3TC-resistant M184V mutant have shown that there is a relatively small change in the ability of the mutant enzyme to bind dNTP (42, 43) and 3TCTP (43) and that the turnover rate of 3TC by M184V HIV-1 RT is substantially decreased relative to the wild-type enzyme. However, another kinetic study with 2′-3′-dideoxy-3′-thia-5-fluorocytidine (FTC) (42), the 5-fluoro analog of 3TC, reported the opposite effect: the M184V mutation affects binding of FTCTP (≈18-fold increase in Kd) rather than the turnover rate of FTC incorporation (unchanged kp). Although the relative resistance of RT to 3TC and to FTC is significantly different (KiM184V RT/KiWT RT is 50 for 3TC and >500 for FTC), the structural similarity of the two inhibitors suggests that these two inhibitors should have a common mechanism of drug resistance. On the basis of the crystal structure of wild-type RT/DNA/dNTP (15), Huang et al. have suggested that the side chain of either a valine or an isoleucine at position 184 would interfere with 3TCTP binding. However, in the absence of structures for 3TC-resistant RTs in complexes with a template-primer and 3TCTP, the structural details of 3TC resistance have not yet been elucidated.
In this study, we compare the crystal structures of the wild-type RT/DNA complex with an HIV-1 RT mutant (M184I) in the presence of an identical oligonucleotide substrate (3.5-Å resolution). The structures of unliganded wild-type and M184I RTs (2.85-Å resolution) were also compared. The structural information was used to develop a model that explains the ability of 3TC-resistant mutant M184I to incorporate dNTPs but not the nucleotide analog 3TCTP. In this model, steric conflict between the oxathiolane ring of 3TCTP and the side chain of β-branched amino acids (Val, Ile, Thr) at position 184 perturbs the stereochemistry of inhibitor binding, leading to a reduced turnover rate. The repositioning of the template-primer seen in the binary complexes may also occur in the catalytic complex and contribute to 3TC resistance. A combination of these factors could interfere with the formation of a catalytically competent closed complex.
MATERIALS AND METHODS
Purification and crystallization of M184I RT/DNA/Fab was done as described previously for the corresponding wild-type complex (13, 14). Specifically, purified HIV-1 RT was mixed with Fab at 1:0.8 mass ratio (total protein concentration: 25 mg/ml) and 0.2 mM 19:18 DNA (19-mer template: 3′-AGGGACAAGCCCGCGGTA-5′, template overhang in bold). Hanging drops were prepared by mixing equal volumes of the complex and crystallization solutions (100 mM cacodylate, pH 5.6/29–31% saturated ammonium sulfate) at 4°C. For unliganded M184I RT, the crystallization solution contained 50 mM Bis⋅Tris, 100 mM (NH4)2SO4, 10% glycerol, and 9% polyethylene glycol 8000, pH 6.8 (16). Diffraction datasets were collected at the Cornell High Energy Synchrotron Source (CHESS) F1 beam line and at the Brookhaven National Light Source X-25 beam line (Table 1).
Table 1.
Summary of crystallographic data and refinement statistics
| M184I RT/DNA/Fab | M184I RT, unliganded | ||
|---|---|---|---|
| Space group | P3212 | P3212 | C2 |
| Temperature, °C | −10 | −165 | −165 |
| Resolution range, Å | 40−3.5 | 40−3.6 | 40−2.85 |
| Cell parameters, a, Å | 169.2 | 167.4 | 238.6 |
| b, Å | 169.2 | 167.4 | 72.7 |
| c, Å | 221.9 | 219.7 | 95.3 |
| β, ° | 90 | 90 | 105.6 |
| Completeness, % | 91.5 (83) | 84.0 (82) | 94.9 (88) |
| (highest shell) | |||
| Rmerge, % | 11.8 | 11.3 | 9.0 |
| Refinement statistics | |||
| Resolution range, Å | 10−3.5 | 8−2.85 | |
| No. of reflections | 39,033 | 31,200 | |
| R-factor | 26.2 | 25.9 | |
| Free R-factor | 33.8 | 34.6 | |
| No. of protein atoms | 11,720 | 7,700 | |
| Luzzati error, Å | 0.53 | 0.48 | |
Structure Determination.
The structures of cooled and frozen M184I RT/DNA/Fab complexes were solved by molecular replacement with wild-type HIV-1 RT/DNA/Fab as a starting model (13, 14). To improve the phase quality and reduce model bias, at the early stages of structure determination we used the program rave and the multiple crystal averaging program dmmulti to average the electron density maps of the frozen and cooled datasets at 3.6-Å resolution. To further reduce model bias, we calculated maps with the DNA and the polymerase active site omitted from the model. Rounds of model building were guided by using both normal and averaged omit maps. The DNA position was clearly indicated in both difference Fourier (Fobs−Fcalc) and the averaged electron density 2Fobs−Fcalc maps computed for M184I RT/DNA/Fab complexes. Both maps were calculated by using phases derived from only the protein model, before any information about bound DNA was included either in the structure refinements or in the map calculations. Stereochemically restrained structure refinement was performed by using the program xplor (44), varying positional and individual isotropic thermal parameters by using a protocol similar to the one used for the corresponding wild-type structure refinement (13). The unliganded M184I structure was solved by using molecular replacement with the unliganded wild-type HIV-1 RT structure (12) as the starting model, with the mutated and the surrounding residues omitted.
RESULTS AND DISCUSSION
Comparison of Structures.
Overall the structures of wild-type HIV-1 RT and the M184I mutant are quite similar either in the presence or in the absence of double-stranded DNA substrate. Specifically, when the unliganded structures of wild-type and mutant HIV-1 RT were compared by superposition of the Cα atoms of amino acid residues 107 to 115 and 151 to 215 of the p66 palm subdomain, the rms deviation was 0.3 Å.
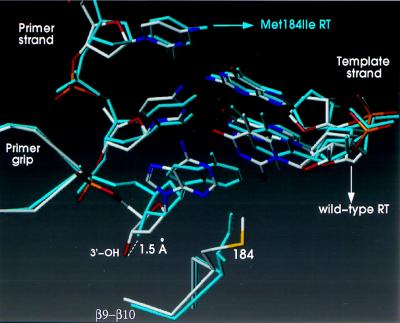
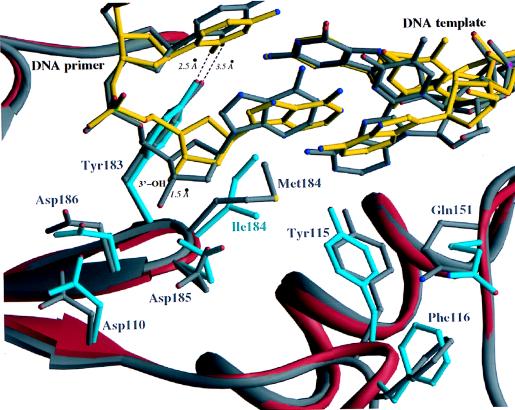
A similar superposition (using the same residue ranges) of the polymerase active sites of the wild-type and mutant RT complexes with DNA gave an rms deviation of 0.4 Å. There is, however, a substantial shift in the position of the last few nucleotides of the primer strand near the polymerase active site (Fig. 2). The largest shift in the primer strand is at the 3′-terminal nucleotide, which is adjacent to the side chain of residue 184 of the p66 subunit, resulting in a displacement of the 3′-OH by approximately 1.5 Å. The hydrogen-bonding interaction between the O2 atom of the penultimate nucleotide of the primer strand and the side chain of Y183 (Fig. 3) is stronger in the M184I complex [the distance between these groups decreases from 3.5 Å in the wild-type RT/DNA complex (13) to 2.5 Å in the M184I RT/DNA complex (Fig. 3)].
Figure 2.
Superposition of the polymerase active sites of wild-type HIV-1 RT/DNA/Fab and M184I HIV-1 RT/DNA/Fab. The wild-type RT complex is shown in white, the mutant RT complex in cyan. The wild-type and M184I RT structures were superimposed on the basis of the core of the p66 palm subdomains (residues 107 to 112 and 151 to 215 of their corresponding p66 subunits).
Figure 3.
Ribbon diagram of the superimposed polymerase active sites of wild-type HIV-1 RT/DNA/Fab and M184I HIV-1 RT/DNA/Fab. The wild-type protein and DNA are shown in gray, the mutant protein in red, and DNA in the mutant RT/DNA complex in yellow.
Smaller changes were observed for complementary nucleotides near the 5′-end of the template strand (Fig. 3). We also observed changes in the position of the unpaired 5′-nucleotide of the template strand (Fig. 3); however, this nucleotide is still stacked over the terminal base pair of the template-primer in a position (relative to the nucleic acid) reminiscent of the arrangement seen in the wild-type RT/DNA complex.
Despite the differences in template-primer position when the wild-type HIV-1 RT is compared with the M184I mutant, the effect of DNA binding on the YXDD motif was similar for the wild-type and mutant enzymes. Specifically, the “punched-down” conformation of the YXDD motif at the tip of β9-β10 that has been previously reported for wild-type HIV-1 RT in the presence of DNA substrate (14) was also observed for the mutant enzyme. The side chain of I184 of the p66 subunit is well ordered in the M184I RT/DNA complex. However, in the unliganded M184I structure the I184 residue in the p66 subunit is disordered. The electron density for I184 in the p51 subunit is well defined in both apo and DNA-bound M184I RT structures.
Changes were also observed in residues proximal to the dNTP-binding site in the M184I RT/DNA complex (Fig. 3). Five of these residues are close to the template-primer (D110, D185, Q151, Y115, and L74). The displacements range up to 2.4 Å when the p66 subunits of the mutant and wild-type RT/DNA complexes are compared (e.g., 1.1 Å for Oδ1 of D110; 2.4 Å for Oγ of D185; 1.2 Å for Oη of Y115; 1.1 Å for Cα of Q151; and 1.8 Å for Cβ of L74). The side chains of D185 and D110 follow the displaced primer terminus of the M184I RT/DNA complex (Fig. 3) and the hydrogen bond between Oδ1 of D185, and 3′-OH of the primer terminus becomes even stronger in the mutant RT/DNA complex (decrease in distance from 3.3 Å to 2.7 Å). However, the hydrogen bond between Oδ2 of D185 and the main-chain carbonyl of V111 is lost (increase in distance from 2.7 to 4.3 Å).
The β12–β13 connecting loop has been previously termed the “primer grip” because it helps position the primer strand at the active site (13, 23, 45–47). A comparison of the DNA-bound structures shows changes (≈1 Å) in the positions of the side chains of residues that are close to the primer DNA strand. These residues include W229 and Y232 of the primer grip, and W266 of the p66 thumb. These changes are associated with the primer repositioning: they are not observed when the unliganded structures are compared.
Resistance to β-l-Nucleosides, Including 3TC, Involves Steric Hindrance.
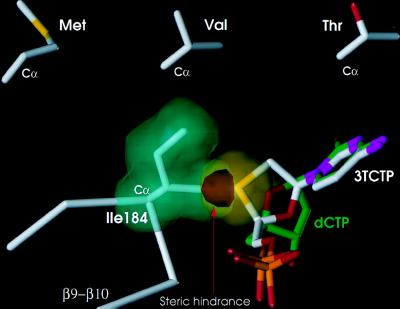
Using the M184I RT/DNA and wild-type RT/DNA crystal structures, we have modeled the interactions of dCTP and 3TCTP at the active site of the M184I enzyme (Fig. 4), assuming similar binding of the triphosphate moieties and similar base-pairing interactions of the incoming inhibitors with a modeled template overhang. The predicted position of the nucleotide with respect to the active site YIDD motif in this model is strongly supported by the structure of the polymerase active site in the recently reported ternary HIV-1 RT/DNA/dTTP complex (15). The 3TCTP and dCTP nucleoside ring configurations are enantiomeric (Fig. 1), so that alignment of the triphosphates and bases would cause the nucleoside ring of the β-l-inhibitor to project 1.5–2.0 Å further toward residue 184 than would the ribose ring of dCTP (Fig. 4). The most significant difference between the interactions of wild-type HIV-1 RT and the M184I mutant with ribose and β-l-oxathiolane nucleoside inhibitors is a very close contact predicted between the Cγ2 methyl of Ile and the protruding oxathiolane ring (Fig. 4). This unfavorably close contact (2.5 Å in M184I vs. 3.9 Å in wild type) would result in severe steric hindrance. The steric conflict would be effectively enforced by the conformational restriction of the β-branched I184 side chain. I184 is constrained both by interactions with the Y115 and Y183 side chains and by interactions with nearby main-chain atoms of the type II′ β turn in an already crowded polymerase active site (Figs. 2 and 3). In contrast to I184, the wild-type M184 side chain not only lacks a β branch but also has greater conformational flexibility, permitting 3TCTP to bind without significant steric conflict. The same mechanism also explains why the only other reported 3TC-resistance mutations are M184V and M184T: valine and threonine are the only other amino acids with β-branched methyl substituents (35, 48).
Figure 4.
Schematic illustration of proposed steric conflict between 3TCTP and 184I. The van der Waals volume of the side chain of I184 (green) is shown to overlap (red) with the sulfur of the β-l-oxathiolane ring of the incoming 3TCTP (yellow). A dCTP molecule (green) is shown superimposed on 3TCTP (white) in a way that maintains the same base pairing with a modeled template strand and proximity to the 3′-OH, but without steric conflict with I184. The model predicts similar steric hindrance with the two other β-branched amino acids (Val and Thr). The side chains of these amino acids are shown in an orientation similar to the I184 observed in our structure. This model is supported by the reported resistance of M184V and M184T RT to 3TCTP. An analogous model can explain resistance of analogous mutants of HBV RT to 3TCTP.
In support of this concept, an analogous complex of M184V RT with DNA reveals that the V184 side chain has its β-methyl group in a similar position (unpublished results). In addition, as predicted by this model, the M184A HIV-1 RT mutant, which lacks a β-methyl substituent at the 184 position, does not discriminate between β-l- and β-d-ring systems (42). RT resistance to 3TC expressed as the Ki/Km ratio for 3TCTP to dTTP is similar for M184A and wild-type HIV-1 RT (42).
Steric hindrance between 3TCTP (or FTCTP) and Ile-184 of M184I RT would not necessarily block the binding of 3TCTP (or FTCTP)—it could shift the binding position of these inhibitors. If the character rather than the extent of binding of β-l-oxathiolane inhibitors is decreased, it is also possible that such changes will affect the interactions that cause the fingers to fold down in the ternary catalytic complex (15). Hence, resistance to 3TC may be the result of perturbed stereochemistry of 3TCTP binding in the M184I RT/DNA/3TCTP complex, potentially leading to an abortive transition state. The nucleoside ring geometry of canonical dNTPs minimizes the steric clash with the β-branched side chain of 3TC-resistant RTs apparently resulting in only small changes in dNTP binding. In the absence of a β-branched chain at the 184 position, wild-type RT is able to incorporate nucleotides with unfavorable stereochemistry such as 3TCTP, albeit less efficiently than normal nucleotide substrates.
In addition to explaining RT resistance to β-l-oxathiolane nucleoside inhibitors (3TC and FTC), this model can be generalized to explain RT resistance to all nucleoside inhibitors with β-l-stereochemistry. In fact, the model correctly predicts that mutants with β-branched amino acids at position 184 have less resistance to β-l-ddCTP than to 3TCTP (42, 49, 50). This is because β-l-ddCTP has a carbon instead of a sulfur at the point of steric conflict. The van der Waals radius of carbon is smaller than that of sulfur, reducing the severity of the steric clash. In addition, as predicted by this model, resistance of M184 mutants to the β-l- version of other nucleoside inhibitors is approximately 50- to 500-fold higher than resistance to the β-d-enantiomers of the same inhibitors (42), as is the case for all reported examples, including 3TCTP, FTCTP, CBVTP (carbovir), d4TTP (stavudine), ddTTP, and ddCTP (42).
4(S)-(6-Amino-9H-purin-9-yl)tetrahydro-2(S)-furanmethanol (IsoddA) is a structurally unique NRTI stereochemically similar to β-d-ddI, but with its base transposed from the natural 1′ position to the 2′ position (Fig. 1). A single Met to Val mutation at codon 184 of RT renders HIV-1 resistant to IsoddA (51). Because of transposition of the base, the ring of IsoddA is expected to project toward the 184 side chain, in a way similar to the β-l-nucleoside inhibitors. Hence, the model can be used to explain resistance of 184 mutants with β-branched side-chains to IsoddA on the basis of the structural similarity between IsoddA and the β-l-nucleoside inhibitors.
The model can also explain the structural basis of 3TC resistance of other polymerases. We have constructed a three-dimensional model of the HBV active site based on the crystal structure of the HIV-1 RT/DNA complex and the sequence homology between HIV-1 RT and HBV polymerase. This model suggests that, like in HIV-1 RT, mutation of the Met of the YMDD motif (M552 in HBV) to a β-branched residue (Ile or Val) would cause steric conflict with the oxathiolane ring of 3TC. However, additional mutations outside the YMDD motif of the HBV polymerase have been reported to confer 3TC resistance, suggesting that other interactions may also affect HBV polymerase sensitivity to 3TCTP (8, 52). Similar to the HIV-1 and HBV enzymes, simian immunodeficiency virus RT develops resistance to 3TC through Met to Ile mutation in the YMDD motif (36), and it was recently reported that the Met to Thr mutation in the YMDD motif of feline immunodeficiency virus RT confers resistance to oxathiolane nucleosides (37, 38). Interestingly, MuLV RT, which has a valine as residue X at the YXDD motif, is naturally resistant to 3TCTP (30).
Possible Role of Template-Primer Repositioning in Resistance to 3TC and Other NRTIs.
In the M184I RT/DNA complex, changes are observed in the position of the template-primer, along with less pronounced changes at residues such as L74, D110, Y115, F116, Q151, and D185, but not in the uncomplexed mutant. When the substrate is a dNTP, these changes may be the cause of the moderate reductions in polymerase activity, processivity, and dNTP binding that have been reported for the mutant enzyme (41–42, 53). M184I retains the ability to incorporate normal nucleotides in a reasonably efficient manner (35, 41, 48), suggesting that the transition state geometry of M184I RT resembles that of the wild-type enzyme.
It is not clear whether the template-primer shift observed in the M184I/DNA complex is maintained in the M184I/DNA/3TCTP ternary complex and to what extent nucleic acid repositioning contributes to 3TC resistance. A shift in the position of the primer would likely contribute to a misalignment of 3TCTP at the transition state and a decrease in the turnover rate of the reaction. Alternatively, shifting the primer back to the position it occupies in a complex with wild-type RT may incur an energetic cost that could also affect formation of the transition state. When a dNTP is the substrate, the effect of primer shift may contribute to the modest changes in polymerization rate, processivity, and dNTP binding seen with the mutant enzyme. When 3TCTP is the incoming substrate, the primer shift may increase the free energy needed to reach the transition state and contribute to 3TC resistance.
Low-level resistance to dideoxynucleoside inhibitors (ddNTPs) has been reported for 184 mutants of HIV-1 RT (2- to 5-fold) (29, 34, 54). This may be related to the distortions of the dNTP-binding pocket caused (i) by changes in the binding of template-primer and/or (ii) by movements of residues near the dNTP-binding pocket (Y115, F116, Q151, D110, D185, and L74). Substitution of two of these residues (L74V or Q151M) (34, 55) causes phenotypic resistance of HIV-1 RT to ddI and ddC. The 3′-hydroxyl of a normal dNTP substrate makes a hydrogen-bond interaction with the backbone NH of Y115 (15); loss of this interaction could alter the position at which a ddNTP is bound. The loss of this hydrogen bond could exacerbate the changes at the M184I dNTP-binding site and result in low-level resistance to ddNTP inhibitors.
Implications for Drug Design.
In conclusion, we have presented a model that explains resistance to 3TC in HIV-1 and other retroviral RTs and in HBV polymerase. We propose that high-level resistance to β-l-nucleoside inhibitors (3TC, FTC) or NRTIs with similar ring orientation (IsoddA) results from a steric conflict between a β-branched amino acid residue at the 184 position of HIV-1 RT and the β-l- or similarly shaped nucleoside ring. Inhibitor molecules designed to have reduced steric conflict with β-branched amino acids at position 184 should not be excluded as effectively by the M184I and M184V mutants. Such inhibitors should include 3TC derivatives that contain in place of the sulfur (radius = 1.80 Å) more compact atoms such as oxygen (radius = 1.52 Å) or carbon (radius = 1.70 Å for methylene). Alternatively, analogs of 3TC containing ring system modifications (e.g., unsaturated, acyclic, or four-membered) may also have less steric clash than 3TC. These inhibitors would potentially have advantages for treating AIDS and viruses whose polymerases have similar active sites, for example hepatitis B.
Acknowledgments
We thank Aaron Shatkin for useful suggestions on the manuscript, Pat Clark for help in protein production, and Yu Hsiou, Karen Lentz, Chris Tantillo, Hemant Yennawar, Wanyi Zhang, and Steve Ealick, Dan Thiel, and coworkers at Cornell High Energy Synchrotron Source and Lonny Berman and coworkers at Brookhaven National Light Source for help with data collection. S.G.S. is supported by a National Institutes of Health–National Institute of Allergy and Infectious Diseases National Research Service Award fellowship (AI 09578) and dedicates his efforts in this work to the memory of George E. Sarafianos. The research in E.A.’s laboratory has been supported by National Institutes of Health Grant AI 36144 and the Keck Foundation. S.H.H.’s laboratory is sponsored in part by the National Cancer Institute, Department of Health and Human Services, under contract with Advanced BioScience Laboratories and by the National Institutes of General Medical Sciences.
ABBREVIATIONS
- 3TC
2′,3′-dideoxy-3′-thiacytidine
- FTC
2′-3′-dideoxy-3′-thia-5-fluorocytidine
- HBV
hepatitis B virus
- MuLV
Moloney murine leukemia virus
- NRTI
nucleoside analog reverse transcriptase inhibitor
- RT
reverse transcriptase
- YXDD motif
Tyr-X-Asp-Asp motif
- 3TCTP
the triphosphate of 3TC
Footnotes
Data deposition: the atomic coordinates have been deposited in the Protein Data Bank, www.rcsb.org [PDB ID code 1C9R (M184I/DNA complex) and 1QE1 (M184I)].
References
- 1.Furman P A, Davis M, Liotta D C, Paff M, Frick L W, Nelson D J, Dornsife R E, Wurster J A, Wilson L J, Fyfe J A, et al. Antimicrob Agents Chemother. 1992;36:2686–2692. doi: 10.1128/aac.36.12.2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kukhanova M, Liu S H, Mozzherin D, Lin T S, Chu C K, Cheng Y C. J Biol Chem. 1995;270:23055–23059. doi: 10.1074/jbc.270.39.23055. [DOI] [PubMed] [Google Scholar]
- 3.Chen C H, Vazquez-Padua M, Cheng Y C. Mol Pharmacol. 1991;39:625–628. [PubMed] [Google Scholar]
- 4.Schinazi R F, McMillan A, Cannon D, Mathis R, Lloyd R M, Peck A, Sommadossi J P, St. Clair M, Wilson J, Furman P A, et al. Antimicrob Agents Chemother. 1992;36:2423–2431. doi: 10.1128/aac.36.11.2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Severini A, Liu X Y, Wilson J S, Tyrrell D L J. Antimicrob Agents Chemother. 1995;39:1430–1435. doi: 10.1128/aac.39.7.1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maynard J E. Vaccine. 1990;8,Suppl.:S18–S20. doi: 10.1016/0264-410x(90)90209-5. [DOI] [PubMed] [Google Scholar]
- 7.Dienstag J L, Perillo R P, Schiff E R, Bartholomew M, Vicary C, Rubin M. N Engl J Med. 1995;333:1657–1661. doi: 10.1056/NEJM199512213332501. [DOI] [PubMed] [Google Scholar]
- 8.Chang C N, Doong S L, Zhou J H, Beach J W, Jeong L S, Chu C K, Tsai C H, Liotta D, Schinazi R F, Cheng Y C. J Biol Chem. 1992;267:13938–13942. [PubMed] [Google Scholar]
- 9.Allen M I, Deslauriers M, Andrews C W, Tipples G A, Walters K A, Tyrrell D L J, Brown N, Condreay L D. Hepatology. 1998;27:1670–1677. doi: 10.1002/hep.510270628. [DOI] [PubMed] [Google Scholar]
- 10.Esnouf R, Ren J, Ross R, Jones Y, Stammers D, Stuart D. Nat Struct Biol. 1995;2:303–308. doi: 10.1038/nsb0495-303. [DOI] [PubMed] [Google Scholar]
- 11.Rodgers D W, Gamblin S J, Harris B A, Ray S, Culp J S, Hellmig B, Woolf D J, Debouck C, Harrison S C. Proc Natl Acad Sci USA. 1995;92:1222–1226. doi: 10.1073/pnas.92.4.1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hsiou Y, Ding J, Das K, Clark A D, Jr, Hughes S H, Arnold E. Structure (London) 1996;4:853–860. doi: 10.1016/s0969-2126(96)00091-3. [DOI] [PubMed] [Google Scholar]
- 13.Jacobo-Molina A, Ding J, Nanni R G, Clark A D, Jr, Lu X, Tantillo C, Williams R L, Kamer G, Ferris A L, Clark P, et al. Proc Natl Acad Sci USA. 1993;90:6320–6324. doi: 10.1073/pnas.90.13.6320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ding J, Das K, Hsiou Y, Sarafianos S G, Clark A D, Jr, Jacobo-Molina A, Tantillo C, Hughes S H, Arnold E. J Mol Biol. 1998;284:1095–1111. doi: 10.1006/jmbi.1998.2208. [DOI] [PubMed] [Google Scholar]
- 15.Huang H, Chopra R, Verdine G L, Harrison S C. Science. 1998;282:1669–1675. doi: 10.1126/science.282.5394.1669. [DOI] [PubMed] [Google Scholar]
- 16.Kohlstaedt L A, Wang J, Friedman J M, Rice P A, Steitz T A. Science. 1992;256:1783–1790. doi: 10.1126/science.1377403. [DOI] [PubMed] [Google Scholar]
- 17.Johnson M S, McClure M A, Feng D-F, Gray J, Doolittle R F. Proc Natl Acad Sci USA. 1986;83:7648–7652. doi: 10.1073/pnas.83.20.7648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Larder B A, Purifoy D J M, Powell K L, Darby G. Nature (London) 1987;327:716–717. doi: 10.1038/327716a0. [DOI] [PubMed] [Google Scholar]
- 19.Doolittle R F, Feng D-F, Johnson M S, McClure M A. Q Rev Biol. 1989;64:1–30. doi: 10.1086/416128. [DOI] [PubMed] [Google Scholar]
- 20.Poch O, Sauvaget I, Delarue M, Tordo N. EMBO J. 1989;8:3867–3874. doi: 10.1002/j.1460-2075.1989.tb08565.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Inouye S, Inouye M. Curr Opin Genet Dev. 1993;3:713–718. doi: 10.1016/s0959-437x(05)80088-7. [DOI] [PubMed] [Google Scholar]
- 22.Boyer P L, Ferris A L, Clark P, Whitmer J, Frank P, Tantillo C, Arnold E, Hughes S H. J Mol Biol. 1994;243:472–483. doi: 10.1006/jmbi.1994.1673. [DOI] [PubMed] [Google Scholar]
- 23.Tantillo C, Ding J, Jacobo-Molina A, Nanni R G, Boyer P L, Hughes S H, Pauwels R, Andries K, Janssen P A J, Arnold E. J Mol Biol. 1994;243:369–387. doi: 10.1006/jmbi.1994.1665. [DOI] [PubMed] [Google Scholar]
- 24.Sarafianos S G, Pandey V N, Kaushik N, Modak M J. Biochemistry. 1995;34:7207–7216. doi: 10.1021/bi00021a036. [DOI] [PubMed] [Google Scholar]
- 25.Shirasaka T, Kavlick M F, Ueno T, Gao W-Y, Kojima E, Alcaide M L, Chokekijchai S, Roy B M, Arnold E, Yarchoan R, et al. Proc Natl Acad Sci USA. 1995;92:2398–2402. doi: 10.1073/pnas.92.6.2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martin-Hernandez A, Domingo E, Menendez-Arias L. EMBO J. 1996;15:4434–4442. [PMC free article] [PubMed] [Google Scholar]
- 27.Sarafianos S G, Das K, Ding J, Boyer P L, Hughes S, H, Arnold E. Chem Biol. 1999;5:257–264. doi: 10.1016/s1074-5521(99)80071-4. [DOI] [PubMed] [Google Scholar]
- 28.Boucher C A, Cammack N, Schipper P, Schuurman R, Rouse P, Wainberg M A, Cameron J M. Antimicrob Agents Chemother. 1993;37:2231–2234. doi: 10.1128/aac.37.10.2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gao Q, Gu Z, Parniak M A, Cameron J, Cammack N, Boucher C, Wainberg M A. Antimicrob Agents Chemother. 1993;37:1390–1392. doi: 10.1128/aac.37.6.1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schinazi R F, Lloyd R J J, Nguyen M-H, Cannon D L, McMillan N, Ilksoy N, Chu C K, Liotta D C, Bazmi H Z, Mellors J W. Antimicrob Agents Chemother. 1993;37:875–881. doi: 10.1128/aac.37.4.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schinazi R F. Perspect Drug Discovery Des. 1993;1:151–180. [Google Scholar]
- 32.Tisdale M, Kemp S D, Parry N R, Larder B A. Proc Natl Acad Sci USA. 1993;90:5653–5656. doi: 10.1073/pnas.90.12.5653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.De Clercq E. Biochem Pharmacol. 1994;47:155–169. doi: 10.1016/0006-2952(94)90001-9. [DOI] [PubMed] [Google Scholar]
- 34.Schinazi R F, Larder B A, Mellors J W. Int Antiviral News. 1997;5:129–142. [Google Scholar]
- 35.Keulen W, Back N K T, Van Wijk A, Boucher C A B, Berkhout B. J Virol. 1997;71:3346–3350. doi: 10.1128/jvi.71.4.3346-3350.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cherry E, Slater M, Salomon H, Rud E, Wainberg M A. Antimicrob Agents Chemother. 1997;41:2763–2765. doi: 10.1128/aac.41.12.2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smith R, Remington K M, Lloyd R M J, Schinazi R F, North T W. J Virol. 1997;71:2357–2362. doi: 10.1128/jvi.71.3.2357-2362.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smith R A, Remington K M, Preston B D, Schinazi R F, North T W. J Virol. 1998;72:2335–2340. doi: 10.1128/jvi.72.3.2335-2340.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhu Y L, Dutschman D E, Liu S H, Bridges E G, Cheng Y C. Antimicrob Agents Chemother. 1998;42:1805–1810. doi: 10.1128/aac.42.7.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Boyer P L, Tantillo C, Jacobo-Molina A, Nanni R G, Ding J, Arnold E, Hughes S H. Proc Natl Acad Sci USA. 1994;91:4882–4886. doi: 10.1073/pnas.91.11.4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Boyer P L, Hughes S H. Antimicrob Agents Chemother. 1995;39:1624–1628. doi: 10.1128/aac.39.7.1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wilson J E, Aulabaugh A, Caligan B, McPherson S, Wakefield J K, Jablonski S, Morrow C D, Reardon J E, Furman P A. J Biol Chem. 1996;271:13656–13662. doi: 10.1074/jbc.271.23.13656. [DOI] [PubMed] [Google Scholar]
- 43.Krebs R, Immendorfer U, Thrall S H, Wöhrl B M, Goody R S. Biochemistry. 1997;36:10292–10300. doi: 10.1021/bi970512z. [DOI] [PubMed] [Google Scholar]
- 44.Brunger A T. x-plorManual Version 3.8: A System for X-Ray Crystallography and NMR. 1996. [Google Scholar]
- 45.Jacques P S, Wöhrl B M, Ottmann M, Darlix J L, Le Grice S F J. J Biol Chem. 1994;269:26472–26478. [PubMed] [Google Scholar]
- 46.Ghosh M, Jacques P S, Rodgers D R, Ottman M, Darlix J-L, Le Grice S F. Biochemistry. 1996;35:8553–8562. doi: 10.1021/bi952773j. [DOI] [PubMed] [Google Scholar]
- 47.Wöhrl B M, Krebs R, Thrall S H, Le Grice S F J, Scheidig A J, Goody R S. J Biol Chem. 1997;272:17581–17587. doi: 10.1074/jbc.272.28.17581. [DOI] [PubMed] [Google Scholar]
- 48.Back N K T, Nijhuis M, Keulen W, Boucher C A B, Oude Essink B B, van Kuilenburg A B P, van Gennip A H, Berkhout B. EMBO J. 1996;15:4040–4049. [PMC free article] [PubMed] [Google Scholar]
- 49.Faraj A, Agrofoglio L A, Wakefield J K, McPherson S, Morrow C D, Gosselin G, Mathe C, Imbach J, Schinazi R F, Sommadossi J P. Antimicrob Agents Chemother. 1994;38:2300–2305. doi: 10.1128/aac.38.10.2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Van Draanen N, Tisdale M, Parry N R, Jansen R, Dornsife R E, Tuttle J V, Averett D R, Koszalka G W. Antimicrob Agents Chemother. 1994;38:868–871. doi: 10.1128/aac.38.4.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nair V, St. Clair M, Reardon J, Krasny H, Hazen R, Paff M, Boone L, Tisdale M, Najera I, Dornsife R E. Antimicrob Agents Chemother. 1995;39:1993–1999. doi: 10.1128/aac.39.9.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fu L, Cheng Y C. Biochem Pharmacol. 1998;55:1567–1572. doi: 10.1016/s0006-2952(98)00050-1. [DOI] [PubMed] [Google Scholar]
- 53.Pandey V N, Kaushik N, Rege N, Sarafianos S G, Yadav N S, Modak M J. Biochemistry. 1996;35:2168–2179. doi: 10.1021/bi9516642. [DOI] [PubMed] [Google Scholar]
- 54.St. Clair M H, Martin J L, Tudor-Williams G, Bach M C, Vavro C L, King D M, Kellam P, Kemp S D, Larder B A. Science. 1991;253:1557–1559. doi: 10.1126/science.1716788. [DOI] [PubMed] [Google Scholar]
- 55.Ueno T, Mitsuya H. Biochemistry. 1997;36:1092–1099. doi: 10.1021/bi962393d. [DOI] [PubMed] [Google Scholar]