Abstract
Transcriptional activation is a key process required for long-term memory formation. Recently, the transcriptional coactivator CREB-binding protein (CBP) was shown to be critical for hippocampus-dependent long-term memory and hippocampal synaptic plasticity. As a coactivator with intrinsic histone acetyltransferase activity, CBP interacts with numerous transcription factors and contains multiple functional domains. Currently, it is not known which transcription factor-binding domain of CBP is essential for memory storage. Using mice that carry inactivating mutations in the CREB-binding (KIX) domain of the coactivator CBP (CBPKIX/KIX mice), we show that the KIX domain is required for long-term memory storage. These results are the first to identify an in vivo function for the KIX domain of CBP in the brain, and they suggest that KIX-interacting transcription factors recruit CBP histone acetyltransferase activity during long-term memory storage. One such KIX-interacting factor is the transcription factor CREB. Using quantitative real-time RT-PCR, we find that the expression of specific CREB target genes is reduced in the hippocampi of CBPKIX/KIX mice during memory consolidation. The recruitment of the transcriptional coactivator CBP via the KIX domain thus imparts target gene-dependent selectivity to CREB-driven transcriptional regulation, thereby activating genes required for the long-term storage of hippocampus-dependent memory.
Recently, we and others have shown that CREB-binding protein (CBP) is critical for hippocampus-dependent long-term memory (Oike et al. 1999; Bourtchouladze et al. 2003; Korzus et al. 2004) and synaptic plasticity (Alarcon et al. 2004; Wood et al. 2005; for review, see Josselyn 2005). CBP is a transcriptional coactivator that regulates transcription via direct interactions with transcription factors and the basal transcription machinery as well as via its intrinsic histone acetyltransferase (HAT) activity, which modifies chromatin structure (Goodman and Smolik 2000; Kalkhoven 2004). CBP interacts with numerous transcription factors and contains multiple functional domains. Studies focusing on memory storage have demonstrated that the CBP HAT domain is necessary for memory storage (Korzus et al. 2004) and suggested that CBP is recruited to DNA by transcription factors interacting with the amino terminus of CBP (Oike et al. 1999; Bourtchouladze et al. 2003; Wood et al. 2005). However, which transcription factor-binding domain of CBP is essential for memory storage remains a key open question.
The KIX domain of CBP provides a critical interface for the recruitment of CBP by specific transcription factors and the subsequent regulation of gene expression. Several transcriptional activators bind to the KIX domain of CBP, but most of these factors do so in a constitutive fashion. The interactions between CREB and CBP, however, are regulated by the phosphorylation state of CREB. Phosphorylation of CREB at Ser 133 in the kinase-inducible domain (KID), as a result of multiple signaling transduction cascades activated by extracellular stimuli, results in the association between phospho-KID and the KIX domain of CBP (Parker et al. 1996). Many models of long-term memory storage suggest that the phosphorylation of a constitutively present factor such as CREB may mediate the induction of gene expression required for memory consolidation. The importance of understanding transcription factor–coactivator interactions is underscored by the numerous studies examining the KID–KIX interaction (Parker et al. 1998, 1999; Radhakrishnan et al. 1998, 1999; Shaywitz et al. 2000; Campbell and Lumb 2002), including a solution structure of the transactivation domain of CREB in a complex with the CBP KIX domain (Radhakrishnan et al. 1997). In contrast to our understanding of the biochemical details regarding the KID-KIX interaction, the function of the CBP KIX domain in vivo is not known.
To address this question, we examined mice that carry a triple point mutation in the KIX domain of CBP (Kasper et al. 2002). These mice express a mutant form of CBP that is impaired in its ability to interact with phospho-CREB and c-Myb, but with the HAT domain of CBP intact. Homozygous CBP KIX mutant mice are viable, enabling us to examine these mice behaviorally. Our results show that the KIX domain is required for long-term memory storage. These results represent the first demonstration of an in vivo function for the CBP KIX domain in the brain and identify this transcription factor-binding domain of CBP as essential for memory processes. Importantly, our findings also show that the recruitment of the transcriptional coactivator CBP via the KIX domain imparts selectivity to CREB-driven transcriptional regulation by activating specific genes required for the long-term storage of hippocampus-dependent memory.
Results
Generation of CBPKIX/KIX mice
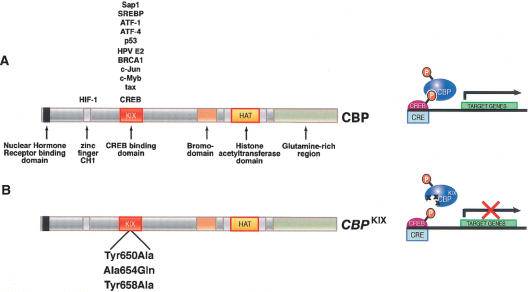
CBPKIX/KIX (current gene symbol is Crebbp) mice carry mutations in three highly conserved residues (Tyr650Ala, Ala654Gln, and Tyr658Ala) within the CBP KIX domain of an otherwise wild-type CBP allele (Fig. 1; Kasper et al. 2002). These residues are critical for the binding surface between the CBP KIX domain and the phosphorylated CREB KID domain (Radhakrishnan et al. 1998) as well as the hematopoietic transcription factor c-Myb (Parker et al. 1999). CBPKIX/KIX mice are essentially normal, apart from a modest decrease in thymus size (Kasper et al. 2002; Koo et al. 2005). Mouse embryonic fibroblast (MEF) lines derived from CBPKIX/KIX mice are compromised in their ability to support CREB-mediated and c-Myb-mediated transcription in transient transfection assays (Kasper et al. 2002). Importantly, there does not appear to be compensation by the increased expression of the CBP paralog p300 in CBPKIX/KIX cells, and the expression of the CBPKIX allele appears similar to that of the wild-type CBP allele (Kasper et al. 2002, and see below).
Figure 1.
Schematic diagram of CREB-binding protein (CBP) showing different functional domains. (A) The endogenous allele of CBP generates a protein product with multiple domains, including several protein–protein interaction motifs as well as the histone acetyltransferase domain. (B) The CBPKIX allele generates a protein product carrying three point mutations in the CREB-binding domain (KIX). These mutations block the interaction between phosphorylated CREB and CBP.
CBP and CBPKIX are expressed similarly in wild-type and CBPKIX/KIX littermate mice, respectively
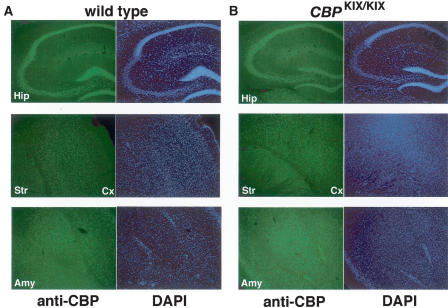
To date, the expression of CBP protein in the mouse brain has not been carefully examined. To determine CBP protein expression patterns, we performed immunohistochemistry on coronal slices of wild-type and CBPKIX/KIX littermate mice using anti-CBP antibodies. As shown in Figure 2A, CBP is expressed throughout the hippocampus, striatum, and amygdala in wild-type mice, and CBP was found to be expressed in all brain regions examined. Similarly, CBPKIX is expressed evenly throughout the same brain regions in CBPKIX/KIX littermate mice (Fig. 2B). These results show that CBPKIX is expressed in a similar manner to CBP.
Figure 2.
CBP and CBPKIX are expressed similarly in wild-type and CBPKIX/KIX littermate mice, respectively. (A) A representative section from a wild-type mouse shows normal distribution of CBP protein in all brain regions examined (Hip, Str, Cx, and Amy shown in left column). DAPI staining is shown in the right column. (B) A representative section from a CBPKIX/KIX homozygous knock-in mouse shows similar expression of CBPKIX protein in the same brain regions as CBP in wild-type sections (Hip, Str, Cx, and Amy shown in left column). DAPI staining is shown in the right column. Hip, hippocampus; Str, striatum; Cx, cortex; Amy, amygdala.
CBPKIX/KIX mice exhibit significantly impaired long-term memory for contextual but not cued fear
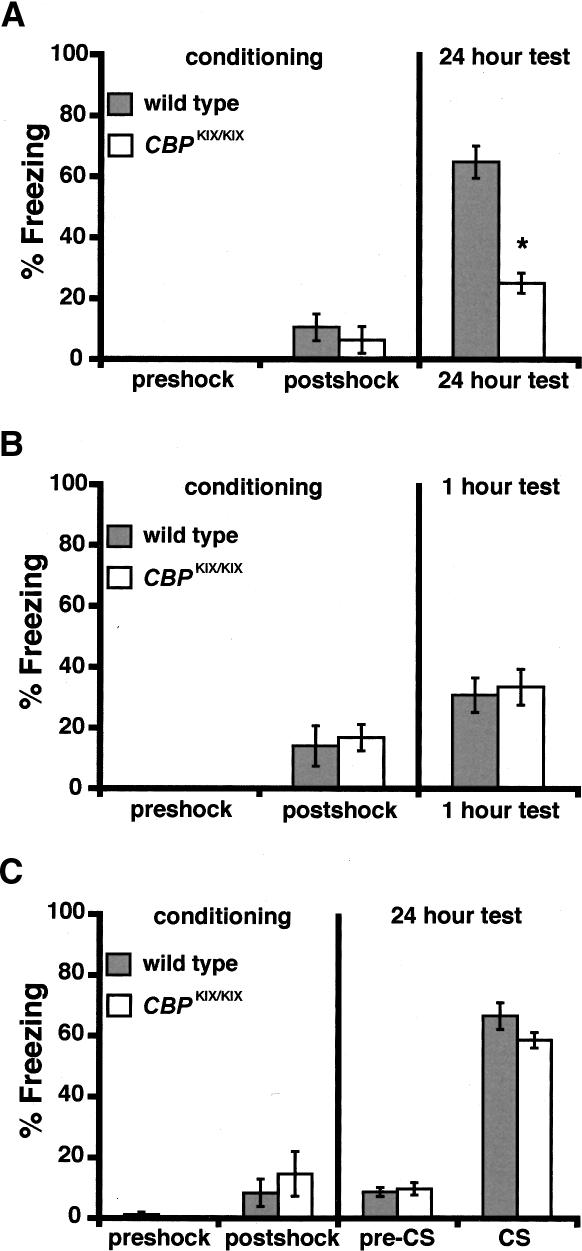
To determine whether the KIX domain of CBP is required for long-term memory storage, we subjected CBPKIX/KIX mice to Pavlovian fear conditioning. In this type of associative learning, a conditioned stimulus (CS) is paired with a noxious stimulus (unconditioned stimulus, US) (Maren 2001). A single conditioning trial is sufficient to elicit a learned fear response (freezing) (Fanselow 1980) and create a long-term memory for the CS. We used two types of fear conditioning, contextual and cued fear conditioning, which require partially distinct, but overlapping, neural systems. Lesions of the hippocampus affect contextual conditioning, whereas lesions of the amygdala affect both contextual and cued conditioning. As shown in Figure 3A, CBPKIX/KIX mice and wild-type littermates showed a similar level of freezing before and after the shock on the training day. In contrast, CBPKIX/KIX mice have significantly decreased freezing in the conditioned context in a 24-h retention test as compared with wild-type littermates (Fig. 3A; 24-h test: wild-type mice exhibited 64.7 ± 5.3% freezing; CBPKIX/KIX mice exhibited 24.9 ± 3.3% freezing, P < 0.0001, Student’s t-test). To assess whether CBPKIX/KIX mice have normal short-term memory for contextual fear, a separate set of CBPKIX/KIX mice and wild-type littermates was tested in the conditioned context 1 h after training (Fig. 3B). CBPKIX/KIX mice and wild-type littermates showed similar levels of freezing 1 h after training (Fig. 3B; wild-type mice exhibited 30.7 ± 5.7% freezing; CBPKIX/KIX mice exhibited 33.3 ± 5.9% freezing, P = NS, Student’s t-test). These results indicate that CBPKIX/KIX mice have normal short-term memory, but impaired long-term memory for the conditioned context.
Figure 3.
CBPKIX/KIX mice exhibit impaired long-term memory for contextual fear conditioning but normal memory for cued fear conditioning. (A) Training and 24-h long-term memory test for contextual fear conditioning. CBPKIX/KIX mice (n = 8) exhibited a significant decrease in freezing in a 24-h retention test, performed in the conditioned context, as compared with wild-type littermates (n = 8). (B) Training and 1-h short-term memory test for contextual fear conditioning. CBPKIX/KIX mice (n = 6) and wild-type littermates (n = 6) showed similar levels of freezing in a 1-h retention test, performed in the conditioned context. (C) Training and 24-h long-term memory test for cued fear conditioning. No differences in freezing behavior were observed between CBPKIX/KIX mice (n = 8) and wild-type littermates (n = 10) during training or the 24-h retention test. The 24-h test was performed in a novel context. Values are mean ±SEM. *P < 0.05.
A different set of CBPKIX/KIX mice and wild-type mice that were naive to fear conditioning was used for a cued fear conditioning experiment. CBPKIX/KIX mice and wild-type littermates showed a similar level of freezing before and after the shock on the training day. No reliable differences were observed in the 24-h retention test, performed in a novel context, between groups either before or after presentation of the cue (Fig. 3C; pre-CS: wild-type mice exhibited 8.6 ± 1.4% freezing; CBPKIX/KIX mice exhibited 9.7 ± 2.0% freezing, P = NS; CS: wild-type mice exhibited 66.6 ± 4.4% freezing; CBPKIX/KIX mice exhibited 58.6 ± 2.6% freezing, P = NS, Student’s t-test). Together, these results demonstrate that the KIX domain of CBP is required selectively in the hippocampal memory system for long-term memory storage of contextual fear conditioning.
CBPKIX/KIX mice exhibit significantly impaired long-term memory for novel object recognition
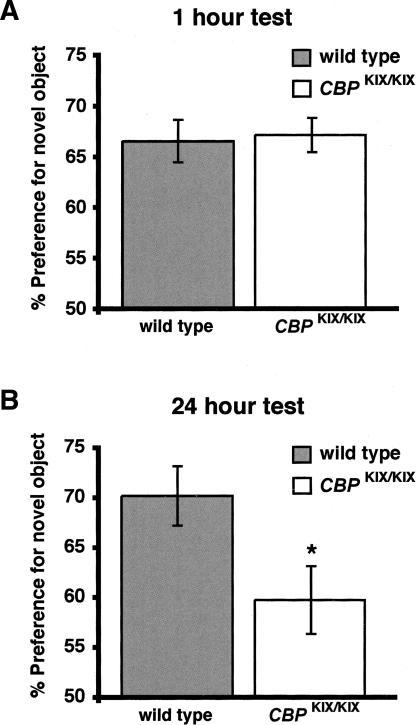
Novel object recognition tasks rely on the natural exploratory activity of rodents toward novel objects, and they provide a non-aversive single-trial learning paradigm. Novel object recognition is a form of visual paired comparison task that can be used to examine the role of the hippocampus in memory storage (Squire et al. 2004). The object recognition task has only recently been used to study genetically modified mice, but evidence suggests that the cAMP pathway and CREB are involved in long-term recognition memory (Bozon et al. 2004; Pineda et al. 2004; Wang et al. 2004). The coactivator CBP and its associated histone acetyltransferase activity also appear to play a role in long-term memory in the object recognition task (Bourtchouladze et al. 2003; Alarcon et al. 2004; Korzus et al. 2004). However, the role of the CREB-binding (KIX) domain of CBP, which meditates PKA-dependent transcriptional regulation, has not been examined. To address this question, we measured long-term memory for recognition memory in CBPKIX/KIX mice. Measurement of the time spent exploring each object was recorded and expressed as the percent time spent exploring the novel object relative to the total time spent exploring both of the objects (% preference for novel object). In a 1-h retention test to examine short-term memory, wild-type mice and CBPKIX/KIX littermates show similar preference for the novel object (Fig. 4A). In a 24-h retention test, CBPKIX/KIX mice have significantly reduced preference for the novel object (Fig. 4B; 24-h test: wild-type mice exhibited 70.1 ± 2.9% preference; CBPKIX/KIX mice exhibited 59.7 ± 3.4% preference; P < 0.05, Student’s t-test), suggesting that long-term recognition memory is significantly impaired in CBPKIX/KIX mice.
Figure 4.
Impaired long-term memory in the novel object recognition task in CBPKIX/KIX mice. (A) Wild-type mice (n = 8) show similar preference for a novel object in a 1-h retention test as compared with CBPKIX/KIX (n = 6) mice. (B) In contrast, wild-type mice (n = 11) show significantly higher preference for a novel object in 24-h retention test than CBPKIX/KIX (n = 10) mice. Values are mean ±SEM. *P < 0.05.
Decreased expression of CRE-containing genes in the hippocampus of CBPKIX/KIX mice after contextual fear conditioning
The triple point mutation carried by CBPKIX/KIX mice was designed to specifically disrupt CREB and c-Myb binding and inhibits CRE- and CREB-mediated transcription in mouse embryo fibroblasts derived from CBPKIX/KIX mice (Kasper et al. 2002). Thus, we predicted that CBPKIX/KIX mice would have impaired CREB-mediated transcription within the hippocampus.
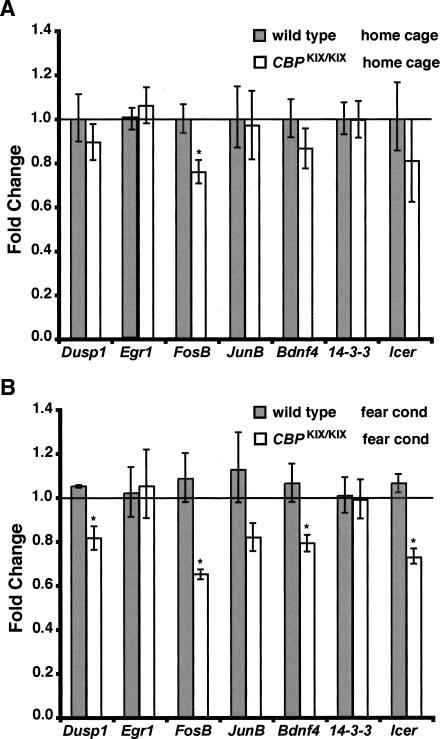
To test this hypothesis, we used quantitative real-time RT-PCR to examine the expression of seven CRE-containing genes, Dusp1, Egr1, Fosb, Junb, Bdnf (Bdnf4 variant), 14-3-3 eta (official symbol is whah), and Icer (a variant of Crem), all of which contain one or more CREB-binding sites (cAMP response-element [CRE] motifs), and many of which have been implicated in memory storage and whose expression is regulated by histone acetylation (Herdegen and Leah 1998; Silva et al. 1998; Conti et al. 2002, 2004; Kida et al. 2002; Conkright et al. 2003; Fass et al. 2003). To determine whether the expression of these genes was altered in basal conditions, we first examined hippocampal samples from CBPKIX/KIX mice and wild-type littermates that only received handling (Fig. 5A). Only the expression of Fosb was significantly decreased in CBPKIX/KIX mice in the basal state (P < 0.05, Mann-Whitney Rank Sum Test).
Figure 5.
Differential regulation of CREB-target genes in the hippocampus of CBPKIX/KIX mice. (A) Samples for quantitative real-time RT-PCR (qRT-PCR) were prepared from the hippocampi of CBPKIX/KIX mice (n = 4) and wild-type (n = 3) littermates that were subject to handling (home cage). qRT-PCR was performed in triplicate. Expression levels were normalized to the house keeping genes Actg1 and Tuba4. Levels of expression for CBPKIX/KIX mice are presented as fold change from wild-type levels, which were set to 1.0. Only Fosb had significantly reduced expression in home cage samples. (B) Samples were prepared from CBPKIX/KIX mice (n = 3) and wild-type (n = 3) littermates that were subject to contextual fear conditioning (fear cond). At 4 h after conditioning, mice were sacrificed, and samples were prepared for qRT-PCR. Levels of expression for CBPKIX/KIX mice and wild-type littermates are presented as a fold change from wild-type (home cage) levels, which were set to 1.0. Dusp1, Fosb, Bdnf4, and Icer had significantly reduced expression. *P < 0.05.
The deficit in long-term memory for contextual fear conditioning that we observed in CBPKIX/KIX mice (Fig. 3A) suggests that the expression of genes required for memory consolidation is affected in CBPKIX/KIX mice. To address this question, CBPKIX/KIX mice and wild-type littermates were subjected to contextual fear conditioning, sacrificed 4 h later, a time point which represents a key period of signaling and gene expression during the consolidation of fear conditioning (Bourtchouladze et al. 1998), and samples were prepared for quantitative real-time RT-PCR. As shown in Figure 5B, the expression of Dusp1, Fosb, Bdnf4, and Icer was significantly reduced in CBPKIX/KIX mice 4 h after training relative to wild-type littermates (P < 0.05, Mann-Whitney Rank Sum Test).
Discussion
CBP is a transcriptional coactivator that interacts with numerous transcription factors that bind to different domains of CBP (Goodman and Smolik 2000), including the KIX domain (Fig. 1). We have shown here that mice carrying a triple point mutation in the KIX domain of CBP exhibit long-term memory deficits. These results are the first to demonstrate an in vivo role for the KIX domain of CBP in the brain, and they suggest that the KIX domain and the transcription factors that bind to it are necessary for long-term memory storage. Recently, we and others have demonstrated that CBP has a role in long-term memory storage (Oike et al. 1999; Bourtchouladze et al. 2003; Alarcon et al. 2004; Korzus et al. 2004; Wood et al. 2005). In these previous studies, CBP mutant mice were examined that either lacked an entire CBP allele (Alarcon et al. 2004), expressed a histone acetyltransferase deficient allele of CBP (Korzus et al. 2004), or expressed a dominant-negative truncation mutant of CBP (Oike et al. 1999; Bourtchouladze et al. 2003; Wood et al. 2005). These studies demonstrated that CBP HAT activity is required for memory storage (Korzus et al. 2004) and implicated the amino terminus of CBP in binding transcription factors involved in synaptic plasticity and long-term memory (Oike et al. 1999; Bourtchouladze et al. 2003; Wood et al. 2005). There is an important difference between the nature of the mutation in the CBPKIX/KIX mice used in this study and the CBP mutant mice generated in the previous studies. The CBPKIX/KIX mice studied here allow us to begin to define which transcription factors interact with CBP for memory storage, which is a critical open question considering the many factors that utilize CBP for the regulation of target gene expression. Importantly, the identification of the transcription factor interface of CBP important for memory storage has enabled us to begin to identify the target genes regulated by CBP during memory consolidation.
Our findings show that the KIX domain of CBP is essential for long-term memory, suggesting that CBP is recruited to DNA by KIX-binding transcription factors during memory consolidation. Many transcription factors bind to the KIX domain of CBP including JUN, c-Myb, p53, SAP1a, SREBP, BRCA1, TAX, HPV E2, and CREB (Goodman and Smolik 2000). Interestingly, all of these transcription factors bind to CBP constitutively, except CREB, which binds to the KIX domain via a phosphorylation-dependent inducible mechanism (Parker et al. 1996). Thus, the CBP KIX domain has the unique ability for both constitutive and inducible interactions. This ability has been attributed to secondary structure characteristics of the binding regions that influence the thermodynamics of protein–protein interactions (Parker et al. 1999). Also, the KIX domain is thought to have one binding surface for JUN, p53, TAX, and SREBP, and a distinctly different binding surface for CREB and c-Myb (Campbell and Lumb 2002; Kasper et al. 2002; Liu et al. 2003).
The inducible nature of the binding of the phospho-KID domain of CREB to CBP KIX may explain our findings of greater changes in the expression of CREB-target genes after conditioning than in the basal state. Only Fosb had significantly reduced expression in the hippocampi of CBPKIX/KIX mice taken from their home cage (Fig. 5A), indicating that KIX interactions are necessary for the basal expression of Fosb. Similarly, transgenic CBP mutant mice expressing CBP with a mutated histone acetyltransferase (HAT) domain have significantly reduced hippocampal Fosb expression (Korzus et al. 2004). Thus, basal Fosb expression may require both the KIX and HAT domains of CBP. However, Alarcon et al. (2004) observed no reduction in basal expression of Fosb in CBP+/− heterozgyous mice. In contrast to the home cage data, we found that the expression of several genes (Fosb, Dusp1, Bdnf4, and Icer) was reduced in the hippocampi of CBPKIX/KIX mice 4 h after conditioning. Thus, fear conditioning may induce the activation of CREB via phosphorylation (Stanciu et al. 2001), thereby recruiting CBP in a KIX-dependent fashion to regulate the expression of a subset of CREB-dependent genes.
Interestingly, the expression of Egr1 and 14-3-3 eta was unchanged regardless of genotype or conditioning. Both Egr1 and 14-3-3 eta have been shown to be involved in memory storage and are at least partially regulated by CREB (Jones et al. 2001; Kida et al. 2002; Davis et al. 2003). There are several possibilities that may explain the discrepancy between our observations and those implicating Egr1 and 14-3-3 eta in memory storage. One explanation is that different genes have different temporal expression profiles, and thus our 4-h time point (after fear conditioning) may have been too late to observe expression changes in immediate early genes, such as Egr1. This 4-h time point may also explain why we did not observe a change in gene expression in home-cage wild-type samples versus fear-conditioned wild-type samples. In an earlier study, we found Dusp1, Egr1, Fosb, and Junb were upregulated 30 min after contextual fear conditioning (Keeley et al. 2006). It will be interesting to perform a more detailed examination of gene expression changes in wild-type and CBPKIX/KIX mice after fear conditioning in future studies to determine how gene profiles regulated by CBP change temporally.
Another possible explanation for the differential regulation of CREB target genes in our study is that Egr1 and 14-3-3 eta may require different CREB coactivators for their expression. For example, the coactivator TORC, and not CBP, was shown to be the crucial mediator of CREB function in the liver, at least for the regulation of genes involved in gluconeogenesis (Koo et al. 2005). TORC interacts with CREB through the basic leucine zipper region, not the KID domain. In conjunction with our data, the findings for the liver suggest that the requirement of the CBP KIX domain for CREB target gene expression not only varies between different genes, but also between tissues; perhaps CREB target genes that are transcribed independently of the KIX domain are instead dependent on TORC function. Another coactivator closely related to CBP, p300, may also play a role in the differential regulation of CREB target genes, as suggested by recent observations of distinct in vivo functions of p300 and CBP during hematopoiesis (Kasper et al. 2002) and in the brain (Oliveira et al. 2006). Recently, a genome-wide analysis of CREB occupancy, phosphorylation, and target gene activation revealed that although ∼4000 promoter sites are occupied by CREB, only a small proportion of CREB target genes was induced by cAMP (Zhang et al. 2005). This activated subset correlated with the preferential recruitment of CBP, suggesting that CREB phosphorylation by itself does not directly predict target gene activation (also see Brindle et al. 1995; Thompson et al. 1995; Chawla et al. 1998; Impey et al. 1998; Hardingham et al. 1999; Hu et al. 1999), but rather that the coactivator CBP mediates the selective induction of particular genes.
The recruitment of CBP may depend not only on the phosphorylation of CREB but on the binding of several other transcription factors, as has been described for the enhanceosome regulating the expression of interferon β (for review, see Merika and Thanos 2001). Thus, it is also reasonable that these genes are regulated by other transcription factors in conjunction with CREB. Comparative genomics has revealed that cis-regulatory sequences within promoter regions of genes are organized with a specific architecture for regulating gene expression (Harbison et al. 2004). Specific activity-dependent stimuli will give rise to unique transcription factor binding patterns in which synergy and occlusion between transcription factors (and associated coactivators) regulates temporal and spatial expression of target genes (Keeley et al. 2006). Indeed, our analysis of transcription factor binding motifs in the genes induced during memory consolidation revealed that both common and brain region-specific regulatory mechanisms likely exist in the hippocampus and the amygdala (Keeley et al. 2006).
The role of other CREB coactivators and other transcription factors in the regulation of gene expression may also explain why we observed a differential role for CBP in memory for contextual and cued fear conditioning. Long-term memory for contextual fear conditioning was impaired in CBPKIX/KIX mice, but memory for cued fear conditioning was unaltered. We observed similar results in a previous study examining the effect of a truncated inhibitory mutation of CBP on memory storage (Wood et al. 2005). Alarcon et al. (2004) also observed impaired memory for contextual fear in mice heterozygous for CBP. Whereas both contextual and cued fear conditioning are dependent on the amygdala, only contextual fear conditioning is also dependent on the hippocampus (Maren 2001). Because CBP is similarly expressed in the hippocampus and amygdala and this expression is not altered in CBP KIX mutant mice (Fig. 2), the most straightforward explanation for the differential role of CBP in contextual and cued fear conditioning is that different memory systems require different molecular machinery with a distinct requirement for CBP. It may be that the hippocampus is part of a memory system that relies more heavily on CBP activity and is thus adversely affected by impaired CBP function. It is tempting to compare the role of CBP in memory for fear conditioning to the findings in CREB mutant mice. Research using CREB mutant mice (Bourtchuladze et al. 1994; Kogan et al. 1997; Gass et al. 1998; Graves et al. 2002; Kida et al. 2002; Pittenger et al. 2002; Balschun et al. 2003; Josselyn et al. 2004) and viral approaches to modifying CREB function (Josselyn et al. 2001; Barco et al. 2002) suggest that this transcription factor is involved in both hippocampus and amygdala function, although the studies are, at times, contradictory, with memory impairments in CREB mutant mice dependent upon gene dosage, genetic background, and training protocol (for reviews, see Lonze and Ginty 2002; Kaplan and Abel 2003). Both CBP and CREB interact with numerous other factors, and it should not be assumed that they always function together during memory storage. It is possible that CBP interacts with other CREB family members, such as ATF-1 or CREM, for the activation of CRE-containing genes in the amygdala. Conversely, it is possible that CREB may be using a different transcriptional coactivator in the amygdala (such as p300, a paralog of CBP, or TORCs). Lastly, as discussed above, different brain regions have common as well as brain region-specific mechanisms that involve the coordinate assembly of multiple transcription factors (known as modules) to regulate gene expression during memory storage (Keeley et al. 2006).
The generation of genetically modified mice carrying point mutations that inactivate individual functional or transcription factor-binding domains of CBP promises to yield a much greater understanding of the in vivo role of CBP in processes ranging from development to memory storage. To more completely understand the function of a particular gene in any process it is necessary to analyze an allelic series of that gene, which will include null mutations, gain-of-function mutations, and loss-of-function mutations that target specific domains of the gene product. To date, the allelic series used to examine the role of CBP in synaptic plasticity and memory storage includes null mutations (Alarcon et al. 2004), a point mutation within the HAT domain (Korzus et al. 2004), and large deletion mutations (Oike et al. 1999; Bourtchouladze et al. 2003; Wood et al. 2005), which have all demonstrated a role for CBP in synaptic plasticity and long-term memory storage. In the current study, we established that a protein surface of the CBP KIX domain that binds specific transcription factors is required for long-term memory storage and is involved in the induction of CREB-target genes during memory consolidation. The identification of specific domains of CBP involved in memory storage not only helps define the molecular mechanisms underlying gene expression required for memory, but also guides the design of much more specific therapeutic agents for cognitive disorders.
Materials and Methods
Subjects
The CBPKIX/KIX mice were generated as previously described (Kasper et al. 2002). Briefly, the targeting vector for CBP contained the point mutations Tyr650Ala, Ala654Gln, and Tyr658Ala. The three mutations were introduced into the CBP locus of 129P2/OlaHsd-derived E14 embryonic stem cells by homologous recombination. Mice carrying the mutant allele of the KIX domain of CBP (designated CBPKIX/KIX for homozygous knock-in mice) have been bred and backcrossed in a heterozygous state on a C57BL/6 genetic background for 10 generations. Mice for experiments were generated from heterozygous matings, and wild-type littermates were used as controls. Mice were 8- to 16-wk-old at the time of the experiment and had free access to food and water in their home cages. Lights were maintained on a 12-h light/12 h-dark cycle, with all behavioral testing carried out during the light portion of the cycle. All experiments were conducted according to National Institutes of Health guidelines for animal care and use and were approved by the Institutional Animal Care and Use Committee of the University of Pennsylvania. The investigator was blind to the genotype of the mice during behavioral testing.
Immunohistochemistry
CBPKIX/KIX mouse and wild-type littermate mice were anesthetized with isofluorane and transcardially perfused with phosphate-buffered saline (PBS) followed by 4% paraformaldehyde in PBS using a peristaltic perfusion pump. Fixed brains were dissected, post-fixed overnight, and then cryoprotected in 30% sucrose at 4°C. Brains were flash frozen in 2-methylbutane on dry ice and mounted on cryostat chucks using OCT. Coronal sections were cut at a thickness of 30 μm and collected in PBS. Floating sections were blocked for 1 h at room temperature in 8% normal goat serum (NGS, Vector Laboratories) with 0.3% Triton-X (Fisher) in PBS and then incubated overnight at 4°C in 2% NGS, 0.3% Triton X-100 in PBS with CBP antibodies (A22 and C20, 1:1000, Santa Cruz Biotechnology) and for 2 h at room temperature with goat anti-rabbit IgG-FITC secondary antibody (1:1000, Santa Cruz Biotechnology). Sections were washed three times for 5 min each in PBS before and after each incubation step. DAPI (Molecular Probes) was added during the last wash. Sections were mounted on slides in 50% glycerol and imaged using a Leica DMRBE fluorescent microscope using a 10× objective and a Hamamatsu CCD camera.
Fear conditioning
Fear conditioning experiments were performed in chambers using methods previously described (Wood et al. 2005). Mice were handled for three consecutive days for 1 min each day. For contextual fear conditioning, mice were placed into the conditioning chamber and received a 2-sec 1.5-mA scrambled footshock 2.5 min after placement into the chamber. Mice were removed from the chamber after 3 min. During testing, mice received one 5-min exposure to the same conditioned context in the absence of shock 24 h after conditioning. For cued fear conditioning, mice were placed into the chamber and the cue (white noise) was presented 2–2.5 min after placement into the chamber and co-terminated with a 2-sec 1.5-mA footshock. Mice were removed from the chamber after a total of 3 min. On the testing day, mice in the cued group received one 5-min exposure to a novel context (another conditioning chamber with a smooth flat floor, altered dimensions, and a novel odorant) for 0–2 min (pre-CS) followed by exposure to the cue from 2–5 min (CS), 24 h after conditioning. Conditioning was assayed by measuring freezing behavior, the complete absence of movement (Fanselow 1980). Freezing was scored during conditioning as well as testing. The behavior of each mouse was sampled at 5-sec intervals, and the percentage of those intervals in which the mouse froze was calculated. Different sets of mice were used for contextual and cued conditioning experiments.
Object recognition
The object recognition task consisted of a training phase and a testing phase (Mansuy et al. 1998). Prior to training, all mice were handled for 1 min a day for 2 d and habituated to the experimental apparatus with 5 min of exploration in the absence of objects. During the training phase (15 min), mice were placed in the experimental apparatus (a white rectangular open field 60 cm × 50 cm × 26 cm) with two identical objects placed in specific locations. After a retention interval of 24 h, mice were placed back into the rectangular environment for the testing phase. Two objects were again present, but one of the familiar objects was now replaced with a novel one. Mice were again allowed to freely explore the environment and the objects for 15 min. All testing and training sessions were videotaped and analyzed by an individual blind to the genotype of the animals examined. A mouse was scored as exploring an object whenever any part of its body other than the tail was touching the object, or whenever it was within 1 cm of the object and facing it. Measurement of the time spent exploring each object was recorded and expressed as the percent time spent exploring the novel object relative to the total time spent exploring both of the objects. The identity of the objects—which one was novel and which one was familiar—as well as the spatial location in which the novel and familiar objects were located were counterbalanced between subjects.
Preparation and purification of mRNA
Hippocampi were harvested from each animal and placed in RNAlater (Ambion). Aerosol Barrier ART tips, DEPC-treated sterile water, and autoclaved microcentrifuge tubes were used for all subsequent steps. Tissue was homogenized in Trizol (Invitrogen) and incubated at room temperature for 5 min. Phenol-chloroform addition was followed by vigorous mixing and room temperature incubation for 3 min. Samples were transferred to phaselock gel tubes (Eppendorf) and were centrifuged at full speed for 15 min. The aqueous phase was transferred to new tubes, where two volumes of ethanol, 1/10 volume NaOAc, and glycogen were added. After gentle mixing, the samples were incubated for 10 min at −20°C. Centrifugation at 4°C and full speed was followed by aspiration of the supernatant. Pellets were washed with 70% ethanol, and then centrifuged for 5 min at 4°C and full speed. The supernatant was aspirated and samples were air-dried for ∼10 min. After resuspension in water, RNA was purified using the RNeasy system (Qiagen) according to the manufacturer’s protocol. Residual DNA was removed by treatment with DNA-free (Ambion). Optical density measurement at 260 and 280 nm by the SmartSpec 3000 (BioRad) spectrophotometer was used to assess the concentration and quality of mRNA in each sample.
Preparation of cDNA
Generation of cDNA was carried out by the RETROscript kit (Ambion). Briefly, each reaction was performed with 1 μg RNA in a total volume of 20 μL composed as follows: 50 mM Tris-HCl pH 8.3, 75 mM KCl, 3 mM MgCl2, 5 mM DTT, 500 μM each dNTP, 5 μM random decamers, 10 units RNase inhibitor, and 100 units MMLV-RT. As controls, additional reaction mixtures were generated that lacked either reverse transcriptase or template mRNA. Subsequent reactions were allowed to proceed for 1 h at 44°C, followed by heat inactivation for 10 min at 100°C.
Quantitative real time RT–PCR
Reactions were carried out in 96-well optical reaction plates (ABI) with optical adhesive covers (ABI) and prepared according to the protocol for using Quantitect SYBRgreen PCR mastermix (Qiagen). Three or four biological (see Fig. 5 legend) and three technical replicates were used in experiments. Reactions were carried out in the ABI Prism 7000 with an initial activation by incubation for 2 min at 50°C followed by incubation for 15 min at 95°C and 40 subsequent cycles of 15 sec at 95°C, 30 sec at 56°C, and 30 sec at 72°C. Primers used for amplification were:
Dusp1 forward 5′-GGAGGATATGAAGCGTTTTCGG-3′
reverse 5′-GGATTCTGCACTGTCAGGCACA-3′
Egr1 forward 5′-CAAGGCCGAGATGCAATTG-3′
reverse: 5′-GACTCTGTGGTCAGGTGCTCA-3′
Fosb forward 5′-CCGAGAAGAGACACTTACCCCA-3′
reverse 5′-AAGTCGATCTGTCAGCTCCCTC-3′
Junb forward 5′-CTTTAAAGAGGAACCGCAGACC-3′
reverse 5′-CGCTTTCGCTCCACTTTGAT-3′
Bdnf4 forward 5′-TGCGAGTATTACCTCCGCCAT-3′
reverse 5′-TCACGTGCTCAAAAGTGTCAG-3′
14-3-3 eta forward 5′-AATGTAGTTGGTGCCAGGCGAT-
reverse 5′-TGCCAGGTAGCGGTAGTAATCG-
Icer forward 5′-GGTGACATGCCAACTTACCAGA-
reverse 5′-TTGCGACTTGCTTCTTCTGC-3′
Actg1 forward 5′-CCGATCGCAATGGAAGAAG-3′
reverse 5′-CGTATGAGTCTTTCTGGCCCA-3′
Tuba4 forward 5′-ATGCGCGAGTGCATTTCAG-3′
reverse 5′-CACCAATGGTCTTATCGCTGG-3′
PCR products were cloned using the TA cloning kit (Invitrogen) following the manufacturer’s protocol. Cloned fragments were then sequenced to confirm the identity of the amplified gene. Data were normalized to Actg1 and Tubulin, alpha 4 (Tuba4) prior to calculation of differences. The Icer primers are the same as previously described (Conti et al. 2004). Relative quantitation of gene expression was performed according to Applied Biosystem’s User Bulletin #2. Fold change was calculated from the ΔCt values with corrections for standard curve data from each gene and housekeeping gene expression levels for each sample based on the relative standard curve method described in the Applied Biosystems manual. For each sample, the ΔCt was calculated against the mean for that gene’s sample set. Next, each of these ΔCt values was corrected with the slope of the standard curve for the relevant primer to account for any variation in primer amplification efficiency. The efficiency-corrected ΔCt value was normalized to the similarly corrected ΔCt from housekeeping genes for each sample to account for variability in mRNA input. The difference between corrected ΔCt for each sample was then grouped according to condition (CBPKIX/KIX or wild-type) and the average ΔCt calculated. Because Ct values are on a logarithmic scale, fold change is equal to two raised to the difference between experimental and control ΔCt values. Because correction has been made for primer efficiency, the fold change of the gene was the most logical expression of the data. The data presented are the calculated means for the biological replicates with n being equal to the number of biological replicates (i.e., the number of mice examined).
Data analysis
Statistics were performed using SigmaStat (version 2.03). Simple planned comparisons were made using a Student's t-test. For the quantitative real-time RT-PCR data, a Mann-Whitney Rank Sum test was used to determine significance, which does not assume a normally distributed data set. Experimenters were blind to genotype, and genotypes were confirmed by Southern blot analysis or PCR following behavioral and molecular experiments.
Acknowledgments
We thank Colleen Brensinger for advice on statistical methods. This research was supported by a postdoctoral Training Program fellowship in Neurodegenerative Diseases (to M.A.W.; V.M.Y.L., Principal Investigator), Nassau Fund Undergraduate Research Award (to M.A.A.), Foundation for Science and Technology fellowship, Portugal (to A.M.M.O.), and by grants from the National Institutes of Health (RO1 MH060244), the Packard Foundation, and the University of Pennsylvania Research Foundation (to T.A.). Also supported by NIH grant R01 DK058199 and by the American Lebanese Syrian Associated Charities of St. Jude Children’s Research Hospital (P.K.B.).
Footnotes
Article published online before print. Article and publication date are at http://www.learnmem.org/cgi/doi/10.1101/lm.213906
References
- Alarcon J.M., Malleret G., Touzani K., Vronskaya S., Ishii S., Kandel E.R., Barco A. Chromatin acetylation, memory, and LTP are impaired in CBP+/− mice: A model for the cognitive deficit in Rubinstein-Taybi syndrome and its amelioration. Neuron. 2004;42:947–959. doi: 10.1016/j.neuron.2004.05.021. [DOI] [PubMed] [Google Scholar]
- Balschun D., Wolfer D.P., Gass P., Mantamadiotis T., Welzl H., Schutz G., Frey J.U., Lipp H.P. Does cAMP response element-binding protein have a pivotal role in hippocampal synaptic plasticity and hippocampus-dependent memory? J. Neurosci. 2003;23:6304–6314. doi: 10.1523/JNEUROSCI.23-15-06304.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barco A., Alarcon J.M., Kandel E.R. Expression of constitutively active CREB protein facilitates the late phase of long-term potentiation by enhancing synaptic capture. Cell. 2002;108:689–703. doi: 10.1016/s0092-8674(02)00657-8. [DOI] [PubMed] [Google Scholar]
- Bourtchuladze R., Frenguelli B., Blendy J., Cioffi D., Schutz G., Silva A.J. Deficient long-term memory in mice with a targeted mutation of the cAMP-responsive element-binding protein. Cell. 1994;79:59–68. doi: 10.1016/0092-8674(94)90400-6. [DOI] [PubMed] [Google Scholar]
- Bourtchouladze R., Abel T., Berman N., Gordon R., Lapidus K., Kandel E.R. Different training procedures recruit either one or two critical periods for contextual memory consolidation, each of which requires protein synthesis and PKA. Learn. Mem. 1998;5:365–374. [PMC free article] [PubMed] [Google Scholar]
- Bourtchouladze R., Lidge R., Catapano R., Stanley J., Gossweiler S., Romashko D., Scott R., Tully T. A mouse model of Rubinstein-Taybi syndrome: Defective long-term memory is ameliorated by inhibitors of phosphodiesterase 4. Proc. Natl. Acad. Sci. 2003;100:10518–10522. doi: 10.1073/pnas.1834280100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozon B., Kelly A., Josselyn S.A., Silva A.J., Davis S., Laroche S. MAPK, CREB and zif268 are all required for the consolidation of recognition memory. Philo. Trans. R. Soc. Lond. B Biol. Sci. 2003;358:805–814. doi: 10.1098/rstb.2002.1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brindle P., Nakajima T., Montminy M. Multiple protein kinase A-regulated events are required for transcriptional induction by cAMP. Proc. Natl. Acad. Sci. 1995;92:10521–10525. doi: 10.1073/pnas.92.23.10521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell K.M., Lumb K.J. Structurally distinct modes of recognition of the KIX domain of CBP by Jun and CREB. Biochemistry. 2002;41:13956–13964. doi: 10.1021/bi026222m. [DOI] [PubMed] [Google Scholar]
- Chawla S., Hardingham G.E., Quinn D.R., Bading H. CBP: A signal-regulated transcriptional coactivator controlled by nuclear calcium and CaM kinase IV. Science. 1998;281:1505–1509. doi: 10.1126/science.281.5382.1505. [DOI] [PubMed] [Google Scholar]
- Conkright M.D., Guzman E., Flechner L., Su A.I., Hogenesch J.B., Montminy M. Genome-wide analysis of CREB target genes reveals a core promoter requirement for cAMP responsiveness. Mol. Cell. 2003;11:1101–1108. doi: 10.1016/s1097-2765(03)00134-5. [DOI] [PubMed] [Google Scholar]
- Conti A.C., Cryan J.F., Dalvi A., Lucki I., Blendy J.A. cAMP response element-binding protein is essential for the upregulation of brain-derived neurotrophic factor transcription, but not the behavioral or endocrine responses to antidepressant drugs. J. Neurosci. 2002;22:3262–3268. doi: 10.1523/JNEUROSCI.22-08-03262.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conti A.C., Kuo Y.C., Valentino R.J., Blendy J.A. Inducible cAMP early repressor regulates corticosterone suppression after tricyclic antidepressant treatment. J. Neurosci. 2004;24:1967–1975. doi: 10.1523/JNEUROSCI.4804-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis S., Bozon B., Laroche S. How necessary is the activation of the immediate early gene zif268 in synaptic plasticity and learning? Behav. Brain Res. 2003;142:17–30. doi: 10.1016/s0166-4328(02)00421-7. [DOI] [PubMed] [Google Scholar]
- Fanselow M.S. Conditioned and unconditional components of post-shock freezing. Pavlov. J. Biol. Sci. 1980;15:177–182. doi: 10.1007/BF03001163. [DOI] [PubMed] [Google Scholar]
- Fass D.M., Butler J.E., Goodman R.H. Deacetylase activity is required for cAMP activation of a subset of CREB target genes. J. Biol. Chem. 2003;278:43014–43019. doi: 10.1074/jbc.M305905200. [DOI] [PubMed] [Google Scholar]
- Gass P., Wolfer D.P., Balschun D., Rudolph D., Frey U., Lipp H.P., Schutz G. Deficits in memory tasks of mice with CREB mutations depend on gene dosage. Learn. Mem. 1998;5:274–288. [PMC free article] [PubMed] [Google Scholar]
- Goodman R.H., Smolik S. CBP/p300 in cell growth, transformation, and development. Genes & Dev. 2000;14:1553–1577. [PubMed] [Google Scholar]
- Graves L., Dalvi A., Lucki I., Blendy J.A., Abel T. Behavioral analysis of CREB αδ mutation on a B6/129 F1 hybrid background. Hippocampus. 2002;12:18–26. doi: 10.1002/hipo.10003. [DOI] [PubMed] [Google Scholar]
- Harbison C.T., Gordon D.B., Lee T.I., Rinaldi N.J., Macisaac K.D., Danford T.W., Hannett N.M., Tagne J.B., Reynolds D.B., Yoo J., et al. Transcriptional regulatory code of a eukaryotic genome. Nature. 2004;431:99–104. doi: 10.1038/nature02800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardingham G.E., Chawla S., Cruzalegui F.H., Bading H. Control of recruitment and transcription-activating function of CBP determines gene regulation by NMDA receptors and L-type calcium channels. Neuron. 1999;22:789–798. doi: 10.1016/s0896-6273(00)80737-0. [DOI] [PubMed] [Google Scholar]
- Herdegen T., Leah J.D. Inducible and constitutive transcription factors in the mammalian nervous system: Control of gene expression by Jun, Fos and Krox, and CREB/ATF proteins. Brain Res. Brain Res. Rev. 1998;28:370–490. doi: 10.1016/s0165-0173(98)00018-6. [DOI] [PubMed] [Google Scholar]
- Hu S.C., Chrivia J., Ghosh A. Regulation of CBP-mediated transcription by neuronal calcium signaling. Neuron. 1999;22:799–808. doi: 10.1016/s0896-6273(00)80738-2. [DOI] [PubMed] [Google Scholar]
- Impey S., Obrietan K., Wong S.T., Poser S., Yano S., Wayman G., Deloulme J.C., Chan G., Storm D.R. Cross talk between ERK and PKA is required for Ca2+ stimulation of CREB-dependent transcription and ERK nuclear translocation. Neuron. 1998;21:869–883. doi: 10.1016/s0896-6273(00)80602-9. [DOI] [PubMed] [Google Scholar]
- Jones M.W., Errington M.L., French P.J., Fine A., Bliss T.V., Garel S., Charnay P., Bozon B., Laroche S., Davis S. A requirement for the immediate early gene zif268 in the expression of late LTP and long-term memories. Nat. Neurosci. 2001;4:289–296. doi: 10.1038/85138. [DOI] [PubMed] [Google Scholar]
- Josselyn S.A. What’s right with my mouse model? New insights into the molecular and cellular basis of cognition from mouse models of Rubinstein-Taybi Syndrome. Learn. Mem. 2005;12:80–83. doi: 10.1101/lm.93505. [DOI] [PubMed] [Google Scholar]
- Josselyn S.A., Shi C., Carlezon W.A., Jr., Neve R.L., Nestler E.J., Davis M. Long-term memory is facilitated by cAMP response element-binding protein overexpression in the amygdala. J. Neurosci. 2001;21:2404–2412. doi: 10.1523/JNEUROSCI.21-07-02404.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josselyn S.A., Kida S., Silva A.J. Inducible repression of CREB function disrupts amygdala-dependent memory. Neurobiol. Learn. Mem. 2004;82:159–163. doi: 10.1016/j.nlm.2004.05.008. [DOI] [PubMed] [Google Scholar]
- Kalkhoven E. CBP and p300: HATs for different occasions. Biochem. Pharmacol. 2004;68:1145–1155. doi: 10.1016/j.bcp.2004.03.045. [DOI] [PubMed] [Google Scholar]
- Kaplan M.P., Abel T. Genetic approaches to the study of synaptic plasticity and memory storage. CNS Spectr. 2003;8:597–610. doi: 10.1017/s1092852900018873. [DOI] [PubMed] [Google Scholar]
- Kasper L.H., Boussouar F., Ney P.A., Jackson C.W., Rehg J., van Deursen J.M., Brindle P.K. A transcription-factor-binding surface of coactivator p300 is required for haematopoiesis. Nature. 2002;419:738–743. doi: 10.1038/nature01062. [DOI] [PubMed] [Google Scholar]
- Keeley M.B., Wood M.A., Isiegas C., Stein J., Hellman K., Hannenhalli S., Abel T. Differential transcriptional response to nonassociative and associative components of classical fear conditioning in the amygdala and hippocampus. Learn. Mem. 2006;13:135–142. doi: 10.1101/lm.86906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kida S., Josselyn S.A., de Ortiz S.P., Kogan J.H., Chevere I., Masushige S., Silva A.J. CREB required for the stability of new and reactivated fear memories. Nat. Neurosci. 2002;5:348–355. doi: 10.1038/nn819. [DOI] [PubMed] [Google Scholar]
- Kogan J.H., Frankland P.W., Blendy J.A., Coblentz J., Marowitz Z., Schutz G., Silva A.J. Spaced training induces normal long-term memory in CREB mutant mice. Curr. Biol. 1997;7:1–11. doi: 10.1016/s0960-9822(06)00022-4. [DOI] [PubMed] [Google Scholar]
- Koo S.H., Flechner L., Qi L., Zhang X., Screaton R.A., Jeffries S., Hedrick S., Xu W., Boussouar F., Brindle P., et al. The CREB coactivator TORC2 is a key regulator of fasting glucose metabolism. Nature. 2005;437:1109–1111. doi: 10.1038/nature03967. [DOI] [PubMed] [Google Scholar]
- Korzus E., Rosenfeld M.G., Mayford M. CBP histone acetyltransferase activity is a critical component of memory consolidation. Neuron. 2004;42:961–972. doi: 10.1016/j.neuron.2004.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y.P., Chang C.W., Chang K.Y. Mutational analysis of the KIX domain of CBP reveals residues critical for SREBP binding. FEBS Lett. 2003;554:403–409. doi: 10.1016/s0014-5793(03)01200-6. [DOI] [PubMed] [Google Scholar]
- Lonze B.E., Ginty D.D. Function and regulation of CREB family transcription factors in the nervous system. Neuron. 2002;35:605–623. doi: 10.1016/s0896-6273(02)00828-0. [DOI] [PubMed] [Google Scholar]
- Mansuy I.M., Mayford M., Jacob B., Kandel E.R., Bach M.E. Restricted and regulated overexpression reveals calcineurin as a key component in the transition from short-term to long-term memory. Cell. 1998;92:39–49. doi: 10.1016/s0092-8674(00)80897-1. [DOI] [PubMed] [Google Scholar]
- Maren S. Neurobiology of Pavlovian fear conditioning. Annu. Rev. Neurosci. 2001;24:897–931. doi: 10.1146/annurev.neuro.24.1.897. [DOI] [PubMed] [Google Scholar]
- Merika M., Thanos D. Enhanceosomes. Curr. Opin. Genet. Dev. 2001;11:205–208. doi: 10.1016/s0959-437x(00)00180-5. [DOI] [PubMed] [Google Scholar]
- Oike Y., Hata A., Mamiya T., Kaname T., Noda Y., Suzuki M., Yasue H., Nabeshima T., Araki K., Yamamura K. Truncated CBP protein leads to classical Rubinstein-Taybi syndrome phenotypes in mice: Implications for a dominant-negative mechanism. Hum. Mol. Genet. 1999;8:387–396. doi: 10.1093/hmg/8.3.387. [DOI] [PubMed] [Google Scholar]
- Oliveira A.M.M., Abel T., Brindle P.K., Wood M.A. Differential role for CBP and p300 CREB-binding domain in motor skill learning. Behav. Neurosci. 2006;120:724–729. doi: 10.1037/0735-7044.120.3.724. [DOI] [PubMed] [Google Scholar]
- Parker D., Ferreri K., Nakajima T., LaMorte V.J., Evans R., Koerber S.C., Hoeger C., Montminy M.R. Phosphorylation of CREB at Ser-133 induces complex formation with CREB-binding protein via a direct mechanism. Mol. Cell. Biol. 1996;16:694–703. doi: 10.1128/mcb.16.2.694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker D., Jhala U.S., Radhakrishnan I., Yaffe M.B., Reyes C., Shulman A.I., Cantley L.C., Wright P.E., Montminy M. Analysis of an activator:coactivator complex reveals an essential role for secondary structure in transcriptional activation. Mol. Cell. 1998;2:353–359. doi: 10.1016/s1097-2765(00)80279-8. [DOI] [PubMed] [Google Scholar]
- Parker D., Rivera M., Zor T., Henrion-Caude A., Radhakrishnan I., Kumar A., Shapiro L.H., Wright P.E., Montminy M., Brindle P.K. Role of secondary structure in discrimination between constitutive and inducible activators. Mol. Cell. Biol. 1999;19:5601–5607. doi: 10.1128/mcb.19.8.5601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pineda V.V., Athos J.I., Wang H., Celver J., Ippolito D., Boulay G., Birnbaumer L., Storm D.R. Removal of G(iα1) constraints on adenylyl cyclase in the hippocampus enhances LTP and impairs memory formation. Neuron. 2004;41:153–163. doi: 10.1016/s0896-6273(03)00813-4. [DOI] [PubMed] [Google Scholar]
- Pittenger C., Huang Y.Y., Paletzki R.F., Bourtchouladze R., Scanlin H., Vronskaya S., Kandel E.R. Reversible inhibition of CREB/ATF transcription factors in region CA1 of the dorsal hippocampus disrupts hippocampus-dependent spatial memory. Neuron. 2002;34:447–462. doi: 10.1016/s0896-6273(02)00684-0. [DOI] [PubMed] [Google Scholar]
- Radhakrishnan I., Perez-Alvarado G.C., Parker D., Dyson H.J., Montminy M.R., Wright P.E. Solution structure of the KIX domain of CBP bound to the transactivation domain of CREB: A model for activator:coactivator interactions. Cell. 1997;91:741–752. doi: 10.1016/s0092-8674(00)80463-8. [DOI] [PubMed] [Google Scholar]
- Radhakrishnan I., Perez-Alvarado G.C., Dyson H.J., Wright P.E. Conformational preferences in the Ser133-phosphorylated and non-phosphorylated forms of the kinase inducible transactivation domain of CREB. FEBS Lett. 1998;430:317–322. doi: 10.1016/s0014-5793(98)00680-2. [DOI] [PubMed] [Google Scholar]
- Radhakrishnan I., Perez-Alvarado G.C., Parker D., Dyson H.J., Montminy M.R., Wright P.E. Structural analyses of CREB–CBP transcriptional activator–coactivator complexes by NMR spectroscopy: Implications for mapping the boundaries of structural domains. J. Mol. Biol. 1999;287:859–865. doi: 10.1006/jmbi.1999.2658. [DOI] [PubMed] [Google Scholar]
- Shaywitz A.J., Dove S.L., Kornhauser J.M., Hochschild A., Greenberg M.E. Magnitude of the CREB-dependent transcriptional response is determined by the strength of the interaction between the kinase-inducible domain of CREB and the KIX domain of CREB-binding protein. Mol. Cell. Biol. 2000;20:9409–9422. doi: 10.1128/mcb.20.24.9409-9422.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva A.J., Kogan J.H., Frankland P.W., Kida S. CREB and memory. Annu. Rev. Neurosci. 1998;21:127–148. doi: 10.1146/annurev.neuro.21.1.127. [DOI] [PubMed] [Google Scholar]
- Squire L.R., Stark C.E., Clark R.E. The medial temporal lobe. Annu. Rev. Neurosci. 2004;27:279–306. doi: 10.1146/annurev.neuro.27.070203.144130. [DOI] [PubMed] [Google Scholar]
- Stanciu M., Radulovic J., Spiess J. Phosphorylated cAMP response element binding protein in the mouse brain after fear conditioning: Relationship to Fos production. Brain Res. Mol. Brain Res. 2001;94:15–24. doi: 10.1016/s0169-328x(01)00174-7. [DOI] [PubMed] [Google Scholar]
- Thompson M.A., Ginty D.D., Bonni A., Greenberg M.E. L-type voltage-sensitive Ca2+ channel activation regulates c-fos transcription at multiple levels. J. Biol. Chem. 1995;270:4224–4235. doi: 10.1074/jbc.270.9.4224. [DOI] [PubMed] [Google Scholar]
- Wang H., Ferguson G.D., Pineda V.V., Cundiff P.E., Storm D.R. Overexpression of type-1 adenylyl cyclase in mouse forebrain enhances recognition memory and LTP. Nat. Neurosci. 2004;7:635–642. doi: 10.1038/nn1248. [DOI] [PubMed] [Google Scholar]
- Wood M.A., Kaplan M.P., Park A., Blanchard E.J., Oliveira A.M., Lombardi T.L., Abel T. Transgenic mice expressing a truncated form of CREB-binding protein (CBP) exhibit deficits in hippocampal synaptic plasticity and memory storage. Learn. Mem. 2005;12:111–119. doi: 10.1101/lm.86605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Odom D.T., Koo S.H., Conkright M.D., Canettieri G., Best J., Chen H., Jenner R., Herbolsheimer E., Jacobsen E., et al. Genome-wide analysis of cAMP-response element binding protein occupancy, phosphorylation, and target gene activation in human tissues. Proc. Natl. Acad. Sci. 2005;102:4459–4464. doi: 10.1073/pnas.0501076102. [DOI] [PMC free article] [PubMed] [Google Scholar]