Abstract
Rapid identification of in vivo affinity ligands would have far-reaching applications for imaging specific molecular targets, in vivo systems imaging, and medical use. We have developed a high-throughput method for identifying and optimizing ligands to map and image biologic targets of interest in vivo. We directly labeled viable phage clones with far-red fluorochromes and comparatively imaged them in vivo by multichannel fluorescence ratio imaging. Using Secreted Protein Acidic and Rich in Cysteine (osteonectin) and vascular cell adhesion molecule-1 as model targets, we show that: 1) fluorescently labeled phage retains target specificity on labeling; 2) in vivo distribution can be quantitated (detection thresholds of ∼ 300 phage/mm3 tissue) throughout the entire depth of the tumor using fluorescent tomographic imaging; and 3) fluorescently labeled phage itself can serve as a replenishable molecular imaging agent. The described method should find widespread application in the rapid in vivo discovery and validation of affinity ligands and, importantly, in the use of fluorochrome-labeled phage clones as in vivo imaging agents.
Keywords: Phage display, screening, drug discovery, molecular imaging, systems biology
Introduction
Molecularly targeted affinity ligands derived from screens play an important role in our further understanding of human disease [1–4], pharmaceutical discovery and development [5–7], nanotechnology sensing applications [8], systems biology [9], and the design of molecular imaging agents [10–13]. Various types of high-throughput screens, including small molecule screens [14,15], yeast two-hybrid screens [16], bacterial display screens [17], and phage display screens [5,10,18,19], have been developed. The latter offers a number of important advantages, such as rapid and economical biologic expansion (rather than the more time-consuming chemical resynthesis), vast peptide diversity, rapid screening process, and the availability of many types of phage clones and libraries (for a review, see Smith and Petrenko [20] and Willats [21]).
A number of modifications to traditional phage screens have been developed over the last decade. Traditional in vitro screens commonly use purified target proteins immobilized on plates [22]. Alternatively, screens have been performed on live cells, with targets expressed in their native environments, which has allowed the development of modified screens that bias toward cell internalized phage [23,24], binding under flow conditions [10] or other biologic processes. One of the most exciting recent developments has been the use of in vivo phage display to yield disease-specific or organ-specific phage clones [18,19]. For example, a number of atherosclerosis-targeted phage have recently been developed [25], and endothelial bed-specific clones have been found in both mice and humans [2,26]. However, irrespective of the method employed, it is common for a given screen to yield tens to hundreds of potential phage clones that subsequently require time-consuming and costly validation. In addition, once a clone is validated, developing, validating, and scaling up an imaging agent based on lead peptides can be challenging and costly.
Given the existing bottlenecks in identifying clones with the highest likelihood of in vivo success and to determine if we could use phage as a targeted imaging agent, we set out to develop comparative in vivo screening tools. We reasoned that such comparative screens could be of value in eliminating in vitro-identified clones that would ultimately fail because of unfavorable in vivo pharmacokinetics, delivery barriers, opsonization, or insufficiently high target-to-background ratios [27,28]. Specifically, we applied far-red and near-infrared fluorescent labeling and newer in vivo fluorescence methods to track phage in vivo to intended targets of interest. Using Secreted Protein Acidic and Rich in Cysteine (SPARC) as a model target for invasive cancer [29,30] and vascular cell adhesion molecule-1 (VCAM-1) for inflammatory endothelium [31], we show how novel targeted peptide sequences can be rapidly developed. We furthermore show that individual fluorescently labeled phage clones can be remarkably efficient as in vivo imaging agents, akin to peptide-decorated nanoparticles [32].
Materials and Methods
Materials
N-hydroxysuccinimide esters of Cy3.5 and Cy5.5 were obtained from Amersham (Piscataway, NJ); fluorescein isothiocyanate (FITC) and tetramethyl rhodamine isothiocyanate (TRITC) were from Sigma Aldrich (St. Louis, MO); N-hydroxysuccinimide esters of Vivotag-680 (VT680) and Angiosense-750 were from Visen Medical (Woburn, MA); and N-hydroxysuccinimide esters of AF750 were from Invitrogen (Eugene, OR). All other chemicals were purchased from Sigma Aldrich or Pierce (Rockford, IL) and used as received.
Phage Libraries
The SPARC-targeted phage used in this study was isolated from the linear 7 mer library (PhD 7), and the VCAM-1-targeted phage was isolated from the disulfide constrained cyclic 7 mer phage display peptide library (PhD C7C; New England Biolaboratories, Cambridge, MA).
Selection of Targeted Phage Clones
To identify SPARC-targeted clones, phages [2 x 1011 plaque-forming units (pfu)] were incubated for 1 hour at 37°C in a well containing purified SPARC protein (Hematologic Technologies, Inc., Essex Junction, VT) that was adsorbed (100 µg/ml) onto MaxiSorp immunoplates (Nalge Nunc, Rochester, NY) and saturated with bovine serum albumin (BSA). Following incubation, unbound phages were removed by 10 washes with Dulbecco's phosphate-buffered saline (DPBS) containing 1% BSA and 0.1% Tween-20. Bound phages were eluted with 100 µl of acidified glycine buffer (0.2 M glycine, pH 2.2) and neutralized with 10 µl of 1 M Tris (pH 9.0) to preserve viability. The eluted phage pool was then amplified, titered, and subjected to three additional rounds of SPARC selection, and individual clones were selected for enzyme-linked immunosorbent assay (ELISA) and sequencing. VCAM-1-targeted phage clones were selected and validated as previously described [10]. A clone expressing the peptide sequence CVHSPNKKC was used for fluorescent labeling.
ELISA Assay
Purified SPARC protein or BSA adsorbed (20 µg/ml) on Nunc MaxiSorp plates (4°C overnight) was incubated at room temperature with phage clones (1012 pfu, 1 hour), washed with DPBS containing 1% BSA and 0.1% Tween-20, incubated with biotinylated anti-M13 antibody (Amersham Pharmacia Biotech, Piscataway, NJ) (1:40, 1 hour), detected with streptavidin-horseradish peroxidase (1:500), and developed with tetramethyl benzidine (Pierce), and absorbance was determined at 650 nm (Emax; Molecular Devices, Sunnyvale, CA). Specificity was determined by dividing SPARC abs 650 nm by BSA abs 650 nm.
Phage Labeling
Phages were labeled with a number of different fluorochromes. Fluorochromes were all conjugated to the phage using standardNHSor isothiocyanate chemistry. Phages (1 x 1012 pfu) were resuspended in 100 ml of a 0.3-M NaHCO3 (pH 8.6) solution containing 1 mg/ml fluorochrome-hydroxy-succinimide ester (Cy3.5, Cy5.5, VT680, and AF750) or 0.25 mg/ml FITC (TRITC). The phage/fluorochrome reaction was allowed to continue for 1 hour at room temperature in the dark. Subsequent to incubation, the volume of the labeled phage was brought up to 1 ml with DPBS, and the phage was purified by polyethyleneglycol precipitation. Fluorochrome-labeled phage was then resuspended in 200 µl of DPBS and titered to determine plaque-forming units, and the concentration of fluorochrome was determined spectrophotometrically (Varian Cary II; Varian, Palo Alto, CA).
In Vitro Characterization of Labeled Phage
Fluorochromes/phage: the number of fluorochromes bound per phage was determined by comparing the concentration of fluorochrome in the sample as determined spectrophotometrically in the phage titer.
Fluorescence as a function of labeling: the fluorescence of phage with increasing dye loading was quantified by fluorescence spectroscopy (Fluorolog2; Jobin Yvon Horiba, Edison, NJ). For in vivo experiments, we chose phage with a dye loading of 800 fluorochromes/phage due to optimized signal versus quenching.
Viability of fluorochrome-labeled phage: phages labeled with increasing amounts of fluorochrome were titered using New England Biolabs (NEB) protocols to determine their ability to transfect and amplify in Escherichia coli.
Mouse Models
Nude mice were injected subcutaneously with 2.5 x 106 Lewis lung carcinoma (LLC) cells in both flanks. Tumors were allowed to grow until they had reached a diameter of 7 to 10 mm before the animals were used for experimentation. To induce microvascular VCAM-1 upregulation, C57Bl/6 mice (n = 10) were injected subcutaneously in the right ear with 5 ng/50 µl mTNFα in normal saline. After 24 hours, animals were anesthetized with inhaled isoflurane and injected intravenously (through the tail vein) with VT680-labeled VCAM-1-targeted phage.
In Vivo Imaging of LLC Tumor-Bearing Mice
For the time course experiment (Figure 3A), mice (n = 10) were coinjected with VT680-labeled SPARC-targeted phage and AF750-labeled wild-type (no insert) phage at equal fluorochrome concentrations and then imaged 2, 4, 6, and 24 hours postinjection. For clone comparison studies (Figure 5A), mice (n = 5 per clone) were also coinjected with VT680-labeled SPARC-targeted phage and AF750-labeled wild-type (no insert) phage through the tail vein then imaged 4 hours postinjection. In Figures 3C and 4, mice (n = 10) were injected with either VT680-labeled SPARC-targeted phage or VT680-labeled wild-type phage (no insert) then imaged 4 hours postinjection with surface reflectance imaging and fluorescent molecular tomography (FMT). Mice were anesthetized by inhalation anesthesia (2% isoflurane, 1 l/min O2). FMT experiments were performed at two wavelengths (at 680/700 nm to detect SPARC-targeted phage and at 750/780 nm to detect wild-type phage excitation/emission) in anesthetized mice [33]. Briefly, mice were positioned and surrounded by matching fluid to simulate tissue properties according to the manufacturer's instructions (Visen Medical). Image data sets were reconstructed using a normalized Born forward model adapted to small mouse models [33]. Image acquisition time per animal was 2 minutes, and reconstruction time was 1 to 2 minutes. Images were displayed as raw data sets (excitation, emission, and masks) and as reconstructed 3D data sets in axial, sagittal, and coronal planes. Fluorochrome concentration in the target was automatically calculated from reconstructed images and expressed as picomoles of fluorochrome per defined target volume. In addition to the 3D FMT described above, we also performed rapid fluorescence screening using reflectance detection only (Figure 3C). This method allowed for a throughput of > 20 mice/hour.
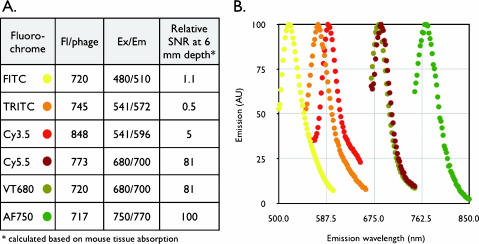
Figure 1.
Phage labeling. (A) Fluorochromes used for phage labeling. Listed are the mean numbers of fluorochromes from a single comparative experiment. The relative fluorescence signal at a 6-mm tissue depth of different fluorochrome-labeled phages was calculated from known spectral tissue absorption. Note the much higher fluorescence of 680- to 750-nm fluorochrome-labeled phages in tissues. (B) Emission spectra of fluorescently labeled phages (excitation and color of data points as listed in A).
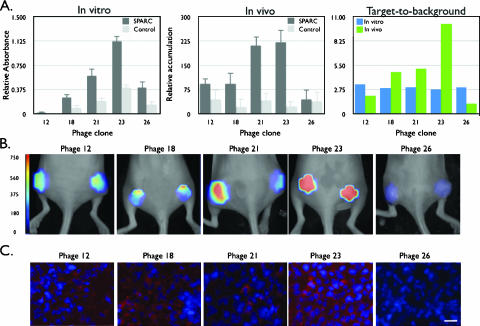
Figure 5.
Screening of SPARC-targeted phage clones. (A) Comparison of the target affinity of different SPARC clones by in vitro (left) or in vivo (middle and right) imaging (control ELISA: BSA; control FMT: wild-type phage). Note the high target-to-background ratio of clone 23 in vivo, whereas most clones are similar by in vitro screening. (B) FMT imaging of mice bearing bilateral LLC tumors injected with different phage clones. Maximum intensity projections are shown for the five phage constructs investigated. All images have been equally leveled. The scale bar is presented as the concentration of fluorochromes in nanomolars. (C) Comparative immunofluorescence of indicated phage clones' tumoral accumulation (red). Nuclei are counterstained with DAPI (blue). Scale bar = 10 µm.
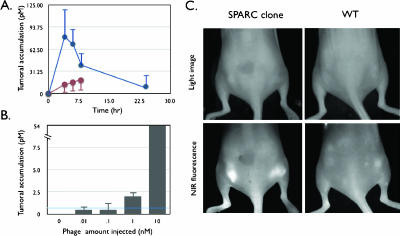
Figure 3.
In vivo behavior of labeled phage. (A) Time course of tumor homing. Mice bearing subcutaneous bilateral LLC-derived tumors were coinjected through the tail vein with VT680-labeled SPARC-targeted phage and AF750-labeled wild-type phage (no insert) and imaged at 0, 2, 4, 6, and 24 hours after injection. Blue line: SPARC-targeted phage clone 23. Brown line: wild-type phage (no insert). (B) Detection threshold. Tumor-bearing mice were injected with increasing log doses of labeled phage and imaged 4 hours after injection. The line indicates detection threshold. (C) Reflectance imaging. Mice bearing subcutaneous bilateral tumors (LLC cells) were injected with either VT680-labeled wild-type phage (right) or VT680-labeled SPARC-targeted phage. Note the brightly fluorescent tumors in the near-infrared fluorescence channel of the SPARC-targeted phage clone [identical white light (WL) settings].
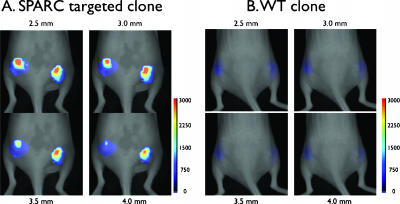
Figure 4.
In vivo imaging of labeled phage. Mice bearing subcutaneous bilateral tumors (LLC cells) were injected with fluorescent phage. (A) Serial coronal tomographic images at different depths of a representative mouse injected with the SPARC-targeted phage clone. Note the distribution of phages throughout the tumors. The scale bar is in nanomolars of fluorescence. (B) Wild-type control phage (identical white light settings as in A).
Intravital Laser Scanning Microscopy
Multichannel fluorescence imaging was performed with a prototypical laser scanning fluorescence microscope (Olympus Corporation, Tokyo, Japan) specifically developed for intravital imaging of mouse organs [34]. Three laser lines at 488, 633, and 748 nm were used. Image acquisition was 1 second. The FluoView 300 software program (Olympus Corporation) was used to control the microscope and to collect images of 512 x 512 pixels with a pixel size of about 5.4 mm/pixel and a total image size of about 2.8 x 2.8 mm. Images were stored as multilayer 16-bit tagged image file format files. Images in the 680-nm channel (660- to 730-nm bandpass filter) and in the 750-nm channel (770-nm longpass filter) were collected concomitantly using custom-made filters and dichroic mirrors (Olympus Corporation). A dry objective (x4 UplanApo NA 0.16; Olympus Corporation) with a field of view of 3.25 mm and a theoretical lateral resolution of about 2.6 µm at 680 nm were used. Wide spectral response photomultiplier tubes (model R928P; Hamamatsu, Bridgewater, NJ) were used as detectors for both visible light and near-infrared signals. After initial images had been obtained, a near-infrared vascular imaging agent (Angiosense-750) was administered to define microvascularity [35].
Histology
Epifluorescence microscopic images were obtained with an upright fluorescence microscope (Eclipse 80i; Nikon Instruments, Melville, NY) with a cooled charge-coupled device camera (Cascade 512B; Photometrics, Tucson, AZ). After imaging by intravital confocal microscopy, immunohistochemical analysis of mouse ears was performed. Frozen sections were stained for the presence of CD31, VCAM-1, or M13. Digital images were taken using a Nikon Eclipse E400 upright microscope (x40 objective; Nikon Instruments) equipped with an Insight color camera (Diagnostics Instruments, Sterling Heights, MI).
Results
A number of commercially available fluorochromes were initially used to test the labeling efficiency of M13 phage coat proteins through hydroxysuccinimide or isothiocyanate chemistry, yielding fluorescent phage clones. Figure 1 summarizes the optical properties of differently-labeled M13 phage. Assuming a 6-mm depth in tissues, near-infrared-labeled phages would be detectable roughly 102-fold more efficiently compared to labeled phages in the visible spectrum. Therefore, we particularly focused on far-red and near-infrared fluorochromes as light penetration in vivo is more efficient at these wavelengths [36]. To better understand the effect of labeling efficiency on phage affinity, fluorescence, and viability, we subsequently performed more in-depth experiments on a phage labeled with a fluorochrome with an excitation wavelength of 680 nm (VT680).
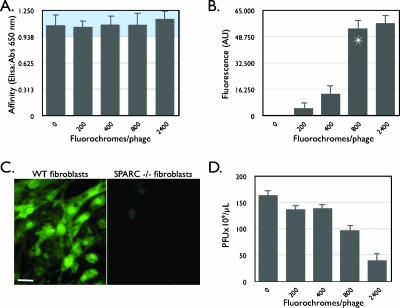
Figure 2 summarizes the effect of the incremental modification of phage coat proteins on the fluorescence, affinity, and viability of isolated phage clones. As a model system, we used one clone (clone 23) isolated from a selection against SPARC protein. Regardless of the extent of phage coat protein modification (up to 2400 fluorochromes/phage particle), derivatized phage bound with the same affinity to SPARC protein, as analyzed through ELISA (Figure 2A). To maximize the net fluorescence per phage and to enhance signal for in vivo imaging, we next measured fluorescence versus the extent of fluorochrome labeling. Beyond 800 fluorochromes per phage, fluorescence did not improve significantly presumably due to dye-dye quenching (Figure 2B). Importantly, at this extent of modification, labeled phages still retained their biologic targeting ability in that they were able to bind to SPARC (Figure 2A). Indeed, in cell-based screens, phages were shown to bind specifically to SPARC-expressing cells but not to fibroblasts isolated from SPARC-/- knockout mice (Figure 2C). Taken together, these results demonstrate an optimum labeling of 800 fluorochrome/phage. Therefore, in subsequent experiments, we used phage with 800 fluorochromes attached, corresponding to ∼ 25% of coat protein modification. The ability of labeled phage to infect host bacteria and to amplify (viability) was affected in a concentration-dependent fashion (Figure 2D). However, at the dose used for in vivo experiments, 50% of phages were viable and still permitted regeneration of phage clones when necessary. Similar experiments, as described above, were also conducted with VT680-labeled wild-type and AF750-labeled SPARC and wild-type phages, showing similar results (data not shown).
Figure 2.
Properties of labeled phage. (A) Effect of increasing fluorochrome labeling (VT680) on phage affinity determined by ELISA (blue region: error range). (B) Effect of increasing labeling on phage fluorescence. Beyond 800 fluorochromes per M13 phage, there is little increase in fluorescence. (C) VT680-labeled phage clone (800 fluorochromes/phage) binds specifically to SPARC-expressing fibroblasts, but not to fibroblasts derived from SPARC-/- mice (scale bar: 10 µm). (D) Effect of successive phage labeling on viability (ability of the phage to infect and amplify in E. coli).
To determine the time course of biologic uptake and system detection levels of fluorochrome-labeled phage, we next tested labeled phage in a mouse tumor model. LLC cells expressing SPARC [37] were grown in mice until tumors had reached a diameter of 7 to 10 mm. Subsequently, mice were injected intravenously with 10 nmol (fluorochrome concentration) of fluorochrome-labeled phage and serially monitored by FMT [38]. Maximum tumor signal occurred 2 to 4 hours after systemic injection of phage (Figure 3A). In dose escalation studies (Figure 3B), tumoral signal intensity was highest with the largest amount of phage injected. At this level of phage labeling, the detection threshold by FMT was calculated to be ∼ 300 phage/mm3. Surface reflectance imaging (rapid screening) demonstrated preferential tumoral accumulation of SPARC-targeted phage over wild-type phage (Figure 3C). To quantitate variability in tumoral distribution and heterogeneity, we also performed tomographic FMT imaging (Figure 4). Screening experiments showed that SPARC-targeted clones accumulated in all bilaterally implanted LLC tumors (Figure 4A), whereas VT680-labeled wild-type phage did not (11,481.92 ± 977 FU in VT680-SPARC phage versus 776.01 ± 24 in VT680-WT phage; P < .0001; specificity ratio = 14.7; n = 10). Serial sections through tumors clearly identified intratumoral phage in a somewhat heterogeneous distribution, which corresponded well with correlative fluorescence microscopy (Figure 5). By FMT measurements, the mean tumoral concentration of SPARC phage was determined to be 23,600 phage/mm3 or 0.297 pM phage. Although a difference between wild-type and SPARC phage accumulation was noted when they were imaged using surface-weighted fluorescence imaging, a larger difference was seen between the wild-type and SPARC phages when they were imaged with FMT. The ability of FMT to detect fluorescent signal throughout the entire depth of the sample allows for a more precise imaging and opens the door for imaging phage-targeted agents in deep tissues where surface-weighted imaging would not be useful.
We next performed comparative in vivo screens of phage clones that had been identified through in vitro screens against SPARC protein, focusing on 5 of 30 ELISA-confirmed clones (Figure 5A). There were considerable in vivo differences in tumoral homing among lead candidates (Figure 5B). Although in vitro analysis demonstrated similar specificity constants for the top five lead candidates, in vivo testing revealed that phage clone 23 (SPPTGIN) demonstrated excellent in vivo targeting ability, whereas clone 12 (NFPQRFL), clone 18 (APTRYNG), clone 21 (HWGMWSY), and clone 26 (GETRAPL) showed much lower in vivo homing to tumors (Figure 5A). These observations held true among different animals in each group, indicating that homing was not due to individual tumor heterogeneity but rather due to in vivo targeting. Furthermore, coinjection of mice with AF750-labeled wild-type phage did not show appreciable targeting (Figure 5A), demonstrating the in vivo tumor targeting of identified peptide sequences. Additional histologic experiments confirmed the imaging findings (Figure 5C).
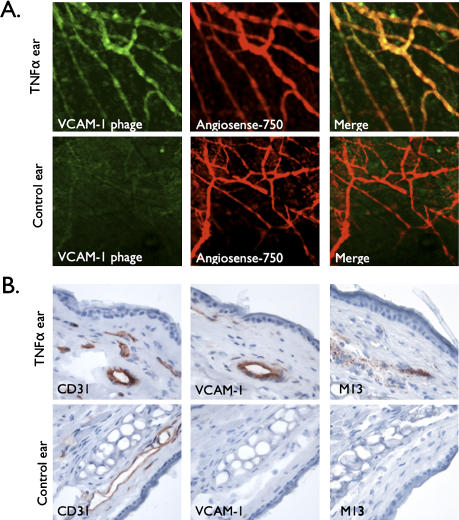
To determine whether fluorochrome-labeled phage could also be used for microscopic imaging and to extend the technique past one in vivo system, we finally performed intravital confocal imaging experiments (Figure 6). Using the TNFα-induced endothelial VCAM-1 upregulation model [10], we performed microvascular imaging in the ear of live mice (Figure 6). Similarly as described above, VCAM-1 internalized phages (bearing the sequence CVHSPNKKC) [10] were labeled with VT680 and injected intravenously 4 hours before imaging. To unequivocally identify microvessels, we coinjected a circulating vascular maker. At the time of imaging, near-infrared fluorescence attributed to phage was highly elevated in the vasculature of the inflamed ear, but was largely absent in the contralateral control ear. There was excellent correlation between local phage homing and signal from the intravascular agent (Figure 6A). Immunohistochemistry confirmed the expression of VCAM-1 in the TNFα-injected ear, but not in the control ear (Figure 6B). In addition, phage particles colocalized with VCAM-1-expressing vessels, confirming the retention of phage specificity for VCAM-1 and the use of this technique for in vivo imaging (Figure 6B).
Figure 6.
In vivo microscopic imaging of phage. (A) Microscopic imaging of bilateral ear vessels of a live mouse injected with labeled VCAM-1-targeted phage (CVHSPNKKC, green) and vascular marker (Angiosense-750, red). Shown are confocal projections of Z stacks from mTNFα-treated ear or control ear at 4 hours after phage injection. Images are representative of different animals in each case and five different fields per ear. (B) Correlative histology: adjacent sections of ears taken from in vivo intravital experiments were stained for CD31, VCAM-1, or phage protein pVIII (M13).
Discussion
The use of phage display libraries is a powerful method for identifying affinity peptides for biologic applications. For example, phage display has been used to identify vascular zip codes [18], markers of angiogenesis [22], targeted therapeutics [39], or affinity ligands for imaging [10,19]. However, the in vivo validation of in vitro hits can remain challenging, time-consuming, and costly. Here, we describe methods to label phages with far-red fluorochromes and to test them in vivo by optical imaging. This approach is universal, fast, cost-effective, and accurate as multiwavelength imaging can incorporate control phage. The described method enhances in vivo phage screening as it allows the direct visualization of injected phage and the identification of cell type-specific clones. In addition, the method is useful for in vivo testing whole phage libraries, subsets, or specific clones. Furthermore, we showthat near-infrared-labeled phages themselves can serve as an economical and replenishable source of molecularly targeted imaging agents.
Labeling of phage can be accomplished with different available fluorochromes that are well established, yielding highly fluorescent targeted phage particles. M13 filamentous phage (6.5 nm in diameter, ∼ 880 nm in length) contains approximately 2700 copies of the major coat protein P8 and 5 copies of three different minor coat proteins (P9, P6, and P3) on the ends. Our results indicate that labeling of coat proteins (800 fluorochromes/phage or ∼ 25% of coat proteins) does not interfere with a displayed peptide's targeting ability or a phage's ability to be internalized into mammalian cells (Figure 2C). Because we only label 25% of phage coat proteins, the probability of labeling the major coat protein (2700 copies/phage)—as opposed to labeling the peptide (5 copies/phage)—that confers targeting ability is high, therefore allowing the retention of selected biologic activity. Indeed, the six phage clones examined (five SPARC and one VCAM-1) all retained their biologic activity. In vivo, we were able to measure the tumor concentration of phage with a threshold of ∼ 300 pfu/mm3. On microscopic resolution, we estimate that the detection threshold is much lower (∼ 10 pfu/mm3) based on confocal laser scanning experiments [34]. The availability of fluorochromes with different spectral properties (Figure 1) allows multicolor imaging (i.e., measurements can be made using a control reference channel) or screening of differently labeled phages simultaneously. These strategies can be exploited to rapidly eliminate clones that would ultimately fail because of unfavorable in vivo pharmacokinetics, delivery barriers, or insufficient target-to-background ratios.
We focused on two different model targets in this research, SPARC and VCAM-1, given their importance in human disease [29,31]. SPARC (osteonectin) is a 32-kDa glycoprotein that interacts with the extracellular matrix and exerts an antiadhesive promigratory effect on cells [29]. In vivo, SPARC expression is restricted to rapidly remodeling tissues, such as invasive cancers [40]. SPARC has recently been used as a biomarker to identify invasive and/or high-metastaticpotential prostate cancer [29], distinguishing it from more indolent forms. We identified one novel phage clone (23-SPPTGIN) that had high affinity and specificity for SPARC and demonstrated excellent in vivo tumoral targeting ability, suggesting that this agent may be useful for the detection and targeting of invasive SPARC-expressing cancers. Conversion of the identified targeted peptide into imaging agents detectable by magnetic resonance imaging and/or positron emission tomography imaging modalities is currently under way. The second example chosen was a phage with high avidity (and endothelial cell internalization) for VCAM-1 expression (CVHSPNKKC). We show that fluorescently labeled clones specifically target VCAM-1-expressing blood vessels in vivo and could be used for the imaging of VCAM-1 expression in high-mortality diseases such as atherosclerosis.
The developed in vivo screening tool could be used to decrease the time between the identification of potential “hits” and the development of in vivo molecularly targeted imaging agents. This could potentially eliminate a number of failed “hits” earlier in the investigation, reducing time and cost. Furthermore, we believe that near-infrared tagged phage could play an important role as a molecularly targeted imaging agent. M13 phage particles have the same width and ability to be functionalized as some synthetic nanomaterials currently being used in vivo [11,41,42] and have been used here as a platform for the development of imaging agents. Infection with filamentous phages is not lethal as they have been injected intravenously [26]. Furthermore, phages have no intrinsic tropism for mammalian cells and are biodegradable. Phages have also been engineered to serve as delivery vehicles, suggesting that the targeted labeled phage can be used for both imaging and therapy [43].
Acknowledgements
The authors acknowledge E. Aikawa for assistance with immunohistochemistry, F. Reynolds for chemistry assistance, M. Pittet for manuscript review and many helpful discussions, R. Anbazhagan for help with phage cultures, and V. Lok for the performance of histology.
References
- 1.Hoffman JA, Giraudo E, Singh M, Zhang L, Inoue M, Porkka K, Hanahan D, Ruoslahti E. Progressive vascular changes in a transgenic mouse model of squamous cell carcinoma. Cancer Cell. 2003;4:383–391. doi: 10.1016/s1535-6108(03)00273-3. [DOI] [PubMed] [Google Scholar]
- 2.Joyce JA, Laakkonen P, Bernasconi M, Bergers G, Ruoslahti E, Hanahan D. Stage-specific vascular markers revealed by phage display in a mouse model of pancreatic islet tumorigenesis. Cancer Cell. 2003;4:393–403. doi: 10.1016/s1535-6108(03)00271-x. [DOI] [PubMed] [Google Scholar]
- 3.Valadon P, Garnett JD, Testa JE, Bauerle M, Oh P, Schnitzer JE. Screening phage display libraries for organ-specific vascular immunotargeting in vivo. Proc Natl Acad Sci USA. 2006;103:407–412. doi: 10.1073/pnas.0506938103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zurita AJ, Arap W, Pasqualini R. Mapping tumor vascular diversity by screening phage display libraries. J Control Release. 2003;91:183–186. doi: 10.1016/s0168-3659(03)00236-0. [DOI] [PubMed] [Google Scholar]
- 5.Ladner RC, Sato AK, Gorzelany J, de Souza M. Phage display-derived peptides as therapeutic alternatives to antibodies. Drug Discov Today. 2004;9:525–529. doi: 10.1016/S1359-6446(04)03104-6. [DOI] [PubMed] [Google Scholar]
- 6.Frank R, Hargreaves R. Clinical biomarkers in drug discovery and development. Nat Rev Drug Discov. 2003;2:566–580. doi: 10.1038/nrd1130. [DOI] [PubMed] [Google Scholar]
- 7.Rudin M, Weissleder R. Molecular imaging in drug discovery and development. Nat Rev Drug Discov. 2003;2:123–131. doi: 10.1038/nrd1007. [DOI] [PubMed] [Google Scholar]
- 8.Perez JM, Josephson L, O'Loughlin T, Hogemann D, Weissleder R. Magnetic relaxation switches capable of sensing molecular interactions. Nat Biotechnol. 2002;20:816–820. doi: 10.1038/nbt720. [DOI] [PubMed] [Google Scholar]
- 9.Hood L, Heath JR, Phelps ME, Lin B. Systems biology and new technologies enable predictive and preventative medicine. Science. 2004;306:640–643. doi: 10.1126/science.1104635. [DOI] [PubMed] [Google Scholar]
- 10.Kelly KA, Allport JR, Tsourkas A, Shinde-Patil VR, Josephson L, Weissleder R. Detection of vascular adhesion molecule-1 expression using a novel multimodal nanoparticle. Circ Res. 2005;96:327–336. doi: 10.1161/01.RES.0000155722.17881.dd. [DOI] [PubMed] [Google Scholar]
- 11.Akerman ME, Chan WC, Laakkonen P, Bhatia SN, Ruoslahti E. Nanocrystal targeting in vivo. Proc Natl Acad Sci USA. 2002;99:12617–12621. doi: 10.1073/pnas.152463399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zitzmann S, Mier W, Schad A, Kinscherf R, Askoxylakis V, Kramer S, Altmann A, Eisenhut M, Haberkorn U. A new prostate carcinoma binding peptide (DUP-1) for tumor imaging and therapy. Clin Cancer Res. 2005;11:139–146. [PubMed] [Google Scholar]
- 13.Mikawa M, Wang H, Guo L, Liu R, Marik J, Takada Y, Lam K, Lau D. Novel peptide ligands for integrin alpha 4 beta 1 overexpressed in cancer cells. Mol Cancer Ther. 2004;3:1329–1334. [PubMed] [Google Scholar]
- 14.Stockwell BR, Haggarty SJ, Schreiber SL. High-throughput screening of small molecules in miniaturized mammalian cell-based assays involving post-translational modifications. Chem Biol. 1999;6:71–83. doi: 10.1016/S1074-5521(99)80004-0. [DOI] [PubMed] [Google Scholar]
- 15.Weissleder R, Kelly K, Sun EY, Shtatland T, Josephson L. Cell-specific targeting of nanoparticles by multivalent attachment of small molecules. Nat Biotechnol. 2005;23:1418–1423. doi: 10.1038/nbt1159. [DOI] [PubMed] [Google Scholar]
- 16.Drees BL. Progress and variations in two-hybrid and threehybrid technologies. Curr Opin Chem Biol. 1999;3:64–70. doi: 10.1016/s1367-5931(99)80012-x. [DOI] [PubMed] [Google Scholar]
- 17.Brown CK, Modzelewski RA, Johnson CS, Wong MK. A novel approach for the identification of unique tumor vasculature binding peptides using an E. coli peptide display library. Ann Surg Oncol. 2000;7:743–749. doi: 10.1007/s10434-000-0743-0. [DOI] [PubMed] [Google Scholar]
- 18.Rajotte D, Arap W, Hagedorn M, Koivunen E, Pasqualini R, Ruoslahti E. Molecular heterogeneity of the vascular endothelium revealed by in vivo phage display. J Clin Invest. 1998;102:430–437. doi: 10.1172/JCI3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Landon LA, Deutscher SL. Combinatorial discovery of tumor targeting peptides using phage display. J Cell Biochem. 2003;90:509–517. doi: 10.1002/jcb.10634. [DOI] [PubMed] [Google Scholar]
- 20.Smith GP, Petrenko VA. Phage display. Chem Rev. 1997;97:391–410. doi: 10.1021/cr960065d. [DOI] [PubMed] [Google Scholar]
- 21.Willats WG. Phage display: practicalities and prospects. Plant Mol Biol. 2002;50:837–854. doi: 10.1023/a:1021215516430. [DOI] [PubMed] [Google Scholar]
- 22.Koivunen E, Gay DA, Ruoslahti E. Selection of peptides binding to the alpha 5 beta 1 integrin from phage display library. J Biol Chem. 1993;268:20205–20210. [PubMed] [Google Scholar]
- 23.Hong FD, Clayman GL. Isolation of a peptide for targeted drug delivery into human head and neck solid tumors. Cancer Res. 2000;60:6551–6556. [PubMed] [Google Scholar]
- 24.Brown KC. New approaches for cell-specific targeting: identification of cell-selective peptides from combinatorial libraries. Curr Opin Chem Biol. 2000;4:16–21. doi: 10.1016/s1367-5931(99)00045-9. [DOI] [PubMed] [Google Scholar]
- 25.Liu C, Bhattacharjee G, Boisvert W, Dilley R, Edgington T. In vivo interrogation of the molecular display of atherosclerotic lesion surfaces. Am J Pathol. 2003;163:1859–1871. doi: 10.1016/S0002-9440(10)63545-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arap W, Kolonin MG, Trepel M, Lahdenranta J, Cardo-Vila M, Giordano RJ, Mintz PJ, Ardelt PU, Yao VJ, Vidal CI, et al. Steps toward mapping the human vasculature by phage display. Nat Med. 2002;8:121–127. doi: 10.1038/nm0202-121. [DOI] [PubMed] [Google Scholar]
- 27.Brown EB, Campbell RB, Tsuzuki Y, Xu L, Carmeliet P, Fukumura D, Jain RK. In vivo measurement of gene expression, angiogenesis and physiological function in tumors using multiphoton laser scanning microscopy. Nat Med. 2001;7:864–868. doi: 10.1038/89997. [DOI] [PubMed] [Google Scholar]
- 28.Massoud TF, Gambhir SS. Molecular imaging in living subjects: seeing fundamental biological processes in a new light. Genes Dev. 2003;17:545–580. doi: 10.1101/gad.1047403. [DOI] [PubMed] [Google Scholar]
- 29.De S, Chen J, Narizhneva NV, Heston W, Brainard J, Sage EH, Byzova TV. Molecular pathway for cancer metastasis to bone. J Biol Chem. 2003;278:39044–39050. doi: 10.1074/jbc.M304494200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Koukourakis MI, Giatromanolaki A, Brekken RA, Sivridis E, Gatter KC, Harris AL, Sage EH. Enhanced expression of SPARC/osteonectin in the tumor-associated stroma of non-small cell lung cancer is correlated with markers of hypoxia/acidity and with poor prognosis of patients. Cancer Res. 2003;63:5376–5380. [PubMed] [Google Scholar]
- 31.Cybulsky MI, Gimbrone MAJ. Endothelial expression of a mononuclear leukocyte adhesion molecule during atherogenesis. Science. 1991;251:788–791. doi: 10.1126/science.1990440. [DOI] [PubMed] [Google Scholar]
- 32.Funovics M, Montet X, Reynolds F, Weissleder R, Josephson L. Nanoparticles for the optical imaging of tumor E-selectin. Neoplasia. 2005;7:904–911. doi: 10.1593/neo.05352. [DOI] [PubMed] [Google Scholar]
- 33.Montet X, Ntziachristos V, Grimm J, Weissleder R. Tomographic fluorescence mapping of tumor targets. Cancer Res. 2005;65:6330–6336. doi: 10.1158/0008-5472.CAN-05-0382. [DOI] [PubMed] [Google Scholar]
- 34.Alencar H, Mahmood U, Kawano Y, Hirata T, Weissleder R. Novel multiwavelength microscopic scanner for mouse imaging. Neoplasia. 2005;7:977–983. doi: 10.1593/neo.05376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Binstadt BA, Patel PR, Alencar H, Nigrovic PA, Lee DM, Mahmood U, Weissleder R, Mathis D, Benoist C. Particularities of the vasculature can promote the organ specificity of autoimmune attack. Nat Immunol. 2006;7:284–292. doi: 10.1038/ni1306. [DOI] [PubMed] [Google Scholar]
- 36.Weissleder R, Ntziachristos V. Shedding light onto live molecular targets. Nat Med. 2003;9:123–128. doi: 10.1038/nm0103-123. [DOI] [PubMed] [Google Scholar]
- 37.Brekken RA, Puolakkainen P, Graves DC, Workman G, Lubkin SR, Sage EH. Enhanced growth of tumors in SPARC null mice is associated with changes in the ECM. J Clin Invest. 2003;111:487–495. doi: 10.1172/JCI16804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ntziachristos V, Ripoll J, Wang LV, Weissleder R. Looking and listening to light: the evolution of whole-body photonic imaging. Nat Biotechnol. 2005;23:313–320. doi: 10.1038/nbt1074. [DOI] [PubMed] [Google Scholar]
- 39.Arap W, Haedicke W, Bernasconi M, Kain R, Rajotte D, Krajewski S, Ellerby HM, Bredesen DE, Pasqualini R, Ruoslahi E. Targeting the prostate for destruction through a vascular address. Proc Natl Acad Sci USA. 2002;99:1527–1531. doi: 10.1073/pnas.241655998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Framson PE, Sage EH. SPARC and tumor growth: where the seed meets the soil? J Cell Biochem. 2004;92:679–690. doi: 10.1002/jcb.20091. [DOI] [PubMed] [Google Scholar]
- 41.Harisinghani MG, Barentsz J, Hahn PF, Deserno WM, Tabatabael S, van de Kaa CH, de la Rosette J, Weissleder R. Noninvasive detection of clinically occult lymph-node metastases in prostate cancer. N Engl J Med. 2003;348:2491–2499. doi: 10.1056/NEJMoa022749. [DOI] [PubMed] [Google Scholar]
- 42.Sipkins DA, Cheresh DA, Kazemi MR, Nevin LM, Bednarski MD, Li KC. Detection of tumor angiogenesis in vivo by alphavbeta3-targeted magnetic resonance imaging. Nat Med. 1998;4:623–626. doi: 10.1038/nm0598-623. [DOI] [PubMed] [Google Scholar]
- 43.Larocca D, Burg MA, Jensen-Pergakes K, Ravey EP, Gonzalez AM, Baird A. Evolving phage vectors for cell targeted gene delivery. Curr Pharm Biotechnol. 2002;3:45–57. doi: 10.2174/1389201023378490. [DOI] [PubMed] [Google Scholar]