Abstract
The progression of organ-confined prostate cancer to metastatic cancer is inevitably fatal. Consequently, identification of structural changes in the genome and associated transcriptional responses that drive this progression is critical to understanding the disease process and the development of biomarkers and therapeutic targets. In this study, whole genome copy number changes in genomes of hormone-naïve lymph node metastases were profiled using array comparative genomic hybridization, and matched primaries were included for a subset. Matched primaries and lymph node metastases showed very similar copy number profiles that are distinct from primary tumors that fail to metastasize.
Keywords: Lymph nodes, prostate cancer, aCGH, metastases, biomarkers
Introduction
An estimated 30% of all prostate cancer patients suffer from recurrent disease after undergoing primary treatment (radical prostatectomy or radiation) [1]. Metastatic prostate cancer inevitably progresses to androgen independency, at which point it is essentially incurable. Due to early detection, the 5-year disease-specific survival rate is close to 100% in men with localized prostate cancer, but is only 34% in men with distant metastasis [1]. It appears that a high proportion of men initially presenting with apparently localized disease have undetectable metastases that eventually progress to life-threatening metastatic tumors. Therefore, developing diagnostic and therapeutic approaches for targeting metastatic disease holds considerable potential for improving the quality of life, treatment modalities, and survival rates of a significant proportion of patients with prostate cancer.
Prostate cancer often migrates through the lymphatic route, depositing tumor cells into pelvic lymph nodes. There have not been many publications to date on the genetic makeup of lymph node metastases of prostate cancer [2,3]. This is partly due to the fact that lymph node metastases, particular those that are hormone-naïve, are hard to obtain. In the United States, a patient diagnosed with prostate cancer with clinical lymph node involvement rarely undergoes radical prostatectomy. We analyzed five hormone-naïve lymph node (HNLN) metastases and two matching sets of primary prostate cancers and their respective HNLN metastases. One matched set consisted of a primary prostate tumor and lymph node metastasis from a unilateral metastatic case, and the second consisted of a primary prostate tumor with bilateral lymph node metastases. Array comparative genomic hybridization (aCGH) was performed on all eight lymph node tumors, as well as on two primaries. We report our genomewide characterization of this set of tumors, highlighting frequent copy number aberrations (CNAs).
Materials and Methods
Tumor Samples
Five lymph nodes were obtained through a collaboration with the University of Ulm (Ulm, Germany). Fifteen-micron sections for each fresh frozen sample were sent. Hematoxylin-eosing stainings were reviewed by one pathologist (M.A.R.). Microdissection was not necessary due to the high percentage of tumors in the sections. Tumor tissue was directly placed into proteinase K (PK) cell lysis buffer (Promega, Madison, WI) with 20 µl of PK and then incubated at 55°C. An additional 20 µl of PK was added twice daily for 2 to 3 days. DNA was extracted using Promega's Wizard DNA purification kit. DNA was cleaned by two phenol/chloroform/isoamyl alcohol (25:24:1) extractions and then purified using 2-ml Eppendorf Phase Lock Gel Light tubes (Eppendorf, Westbury, NY). Finally, DNA was ethanol-precipitated with ammoniumacetate, resuspended in 25 to 50 µl of 1x Tris-EDTA (TE), and stored at 4°C. RNA was isolated by macrodissection for matched primary and HNLN cases (PR1263 and PR1264) using the RNeasy kit (Qiagen, Valencia, CA). TaqMan assay was carried out at the University of California at San Francisco (UCSF) Comprehensive Cancer Center's Genome Core.
aCGH
Human version 2.0 BAC arrays were purchased from the UCSF Array Core. Each array consists of 2460 BAC clones on chromium slides spotted in triplicate [4]. The resolution is approximately 1.4 Mb. We followed our published hybridization protocol [5].
aCGH Statistical Analysis
Tumor/reference fluorescence intensity ratios were converted to log2 domain, and replicate spots were averaged. Observed log2 ratios were excluded if there were fewer than two (of three) usable replicate spots, or if the standard deviation of replicates was > 0.2. Each array was normalized to have a median log2 ratio of 0. To identify gained and lost clones in individual samples, sample-specific thresholds were constructed [6,7]. GEMCaP clones with log2 ratios above or below (±) a tumor's threshold were considered gained or lost, respectively. A Microsoft Excel (Seattle, WA) macro written in our laboratory, was used to determine the percentage of aberrant GEMCaP loci based on inputted aCGH data and corresponding tumor-based threshold (TBT) for a given patient. All tumor aCGH data were analyzed with this macro. Frequent CNAs were plotted only using clones that were present in ≥ 50% of samples.
Results
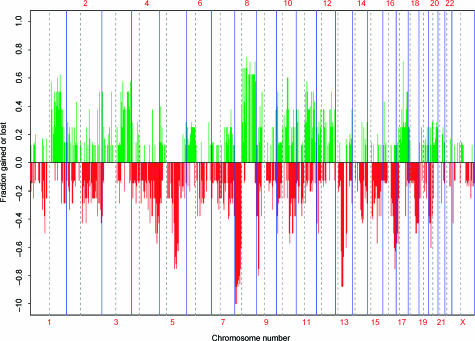
Whole genome aCGH was performed on eight HNLN tumors. Figure 1 displays the frequency of copy number changes across the whole genome for all eight hormone-naïve tumors. The numerical frequency for aberration, along with BAC clone names for this set of tumors, is provided in Table W1. Sixty-three percent of the clones with increased copy number that were aberrant at a frequency of ≥ 50% resided on chromosome 8. Similarly, 57% of frequently deleted clones (frequency of > 50%) mapped to chromosome 8. A summary highlighting some of the frequent (frequency of ≥ 50%) CNAs identified by aCGH is shown in Table 1. In addition, a list of candidate genes that directly map to, or near, an aberrant clone is provided in Table 1.
Figure 1.
Frequency plot for eight HNLN tumors. Green bars represent gains; red bars represent losses.
Table 1.
Selection of Frequent (≥ 50%) CNAs in HNLN Tumors.
| (A) | |||
| Clone | Frequency of CNA (Gain) (%) | Locus | Candidate Genes |
| RP11-214E11 | 75 | 8q21.13 | TPD52 |
| RP5-1071I14* | 71 | 17q21.33 | NGFR |
| RP11-80C11 | 71 | 8q21.13 | FABP5 |
| RP11-237F24 | 62 | 8q24.21 | MYC |
| RP11-30I17 | 57 | 1q21.1 | PIAS3 |
| RP11-179B9 | 50 | 3p14.1 | Prickle2 |
| RP11-1146E5* | 50 | 3q26.2 | EVI1 |
| RP11-114M1* | 50 | 3q26.32 | IRA1 (∼ 500 kb away) |
| RP11-548G17 | 50 | 11q13.1 | ARL2, SNX15, ZFPL1 |
| RP11-168B13 | 50 | 11q13.4-13.5 | RPS3 |
| RP11-46E14* | 50 | 17q25.3 | CBX8, CBX4 |
| (B) | |||
| Clone | Frequency of CNA (Loss) (%) | Locus | Candidate Genes |
| RP11-246G24 | 100 | 8p23.2 | MYOM2 (∼ 200 kb away) |
| GS1-77L23 | 88 | 7q32.1 | LEP |
| RP11-92C1 | 88 | 8p22 | DLC1 |
| RP11-252K12 | 88 | 8p23.1 | C8orf21 |
| RP11-122N11 | 88 | 8p23.1 | SPAG11 |
| RP11-287P18 | 88 | 8p23.1 | SPAG11, DEFBs |
| RP11-112G9 | 88 | 8p23.1 | MSRA |
| RP11-17I11 | 88 | 13q14.11 | DNAJD1 |
| RP11-21H9 | 88 | 13q14.11 | TNFSF11 |
| RP11-217H23* | 88 | 13q14.3 | GTF2F2 |
GEMCaP clone.
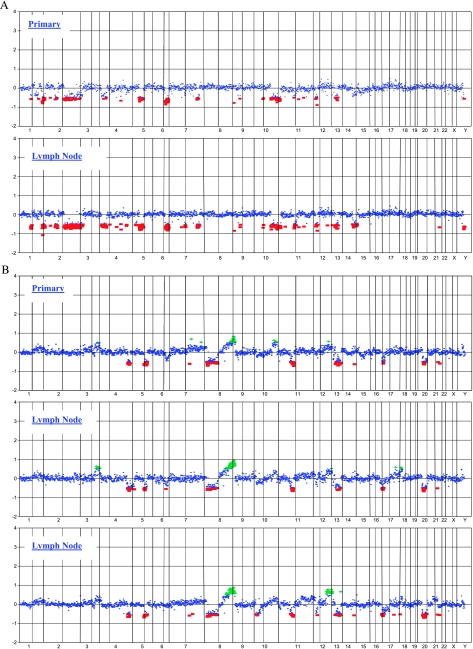
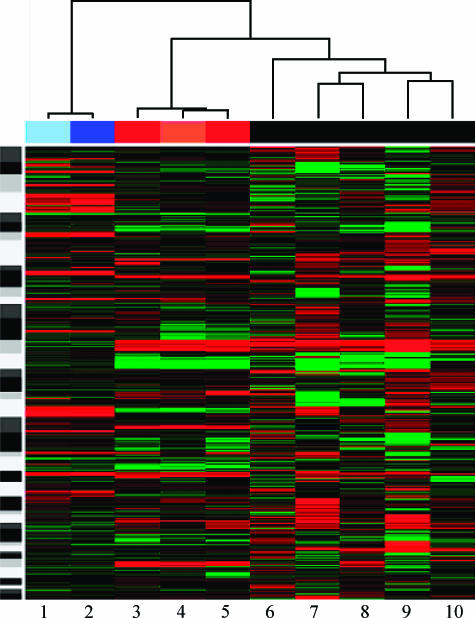
aCGH was also performed on two primary tumors possessing matched HNLN tumors. The copy number profiles of matched primary and lymph node cases are shown in Figure 2. The median Pearson correlation between primaries and their matched metastasis was 0.88, as opposed to 0.21 for unrelated samples. The smallest correlation among related samples was 0.85, and the largest correlation among unrelated samples was 0.49. Note that the direct test comparing distributions of correlations between matched and unmatched samples leads to P < 1e-04 (Wilcoxon rank sum test). Figure 3 shows a heat map of copy number changes across the genome of each tumor studied. The corresponding dendrogram also shows clustering of related samples. Aggregated hierarchical clustering with Ward method and Pearson correlation was used as a measure of similarity.
Figure 2.
aCGH whole genome CNA profiles for the two matched primary and lymph node tumors. The primary case is displayed first for each set. (A) Unilateral case. (B) Bilateral case.
Figure 3.
Heat map for all eight HNLN tumors and two primary prostate tumors. Red represents loss; green denotes copy number gain. 1 = PR1263- primary; 2 = PR1263HNLN; 3 = PR1264leftHNLN; 4 = PR1264primary; 5 = PR1264rightHNLN; 6 = HNLN13; 7 = HNLN10; 8 = HNLN12; 9 = HNLN8; 10 = HNLN11.
We compared the genomic profiles of the eight HNLN metastases to our existing aCGH database, which consists of 19 distant metastatic tumors, 12 primary tumors that progressed by prostate-specific antigen failure and confirmed bone metastases, and 32 primary tumors that did not relapse biochemically. In regards to gains, four clones were gained with statistical significance (P < .05) in > 20% of HNLN metastases, distant metastases, and primaries that progressed, but were gained in < 20% of nonprogressors. Fourteen clones met this criterion for losses. Clone names and their corresponding CNA frequencies are listed in Table 2.
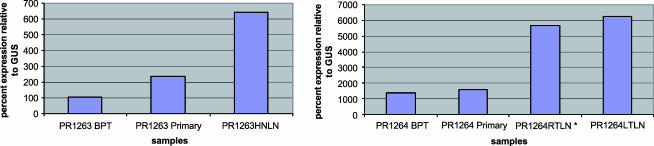
To evaluate some of the candidate genes identified in this study, existing and unpublished Affymetrix gene expression results for three lymph node tumors (HNLN8, HNLN12, and HNLN13) were queried. Gene expression for unmatched primary tumors was used for comparison. The following genes mapping to regions of copy number gain listed in Table 1 were represented on Affymetrix array: fatty acid-binding protein 5 (FABP5), tumor protein D52 (TPD52), nerve growth factor receptor (NGFR), myelocytomatosis viral oncogene homolog (MYC), protein inhibitor of activated STAT protein 3 (PIAS3), ADP ribosylation factor-like 2 (ARL2), sorting nexin 15 (SNX15), multiple endocrine neoplasia 1 (MEN1), ribosomal protein S3 (RPS3), chromobox homolog 8 (CBX8), and chromobox homolog 4 (CBX4). Table 3 shows the combined aCGH and expression results for HNLN8, HNLN12, and HNLN13. We further evaluated FABP5 with TaqMan using RNA from two matched primary HNLN cases (PR1263 and PR1264) that were not included in the Affymetrix gene expression study. FABP5 was selected because it was differentially and highly expressed in HNLN metastases compared to primaries (Table 3). There was a trend toward the upregulation of FABP5 in HNLNs in the case with concordant copy number change (PR1264HNLN), as well as in the case without it (PR1263) (Figure 4).
Table 3.
Expression Levels in a Subset of HNLN Metastases for Genes Mapping to Frequent CNAs in HNLN Tumors.
| Gene | Mean | |||
| Primaries (n = 15) | HNLN8 | HNLN12 | HNLN13 | |
| TPD52 | 408.76 | 338.43 | 284.87 | 265.83 |
| NGFR | 38.72 | 42.54 | 28.30 | 35.25 |
| FABP5 | 671.08 | 2990.93 | 1776.89 | 35.46 |
| MYC | 231.68 | 342.6 | 1232.99 | 263.41 |
| PIAS3 | 93.93 | 55.54 | 38.79 | 76.69 |
| ARL2 | 53.46 | 186.49 | 92.27 | 32.53 |
| SNX15 | 84.2 | 95.27 | 86.63 | 62.86 |
| MEN1 | 113.92 | 225.55 | 155.01 | 120.52 |
| RPS3 | 5060.5 | 5855.31 | 5719.49 | 5644.25 |
| CBX4 | 51.82 | 59.67 | 43.51 | 60.83 |
| CBX8 | 26.35 | 43.25 | 28.32 | 35.30 |
Corresponding copy number data are also codified for reference. Gray represents copy number gain, and white indicates that normal copy number was observed. An expression level of ∼ 50 is considered normal.
Figure 4.
FABP5 TaqMan results for matched primary and HNLN cases. BPT = benign prostatic tissue. *A sample with aCGH copy number gain.
Discussion
Zitzelsberger et al. [3] performed CGH on a set of five HNLN metastases. A direct comparison with our aCGH results could not be made because CGH has a lower resolution. However, based on their published ideogram, they also observed gains involving 2p, 8q, 9q, 17, 20, and X, and losses involving 2p, 2q, 3p, 3q, 4q, 8p, 11q, 13q, and X (Figure 1).
The majority of frequently deleted metastasis-associated regions in our tumor set map to chromosome 8p, which has been documented to be involved in prostate cancer [6,8–12]. In a study of 64 primary tumors, we found 8p loss to be associated with advanced stage disease (pT ≥ 3) [6]. Tumor-node-metastasis stage ≥ 3 indicates extracapsular extension and invasion of seminal vesicles or adjacent structures [13].
When comparing expression levels in HNLNs to those in primary tumors, assuming that normal expression corresponds to a value of ∼ 50, copy number gain corresponded to increased expression for all genes except NGFR and chromobox homologs (Table 3). SNX15 showed only a slight increase in expression for the HNLN sample with copy number gain at that locus. Some genes were upregulated in primary tumors and HNLN metastatic tumors, namely, TPD52, FABP5, MYC, MEN1, and RPS3. MYC and FABP5 were the only genes queried that showed a large increase in expression for the HNLNs compared to high levels in primary tumors. We further validated FABP5 upregulation in matched primary HNLN cases (PR1263 and PR1264) using TaqMan (Figure 4). The HNLNs (HNLN8 and HNLN12) with high expression of TPD52 also had increased copy number at that locus. Interestingly, the nodal tumor HNLN13 did not show copy number gain at the TPD52 locus, but had increased expression of TPD52, suggesting an independent mechanism leading to aberrant TPD52 gene expression. This provides further evidence of the important role that TPD52 may play in cancer [14]. The trend of expression change without corresponding copy number change was also observed for RPS3, which has been shown to be upregulated in colon cancer [15], and for the ARL2 and SNX15 genes, which both map to the 11q13 CNA locus.
Several putative metastasis-related genes that were identified in this study and validated by expression studies have been previously reported to be involved in prostate cancer. Two independent expression studies focusing on FABP5 found it to be upregulated in prostate cancer relative to normal prostate [16,17]. In addition, a query of two whole transcriptome studies showed FABP5 to be upregulated in metastatic tumors (including lymph nodes) compared to primary and normal prostate tissues [2,18]. FABP5 has been proposed to be a metastasis-inducing gene [17,19]. Previous work has shown that FABP5 acts by upregulating vascular endothelial growth factor to induce invasion in vitro and metastasis in vivo [16,19]. The candidate oncogene TPD52 has been found to be upregulated in prostate cancer at DNA, RNA, and protein levels [14]. The ARL2 and SNX15 genes map ∼ 200 kb from MEN1. We previously reported MEN1 as a candidate oncogene in prostate cancer [6]. More specifically, copy number gain at the 11q13.1 locus associates with cases that fail biochemically following radical prostatectomy. MEN1 was found, with expression data, to be upregulated in three lymph nodes (HNLN8, HNLN12, and HNLN13), as well as in primary tumors. Only one of three nodes (HNLN8) had corresponding copy number gain. This stresses the importance of the role that MEN1 might play in prostate cancer. In addition, a query in the Oncomine expression database (http://www.oncomine.org) for upregulated genes provided similar results in Table 3. The genes FABP5, TPD52, MYC, RPS3, and SNX15 had expression data available for HNLNs. Similar to our findings, these genes showed a trend toward increased expression in HNLNs [2].
We recently reported a set of 39 biomarkers, which we termed GEMCaP, that corresponds to regions of the human genome that are more frequently altered in primary prostate tumors recurring after surgical removal, as well as in actual metastases [6]. GEMCaP consists of 16 amplified loci and 23 deleted loci. Four of the amplified GEMCaP loci were among the frequent CNAs in this study (Table 1). We have previously demonstrated that when > 20% of the GEMCaP loci are altered, patients are at high risk for recurrence following radical prostatectomy [20]. The eight HNLNs had copy number changes at 5%, 27%, 34%, 45%, 46%, 54%, 58%, and 60% of the GEMCaP loci. Therefore, seven of eight HNLNs fitted our criteria for an aggressive genotype. The entire GEMCaP results are supplied as Table W1. The whole genome aCGH profile for the HNLN sample with only a 5% GEMCaP score possessed primarily deletions. We have observed this genotype in other sets of primary and metastatic tumors (data not shown). This deletion genotype appears to represent a small fraction (< 10%) of prostate tumors. In our aCGH database query (Table 2), all four clones that were more frequently gained in metastatic tumors and primaries that progressed were GEMCaP clones. A similar analysis for deletions resulted in seven GEMCaP clones.
Table 2.
CNAs More Frequently Found in Primaries That Progressed, and Metastases Versus Nonprogressors.
| (A) | |||||||
| Clone Mapt 5.0 | Locus | Gene | Gains (%) | P | |||
| Primary Progressors (n = 12) | Nonprogressors (n = 32) | Harvard Metastases (n = 19) | HNLN (n = 8) | ||||
| RP11-1146E5* | 3q26.2 | EVI1 | 20 | 0 | 39 | 50 | .0003 |
| RP4-693L23* | 11p15.5 | p57 | 45 | 14 | 53 | 38 | .008 |
| RP11-46E14* | 17q25.3 | CBX8, CBX4 | 27 | 15 | 56 | 38 | .03 |
| RP11-114M1* | 3q26.32 | IRA1 | 45 | 13 | 42 | 50 | .01 |
| (B) | |||||||
| Clone Mapt 5.0 | Locus | Gene | Deletions (%) | P | |||
| Primary Progressors (n = 12) | Nonprogressors (n = 32) | Harvard Metastases (n = 19) | HNLN (n = 8) | ||||
| RP11-232J22* | 8p21.2 | BNIP3L | 27 | 11 | 53 | 75 | .0013 |
| CTD-2015D3* | 8p21.2 | ADRA1A | 25 | 17 | 63 | 57 | .0013 |
| RP11-57I3* | 8p12 | NRG1 | 36 | 6 | 56 | 71 | .0001 |
| RP11-262I23 | 8p11.22-p11.21 | INDO | 27 | 3 | 24 | 50 | .0038 |
| RP11-28L24* | 6q21 | LAMA4 | 36 | 7 | 35 | 25 | .015 |
| RP11-262B7 | 8p23.1 | Not in database | 25 | 13 | 35 | 88 | .0145 |
| CTD-2173J2* | 13q14.2 | RB | 25 | 3 | 61 | 38 | .0001 |
| CTD-2079J2* | 5q21.3 | FER | 45 | 13 | 29 | 71 | .0133 |
| RP11-73N22 | 5q23.1 | No gene | 36 | 8 | 21 | 63 | .0185 |
| RP11-115L24 | 5q21.2 | No gene | 45 | 14 | 33 | 75 | .0081 |
| RP11-76B12* | 8p21.2 | DOCK5 | 25 | 13 | 50 | 50 | .015 |
| RP11-70L1 | 8p21.2 | GNRH1, KCTD9 | 25 | 19 | 53 | 63 | .0228 |
| RP11-217H23 | 13q14.13 | GTF2F2 | 27 | 3 | 53 | 88 | .0001 |
| RP11-241I4 | 8p23.1 | MSRA | 30 | 12 | 37 | 88 | .0059 |
GEMCaP clone.
Our results show a high degree of concordance between the genomic copy number profiles of matched primary and metastasis cases. This is similar to what we observed in a cohort of unmatched primaries and metastatic tumors [6]. A CGH study by Zitzelsberger et al. [3] showed common changes for matched primaries and HNLN cases, but it only listed gains of 9q and 16 and loss of 13q as CNAs in common. Two previous reports that studied primaries and matched lymph node metastases by fluorescence in situ hybridization (FISH) reported that chromosomal changes in lymph node tumors resembled those from primary tumors [21,22]. Even though FISH studies included only selected regions on chromosomes 7, 8, 10, 12, and 17, this is in line with our observations.
This study emphasizes the importance of studying metastatic tumors, as well as primary tumors. Biomarkers of aggressive disease were found in HNLN metastatic lesions. Genomic profiles of matched primaries and lymph node metastases were found to be very similar. Studies such as these can highlight genomic changes in primary tumors that can be used for the early detection of aggressive tumors and, conversely, for the identification of patients who can safely elect active surveillance. In addition, these types of study can identify drug targets with associated biomarkers.
Supplementary Material
Acknowledgements
We would like to thank Lars Schmitt (UCSF Pathology Department) for assistance with UCSF cases, and Shivaranjani Sridharan and Yasuko Kobayashi for help with manuscript revisions.
Abbreviations
- FABP5
fatty acid-binding protein 5
- TPD52
tumor protein D52
- NGFR
nerve growth factor receptor
- MYC
myelocytomatosis viral oncogene homolog
- PIAS3
protein inhibitor of activated STAT protein 3
- ARL2
ADP ribosylation factor-like 2
- SNX15
sorting nexin 15
- MEN1
multiple endocrine neoplasia 1
- RPS3
ribosomal protein S3
- CBX8
chromobox homolog 8
- CBX4
chromobox homolog 4
Footnotes
This work was supported by the University of California at San Francisco Prostate Cancer SPORE, National Institutes of Health grant P50CA89520.
This article refers to supplementary material, which is designated by W (i.e., Table W1) and is available online at www.bcdecker.com.
References
- 1.Jemal A, Tiwari RC, Murray T, Ghafoor A, Samuels A, Ward E, Feuer EJ, Thun MJ. Cancer statistics, 2004. CA Cancer J Clin. 2004;54:8–29. doi: 10.3322/canjclin.54.1.8. [DOI] [PubMed] [Google Scholar]
- 2.Lapointe J, Li C, Higgins JP, van de Rijn M, Bair E, Montgomery K, Ferrari M, Egevad L, Rayford W, Bergerheim U, et al. Gene expression profiling identifies clinically relevant subtypes of prostate cancer. Proc Natl Acad Sci USA. 2004;101:811–816. doi: 10.1073/pnas.0304146101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zitzelsberger H, Engert D, Walch A, Kulka U, Aubele M, Hofler H, Bauchinger M, Werner M. Chromosomal changes during development and progression of prostate adenocarcinomas. Br J Cancer. 2001;84:202–208. doi: 10.1054/bjoc.2000.1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pinkel D, Segraves R, Sudar D, Clark S, Poole I, Kowbel D, Collins C, Kuo WL, Chen C, Zhai Y, et al. High resolution analysis of DNA copy number variation using comparative genomic hybridization to microarrays. Nat Genet. 1998;20:207–211. doi: 10.1038/2524. [DOI] [PubMed] [Google Scholar]
- 5.Paris PL, Albertson DG, Alers JC, Andaya A, Carroll P, Fridlyand J, Jain AN, Kamkar S, Kowbel D, Krijtenburg PJ, et al. High-resolution analysis of paraffin-embedded and formalin-fixed prostate tumors using comparative genomic hybridization to genomic microarrays. Am J Pathol. 2003;162:763–770. doi: 10.1016/S0002-9440(10)63873-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Paris PL, Andaya A, Fridlyand J, Jain AN, Weinberg V, Kowbel D, Brebner JH, Simko J, Watson JE, Volik S, et al. Whole genome scanning identifies genotypes associated with recurrence and metastasis in prostate tumors. Hum Mol Genet. 2004;13:1303–1313. doi: 10.1093/hmg/ddh155. [DOI] [PubMed] [Google Scholar]
- 7.Fridlyand J, Snijders AM, Pinkel D, Albertson DG, Jain A. Application of Hidden Markov Models to the analysis of the array CGH data. J Multivar Anal (Spec Genomic Issue) 2004;90:132–153. [Google Scholar]
- 8.Swalwell JI, Vocke CD, Yang Y, Walker JR, Grouse L, Myers SH, Gillespie JW, Bostwick DG, Duray PH, Linehan WM, et al. Determination of a minimal deletion interval on chromosome band 8p21 in sporadic prostate cancer. Genes Chromosomes Cancer. 2002;33:201–205. doi: 10.1002/gcc.10015. [DOI] [PubMed] [Google Scholar]
- 9.Oba K, Matsuyama H, Yoshihiro S, Kishi F, Takahashi M, Tsukamoto M, Kinjo M, Sagiyama K, Naito K. Two putative tumor suppressor genes on chromosome arm 8p may play different roles in prostate cancer. Cancer Genet Cytogenet. 2001;124:20–26. doi: 10.1016/s0165-4608(00)00248-x. [DOI] [PubMed] [Google Scholar]
- 10.Washburn JG, Wojno KJ, Dey J, Powell IJ, Macoska JA. 8pter-p23 deletion is associated with racial differences in prostate cancer outcome. Clin Cancer Res. 2000;6:4647–4652. [PubMed] [Google Scholar]
- 11.Macoska JA, Paris P, Collins C, Andaya A, Beheshti B, Chaib H, Kant R, Begley L, MacDonald JW, Squire JA. Evolution of 8p loss in transformed human prostate epithelial cells. Cancer Genet Cytogenet. 2004;154:36–43. doi: 10.1016/j.cancergencyto.2004.02.013. [DOI] [PubMed] [Google Scholar]
- 12.Tsuchiya N, Slezak JM, Lieber MM, Bergstralh EJ, Jenkins RB. Clinical significance of alterations of chromosome 8 detected by fluorescence in situ hybridization analysis in pathologic organ-confined prostate cancer. Genes Chromosomes Cancer. 2002;34:363–371. doi: 10.1002/gcc.10064. [DOI] [PubMed] [Google Scholar]
- 13.Hermanek P, Hutter RVP, Sobin LH. TNM Atlas IUAC. New York: Springer; 1997. [Google Scholar]
- 14.Rubin MA, Varambally S, Beroukhim R, Tomlins SA, Rhodes DR, Paris PL, Hofer MD, Storz-Schweizer M, Kuefer R, Fletcher JA, et al. Overexpression, amplification, and androgen regulation of TPD52 in prostate cancer. Cancer Res. 2004;64:3814–3822. doi: 10.1158/0008-5472.CAN-03-3881. [DOI] [PubMed] [Google Scholar]
- 15.Pogue-Geile K, Geiser JR, Shu M, Miller C, Wool IG, Meisler AI, Pipas JM. Ribosomal protein genes are overexpressed in colorectal cancer: isolation of a cDNA clone encoding the human S3 ribosomal protein. Mol Cell Biol. 1991;11:3842–3849. doi: 10.1128/mcb.11.8.3842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Adamson J, Morgan EA, Beesley C, Mei Y, Foster CS, Fujii H, Rudland PS, Smith PH, Ke Y. High-level expression of cutaneous fatty acid-binding protein in prostatic carcinomas and its effect on tumorigenicity. Oncogene. 2003;22:2739–2749. doi: 10.1038/sj.onc.1206341. [DOI] [PubMed] [Google Scholar]
- 17.Jing C, Beesley C, Foster CS, Rudland PS, Fujii H, Ono T, Chen H, Smith PH, Ke Y. Identification of the messenger RNA for human cutaneous fatty acid-binding protein as a metastasis inducer. Cancer Res. 2000;60:2390–2398. [PubMed] [Google Scholar]
- 18.Yu YP, Landsittel D, Jing L, Nelson J, Ren B, Liu L, McDonald C, Thomas R, Dhir R, Finkelstein S, et al. Gene expression alterations in prostate cancer predicting tumor aggression and preceding development of malignancy. J Clin Oncol. 2004;22:2790–2799. doi: 10.1200/JCO.2004.05.158. [DOI] [PubMed] [Google Scholar]
- 19.Jing C, Beesley C, Foster CS, Chen H, Rudland PS, West DC, Fujii H, Smith PH, Ke Y. Human cutaneous fatty acid-binding protein induces metastasis by up-regulating the expression of vascular endothelial growth factor gene in rat Rama 37 model cells. Cancer Res. 2001;61:4357–4364. [PubMed] [Google Scholar]
- 20.Paris PL, Weinberg V, Simko J, Andaya A, Albo G, Rubin MA, Carroll PR, Collins C. Preliminary evaluation of prostate cancer metastatic risk biomarkers. Int J Biol Markers. 2005;20:141–145. doi: 10.1177/172460080502000301. [DOI] [PubMed] [Google Scholar]
- 21.Qian J, Hirasawa K, Bostwick DG, Bergstralh EJ, Slezak JM, Anderl KL, Borell TJ, Lieber MM, Jenkins RB. Loss of p53 and c-myc overrepresentation in stage T(2–3)N(1–3)M(0) prostate cancer are potential markers for cancer progression. Mod Pathol. 2002;15:35–44. doi: 10.1038/modpathol.3880487. [DOI] [PubMed] [Google Scholar]
- 22.Alcaraz A, Corral JM, Ribal MJ, Mallofre C, Mengual L, Carrio A, Gil-Vernet Sedo JM, Villavicencio H. Fluorescence in situ hybridization analysis of matched primary tumour and lymph-node metastasis of D1 (pT2–3pN1M0) prostate cancer. BJU Int. 2004;94:407–411. doi: 10.1111/j.1464-410X.2004.04829.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.