Abstract
The importance of genetic mutations in carcinogenesis has been recognized, and it has been proposed that aberrant mutation of mRNA may represent a novel oncogenic principle. Here we report that the mRNA of a homeobox gene prox1, a candidate tumor suppressor, suffers adenosine-to-inosine nucleotide conversion and loses tumor-suppressive functions in a subset of human cancers. Expression of Prox1 was reduced in pancreatic cancers, and the extent of reduction correlated with progression of tumor differentiation. A-to-G base change was found in prox1 cDNA taken from human cancer cells, but not in corresponding genomic DNA. We mapped four common mutation sites in prox1 gene, and the same four sites were mutated in human clinical samples from several cancers. Tetracycline-induced wild-type (wt) Prox1 in tumor cells inhibited transforming activity and cellular proliferation. However, mutant Prox1 with the four common sites altered from A to G lost these inhibitory functions. In mice, xenografts of tumor cells with tetracycline-induced wt-Prox1 formed tumor masses significantly more slowly than control tumors, whereas mutated Prox1 had no effect. These findings may point to a pivotal role of the RNA mutation of prox1 gene in the pathogenesis of human cancer progression.
Keywords: RNA mutation, tumor-suppressor gene, prox1, cancer, asymmetrical cell division
Introduction
Cancer is essentially a genetic disease of somatic cells [1]. According to the currently accepted model of carcinogenesis, a series of mutations in coding regions of oncogenes or tumor-suppressor genes is required for cancer development. In the model, genomic DNA sequence with mutations is transcribed to mRNA that is finally translated into a functionally aberrant protein, leading to deregulation of fundamental cellular processes. Besides genetic mutations, several epigenetic events, such as methylation of promoters and histone acetylation, are known to affect targeted gene expression and, thus, to quantitatively modify the level of functional proteins [2]. In all these scenarios, mRNA is viewed as an intermediate between DNA and protein [3]. However, it has been recently revealed that mRNA is actively regulated by a variety of machineries playing roles at the level of mRNA processing, mRNA stabilization, and gene transcription, such as nonsense-mediated decay (NMD), RNA interference, and RNA mutation, as well as alternative splicing. Recent reports suggest that functional deregulation of these RNA-based mechanisms may be involved in carcinogenesis. NMD is shown to degrade BRCA1 mRNA bearing nonsense mutations [4]. Ras genes contain multiple binding sites for the let-7 family of microRNA, and the expression of let-7 and Ras is inversely correlated in human lung cancers [5].
The homeobox gene prox1 is related to the Drosophila prospero gene, which mediates cell fate decisions of neuroblasts [6]. Analysis of Prox1-deficient mice indicated its various role in lens fiber elongation, hepatocyte migration, development of lymphatic vessels, retinal cell differentiation, and pancreatic development. In pancreatic development, lack of Prox1 activity disrupts epithelial pancreatic morphology, hinders pancreatic growth before E11.5, and decreases the formation of islet cell precursors after E13.5 [7]. Prox1 is considered to contribute to the allocation of an adequate supply of islet cells throughout pancreatic ontogeny by preventing exocrine cell differentiation of multipotent pancreatic progenitors, whereas the role of Prox1 in the pancreas of adults remains unknown. Prox1 is also required for the proliferation and migration of hepatoblasts in liver development [8]. However, cells lacking Prox1 are less likely to stop dividing, and ectopic expression of Prox1 forces progenitor cells to exit the cell cycle in the retina [9] and causes abnormal cellular proliferation by a downregulated expression of cell cycle inhibitors in lens fibers [10]. The role of Prox1 is very multifunctional, and its physiological functions may change according to developmental stage, organ, or type of cancer.
We have recently reported that there is a significant correlation between Prox1 expression and the differentiation scores of hepatocellular carcinoma (HCC) [11]. Low expression of Prox1 in tumors is closely associated with poor prognosis. Specific knockdown of prox1 by RNA interference strongly accelerates in vitro cell growth, whereas overexpression of Prox1 greatly suppresses growth. These results suggest that Prox1 is involved in the differentiation and progression of HCC and, thus, may be a candidate tumor-suppressor gene for HCC.
In this study, we report our finding that prox1 suffers adenosine-to-inosine (A-to-I) RNA mutation without genomic mutation in a subset of human cancer cells. RNA mutation is a mechanism capable of changing genetic information without affecting genomic DNA. A-to-I nucleotide conversion might change the coding potential of mRNA and might result in the synthesis of an isoform of the protein, which is not predictable from unaffected genomic DNA. Analyzing the effect of RNA mutation on protein function, we found that wild-type (wt) Prox1 suppressed tumor cell proliferation in vitro and in vivo, and that the mutant form of Prox1 lost its suppressive effect. prox1 genomic mutation and methylation have been identified in hematologic cell lines [12], and our study suggests that RNA mutation, as a new mechanism of tumorigenesis, may induce or alter tumor progression.
Materials and Methods
Cells and cDNA Libraries
Miapaca2 and Panc1 are human pancreatic cancer cell lines with relatively poor differentiation, HCT116 is a human colon cancer cell line, and human embryonic kidney 293 cells are used as controls. All four cell lines were cultured in DMEM supplemented with 10% fetal calf serum (JRH Biosciences, Lenexa, KS). Human cDNA libraries of the brain, cerebellum, lung, heart, esophagus, small intestine, colon, liver, pancreas, spleen, kidney, testis, and ovary were purchased from Biochain Institute (Hayward, CA).
Patient Samples
Sixty paraffin-embedded formalin-fixed tissues of pancreatic cancers were retrieved from the 2000 to 2004 surgical pathology files of Kyoto University Hospital (Kyoto, Japan). Frozen surgical tissue samples from 9 cases of esophageal cancer, 24 cases of pancreatic cancer, and 5 cases of colon cancer were obtained for the preparation of total RNA and genomic DNA. Total RNA and genomic DNA were isolated sequentially using ISOGEN (Nippon Gene, Tokyo, Japan) according to the manufacturer's instructions. All samples were obtained with informed consent, and their use was approved by the ethics committee of the hospital.
Immunohistochemistry
Paraffin-embedded sections were deparaffinized and hydrated by standard methods. After antigen retrieval with microwave (600 W for 5 minutes; 200 W for 10 minutes) in sodium citrate, endogenous peroxidase was quenched with 3% H2O2 in methanol for 10 minutes. The sections were placed in TNB buffer (TSA Biotin System kit/NEL700; Perkin Elmer Life Science, Wellesley, MA) for 30 minutes to block nonspecific hybridization, after which they were exposed to 1:100 rabbit anti-Prox1 antibody (ReliaTech GmbH, Braunschweig, Germany) in TNB for 16 hours at 4°C, washed with TNT buffer, and incubated for 30 minutes at room temperature with biotinylated secondary antibodies (Dako, Carpinteria, CA) diluted in TNB. Tyramide signal amplification was used to enhance staining. Peroxidase activity was developed with EnVision kit/HRP (DAB; Dako), and sections were counterstained with hematoxylin. In negative control stains, primary antibodies were omitted. Staining intensity was estimated for each cell type on a three-step scale (-, +, and ++) by three investigators.
Genomic DNA and RNA Isolation, and Reverse Transcription-Polymerase Chain Reaction (RT-PCR)
Genomic DNA were isolated using QIAamp DNA Mini Kit (Qiagen, Valencia, CA) according to the manufacturer's instructions. Total RNA from cultured cell lines was isolated with TRIzol Reagent (Gibco BRL, Grand Island, NY), subsequently digested with RNase-free DNase I (Roche, Indianapolis, IN), and tested for integrity. Ten micrograms of total RNA was used to synthesize cDNA by using SuperScript II reverse transcriptase (Invitrogen, Carlsbad, CA) and oligo-dT primers. Four sets of PCR primers intended to cover the entire coding region of prox1 gene were used for sequencing. Primer sequences were as follows: set 1, 5′-ATGCCTGACCATGACAGCAC-3′ (forward) and 5′-TTTCATTGCCCCTTAATGCC-3′ (reverse); set 2, 5′-TAATTCGGGGTATGAGCCAT-3′ (forward) and 5′-TCTGGCCTGGGGGATCTG-3′ (reverse); set 3, 5′-CAGGTTCCTCAGGTCTTC-3′ (forward) and 5′-CTTCCTGCATTGCACTTCC-3′ (reverse); set 4, 5′-CATCTCACCACCTGAGCC-3′ (forward) and 5′-CTACTCATGAAGCAGCTCTTG-3′ (reverse). PCR conditions were 94°C for 3 minutes, followed by 30 cycles of 94°C for 30 seconds, 60°C for 30 seconds, and 72°C for 1 minute; and a final extension step of 72°C for 7 minutes. For analysis of Prox1 expression, its 3′ region was amplified with primers 5′-CAGATGGAGAAGTACGCAC-3′ (forward) and 5′-CTACTCATGAAGCAGCTCTTG-3′ (reverse) and, as control, glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA was amplified with primers 5′-GACAACAGCCTCAAGATCATCA-3′ (forward) and 5′-GGTCCACCACTGACACGTTG-3′ (reverse). Both reactions involved initial denaturation at 94°C for 3 minutes, followed by 25 to 30 cycles at 94°C for 30 seconds, 57°C for 30 seconds, and 72°C for 30 seconds (for prox1); or 18 cycles at 94°C for 30 seconds, 58°C for 30 seconds, and 72°C for 30 seconds (for GAPDH), with a final extension step of 72°C for 7 minutes.
Sequence Analysis
All PCR products were first directly sequenced without cloning using the ABI PRISM Big Dye Terminator Cycle Sequencing Ready Reaction (Applied Biosystems, Foster City, CA) by ABI PRISM 3100 sequence analyzer (Applied Biosystems). PCR products were run on 2% agarose gels and, if a single clear band of the correct approximate size was obtained, it was excised or purified by QIAquick PCR purification kit (Qiagen). When PCR products were directly sequenced, the extent of mutation at each site was determined with electropherograms from sequencing reactions to estimate the relative amounts of A and G semiquantitatively: (-) no trace of G; (+) presence of a trace of G but less than A; (++) trace of G more than A; (+++) no trace of A. Subsequently, PCR products were cloned using TOPO TA cloning kits (Invitrogen), and editing efficiency was estimated by evaluating the sequence of at least 10 clones.
Plasmids, Transfection, and Cell Proliferation
cDNA of wt-prox1 or mutant (mut) prox1 were subcloned into the mammalian expression vector pcDNA6/TO (tet operator; Invitrogen). Both 293 and Miapaca2 cells expressing tet repressor were transfected with wt-prox1 or mut-prox1 construct by using Lipofectamine (Gibco BRL) following the manufacturer's instructions. Stable clones were obtained by selecting for blasticidin and zeocin resistance and were further confirmed to induce wt-prox1 or mut-prox1 expression in the presence of tetracycline (0.01 µg/ml for 293 cells; 0.2 µg/ml for Miapaca2 cells). Cell proliferation assays were performed by cell counts and colony formation assay. For colony formation assay, 293 cells were seeded into 10-cm culture plates at 5 x 105 cells/plate, cultured for 10 days in the presence of tet, and stained with Giemsa solution. Tet-inducible 293/LacZ or Miapaca2/LacZ cells were used as negative controls.
Transformation Assay
For cell transformation assay, tet-inducible Miapaca2 cells were seeded into 0.4% top agar layer of 10-cm culture plates with 0.8% base agar layer at 10,000 cells/plate and cultured for 21 days in the presence of 0.2 µg/ml tet. The cells were stained with cell stain solution according to the manufacturer's instructions.
In Vivo Study
Mice were injected subcutaneously with tet-inducible Miapaca2 cells at 1 x 107 cells/mouse. They were maintained under a continuous administration of 0.4 mg/ml doxycycline (Dox) in drinking water for 7 weeks. Tumor size was measured weekly using the formula width2 x length/2 and was finally weighed.
Results
Expression of Prox1 Is Downregulated in Human Pancreatic Cancer and Correlates with Its Differentiation
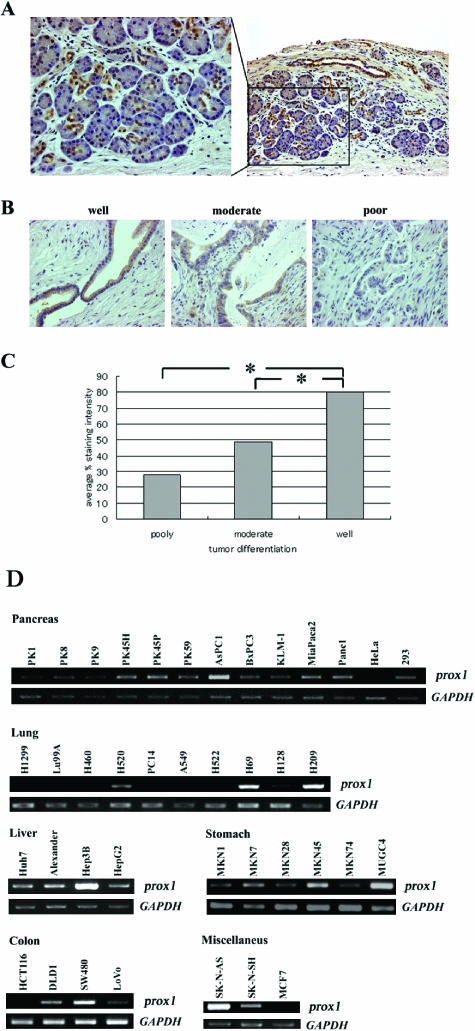
To test whether Prox1 protein is associated with pancreatic tumor progression, we evaluated Prox1 expression in 60 cancerous pancreatic tissues of various grades by immunostaining. Prox1 was expressed exclusively by ductal cells in normal tissues adjacent to cancerous regions (Figure 1A), and staining levels were in proportion to the differentiation grades of cancer cells (Figure 1, B and C). On screening various types of human cancer cell lines for prox1 expression by RT-PCR, although the distribution of prox1 expression reported previously was restricted, detectable levels were found in most types of tumor cell lines in various organs: 11, pancreatic cancer; 3, small cell lung cancer; 4, HCC; 6, gastric cancer; 4, colon cancer; 2, neuroblastoma; 1, breast cancer (Figure 1D).
Figure 1.
Expression of Prox1 is downregulated in human pancreatic cancer and correlates with its differentiation. (A) Immunostaining of normal pancreatic tissue. Prox1 expression is localized in ductal cells. (B) Immunostaining of pancreatic cancers of various grades: well-differentiated (well), moderately differentiated (moderate), and poorly differentiated (poor). (C) Staining intensity for Prox1 was estimated for each cell type. Columns show the relationship between tumor differentiation level and the average percent staining intensity for prox1. *P < .05. (D) Expression and mRNA editing of prox1 in diverse types of human cancer cell lines. RT-PCR analyses of prox1 expression in human pancreatic cancer cell lines: HeLa and 293 cells (pancreas), human lung cancer cell line (lung), human HCC (liver), human gastric cancer cell lines (stomach), human colon cancer cell lines (colon), and other types of human cancer cell lines (miscellaneous). GAPDH was used as internal control.
prox1 mRNA Mutation in Human Cancer Cells
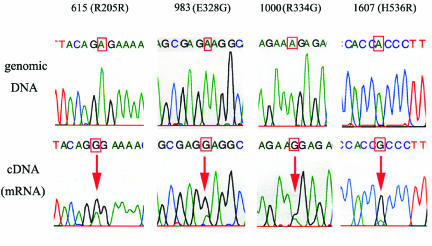
To investigate mutations in coding regions of prox1, cDNA obtained from cancer cell lines were amplified by PCR and sequenced as a population without cloning. We compared the sequences with the published human genomic sequence and found that electropherograms revealed the presence of an unambiguous trace of guanosine in positions for which published data clearly indicated the presence of an adenosine in Panc1, Miapaca2, and HCT116 cells at nucleotide positions 615, 983, 1000, and 1607 (NM002763; CDS 273-2486) (Figure 2). Three of four mutations identified, at nucleotide positions 983 (E328G), 1000 (R334G), and 1607 (H536R), generate amino acid changes in translation products. The four sites did not correspond with known single-nucleotide polymorphisms (SNPs) of genomic origin. We then sequenced matching genomic DNA samples retrieved from the same tumor cell lines and confirmed that genomic DNA sequences exhibit only adenosine signals (Figure 2). The RNA mutations identified are not seen in any cDNA library and publicly available expressed sequence tags covering this gene. Finally, we analyzed human cDNA libraries obtained from the brain, cerebellum, lung, heart, esophagus, small intestine, colon, liver, pancreas, spleen, kidney, testis, and ovary, but found no signs of A-to-G mutation (data not shown).
Figure 2.
Matching genomic DNA and cDNA sequences for Miapaca2 cells. The mutated site is characterized by a trace of guanosine in cDNA sequence, where genomic DNA sequence exhibits only adenosine signals (indicated by arrows).
Characterization of Mutation Sites in prox1 Transcript
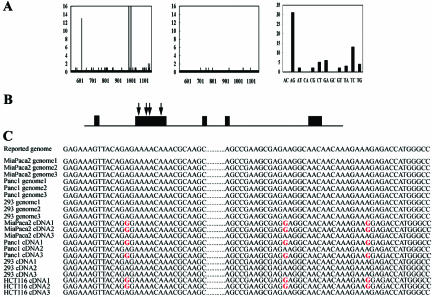
To further analyze mutation sites, PCR products were cloned and at least 10 individual clones were sequenced. Notably, inosine is recognized by translational apparatus as guanosine, so that the change of A-to-G in cDNA implicates A-to-I conversion in mRNA. The frequency of A-to-I mutation was validated for Panc1, Miapaca2, HCT116, and 293 cells. Set 2 primers (see Materials and Methods section) were used to amplify the region covering the first three mutations (R205R, E328G, and R334G). Comparing cDNA sequences with the published genomic sequence revealed a high number of nucleotide discrepancies in three restricted sites (Figure 3A, left). However, the sequences of cloned genomic fragments harbored nucleotide mismatches at random sites, most probably due to sequencing error (Figure 3A, center). The number of A-to-G mismatches between the cDNA sequence and the published genomic sequence is higher than all the other 11 types of nucleotide discrepancies (Figure 3A, right). Excess exclusively in the number of A-to-G discrepancies over other base changes most probably reflects cases of bona fide A-to-I RNA mutation. The same results were found in the other mutation (H536R) covered by set 3 primers (data not shown). Consequently, no A-to-G conversion was found in 293 cells, whereas four common mutation sites were found in Panc1, Miapaca2, and HCT116 cells (Figure 3, B and C). All four sites were within the N-terminal domain of the prox1 gene, which has not yet been well characterized (Figure 3B), but interestingly, adenosine residues at these positions are all conserved in a number of prox1 homologues from different species (data not shown). Common multiple base changes within a stretch of several hundred nucleotides, all being of the A-to-G type, are not likely accounted for by SNPs or sequencing errors. They are flanked by evolutionarily conserved regions, and these nucleotides may have been passed on to all descendants in the course of evolution because they were somehow important for the function of protein products.
Figure 3.
Characterization of mutation sites in prox1. (A) Evaluation of the sequences of cloned PCR products for the prox1 gene by set 2 primers (see Materials and Methods section) from Panc1, Miapaca2, and HCT116 cells. The number of nucleotide discrepancies between published human genomic sequences and the sequences of cloned PCR products of cDNA (left) or genomic DNA (center) obtained from Panc1, Miapaca2, and HCT116 cells. Distribution of mismatches between cDNA sequences and published genomic sequences (right). (B) Schematic view of prox1 gene with predicted mutation sites (indicated by arrows) in the coding region (box: exon). (C) Sequences of individually cloned fragments were aligned to the published human genomic sequence. Mutations identified are indicated in red.
Frequency of the RNA Mutation of prox1 in Cancer Cell Lines and Clinical Samples
The overall frequency of mutation in Panc1, Miapaca2, and HCT116 cells was 46%, 56%, and 62%, respectively, as evaluated from cloned sequences, whereas the mutation levels of other cancer cells were negative, as evaluated semiquantitatively by electropherograms (data not shown). A-to-G base change was not found in corresponding genomic DNA from any of the clones sequenced. Furthermore, we found that the same four sites mutated in human clinical samples taken from pancreatic, esophageal, and colon cancers (Table 1). RNA mutation events were observed in four of nine cases of esophageal cancers (44%), although the frequency of mutation in each case varied. A-to-G conversion was also identified in 2 of 24 cases of pancreatic cancer (8%) and in 2 of 5 cases of colon cancer (40%).
Table 1.
The Frequency of A-to-I RNA Mutation in cDNA and Genomic DNA Obtained from Human Samples of Pancreatic, Esophageal, and Colon Cancers.
| Patient Number | cDNA | Genomic DNA |
| Pancreatic cancer | ||
| 5 | 50% (2/4) | 0% (0/4) |
| 24 | 100% (4/4) | 0% (0/4) |
| Esophageal cancer | ||
| 5 | 29% (4/14) | 0% (0/7) |
| 6 | 30% (10/33) | 0% (0/8) |
| 7 | 7% (1/14) | 0% (0/8) |
| 8 | 75% (3/4) | 0% (0/8) |
| Colon cancer | ||
| 1 | 11% (1/9) | ND |
| 2 | 11% (1/9) | ND |
ND, not determined.
Prox1 Functions as Tumor Suppressor, and Mutated Prox1 Loses Its Function In Vitro
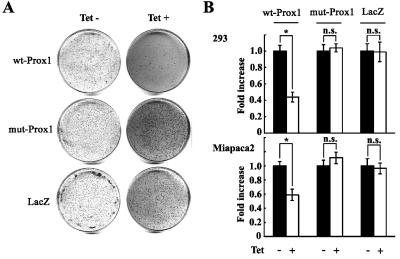
To explore the impact of RNA mutations on tumorigenesis, we expressed wt-Prox1 or mut-Prox1 with the four common sites mutated in 293 and Miapaca2 cells using the tet-on gene expression system and then assessed cellular behavior. We found, by transformation assay, that the suppression of colony formation in soft agar by tetracycline-induced wt-Prox1 was evident in Miapaca2 cells (Figure 4A). wt-Prox1 in both 293 and Miapaca2 cells also inhibited cell proliferation (Figure 4B). However, overexpression of mut-Prox1 lost these suppressive functions (Figure 4, A and B).
Figure 4.
Prox1 functions as a tumor suppressor, and mutated Prox1 loses its function in vitro. (A) Cell transformation assay in Miapaca2 cells with tet-induced wt-Prox1, mut-Prox1, and LacZ. Cells from day 21 after incubation without (left) or with (right) tet are shown. (B) Cell proliferation was suppressed significantly by overexpression of wt-Prox1, but not of mut-Prox1. Graphs show the fold increase of cell numbers (means and SD) for 293 and Miapaca2 cells with tet-induced wt-Prox1, mut-Prox1, and LacZ compared with control cells without tet induction.
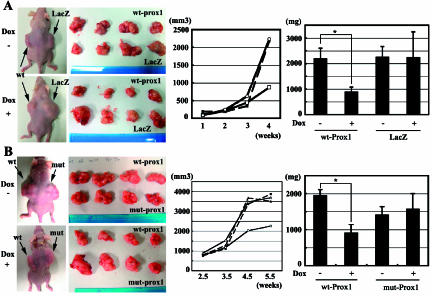
Prox1, But Not mut-Prox1, Suppresses the Growth of Xenografted Tumors in Mice
To investigate Prox1's suppressive effect on tumor formation and the loss of function by mutation, we grafted tetinducible derivatives of Miapaca2 cells (wt-Prox1, mut-Prox1, and LacZ) subcutaneously at both sides of the back of nude mice (Figure 5A, left). wt-Prox1-overexpressing tumors in mice treated with Dox grew more slowly than both LacZ controls in mice with Dox and wt-Prox1 tumors in mice without Dox (Figure 5). However, mut-Prox1 tumors in mice treated with Dox completely lost their suppressive activity on tumorigenesis (Figure 5B).
Figure 5.
Prox1, but not mut-Prox1, suppresses the growth of xenografted tumors in mice. (A) Macroscopic views reveal that Miapaca2 cells with tet-induced wt-Prox1 grow more slowly than control tumors (left). Growth dynamics of Miapaca2 cells estimated by the formula: tumor width2 x length, expressing wt-Prox1 or LacZ (center). (■) wt-Prox1, Dox-; (□) wt-Prox1, Dox+; (●) LacZ, Dox-; (○) LacZ, Dox+. Tumor weight (mg) measured after the sacrifice of mice proves that Dox-induced wt-Prox1 suppresses tumor growth (right). *P < .05. (B) Dox-induced mut-Prox1 loses the suppressive effect of Prox1 on tumorigenesis (left). Growth dynamics of Miapaca2 cells with Dox-induced mut-Prox1 shows loss of the suppressive function of tumor formation (center). (■) wt-Prox1, Dox-; (□) wt-Prox1, Dox+; (▲) mut-Prox1, Dox-; (△) mut-Prox1, Dox+. Tumor weight (mg) shows that mut-Prox1 loses the suppressive function of tumor formation (right). *P < .05.
Discussion
Based on discrepancies between mRNA (cDNA) and genomic sequences, we have identified RNA mutations of a novel candidate tumor-suppressor gene prox1 in a subset of human cancer cells. wt-Prox1 suppressed tumor cell proliferation in vitro and in vivo, and the mutant form of Prox1 lost its function, implicating that RNA mutations, rather than genomic mutations, could induce or alter tumor progression. To our knowledge, this is the first demonstration of RNA mutation of a transcriptional factor that may be involved in oncogenesis. Although RNA mutation found in malignant tissues may be a cause or a consequence of carcinogenesis, mutations of the recoding process of RNA processing, particularly when it targets functionally critical residues in proteins, may be a source of diversity in disease. As an unfortunate side effect of this diversity, aberrant RNA mutation might lead to unpredicted base modifications and might cause epigenetic instability in cancer.
We have recently reported that Prox1 negatively regulates tumor progression in HCC [11]. A transient knockdown of prox1 by siRNA significantly accelerated the growth of HCC cell lines in vitro, and we also demonstrated that overexpression of Prox1 resulted in suppression of cell proliferation. Clinically, a lower expression level of Prox1 corresponded to poorer differentiation of HCC and to poor prognosis for patients with HCC. In this study, we have demonstrated that Prox1 overexpression using tet-on system conferred a slower growth phenotype to pancreatic cancer cell lines and enabled them to form much smaller tumors in nude mice. Expression of Prox1 is downregulated in human pancreatic cancer and correlates with its differentiation. These data support novel functions for Prox1 as a tumor suppressor or as a factor to regulate the progression of the malignant character of cancer cells.
Drosophila neuroblasts divide asymmetrically to produce differentiated cells as a result of the asymmetrical localization of cell fate determinants and cortical cell polarity determinants, including the Prox1 homologue prospero, as well as Numb, Partner of Inscuteable (PINS), and aPKC [13]. It is known that the disruption of the machinery regulating asymmetrical cell division in neuroblasts leads to symmetrical cell division and, consequently, to tumor formation. Cells lacking PINS, Numb, or prospero are tumorigenic, and neuronspecific expression of a constitutive active variant of aPKC causes an increase in dividing neuroblasts [14]. Consistent with the tumorigenic potential in Drosophila, aPKC has been identified as a tumor suppressor in human lung cancers [15]. In fact, these molecules could protect against tumorigenesis through mechanisms independent of their roles in cell polarity in humans. Although it is not known whether Prox1 regulates asymmetrical division by stem cells in the liver or in the pancreas, it is intriguing that Prox1 has been also identified as a tumor suppressor in human cancers.
Posttranscriptional RNA editing is a process by which the nucleotide sequence of a nuclear mRNA is changed from that encoded in genomic DNA. RNA editing occurs through base modification, by deamination of cytidine (C) to uridine (U) or by deamination of adenosine (A) to inosine (I), in nuclear mRNA. Uridine and inosine are recognized by translational apparatus as thymidine and guanosine, respectively, so the net effects are changes of C-to-T and A-to-G. A-to-I RNA editing is mediated by members of the ADAR (adenosine deaminases acting on RNA) family [16]. Until recently, only a limited number of human editing substrates with an editing site in coding regions were known, including brain-specific glutamate receptor and G-protein-coupled serotonin receptors (5-HT2CR) [17]. Abundant A-to-I changes in mRNA prompted us to validate prox1 as a potential substrate of A-to-I RNA editing. We investigated whether prox1 could be a new target for ADARs, but there was no relationship between ADAR1 or ADAR2 expression and A-to-I mutations for prox1 gene in cancer cell lines (unpublished data). Auxiliary protein may be needed for supporting the full activity of ADARs. Alternatively, a different machinery may be responsible for A-to-I RNA mutation of the prox1 gene because the cluster of editing sites lacks Alu repeats, where A-to-I editing in humans has been known primarily to occur. Indeed, a recent report suggests that the transcription repressor gene PRDM1 is a target of RNAmutation, but the type of nucleotide conversions (G→A, U→A) identified in their studies does not involve the deamination process involved in A-to-I conversions mediated by ADARs [18]. Another example of G→A RNA mutation has been reported in hnRNP K protein mRNA in colorectal adenocarcinoma, and the mechanism of RNA mutation is believed to be carried out by yet unknown factors [19]. In human immunovirus-1 mRNA, which undergoes G→A mutation isolated from chronically infected H9 cells, no consensus was detected in the sequence surrounding mutation sites, and there were no consistent secondary structures evident in these regions [20].
Evidence of RNA editing as a potential oncogenic principle has come from limited tumor types, such as malignant neurofibromas in neurofibromatosis type I (NF1) and Wilms tumors [21]. C-to-U RNA editing of the tumor-suppressor NF1 gene and of the Wilms tumor susceptibility gene WT1 was found in tumor samples of patients. A-to-I RNA hyperediting of the hematopoietic cell phosphatase (PTPH6) gene in acute myeloid leukemia may contribute to the decrease in wt protein levels, suggesting its involvement in leukemogenesis [22]. In transgenic animals of APOBEC-1, the catalytic subunit of the apoB mRNA editing enzyme complex, the hepatic overexpression of APOBEC-1 mediated by the promoter of apolipoprotein E causes hepatocellular dysplasia and carcinoma [23]. NAT1 (novel APOBEC-1 target 1), whose hyperediting was identified in APOBEC-1 transgenic animals, was postulated to contribute to oncogenesis [24]. However,RNA editing of NAT1 mRNA or NF1 has not yet been identified in any natural human carcinoma [25].
In conclusion, the finding of RNA mutation in prox1 may challenge the currently accepted model of carcinogenesis requiring mutations in DNA. RNA editing (of which cellular mechanisms are currently being discovered) or RNA mutation could be an important prerequisite for potential deregulation during cancer development. Because the genome is intact, RNA mutations may be repaired if the mechanisms underlying mutation are identified, and this report would greatly contribute to the provision of new drugs and novel therapies for cancer patients.
Acknowledgements
We thank J. Yamashita for helpful discussions and M. Mizutani for technical assistance.
Footnotes
This work was supported by grants from the Ministry of Education, Culture, Sports, Science, and Technology, the Special Coordination Funds for Promoting Science and Technology, the Ichiro Kanehara Foundation, the Takeda Science Foundation, and the Princess Takamatsu Cancer Research Fund.
Meiko Takahashi and Takanobu Yoshimoto contributed equally to this work.
References
- 1.Vogelstein B, Kinzler KW. Cancer genes and the pathways they control. Nat Med. 2004;10:789–799. doi: 10.1038/nm1087. [DOI] [PubMed] [Google Scholar]
- 2.Murrel A, Rakyan VK, Beck S. From genome to epigenome. Hum Mol Genet. 2005;14:R3–R10. doi: 10.1093/hmg/ddi110. [DOI] [PubMed] [Google Scholar]
- 3.Scholzova E, Malik R, Sevcik J, Kleibl Z. RNA regulation and cancer development. Cancer Lett. 2006 doi: 10.1016/j.canlet.2006.03.021. May 2; [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 4.Perrin-Vidoz L, Sinilnikova OM, Stoppa-Lyonnet D, Lenoir GM, Mazoyer S. The nonsense-mediated mRNA decay pathway triggers degradation of most BRCA1 mRNAs bearing premature termination codons. Hum Mol Genet. 2002;11:2805–2814. doi: 10.1093/hmg/11.23.2805. [DOI] [PubMed] [Google Scholar]
- 5.Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, Labourier E, Reinert KL, Brown D, Slack FJ. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 6.Hong YK, Detmar M. Master regulator of the lymphatic vasculature phenotype. Cell Tissue Res. 2003;314:85–92. doi: 10.1007/s00441-003-0747-8. [DOI] [PubMed] [Google Scholar]
- 7.Wang J, Kilic G, Aydin M, Burke Z, Oliver G, Sosa-Pineda B. Prox1 activity controls pancreas morphogenesis and participates in the production of “secondary transition” pancreatic endocrine cells. Dev Biol. 2005;286(1):182–194. doi: 10.1016/j.ydbio.2005.07.021. [DOI] [PubMed] [Google Scholar]
- 8.Sosa-Pineda B, Wigle JT, Oliver G. Hepatocyte migration during liver development requires Prox1. Nat Genet. 2000;25:254–255. doi: 10.1038/76996. [DOI] [PubMed] [Google Scholar]
- 9.Dyer MA, Livesey FJ, Cepko CL, Oliver G. Prox1 function controls progenitor cell proliferation and horizontal cell genesis in the mammalian retina. Nat Genet. 2003;34(1):53–58. doi: 10.1038/ng1144. [DOI] [PubMed] [Google Scholar]
- 10.Wigle JT, Chowdhury K, Gruss P, Oliver G. Prox1 function is crucial for mouse lens-fibre elongation. Nat Genet. 1999;21:318–322. doi: 10.1038/6844. [DOI] [PubMed] [Google Scholar]
- 11.Shimoda M, Takahashi M, Yoshimoto T, Kono T, Ikai I, Kubo H. A homeobox protein prox1 is involved in the differentiation, proliferation and prognosis in hepatocellular carcinoma. Clin Cancer Res. 2006;12:6005–6011. doi: 10.1158/1078-0432.CCR-06-0712. [DOI] [PubMed] [Google Scholar]
- 12.Nagai H, Li Y, Hatano S, Toshihito O, Yuge M, Ito E, Utsumi M, Saito H, Kinoshita T. Mutations and aberrant DNA methylation of the PROX1 gene in hematologic malignancies. Genes Chromosomes Cancer. 2003;38(1):13–21. doi: 10.1002/gcc.10248. [DOI] [PubMed] [Google Scholar]
- 13.Morrison S, Kimble J. Asymmetric and symmetric stem-cell divisions in development and cancer. Nature. 2006;441:1068–1074. doi: 10.1038/nature04956. [DOI] [PubMed] [Google Scholar]
- 14.Caussinus E, Gonzalez C. Induction of tumor growth by altered stem-cell asymmetric division in Drosophila melanogaster. Nat Genet. 2005;37:1125–1129. doi: 10.1038/ng1632. [DOI] [PubMed] [Google Scholar]
- 15.Regala RP, Weems C, Jamieson L, Khoor A, Edell ES, Lohse CM, Fields AP. Atypical protein kinase C iota is an oncogene in human non-small cell lung cancer. Cancer Res. 2005;65:8905–8911. doi: 10.1158/0008-5472.CAN-05-2372. [DOI] [PubMed] [Google Scholar]
- 16.Wang Q, Khillan J, Gadue P, Nishikura K. Requirement of the RNA editing deaminase ADAR1 gene for embryonic erythropoiesis. Science. 2000;290:1765–1768. doi: 10.1126/science.290.5497.1765. [DOI] [PubMed] [Google Scholar]
- 17.Higuchi M, Single FN, Kohler M, Sommer B, Sprengel R, Seeburg PH. RNA editing of AMPA receptor subunit GluR-B: a basepaired intron-exon structure determines position and efficiency. Cell. 1993;75:1361–1370. doi: 10.1016/0092-8674(93)90622-w. [DOI] [PubMed] [Google Scholar]
- 18.Tam W, Gomez M, Chadburn A, Lee JW, Chan WC, Knowles DM. Mutational analysis of PRDM1 indicates a tumor suppressor role in diffuse large B-cell lymphomas. Blood. 2006;107:1090–1100. doi: 10.1182/blood-2005-09-3778. [DOI] [PubMed] [Google Scholar]
- 19.Klimek-Tomczak K, Mikula M, Dzwonek A, Paziewska A, Karczmarski J, Hennig E, Bujnicki JM, Bragoszewski P, Denisenko O, Bomsztyk K, et al. Editing of hnRNP K protein mRNA in colorectal adenocarcinoma and surrounding mucosa. Br J Cancer. 2006;94:586–592. doi: 10.1038/sj.bjc.6602938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bourara K, Litvak S, Araya A. Generation of G-to-A and C-to-U changes in HIV-1 transcripts by RNA editing. Science. 2000;289:1564–1566. doi: 10.1126/science.289.5484.1564. [DOI] [PubMed] [Google Scholar]
- 21.Sharma PM, Bowman M, Madden SL, Rauscher FJ, III, Sukumar S. RNA editing in the Wilms tumor susceptibility gene, WT1. Genes Dev. 1994;8:720–731. doi: 10.1101/gad.8.6.720. [DOI] [PubMed] [Google Scholar]
- 22.Beghini A, Ripamonti CB, Peterlongo P, Roversi G, Cairoli R, Morra E, Larizza L. RNA hyperediting and alternative splicing of hematopoietic cell phosphatase (PTPN6) gene in acute myeloid leukemia. Hum Mol Genet. 2000;9:2297–2304. doi: 10.1093/oxfordjournals.hmg.a018921. [DOI] [PubMed] [Google Scholar]
- 23.Yamanaka S, Balestra ME, Ferrell LD, Fan J, Arnold KS, Taylor S, Taylor JM, Innerarity TL. Apolipoprotein B mRNA-editing protein induces hepatocellular carcinoma and dysplasia in transgenic animals. Proc Natl Acad Sci USA. 1995;92:8483–8487. doi: 10.1073/pnas.92.18.8483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yamanaka S, Poksay KS, Arnold KS, Innerarity TL. A novel translational repressor mRNA is edited extensively in livers containing tumors caused by the transgene expression of the apoB mRNA-editing enzyme. Genes Dev. 1997;11:321–333. doi: 10.1101/gad.11.3.321. [DOI] [PubMed] [Google Scholar]
- 25.Greeve J, Lellek H, Apostel F, Hundoegger K, Barialai A, Kirsten R, Welker S, Greten H. Absence of APOBEC-1 mediated mRNA editing in human carcinomas. Oncogene. 1999;18:6357–6366. doi: 10.1038/sj.onc.1203039. [DOI] [PubMed] [Google Scholar]