Abstract
Upregulated gene 4 (URG4), a novel gene located on 7 chromosome (7p13), was found to contribute to hepatocarcinogenesis. However, the role of URG4 in the gastric carcinogenesis still remains unclear. In the present study, URG4 was found by immunohistochemistry to be upregulated in human gastric cancer tissues compared with matched adjacent nonneoplastic tissues. The proliferating cell nuclear antigen index is higher in gastric cancer tissues with high URG4 expression than in those with low URG4 expression. The growth of GES-1 cells, which are immortalized human gastric epithelial mucosa cells with baseline URG4 expression, was accelerated by URG4 induction. Downregulation of URG4 through URG4 small interfering RNA (siRNA) in SGC7901 and MKN28 cells, which had high endogenous URG4 expression, suppressed cell proliferation in both of these cells. URG4-siRNA also inhibited the proliferation of SGC7901 and MKN28 cells in soft agar and tumor formation in nude mice. Overexpression of URG4 in GES cells upregulated cyclin D1, whereas repression of URG4 in SGC7901 and MKN28 cells downregulated cyclin D1. The data suggested that URG4 played an important role in the development of human gastric cancer by regulating the expression of cyclin D1 and might be used as a potential therapeutic target for gastric cancer.
Keywords: URG4, gastric cancer, carcinogenesis, cell cycle, oncogene
Introduction
Cancer is the result of an accumulation of multiple molecular alterations in the same cell or its descendents [1,2]. Alterations in two groups of genes, proto-oncogenes and tumorsuppressor genes, play a particularly important role in this process [3,4]. In the past decade, a very large number of proto-oncogenes and tumor-suppressor genes have been found. In spite of the sizable number of genes already described, new genes with oncogenic potential or tumorsuppressing activity are still being identified. Recently, upregulated gene 4 (URG4), a novel gene upregulated by HBxAg in human hepatocellular carcinoma, has been identified (GenBank accession no. NM_017920) [5]. URG4 was located on 7 chromosome (7p13). Previous data suggested that overexpression of URG4 in HepG2 cells promoted hepatocellular growth and survival in tissue culture and nude mice. Hence, URG4 may be an oncogene operating in hepatocarcinogenesis [5].
Gastric cancer is one of the most common malignancies in the world, particularly in Eastern Asian countries such as China, Korea, and Japan [6]. The molecular mechanisms of gastric carcinogenesis remain unclear. Whether or not URG4 is involved in gastric carcinogenesis is still unknown. To gain insight into these issues, the expression of URG4 in malignant gastric tissues and its corresponding adjacent counterparts was detected, and the effects of the modulation of URG4 gene expression on the phenotype of gastric cancer cells and on the immortalized human gastric epithelial mucosa cell line GES-1 were also explored.
Materials and Methods
Tissue Specimens and Cell Lines
Serial sections of paraffin-embedded tissues were collected from 100 patients with gastric cancer who underwent gastrectomy in our hospital between January 2004 and June 2005. None of the patients had received preoperative radiation therapy or chemotherapy. Data on sex, age, tumor size, histologic type of neoplasm, and tumor-node-metastasis (TNM) stage were obtained from surgical and pathological records, with the patients' consent. The human gastric cancer cell lines SGC7901, MKN28, MKN45, AGS, and BGC823, and the immortalized human gastric epithelial mucosa cell line GES-1 were preserved in our institute [7,8]. All cell lines were cultured in RPMI 1640 (Invitrogen, Carlsbad, CA) supplemented with 10% heat-inactivated fetal calf serum (FCS) at 37°C with 5% CO2 in a humidified incubator (Forma Scientific, Marietta, OH).
Immunohistochemical Staining
All sections (4 µm) were cut from original paraffin blocks, which were deparaffinized in xylene and rehydrated in graded alcohols. After the inactivation of endogenous peroxidase activity with 0.3% hydrogen peroxide in methanol for 30 minutes, the sections immersed in citrate buffer were heated in a microwave oven for epitope retrieval. Then the sections were blocked with 10% normal goat serum (Biological Technology Co. Ltd., Wuhan, China) for 40 minutes and incubated overnight at 4°C with rabbit anti-human URG4 polyclonal antibody (diluted 1:500; kindly provided by Dr. Mark Feitelson, Department of Pathology, Anatomy, and Cell Biology, Thomas Jefferson University, Philadelphia, PA) or mouse anti-human proliferating cell nuclear antigen (PCNA) monoclonal antibody (diluted 1:200; DAKO, Carpinteria, CA). The primary antibody was detected with the DAKO EnVision+ Kit (DAKO). Reaction products were visualized with the DAKO Liquid DAB+ Substrate-Chromagen System (DAKO) and then counterstained with hematoxylin. Negative control sections were incubated with preimmune rabbit serum or normal mouse serum instead of the primary antibody, respectively. All sections were examined microscopically and scored by two independent pathologists in a blinded manner. URG4 scoring was based on both intensity and extensity, according to previous reports [9,10]. The ratio of positive cells per specimen was evaluated quantitatively and was scored as follows: 0 = staining of < 1%; 1 = staining of 2% to 25%; 2 = staining of 26% to 50%; 3 = staining of 51% to 75%; and 4 = staining of > 75%. Intensity was graded as follows: 0 = no staining; 1 = weak staining; 2 = moderate staining; and 3 = strong staining. Total score (0–12) was calculated and graded as negative (I; score of 0–1), weak (II; score of 2–4), moderate (III; score of 5–8), and strong (IV; score of 9–12).
For PCNA analysis, the PCNA index was examined. It was calculated as the percentage of PCNA-positive cells per 1000 cells, counted at random in each section. This counting was performed under a x400 magnification [11].
RNA Extraction and Semiquantitative Reverse Transcription-Polymerase Chain Reaction (RT-PCR)
Total RNA were isolated from cells with the TRIZOL Reagent (GIBCO BRL, Grand Island, NY). RT reaction was performed using the First-Strand cDNA Synthesis Kit (MBI Fermentas, Vilnius, Lithuania) according to the manufacturer's protocol. Appropriate cycles were chosen to ensure the termination of PCR amplification before reaching a stable stage in each reaction. Gene expression was presented as the yield of PCR products from target sequences relative to the yield of PCR products from the β-actin gene. PCR primers and reaction parameters are listed in Table 1. Each experiment was repeated at least thrice.
Table 1.
Primers and Reaction Parameters for PCR.
| Products | Sequence | Annealing Temperature (°C) | Number of Cycles | Size (bp) |
| URG4 | 5′-CTTCATCCTGAGTCCCTACCG-3′, 5′-GCCGTTCTGCTGCATTCG-3′ | 55 | 32 | 472 |
| β-Actin | 5′-AGCGGGAAATCGTGCGTG-3′, 5′-CAGGGTACATGGTGGTGCC-3′ | 54 | 18 | 309 |
Plasmid Construction and Transfection
Three pairs of hairpin small interfering RNA (siRNA) oligos for URG4 were designed according to the siRNA design guidelines of Ambion, Inc. (Austin, TX) Compare target sequences to the human genome database in a BLASTsearch to eliminate from consideration any target sequence with more than 16 to 17 base pairs of homology contiguous to other coding sequences. For oligo-1, sense: 5′-gatccgtgctgatgccaggaataccttcaagagaggtattcctggcatcagcattttttggaaa-3′, antisense: 5′-agcttttccaaaaaatgctgatgccaggaatacctctcttgaaggtattcctggcatcagcacg-3′; for oligo-2, sense: 5′-gatccgtagaccactcacatgtcctttcaagagaaggacatgtgagtggtctattttttggaaa-3′, antisense: 5′-agcttttccaaaaaatagaccactcacatgtccttctcttgaaaggacatgtgagtggtctacg-3′; for oligo-3, sense: 5′-gatccgcttcgaatgcagcagaacgttcaagagacgttctgctgcattcgaagttttttggaaa-3′, antisense: 5′-agcttttccaaaaaacttcgaatgcagcagaacgtctcttgaacgttctgctgcattcgaagcg-3′. Their target sequences were homologous to nt 349–369, 1200–1220, and 1574–1594 of the URG4 cDNA sequence, respectively. For annealing to form DNA duplexes, 0.01 M each of sense and antisense oligos was used. The duplexes were diluted and then ligated with pSilencer3.1-H1 neo vector (Ambion, Inc.). The products were transformed into DH5a-competent cells. Ampicillin-resistant colonies were chosen, identified by restriction digestion, and further confirmed by DNA sequencing. According to the manufacturer's instructions, siRNA plasmids of URG4 were transfected into SGC7901 and MKN28 cells, and pcDNA3 containing the full-length human URG4 cDNA (which was also kindly provided by Dr. Mark Feitelson) was transfected into GES-1 cells using Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA). The cells transfected with pSilencer3.1-H1 neo vector [which was provided in the siRNA kit (Ambion, Inc.) and expressed hairpin siRNA with limited homology to any known sequence in human genomes] or pcDNA3 vector (Invitrogen) alone served as negative control, respectively. Cells were selected in a growth medium containing G418 (Invitrogen). The expression levels of URG4 in G418-resistant clones were evaluated by Western blot analysis.
Cell Proliferation Assays
MTTassay was used to determine the effect of URG4 on cellular proliferation. Briefly, cells were plated in 96-well plates (2 x 103 cells/well in a final volume of 200 µl) in replicates of three. After 1, 2, 3, 4, 5, 6, and 7 days of incubation, 20 µl of MTT (5 mg/ml; Sigma, St. Louis, MO) was added and incubated for 4 hour. Supernatant was then removed, and 150 µl of dimethyl sulfoxide was added. Culture plates were surged for 10 minutes at room temperature to dissolve MTT crystals. Absorbance values were determined by an ELISA reader (Bio-Rad Laboratories, Richmond, CA) at a wavelength of 490 nm. Each experiment was repeated at least thrice.
Flow Cytometry Assay
For DNA content analysis, cells were harvested and washed twice with ice-cold phosphate-buffered saline (PBS). Cell pellets were fixed in 70% ethanol, treated with RNase A (Boehringer Mannheim, Indianapolis, IN), and stained with propidium iodide (Sigma-Aldrich, St. Louis, MO). DNA contents were measured with a flow cytometer (FACScan; Becton Dickinson, San Jose, CA). The proliferation index (PI) was calculated as: PI = (S + G2)/(S + G2 + G1).
Western Blot Analysis
Cells were lysed with lysis buffer (50 mM Tris pH 7.2, 1% Triton X-100, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate, 500 mM NaCl, and 10 mM MgCl2 with 10 µg/ml leupeptin, 10 µg/ml aprotinin, and 1 mM PMSF) and quantified by the Bradford method. Fifty to sixty micrograms of cellular proteins was separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (10% gel) and transferred to nitrocellulose membranes (Immobilin-P; Millipore, Bedford, MA). Membranes were blocked with 5% nonfat milk at room temperature for 2 hours and incubated overnight with rabbit anti-human URG4 polyclonal antibody (diluted 1:800) or anti-β-actin monoclonal antibody (1:4000; Sigma) at 4°C. After incubation with horseradish peroxidase-conjugated anti-rabbit IgG or anti-mouse IgG (Sigma), respectively, the specific protein band was visualized by enhanced chemiluminescence (Amersham-Pharmacia Biotech, Beijing, China). Autoradiograms were quantified by densitometry (software: Bio Image IQ, BioImage, Ann Arbor, MI). The same membrane was reprobed with β-actin-specific antibody to ensure equal control. Each experiment was repeated at least thrice.
Soft Agar Colony Formation Assays
Anchorage-independent cell growth was determined by analyzing the formation of colonies in soft agar. Cells (2 x 102 cells/well) from each cell line were suspended in 0.3% agar in RPMI 1640 containing 10% FCS and plated on solidified agar (0.5%) in 24-well dishes. Cells were incubated for 3 weeks at 37°C in 5% CO2 before counting colonies under a code. Each assay was performed in triplicate.
Tumorigenicity Test in Nude Mice
Logarithmically growing cells were trypsinized and resuspended in PBS after washing twice with a serum-free medium. About 107 cells in 0.2 ml were injected subcutaneously into 4-week-old female BALB/c nu/nu mice. After 4 weeks of observation, the mice were sacrificed and the tumors were recovered for further analysis. Experimental and control groups had at least five mice each. Tumor volume was measured by vernier caliper, and tumor volumes were calculated according to the formula [12]: Tumor volume (cm3) = (a/2)(b/2)hπ, where a, b, and h are the minor dimension, major dimension, and height of the tumor, respectively (π = 3.1416).
Statistical Analysis
All data were analyzed using the SPSS software package (SPSS, Chicago, IL), and P < .05 was considered statistically significant. The significance of the difference in the frequency of URG4-positive staining between normal samples and tumors, and the difference in cell cycle were analyzed by chi-square test. Mann-Whitney U test for two groups and Kruskal-Wallis H test followed by Nemenyi test for multigroups were used to compare differences among groups on the immunohistochemistry of URG4 with various clinical pathological parameters. Student's t test and one-way analysis of variance followed by Dunnett's multiple comparison tests were adopted for other data.
Results
Immunohistochemical Analysis of URG4 Expression in Human Gastric Cancer Specimen and Matched Adjacent Nonneoplastic Tissues
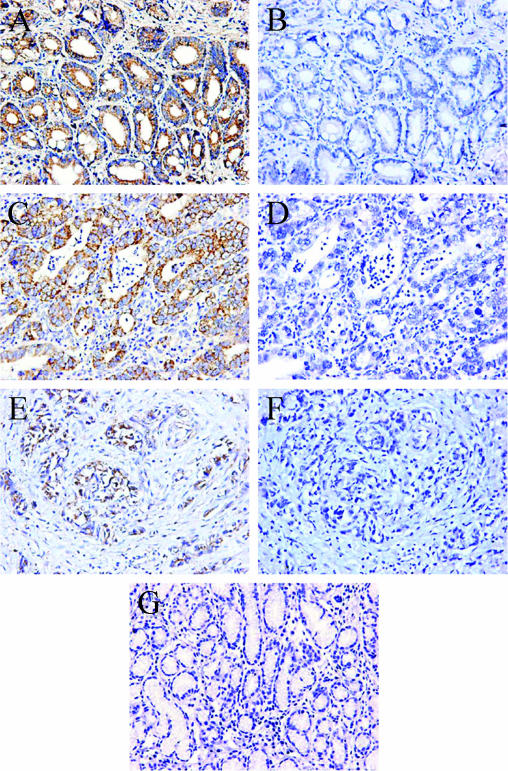
The expression of URG4 in human malignant gastric tissues and matched adjacent nonneoplastic tissues was analyzed by immunohistochemistry. The results showed that URG4 was mainly expressed in the cytoplasm of epithelial cells and only occasionally in nuclei (Figure 1, A–G). In gastric cancer tissues, 65% (65 of 100 patients) had positive staining for URG4 (Figure 1, A–F), which was significantly higher than 30% (30 of 100 patients) in adjacent nonneoplastic tissues (Figure 1G) (P < .001; Table 2). Further analysis of the clinicopathological features of 100 gastric cancer specimens revealed a positive association of URG4 expression with degree of tumor differentiation. In poorly differentiated tumor cells, the average expression of URG4 was lower, whereas URG4 was detected in most epithelial cells, with a higher expression in moderately differentiated and well-differentiated tumor cells (P < .001 and P < .001, respectively) (Table 3). With respect to TNM stage, the expression of URG4 was not significantly different between patients at stages III + IV and patients at stages I + II (P = .133; Table 3). There was no significant difference in URG4 expression between tumors with nodal metastasis and those without (P = .100; Table 3).
Figure 1.
Immunohistochemical staining of URG4 in normal gastric tissues and gastric cancer with different stages of differentiation. (A) Anti-URG4 staining of well-differentiated gastric carcinoma tissue. (B) Preimmune rabbit serum used to stain a consecutive section from the same patient as in (A). (C) Anti-URG4 staining of moderately differentiated gastric carcinoma tissue. (D) Preimmune rabbit serum used to stain a consecutive section from the same patient as in (C). (E) Anti-URG4 staining of poorly differentiated gastric carcinoma tissue. (F) Preimmune rabbit serum used to stain a consecutive section from the same patient as in (E). (G) Normal epithelium exhibited negative URG4 immunostaining. (A–G) Original magnification, x200.
Table 2.
Expression of URG4 in Gastric Cancer Tissues and Adjacent Nonneoplastic Tissues.
| Total (n) | Positive | Negative | P | |
| Nonneoplastic tissues | 100 | 30 | 70 | < .001* |
| Gastric cancer tissues | 100 | 65 | 35 |
P < .05 was considered statistically significant.
Table 3.
Clinicopathological Associations of URG4 Expression in Patients with Gastric Cancer.
| Characteristics | n | URG4 (n) | P | |||
| I | II | III | IV | |||
| Age (years) | ||||||
| < 50 | 22 | 7 | 8 | 6 | 1 | .455 |
| ≥ 50 | 78 | 28 | 25 | 22 | 3 | |
| Gender | ||||||
| Male | 76 | 29 | 26 | 19 | 2 | .103 |
| Female | 24 | 6 | 7 | 9 | 2 | |
| Differentiation | ||||||
| Well-differentiated | 17 | 3 | 4 | 7 | 3 | > .100 (well-differentiated versus moderately differentiated) |
| Moderately differentiated | 37 | 8 | 12 | 16 | 1 | < .001* (well-differentiated versus poorly differentiated) |
| Poorly differentiated | 46 | 24 | 17 | 5 | 0 | < .001* (moderately differentiated versus poorly differentiated) |
| TNM stage | ||||||
| I + II | 53 | 16 | 16 | 18 | 3 | .133 |
| III + IV | 47 | 19 | 17 | 10 | 1 | |
| Metastasis | ||||||
| With | 34 | 15 | 11 | 7 | 1 | .1 |
| Without | 66 | 20 | 22 | 21 | 3 | |
P < .05 was considered statistically significant.
Immunohistochemical Analysis of the PCNA Index in Human Gastric Cancer Tissues
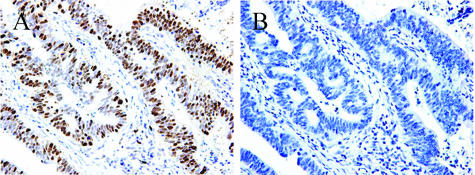
PCNA has been demonstrated to be a useful tool in evaluating cell proliferation [13]. To determine the gastric cancer cell PI, the sections were immunohistochemically stained with PCNA antibody. PCNA protein expression was observed in the nuclei of cancer cells (Figure 2A). The PCNA index of gastric cancer tissues with high URG4 expression is 64.04 ± 11.56, which was higher than 49.84 ± 9.68 in gastric cancer tissues with low URG4 expression. (P < .01; Table 4)
Figure 2.
Immunohistochemical analysis of the PCNA index in human gastric cancer tissues. (A) Anti-PCNA staining of gastric carcinoma tissue. (B) Normal mouse serum used to stain a consecutive section from the same patient as in (A). (A and B) Original magnification, x200.
Table 4.
Associations between PCNA Expression and URG4 Expression in Gastric Cancer Tissues.
| Grade of URG4 Expression | n | PCNA Index (mean ± SD) | P |
| I + II | 32 | 49.84 ± 9.68 | < .01* |
| III + IV | 68 | 64.04 ± 11.56 |
P < .05 was considered statistically significant.
URG4 mRNA and Protein Expression in Gastric Cancer Cell Lines
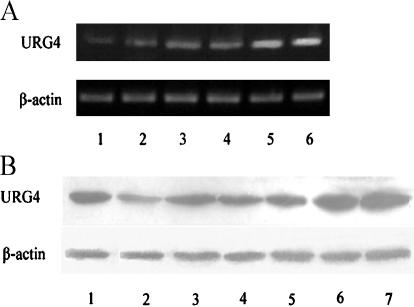
To identify the expression of URG4 in cell lines, the mRNA and protein levels of URG4 were determined in five gastric cancer cell lines and an immortalized gastric epithelial mucosa cell (GES-1). As shown in Figure 3A, a higher expression of URG4 mRNA was detected in the well-differentiated gastric cancer cell line MKN28 and in the moderately differentiated gastric cancer cell line SGC7901, whereas the expression of URG4 mRNA was lower in the three poorly differentiated gastric cell lines BGC823, MKN45, and AGS, and was lowest in GES-1. The protein expression of URG4 displayed a pattern similar to that in the mRNA level, and a 104-kDa band was detected in all cell lines (Figure 3B).
Figure 3.
The URG4 expression level of five gastric cancer cell lines and an immortalized gastric epithelial mucosa cell (GES-1). (A) RT-PCR analysis of the expression level of URG4 mRNA inGES-1 (lane 1), AGS (lane 2), MKN45 (lane 3), BGC823 (lane 4), MKN28 (lane 5), and SGC7901 (lane 6). β-Actin was used as internal control. (B) Western blot analysis of the expression of URG4 protein in HepG2 cells (lane 1, positive control) [5], GES-1 (lane 2), AGS (lane 3), MKN45 (lane 4), BGC823 (lane 5), MKN28 (lane 6), and SGC7901 (lane 7).
Overexpression of URG4 Protein Promotes GES-1 Cell Growth
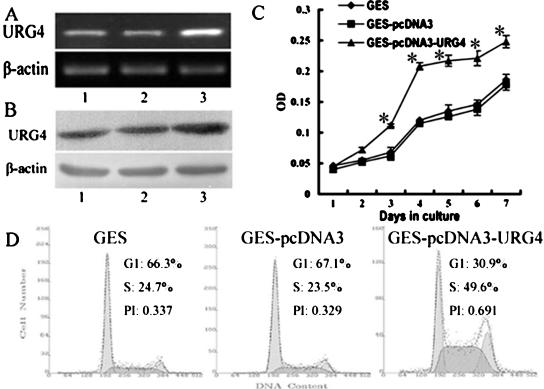
To determine whether URG4 could stimulate the cell growth and survival of the immortalized human gastric epithelial mucosa cell line GES-1, which had a lower expression of URG4, pcDNA3-URG4 was transfected into GES-1 cells. Several G418-resistant clones were obtained after 4 weeks of selection and then tested for URG4 gene expression at both mRNA and protein levels. It was found that pcDNA3 URG4 could significantly upregulate the level of URG4 mRNA (Figure 4A) and protein (Figure 4B) compared with those in controls. Figure 4C showed that GES-1 cells with increased URG4 expression (GES-pcDNA3-URG4) proliferated at a faster rate than did control cells, and statistical analysis showed a significant difference in a medium containing 10%FCS on the third day. The cell cycle of these cells was then measured with flow cytometry. The results showed that 24.7% of GES-1 cells and 23.5% of GES-pcDNA3 cells were in S-phase, whereas 49.6% of GES-pcDNA3-URG4 cells were in S-phase (P < .01; Figure 4D), suggesting that URG4 promotes the entry of cells into S-phase. In addition, this difference became more striking when the cells were cultured in a medium supplemented with low concentrations of FCS (data not shown). Taken together, our data strongly suggest that upregulation of URG4 promotes cell cycle and, therefore, enhances the proliferation of GES-1 cells.
Figure 4.
Overexpression of URG4 protein promotes GES-1 cell growth. (A) Expression of URG4 mRNA in GES-1 (lane 1), GES-pcDNA3 (lane 2), and GES-pcDNA3-URG4 (lane 3) by RT-PCR. (B) Expression of URG4 protein in GES-1 (lane 1), GES-pcDNA3 (lane 2), and GES-pcDNA3-URG4 (lane 3) by Western blot analysis. (C) Growth curves for GES-1, GES-pcDNA3, and GES-pcDNA3-URG4 cells by MTT assay. The value shown is the mean of three determinations. *Statistical significance. (D) Cell cycle distribution and the PI of GES-1, GES-pcDNA3, and GES-pcDNA3-URG4 cells.
Downregulation of URG4 Expression Repressing the Growth and Tumorigenicity of Gastric Cancer Cells In Vitro and In Vivo
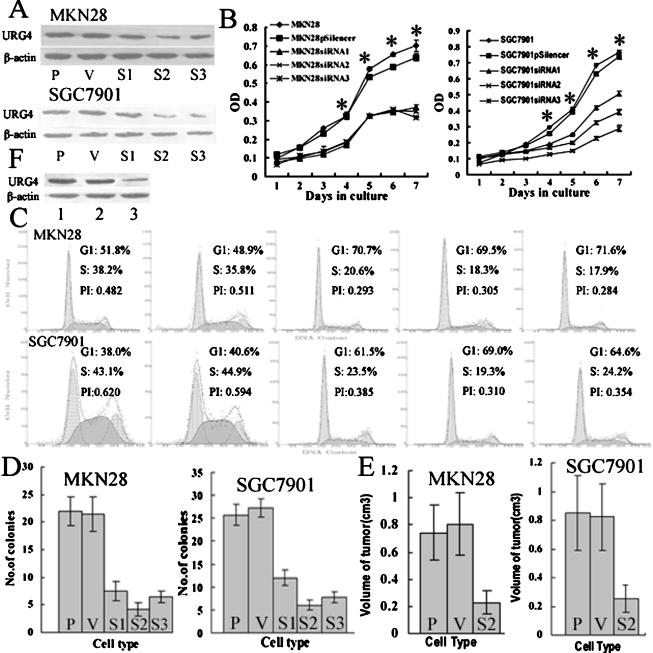
To determine whether inhibition of URG4 expression could suppress the cell growth of gastric cancer cells, three siRNA (URG4-siRNA1, URG4-siRNA2, and URG4-siRNA3) that specifically target URG4 were constructed and transfected into SGC7901 and MKN28 cells. As seen in Figure 5A, the expression level of URG4 in MKN28 and SGC7901 cells was significantly reduced by URG4-siRNA compared to the expression level of cells transfected with pSilencer3.1-H1 neo vector alone or parental cells, and URG4-siRNA2 could most effectively decrease the endogenous level of URG4 protein compared to URG4-siRNA1 and URG4-siRNA3. Significant growth inhibition of URG4-siRNA on MKN28 and SGC7901 cells, compared to control cells, was observed from the fourth day onward (Figure 5B), suggesting that the effects of these URG4-siRNA on proliferation and viability are likely caused by the repression of URG4 protein expression. Then, the effect of URG4-siRNA on the cell cycle of SGC7901 and MKN28 cells was evaluated by flow cytometry. As seen in Figure 5C, the percentages for MKN28 and MKN28-pSilencer cells were 38.2% and 35.8% in S-phase, whereas 20.6% of MKN28-siRNA1, 18.3% of MKN28-siRNA2, and 17.9% of MKN28-siRNA3 cells were in S-phase, respectively (P < .001). In addition, 43.1% of SGC7901 and 44.9% of SGC7901-pSilencer cells were in S-phase compared to 23.5% of SGC7901-siRNA1, 19.3% of SGC7901-siRNA2, and 24.2% of SGC7901-siRNA3 cells (P < .001). Together, the results showed that URG4-siRNA could repress cell proliferation by lengthening the cell cycle.
Figure 5.
Suppression of gastric cancer cell growth by downregulation of URG4 expression in vitro and in vivo. (A) Western blot analysis of URG4 in parental cells (P); control vector transfectants (V); and URG4-siRNA1, URG4-siRNA2, and URG4-siRNA3 transfectants (S1, S2, and S3). β-Actin was used as loading control. (B) Growth curves for P, V, S1, S2, and S3 cells by MTT assay. The value shown is the mean of three determinations. *Statistical significance. (C) Cell cycle distribution and the PI of P, V, S1, S2, and S3 cells by flow cytometry. (D) Colony numbers of P, V, S1, S2, and S3 cells in soft agar. Each soft agar assay was performed in triplicate, and the results were expressed as the mean number of colonies ± SD. (E) Tumor size of P, V, and S2 cells in BALB/c nu/nu mice. (F) URG4 expression in whole tumor tissue extracts by Western blot analysis. Lane 1, tumor tissue of parental SGC7901. Lane 2, tumor tissue of SGC7901-pSilencer. Lane 3, tumor tissue of SGC7901-siRNA2.
Then, the colony formation assay of parental and transfected cells was measured with plating efficiency in soft agar. As shown in Figure 5D, MKN28-siRNA1, MKN28-siRNA2, and MKN28-siRNA3 cells yielded 7.5 ± 1.81, 4.25 ± 1.89, and 6.5 ± 2.51 colonies, whereas MKN28 and MKN28-pSilencer cells yielded 22 ± 2.58 and 21.5 ± 3.41 colonies after 21 days, respectively (P < .01). SGC7901-siRNA1, SGC7901-siRNA2, and SGC7901-siRNA3 cells yielded 12 ± 1.63, 6 ± 1.12, and 7.75 ± 1.26 colonies, whereas SGC7901 and SGC7901-pSilencer cells yielded 25.75 ± 2.21 and 27.25 ± 2.06 colonies after 21 days, respectively (P < .01). Hence, the results showed that there was marked reduction in anchorage-independent growth among cells treated with URG4-siRNA in comparison with controls. The colonies that formed in cells treated with URG4-siRNA were considerably smaller than those in controls (results not shown). Furthermore, the repression potential of URG4-siRNA on the growth of SGC7901 and MKN28 cells in nude mice was also performed. The result showed that tumor size was dramatically smaller in SGC7901 and MKN28 cells transfected with URG4-siRNA2 than in control cells (P < .05; Figure 5E), suggesting that repression of URG4 was directly involved in the inhibition of tumor growth in nude mice. In addition, URG4 protein is much lower in recovered tumors formed in nude mice injected with SGC7901-siRNA2 than in those with SGC7901-pSilencer3.1-H1 or parental SGC7901 cells (Figure 5F). In conclusion, the data here suggested that URG4 played a promoter role in enhancing the malignant growth potential of gastric cancer cells in vitro and in vivo.
URG4-Stimulating Cell Proliferation through Cyclin D1
Because URG4 promotes the entry of cells into S-phase, cyclin D1, a key factor in cell passage through the check point between G0/G1-phase and S-phase, was further detected in cell lines with high or low URG4 expression. The result showed that cyclin D1 was upregulated in URG4-overexpressed GES-pcDNA3-URG4 cells compared with GES-pcDNA3 cells and parental cells (Figure 6A). However, the expression level of cyclin D1 in MKN28 and SGC7901 cells was significantly reduced by URG4-siRNA compared to cells transfected with pSilencer3.1-H1 neo vector alone or parental cells, and URG4-siRNA2 could most effectively decrease the endogenous level of cyclin D1 protein than could URG4-siRNA1 and URG4-siRNA3 (Figure 6, B and C). Moreover, cyclin D1 is lower in recovered tumors formed in nude mice injected with SGC7901-siRNA2 than in those injected with SGC7901-pSilencer3.1-H1 or parental SGC7901 cells (Figure 6D).
Figure 6.
URG4-stimulating cell proliferation through cyclin D1. (A) Expression of cyclin D1 protein in GES-1 (lane 1), GES-pcDNA3 (lane 2), and GES-pcDNA3-URG4 cells (lane 3) by Western blot analysis. (B) Western blot analysis of cyclin D1 in MKN28 cells (lane 1), control MKN28-pSilencer cells (lane 2), MKN28-siRNA1 (lane 3), MKN28-siRNA2 (lane 4), and MKN28-siRNA3 (lane 5). β-Actin was used as loading control. (C) Western blot analysis of cyclin D1 in SGC7901 cells (lane 1), control SGC7901-pSilencer cells (lane 2), SGC7901-siRNA1 (lane 3), SGC7901-siRNA2 (lane 4), and SGC7901-siRNA3 (lane 5). β-Actin was used as loading control. (D) Cyclin D1 expression in whole tumor tissue extracts by Western blot analysis. Lane 1, tumor tissue of parental SGC7901. Lane 2, tumor tissue of SGC7901-pSilencer. Lane 3, tumor tissue of SGC7901-siRNA2.
Discussion
The identification and characterization of genes that are differentially expressed in gastric cancer tissues and matched adjacent nonneoplastic tissues provide important information with regard to understanding the mechanisms responsible for carcinogenesis. In the present study, URG4 expression was found to be upregulated in gastric cancer tissues compared with matched adjacent nonneoplastic tissues. This coincides with our in vitro observation that URG4 is upregulated in gastric cancer cell lines compared with normal gastric epithelial cell lines, suggesting that URG4 might play an oncogenic role in the development of gastric cancer. PCNA, an auxiliary protein of DNA polymerase δ, is a proliferation-associated marker. Its maximal expression peaks in the late G1-phase and S-phase of the cell cycle [14]. PCNA has been used as a proliferation marker in different neoplasms. In the present study, the PCNA index of gastric cancer tissues with high URG4 expression was higher than those with low URG4 expression, suggesting that URG4 was related to the proliferative activity of cancer cells and might promote cell growth in gastric cancer tissues. Moreover, our findings demonstrated that induction of URG4 could promote the proliferation of GES-1 cells and could stimulate cell cycle progression by shortening the emergence of cells from quiescence (G0) and entry into S-phase. In addition, reduction of URG4 with URG4-siRNA inhibited the proliferation of SGC7901 and MKN28 cells, and repressed cell cycle progression. Given that oncogene suppression often leads to inhibition of the anchorage-independent growth of tumor cells in soft agar [15], URG4-reducing SGC7901 and MKN28 cells through siRNA were also tested for anchorage-independent growth, and the results showed that reduced URG4 could repress the anchorage-independent growth of both SGC7901 and MKN28 cells in soft agar (Figure 5D), also suggesting that URG4 might be an oncogene operating in gastric carcinogenesis. This was further supported by the finding that reducing URG4 represses tumor formation and growth in nude mice (Figure 5, E and F).
Cyclin D1 is a periodic regulatory protein that is believed to govern cell cycle transit from G1-phase into S-phase, and has been found to be abnormally expressed in many human cancers [16]. Overexpression of cyclin D1 leads to abnormal cellular proliferation, which underlies the process of tumorigenesis. Thus, cyclin D1 can function as a cooperative oncogene in cell transformation [17]. In the present study, URG4 could upregulate the expression of cyclin D1, indicating that cyclin D1 was involved in the contribution of URG4 to cancer cell growth.
In conclusion, the present study showed that URG4 was upregulated in human gastric cancer tissues and also in gastric cancer cell lines, and overexpression of URG4 could promote cell proliferation, whereas downregulation of URG4 in gastric cancer cells repressed cell proliferation and tumor formation potential. Moreover, URG4 might promote cell growth partly through cyclin D1. Our result strongly indicated that URG4 might be an oncogene involved in the development of gastric cancer and also a promising therapeutic target in cancer treatment.
The mechanism of URG4 biologic activity in normal and malignant cells is not yet fully understood. Despite experimental evidence on the oncogenic potential of URG4, preliminary data from our laboratory concerning URG4 suggested that the role of URG4 might be played through the cell cycle-related protein cyclin D1. Further work should be pursued in our laboratory to expand these recent findings and to obtain more information on the function and potential molecular mechanism(s) of URG4 protein.
Acknowledgements
We thank technician Taidong Qiao and Zhen Chen for excellent technical assistance.
Abbreviations
- URG4
upregulated gene 4
- RT-PCR
reverse transcription-polymerase chain reaction
- siRNA
small interfering RNA
- MTT
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- PCNA
proliferating cell nuclear antigen
Footnotes
This work was supported by the Program for New Century Excellent Talents in University and grants 30371585 and 30570835 from the National Natural Science Foundation of China to Jie Liu.
Jiugang Song and Huahong Xie contributed equally to this study.
References
- 1.Vogelstein B, Kinzler KW. The multistep nature of cancer. Trends Genet. 1993;9:138–141. doi: 10.1016/0168-9525(93)90209-z. [DOI] [PubMed] [Google Scholar]
- 2.Weinstein IB. Disorders in cell circuitry during multistage carcinogenesis: the role of homeostasis. Carcinogenesis. 2000;21:857–858. doi: 10.1093/carcin/21.5.857. [DOI] [PubMed] [Google Scholar]
- 3.Weinberg RA. Oncogenes and tumor suppressor genes. CA Cancer J Clin. 1994;44:160–170. doi: 10.3322/canjclin.44.3.160. [DOI] [PubMed] [Google Scholar]
- 4.Munger K. Disruption of oncogene/tumor suppressor networks during human carcinogenesis. Cancer Invest. 2002;20:71–81. doi: 10.1081/cnv-120000369. [DOI] [PubMed] [Google Scholar]
- 5.Tufan NL, Lian Z, Liu J, Pan J, Arbuthnot P, Kew M, Clayton MM, Zhu M, Feitelson MA. Hepatitis Bx antigen stimulates expression of a novel cellular gene, URG4, that promotes hepatocellular growth and survival. Neoplasia. 2002;4:355–368. doi: 10.1038/sj.neo.7900241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alberts SR, Cervantes A, van de Velde CJ. Gastric cancer: epidemiology, pathology and treatment. Ann Oncol. 2003;14:31–36. doi: 10.1093/annonc/mdg726. [DOI] [PubMed] [Google Scholar]
- 7.Pan Y, Bi F, Liu N, Xue Y, Yao X, Zheng Y, Fan D. Expression of seven main Rho family members in gastric carcinoma. Biochem Biophys Res Commun. 2004;315:686–691. doi: 10.1016/j.bbrc.2004.01.108. [DOI] [PubMed] [Google Scholar]
- 8.Hong L, Zhang Y, Liu N, Liu C, Zhi M, Pan Y, Lan M, Sun L, Fan D. Suppression of the cell proliferation in stomach cancer cells by the ZNRD1 gene. Biochem Biophys Res Commun. 2004;321:611–616. doi: 10.1016/j.bbrc.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 9.Wang J, Wu K, Zhang D, Tang H, Xie H, Hong L, Pan Y, Lan M, Hu S, Ning X, et al. Expressions and clinical significances of angiopoietin-1, -2 and Tie2 in human gastric cancer. Biochem Biophys Res Commun. 2005;337:386–393. doi: 10.1016/j.bbrc.2005.09.051. [DOI] [PubMed] [Google Scholar]
- 10.Maaser K, Daubler P, Barthel B, Heine B, von Lampe B, Stein H, Hoffmeister B, Scherer H, Scherubl H. Oesophageal squamous cell neoplasia in head and neck cancer patients: upregulation of COX-2 during carcinogenesis. Br J Cancer. 2003;88:1217–1222. doi: 10.1038/sj.bjc.6600865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Generali D, Berruti A, Brizzi MP, Campo L, Bonardi S, Wigfield S, Bersiga A, Allevi G, Milani M, Aguggini S, et al. Hypoxiainducible factor-1alpha expression predicts a poor response to primary chemoendocrine therapy and disease-free survival in primary human breast cancer. Clin Cancer Res. 2006;12:4562–4568. doi: 10.1158/1078-0432.CCR-05-2690. [DOI] [PubMed] [Google Scholar]
- 12.Lew YS, Brown SL, Griffin RJ, Song CW, Kim JH. Arsenic trioxide causes selective necrosis in solid murine tumors by vascular shutdown. Cancer Res. 1999;59:6033–6037. [PubMed] [Google Scholar]
- 13.Rokkas T, Liatsos C, Karameris A, Petridou E, Lazaris A, Antoniades D, Kalafatis E. Proliferating cell nuclear antigen (PCNA) immunostaining in Helicobacter pylori infection: impact of eradication. Pathol Oncol Res. 1999;5:304–308. doi: 10.1053/paor.1999.0215. [DOI] [PubMed] [Google Scholar]
- 14.Russo G, Zamparelli A, Howard CM, Minimo C, Bellan C, Carillo G, Califano L, Leoncini L, Giordano A, Claudio PP. Expression of cell cycle-regulated proteins pRB2/p130, p107, E2F4, p27, and pCNA in salivary gland tumors: prognostic and diagnostic implications. Clin Cancer Res. 2005;11:3265–3273. doi: 10.1158/1078-0432.CCR-04-2508. [DOI] [PubMed] [Google Scholar]
- 15.Joseph P, Lei YX, Whong WZ, Ong TM. Molecular cloning and functional analysis of a novel cadmium-responsive proto-oncogene. Cancer Res. 2002;62:703–707. [PubMed] [Google Scholar]
- 16.Segas JV, Lazaris AC, Nikolopoulos TP, Kavantzas NG, Lendari IE, Tzagkaroulakis AM, Patsouris ES, Ferekidis EA. Cyclin D1 protein tissue detection in laryngeal cancer. ORL J Otorhinolaryngol Relat Spec. 2005;67:319–325. doi: 10.1159/000090041. [DOI] [PubMed] [Google Scholar]
- 17.Uhlman DL, Adams G, Knapp D, Aeppli DM, Niehans G. Immunohistochemical staining for markers of future neoplastic progression in the larynx. Cancer Res. 1996;56:2199–2205. [PubMed] [Google Scholar]