Abstract
Transcriptional cofactors are integral to the proper function and regulation of nuclear receptors. Members of the peroxisome proliferator-activated receptor (PPAR) family of nuclear receptors are involved in the regulation of lipid and carbohydrate metabolism. They modulate gene transcription in response to a wide variety of ligands, a process that is mediated by transcriptional coactivators and corepressors. The mechanisms by which these cofactors mediate transcriptional regulation of nuclear receptor function are still being elucidated. The rapidly increasing array of cofactors has brought into focus the need for a clear understanding of how these cofactors interact in ligand- and cell-specific manners. This review highlights the differential effects of the assorted cofactors regulating the transcriptional action of PPARγ and summarizes the recent advances in understanding the physiological functions of corepressors and coactivators.
1. INTRODUCTION
Peroxisome proliferator-activated receptors (PPARs) are a subfamily of structurally similar members of the nuclear hormone receptor superfamily [1]. However, unlike classical nuclear hormone receptors, PPARs do not bind their ligands with high affinity, but possess a relatively low binding affinity for unsaturated fatty acids and a broad range of compounds that includes eicosanoids and their metabolites (notably prostaglandin PGJ2 and leukotriene LTB4) and synthetic ligands such as fibrates (a drug for treatment of hyperlipidemia) and thiazolidinediones (TZDs, antidiabetic drugs). Thus, these receptors are considered to be nutrient sensors that regulate lipid and glucose metabolism in adipocytes and other metabolically active tissues. PPARs have also been shown to be involved in a diverse array of nonmetabolic functions including inflammation, tissue repair, atherosclerosis, and cancer [2–4].
PPARγ is the most highly characterized member of this subfamily and its regulation by nuclear receptor cofactors will be the focus of this review. Two major splice variants have been found; PPARγ1 is expressed in adipocytes, skeletal muscle, liver and heart tissue, while PPARγ2 is almost exclusively found in adipose tissue [5]. Although PPARγ2 may be more adipogenic than PPARγ1 [6, 7], both isoforms are thought to be essential regulators of adipogenesis [8–10]. A common model for adipogenesis 3T3-L1 cell differentiation into adipocytes is mediated by PPARγ2 [11]. This model has been used extensively to define the relationship between PPARγ and its cofactors. In addition to adipogenesis, PPARγ has been shown to play a role in insulin sensitivity, atherosclerosis, inflammation, and cancer [12, 13].
1.1. Overview of cofactors involved in transcriptional regulation of PPARγ
PPAR transactivation is induced by ligand-dependent and independent mechanisms. Ligand-dependent transactivation is induced by ligand binding to the C-terminal activation function (AF-2) domain [14]. The role of transcriptional cofactors in ligand-independent transactivation is poorly understood and outside of the scope of this review. PPARs form heterodimers with the retinoid X receptor (RXR) and bind to PPAR response elements (PPREs) in enhancer sites of regulated genes [15]. In the absence of ligand, nuclear receptor corepressors bind to these heterodimers and recruit histone deactylases (HDACs) to repress transcription. Ligand binding induces a conformational change in the receptor dimer which excludes corepressors from the complex [16]. Ligand binding also increases PPAR's affinity for a number of coactivators, whose binding facilitates chromatin remodeling by histone modification and nucleosome mobilization, leading to the recruitment of the basal transcription machinery to PPAR target genes [17–19]. The short motif LXXLL, where L is leucine and X is any amino acid, is necessary for many coactivators to bind to nuclear receptors [20]. This “NR box” is found in the majority of nuclear receptor coactivators and binds to a hydrophobic pocket in the nuclear receptor binding domain [21].
Cofactors that have been shown to interact directly with PPARγ to initiate its transactivation include members of the p160 family of coactivators, which includes SRC-1/NCoA1, TIF2/GRIP1/NCoA2/SRC-2, and pCIP/ACTR/AIB1/SRC-3 [22]. While having weak histone acetyltransferase (HAT) activities, the C-terminal activation domains of p160 proteins appear to primarily serve as foundations upon which coactivator complexes are assembled. The p160 family of coactivators contains functional activation domains that recruit factors such as cAMP responsive element binding protein (CREB) binding protein (CBP)/p300 via activation domain 1 (AD1). The CBP/p300 complex possesses promiscuous HAT activity, which aids in remodeling chromatin to allow transcriptional activation [23].
The prominent ATP-dependent chromatin remodeling complex SWI/SNF includes components such as BAF250, BAF57, BAF60a, and BRG1 [24]. The SWI/SNF complex is thought to be targeted to nuclear receptor target genes upon ligand induction by interaction with receptors, coactivators, or the general transcription machinery [23]. This complex has also been implicated in chromatin remodeling leading to activation of the PPARγ promoter, thus regulating its expression and adipogenesis [25, 26].
The thyroid receptor associated protein (TRAP)/vitamin D receptor interacting proteins (DRIP)/Mediator complex contains subunits which interact with a variety of transcription factors and serve as a bridge between the basal transcriptional machinery and DNA-bound nuclear receptor cofactors [27, 28]. The TRAP complex interacts with PPARγ in a ligand-dependent fashion. This complex acts more directly on the general transcription machinery, as is evident by its ability to transactivate transcription on naked DNA templates [29]. Furthermore, the TRAP complex interacts with nuclear receptors through PPAR binding protein (PBP)/TRAP220/DRIP205 [30]. Thus, TRAP220 is a critical component of this complex and is required for transcriptional activation of PPARγ [31].
The PPAR-gamma coactivator-1α(PGC-1α) is a unique PPAR coactivator, which serves as a scaffolding protein to integrate a variety of coactivator [32]. Upon docking to PPARγ, PGC-1α recruits HATs such as CBP/p300 and steroid receptor coactivator 1 (SRC-1) to remodel chromatin and initiate transcription [32, 33]. However, interaction of PGC-1α and HAT proteins is not sufficient to activate gene transcription; the C-terminal domain of PGC-1α also interacts with the TRAP complex through direct association with PBP/TRAP220 to induce transcription (Wallberg et al. [33]). PGC-1α has several RNA recognition motifs (RRM), which function in the coupling of transcription to mRNA splicing [34]. The modes of regulation of PPARγ by PGC-1α have been reviewed [35, 36].
Although much is known about the mechanisms by which PPARγ recruits coactivators to initiate transcription, considerably less has been demonstrated with regard to transcriptional repression by corepressors. Both NCoR (nuclear receptor corepressor protein) [37] and SMRT (silencing mediator of retinoid and thyroid hormone receptors) [38] directly interact with PPARγ in vitro [39–41]. It may be noted that PPARγ does not appear to be a strong repressor, however, increasing evidence suggests that NCoR and SMRT do repress PPARγ-modulated gene expression during adipogenesis [42, 43].
The exchange of cofactors may be facilitated by nuclear corepressor exchange factors (NCoEx), namely, transducin β-like 1 (TBL1) and the related protein TBLR1 [44]. TBL1 and TBLR1 are components of the NCoR corepressor complex [45]. However, they activate PPARγ-dependent transcription in response to rosiglitazone. Moreover, embryonic stem cells with a TBL1 deletion fail to differentiate into adipocytes [46] suggesting that TBL1 is necessary for PPARγ activation. The mechanism of TBL1/TBLR1 activation of PPARγ remains elusive, but is probably linked to the proteasome-dependent degradation of corepressors [46].
1.2. Physiological functions of cofactors in adipogenesis
The molecular modes of regulation of nuclear receptor signaling by cofactors have been extensively reviewed [16, 17, 23, 47–49]. Herein we focus on the recent advances in understanding the physiological functions of cofactors in PPARγ-modulated processes, in particular, adipogenesis and energy metabolism. The diversified functions of PPARγ cofactors are studied in cell-based system and/or mice models, which are summarized in Table 1.
Table 1.
Loss-of-function studies on PPARγ cofactors in adipogenesis and energy metabolism
| PPARγ cofactor | Phenotype in the absence of the cofactor | |
|
| ||
| Cell-based studies | Mouse studies | |
|
| ||
| Brg1, hBrm (SWI/SNF components) | Blocked adipogenesis (Salma et al. [25]) | — |
| Reduced presence of Pol II and GTFs on the promoter (Salma et al. [25]) | ||
| Decreased PPARγ transcription (Salma et al. [25]) | ||
|
| ||
| TIF2 | Increased lipolysis (Picard et al. [50]) | Enhanced adaptive thermogenesis (Picard et al. [50]) |
| Protection against obesity (Picard et al. [50]) | ||
| Increased insulin-sensitivity (Picard et al. [50]) | ||
| Improved metabolic profile. Increased lipolysis (Picard et al. [50]) | ||
| Decreased presence of PPARγ | ||
|
| ||
| SRC-1 | — | Predisposition to obesity (Picard et al. [50]) |
| Reduced energy expenditure (Picard et al. [50]) | ||
| Reduced fatty acid oxidation in brown adipose tissue (Picard et al. [50]) | ||
| Decreased energy expenditure, attenuated fatty acid oxidation (Picard et al. [50]) | ||
|
| ||
| SRC-1/pCIP double knockout | Abrogated preadipocyte differentiation (Wang et al. [51]) | Diminished lipid storage in brown fat; increased caloric intake on both chow and high-fat diet due to increased leptin levels; resistance to diet-induced obesity; increased basal metabolic rate and energy expenditure (Wang et al. [51]) |
| Reduced expression of PPARγ-target genes, including UCP-1, due to corepressor recruitment and decreased PPARγ recognition of PPREs (Wang et al. [51]) | ||
|
| ||
| PGC-1α | Impaired induction of thermogenic genes in BAT (Uldry et al. [52]) | Reduced mitochFondrial function (Lin et al. [53]) |
| Decreased number and impaired function of mitochondria (Uldry et al. [52]) | Resistance to obesity and hyperactivity (Lin et al. [53]) | |
|
| ||
| TRAP220/DRIP205/PBP | Defective PPARγ-stimulated adipogenesis (Ge et al. [31]) | Defective vascular development similar to that seen in PPARγ-null mice (Barak et al. [54]; Zhu et al. [55]) |
|
| ||
| PRIP/NRC/RAP250/TRBP | Decreased PPARγ-mediated transcriptional activation (Antonson et al. [56]; Zhu et al. [57]) | — |
|
| ||
| RIP140 | Upregulation of genes involved in energy dissipation (Poweka et al., 2006) | Increased oxygen consumption and resistance to high-fat diet-induced obesity (Leonardsson et al. [58]) |
| Increased PGC-1α expression (Poweka et al., 2006) | Expression of lipgenic enzymes is decreased. UCP-1 (involved in energy dissipation in BAT) expression is increased (Leonardsson et al. [58]) | |
|
| ||
| NCoR and SMRT | Increased adipocyte differentiation (Yu et al. [42]) | — |
|
| ||
| Sirt1 | Decreased NCoR levels (Picard et al. [43]) | — |
2. COACTIVATORS
2.1. PGC-1α a master regulator of adaptive thermogenesis in brown adipose tissue
The thermogenic effect of PPARγ in brown adipose tissue (BAT) is mediated by PGC-1α, which is induced by cold and highly expressed in BAT [35, 36]. PGC-1α regulates the action of PPARγ on adaptive thermogenesis and fatty acid oxidation by interacting with the PPARγ/RXRα heterodimer. This interaction stimulates expression of uncoupling protein 1 (UCP-1), which is responsible for uncoupling β-oxidation from ATP synthesis in oxidative phosphorylation, ultimately resulting in the loss of energy as heat [32].
PGC-1α is unique in that, in addition to its ligand-dependent binding to the PPARγ ligand-binding domain (LBD), it can also bind to the DNA-binding domain (DBD) and the hinge region of nuclear receptors in a ligand-independent fashion [59]. The ligand-independent binding of PGC-1α to PPARγ is mediated by the PGC-1α N-terminal domain and results in the expression of enzymes involved in the mitochondrial respiratory chain to activate adaptive thermogenesis [32, 60]. Chromatin immunoprecipitation (ChIP) analyses revealed that the presence of PGC-1α decreases the association of corepressors on a PPRE-containing gene in the absence of exogenous ligand without altering the binding of PPARγ, and PGC-1α is sufficient to recruit SRC-1, p300, and RNA polymerase II to the PPRE-containing gene in the absence of rosiglitazone [61].
The ectopic expression of PGC-1α in white adipose tissue (WAT) in vitro causes induction of the genes associated with the brown fat phenotype, such as UCP-1 and components of the electron transport chain [62, 63]. The presence of UCP-1 in WAT is associated with a more brown-fat like phenotype, enhanced metabolic rate and insulin sensitivity, and resistance to obesity [64–66], which could indicate a potential therapeutic role for PGC-1α and UCP-1.
The function of PGC-1α in adaptive energy metabolism is reinforced in the PGC-1α knockout mouse model [53]. PGC-1α null mice are born with no obvious defects during embryonic development but have reduced mitochondrial function. Intriguingly, null mice are lean and resistant to diet-induced obesity. The lean phenotype is largely due to hyperactivity caused by lesions in the striatal region of the brain which controls movement [53]. The closely related family member PGC-1β has been less studied, but it appears to induce mitochondrial biogenesis and fatty acid oxidation in several cell types [67–69]. Thus, PGC-1β can regulate some but not all activities of PGC-1α. The most recent PGC-1β knockdown studies in immortal preadipocyte lines derived from PGC-1α null mice reveal complementary actions of the two PGC-1 proteins [52]. Loss of PGC-1α alone severely impairs the induction of thermogenic genes but does not affect brown fat differentiation (Figure 1). Loss of either PGC-1α or PGC-1β exhibits a small decrease in the differentiation-induced mitochondrial biogenesis; however, double knockdown results in a reduced number of mitochondria and functional defects [52]. This study implicates that PGC-1β plays a role in brown fat differentiation, and is at least as important as PGC-1α in this process (Figure 1).
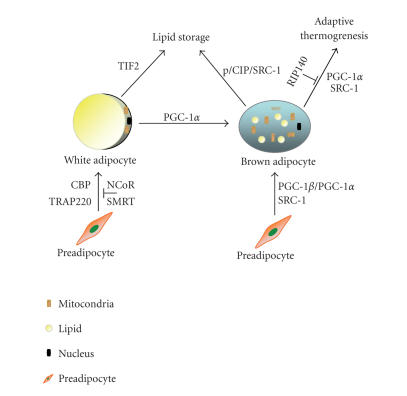
Figure 1.
Putative functions of PPARγ cofactors in white adipose- and brown adipose-modulated lipid and energy metabolism. Positive regulators are highlighted in red. Preadipocytes can be differentiated into white adipocytes via transcriptional regulation of PPARγ by coactivators CBP and TRAP220, or differentiated into brown adipocytes via transactivation by PGC-1β, PGC-1α, and SRC-1. TIF2 plays roles in lipid storage from white adipocytes, while p/CIP and SRC-1 function to promote lipid storage in brown fat. PGC-1α is not only involved in adaptive thermogenesis but it also promotes the conversion of white adipocytes into brown adipocytes. SRC-1 is the only member of p160 proteins that show clear function in energy expenditure.
2.2. Effects of the p160 coregulators SRC-1, TIF2/SRC-2, and p/CIP/SRC-3 on energy metabolism and homeostasis
Members of the 160 kd protein family of coactivators are able to interact directly with the AF2 domain of PPARγ to allow nuclear receptor transactivation function in a ligand-dependent manner via an α-helical LXXLL motif on p160 protein's N-terminal domain. Furthermore, CBP/p300 interacts with p160 cofactors and directly with PPARγ, possibly providing additional stability to the complex through an increased number of contact points [70]. However, although CBP/p300 binding is required for maximal PPARγ activity in vitro, minimal data exists showing a requirement for these cofactors in adipogenesis [71].
Mice deficient in p160 family members exhibit very different phenotypes, providing insights into their physiological functions in adipogenesis and energy metabolism [50]. TIF2−/− mice exhibit enhanced adaptive thermogenesis and protection against obesity, whereas SRC-1−/− mice are predisposed to obesity with accompanying reduced energy expenditure [50]. TIF2−/− mice also show improved metabolic profiles and increased whole-body insulin sensitivity [50]. TIF2 seems to have a greater influence on the p300/PPARγ complex than does the SRC-1 complex, which could possibly be attributed to a weaker capacity of SRC-1 to interact with other coregulators such as p300/CBP and TRAP220, as these coregulators have been shown to have roles in adipogenesis [31, 71]. An increase in lipolysis is observed in TIF2−/− cells, indicating a reduced potential for the storage of fatty acids. Furthermore, a TIF2 dose-dependent attenuation of the PGC-1α/PPARγ activation complex in the presence of SRC-1 suggests that TIF2 competes with SRC-1 for the formation of PGC-1α/PPARγ complexes. However, TIF2 does not significantly enhance PPARγ transactivation mediated by PGC-1α, and an increase in PGC-1α expression level was observed in BAT of TIF2−/− mice [50]. Thus, TIF2 appears to be linked to WAT differentiation and fat storage by potentiating PPARγ activity (Figure 1). In contrast, SRC-1−/− mice displayed increased fat mass and plasma leptin levels. Moreover, the mRNA of UCP-1, PGC1α, and AOX were decreased in BAT, suggesting that the thermogenic machinery in BAT is diminished in the absence of SRC-1. Thus, SRC-1 largely contributes to brown fat differentiation and energy expenditure in brown fat (Figure 1).
A recent study involving p/CIP−/− SRC-1−/− double knockout (DKO) mice revealed that p/CIP and SRC-1 are required for induction of genes necessary for adaptive thermogenesis and lipid storage in BAT [51]. These DKO mice consume more food, both on chow and high fat diets, as a result of decreased blood leptin levels; however, the DKO mice are resistant to diet-induced obesity and remain lean when compared to single knockout and wild type littermates. Furthermore, these mice are more physically active and have increased basal metabolic rates. This phenotype appears to be the result of failed induction of PPARγ target genes, resulting in increased basal metabolism and decreased adipogenesis [51]. Although p/CIP single knockout mice do not exhibit a strong phenotype in adipogenesis, p/CIP appears to potentiate SRC-1-mediated fat storage in BAT and perhaps adaptive thermogenesis (Figure 1).
2.3. The SWI/SNF chromatin remodeling complex is required for induction of the PPARγ promoter and adipogenesis
The mammalian SWI/SNF (mating type switching/sucrose nonfermenting) family of ATP-dependent chromatin remodeling enzymes plays critical roles in the activation of PPARγ transcription for adipogenesis. The core components of the complex include either the Brg1 or Brm ATPase and several Brg1/Brm-associated factors (BAFs). Although in vitro analyses of SWI/SNF complexes containing Brg1 or Brm reveal similarities in chromatin remodeling [72], differences in their functions have been observed in vivo. Brg1 knockout mice are embryonically lethal, and heterozygotes show a predisposition for tumor development [73]. In contrast, Brm knockout mice and cells show only a slight difference in proliferation from wild type [74].
PBAF, a multisubunit complex containing Brg1 and BAF180 subunit was shown to activate PPARγ transcription in an in vitro chromatin-based system [75]. The necessity of the SWI/SNF chromatin remodeling complex is illustrated by experiments revealing that Pol II and general transcription factors are dissociated from the PPARγ promoter when cells are transfected with dominant negative components of the SWI/SNF complex [25]. This suggests that function of the SWI/SNF complex is essential to formation of the preinitiation complex (PIC) on the PPARγ2 promoter and subsequent transcription initiation. Expression of dominant negative Brg1 or hBrm leads to blocked induction of the PPARγ activator and adipogenesis, which was measured both morphologically and by expression of two adipogenic marker genes, aP2 and adipsin [25]. Because Brg1 and hBrm are both crucial members of the SWI/SNF chromatin remodeling complex, this evidence suggests that the SWI/SNF enzymes are required for the activation of PPARγ and adipogenesis [25].
BAF60c, another component of the SWI/SNF complex, serves to anchor the SWI/SNF complex to PPARγ. GST pull-down experiments as well as co-IP confirmed the ability of BAF60c to interact with PPARγ. Moreover, BAF60c interacts with PPARγ in a ligand-dependent fashion to enhance the transcriptional activity of the receptor [26]. However, BAF60c was not shown to affect adipocyte differentiation in these experiments suggesting that BAF60c is not the only factor docking SWI/SNF to PPARγ [26].
2.4. TRAP220/DRIP205/PBP is required for transactivation of PPARγ2 and adipogenesis
The TRAP complex has been implicated as a general transactivator of nuclear receptors [76], apparently functioning by direct interaction with DNA-bound activators and RNA polymerase II [30]. Appreciable evidence for the TRAP complex serving as a coactivator for PPARγ is derived from an in vitro transcription assay in which purified TRAP complex significantly enhanced the transcriptional activity of PPARγ2 on a PPRE-template. GST pull-down assays confirmed the ability of the TRAP complex to bind PPARγ2 only in the presence of TRAP220 [31]. Thus, TRAP220, also known as DRIP205 and PBP [77], anchors the TRAP complex to PPARγ target promoters. A TRAP220−/− mutation is embryonically lethal at day 11.5, showing defects in vascular development similar to those in PPARγ −/− mice, indicating that TRAP220 function is nonredundant and essential for development [54, 78]. Studies using immortalized TRAP220−/− MEFs reveal that TRAP220 acts as a coactivator for PPARγ2 and is an essential mediator of adipogenesis [31]. TRAP220−/− cells exhibit defective PPARγ2-stimulated adipogenesis and expression of adipogenic marker genes. These adipogenic defects can be rescued by ectopic expression of TRAP220 [31]. These data support the model that TRAP220 acts as an anchor in TRAP complex binding, and may also play a role in binding to the CBP-associated complex.
2.5. Evidence of a megacomplex in PPAR transactivation
PPAR interacting protein PRIP/NRC/RAP250/TRBP is ubiquitously expressed in adult mice, and binds to PPARγ enhancing ligand-dependent transcription [55, 56, 79]. PRIP is also necessary for embryonic vascular development, as well as normal cardiac and neural development, as shown by a lethal null mutation [56, 57]. Mouse embryonic fibroblasts isolated from these PRIP null mice exhibited a decreased capacity for ligand-dependent transcriptional activation of PPARγ [56, 57]. PRIP interacting protein with methyltransferase domain (PIMT) was isolated in a yeast two-hybrid screen using PRIP as bait and enhances PRIP-mediated PPARγ transactivation [80]. Interestingly, PIMT binds to CBP/p300 and TRAP220 supporting a model in which the TRAP complex anchored by TRAP220 is bound to PPAR at the same time as the CBP/p300-associated complex [81].
The isolation of PPARα-interacting cofactor (PRIC) complex which enhances the transcription of PPARα further supports the existence of megacomplex on PPAR-target gene promoters [82]. Of the 25 polypeptides comprising PRIC complex, 18 contained one or more LXXLL motifs. Recognized proteins identified in the PRIC complex include SRC-1, CBP, TRAP220, PRIP, PIMT, TRAP100, and PGC-1, suggesting that CBP-associated complex and TRAP220 bound basal transcription factors may be bound simultaneously. PRIC285, a novel member of the PRIC complex renamed PPAR DNA-binding domain interacting protein (PDIP-1), was shown to bind to the DBD of PPARγ in a yeast two-hybrid assay. Two splice variants, PDIP-1a and PDIP-1b, were identified, and both were shown to transactivate all three isotypes of PPAR and thyroid receptor, whereas PDIP-1a but not PDIP-1b transactivates estrogen receptor (ER) α and androgen receptor (AR), indicating some receptor specificity [82].
3. COREPRESSORS
3.1. Corepressor RIP140 regulates energy metabolism but not adipogenesis
RIP140 was originally identified as a corepressor of ligand-dependent ER function by binding to the AF-2 domain [83]. It was later shown to bind to PPARα in a yeast two-hybrid screen [84]. Although PPARγ and RXR ligands promote the interaction of RIP140 with rat PPARγ in solution, RIP140 interaction with PPARγ/RXR heterodimers does not occur on DNA. This cofactor downregulates the activity of several nuclear receptors specifically by attenuating transactivation mediated by SRC-1. For instance, RIP140 competes with the coactivator SRC-1 for binding to PPARγ [84]. This evidence is suggestive of a model in which RIP140 indirectly regulates the activity of PPARγ by competing with coactivators such as SRC-1. RIP140−/− mice exhibit upregulation of energy metabolic genes UCP-1 and carnitine O-palmitoyl transferase I (CPT-I) and increased β-oxidation in adipocytes, albeit adipogenesis is unaffected [58]. This data suggests that a highly specific set of PPARγ mediated functions is modulated by RIP140 repression while other PPARγ functions such as adipogenesis remain unaltered.
3.2. Transcriptional corepressors for PPARγ: NCoR and SMRT
NCoR and SMRT function to recruit HDAC (histone deacetylase) complexes, which covalently modify nucleosomes to compact DNA and repress transcription [47]. Binding of NCoR and SMRT to NRs is mediated by the corepressor nuclear receptor box (CoRNR) [85]. This motif is very similar to the NR box with a consensus sequence of hydrophobic residues including leucine and isoleucine [86, 87]. The α-helix that contains the CoRNR box is predicted to be longer than the helix containing the NR box in coactivators [87], presenting a possible mechanism for cofactor selection via the ligand-induced conformational change of the NR. Thus, conformational change may exclude corepressors from the AF-2 binding pocket.
Evidence exists suggesting that in the absence of ligand, PPARγ recruits the transcriptional corepressors NCoR and SMRT to downregulate PPARγ-mediated transcriptional activity. Gene silencing of NCoR or SMRT in 3T3-L1 preadipocytes has been shown to increase adipocyte differentiation, a classical PPARγ2 function [42]. Moreover, treatment with the synthetic PPARγ ligand pioglitazone decreases both PPARγ-SMRT and PPARγ-NCoR interactions, although the PPARγ-SMRT interaction decrease is much more prominent. Furthermore, in a separate study by Krogsdam et al., repression of PPARγ-mediated transcription by NCoR exists even in the presence of ligand [88]. These studies underscore the transcriptional repression of PPARγ by NCoR and SMRT in vivo.
It appears that gene-specific factors may affect the conformation of PPARγ, further complicating the ligand-receptor-repressor interaction. One example of this variability is the differential activation of glycerol kinase (GyK) and aP2 transcription. Although both contain PPREs, PPARγ recruits corepressor NCoR to the GyK gene while recruiting coactivators to the aP2 gene [89]. The addition of TZD results in the activation of GyK by recruiting PGC-1α and displacing NCoR, while TZD treatment has little effect on transcription of aP2 and does not recruit PGC-1α to the aP2 promoter [89]. These data suggest that gene-specific PPARγ receptor conformation leads to the recruitment of different cofactor complexes.
Another corepressor, Sirt1, has also been shown to effectively inhibit PPARγ-mediated transcription [90]. This NAD-dependent deacetylase binds to NCoR and SMRT, presenting a model where Sirt1 is recruited to PPARγ via interactions with NCoR and/or SMRT. This was further supported by loss of Sirt1-mediated repression when NCoR levels were decreased via RNAi [90].
3.3. Summary of coactivators and corepressors in lipid and energy metabolism
Cellular energy metabolism is maintained through a delicate balance between energy intake and energy expenditure. When energy intake exceeds energy expenditure, excess energy is stored as lipid in WAT. Although BAT also allows storage of small amount of lipids, it is mainly responsible for energy dissipation. As PPARγ plays an essential role in lipid homeostasis, it is not surprising that multiple PPAR cofactors are involved in lipid and energy metabolism; namely, processes including adipocyte differentiation, lipid storage, and adaptive thermogenesis (Figure 1). PPARγ/RXR heterodimers are master regulators of preadipocyte differentiation into brown and white adipocytes. Multiple lines of evidence support the model that CBP/p300 and TRAP220 participate in white adipocyte differentiation, and this process is reversibly regulated by corepressors NCoR and SMRT [31, 42, 71]. On the contrary, differentiation of preadipocytes into BAT is regulated by a different set of coactivators such as PGC-1β/PGC-1α and SRC-1 [50, 52]. Conversion of white adipocyte to brown adipocyte-like cells can be at least partially catalyzed by ectopically expressed PGC-1α [62]. TIF2 plays important functions in the storage of fatty acids in WAT as evident by the fact that TIF2−/− mice are protected from obesity and TIF2−/− cells show an increase in lipolysis [50]. Brown adipocytes are enriched in mitochondria and the major function is adaptive thermogenesis in rodents. PGC-1α and SRC-1 are positive regulators of the thermogenic capacity of BAT [50, 52, 53], whereas the corepressor RIP140 appears to negatively regulate this process [58]. Lipid storage in brown adipocytes can be regulated by coactivators p/CIP and SRC-1 [51]. Figure 1 summarizes some of the major players in lipid and energy homeostasis based on current literature. It is worthy to note that some cellular processes require more stringent regulation than others, such that more than one member of the closely related proteins are simultaneously involved. For example, complementary actions of p/CIP and SRC-1 in lipid storage of brown adipocytes and two PGC-1 coactivators in brown fat differentiation are absolutely essential.
3.4. Ligand- and promoter-specific coregulator recruitment in PPARγ transactivation
A comparison of natural and synthetic PPARγ ligands reveals a distinct differential recruitment of transcriptional coactivators. 15d-PGJ2, an endogenous PPARγ ligand, is capable of inducing interactions between the PPARγ/RXR heterodimer and SRC-1, TIF2, p/CIP, p300, and TRAP220 [91]. However, the synthetic PPARγ ligand troglitazone did not induce interaction between the PPARγ/RXR heterodimer and any of these coactivators. Furthermore, the transactivation function of PPARγ was shown to be increased by these coactivators in the presence of 15d-PGJ2 and 9-HODE, but not troglitazone. FK614, a non-TZD synthetic PPARγ ligand, and two TZDs, rosiglitazone and pioglitazone, induce recruitment of SRC-1, CBP, and PGC-1α when bound to PPARγ. However, the level to which SRC-1 and CBP are recruited by FK614-bound PPARγ is altered in comparison to rosiglitazone- and pioglitazone-bound receptor (Fujimura, 2005) while PGC-1α showed similar levels of recruitment. These data suggest specific ligands can differentially define the coactivator complex, and that similar coactivators might have distinct in vivo functions.
4. CONCLUSIONS
The race to find new nuclear receptor coactivators and corepressors has resulted in a rapid increase in the number of known cofactors accompanied by insufficient knowledge as to their mechanisms of interaction and transcriptional mediation. Initial investigation has shown that seemingly redundant or promiscuous cofactors have a high amount of context specificity. Gene sequence- and ligand-specific nuclear receptor conformation appears to affect cofactor complex recruitment. The relative expression levels of coactivators and corepressors modulate nuclear receptor transactivation. In the case of PPARγ, there are only a few examples of these differential conditions thus far. Further investigation of these interactions may eventually allow for a better comprehension of context-specific expression profiles. Partial PPARγ agonists, such as FK614, that differentially activate PPARγ target genes may be effective in treating metabolic disease while reducing the side effects (e.g., promoting obesity) caused by current TZD-based treatments. The ability to target unique expression profiles may also lead to a more widespread ability to treat illnesses related to nuclear receptor function.
ACKNOWLEDGMENTS
We thank Chih-Hao Lee and Weimin He for critical reading of the manuscript. The third author is supported by a Susan Komen Breast Cancer Foundation Grant BCTR95306 and UWCCC core grant. The second author was supported by NIH Grant T32 CA009135.
LIST OF ABBREVIATIONS
- 15dPGJ2:
15-deoxy-Δ 12, 14-prostaglandin J2
- 9-HODE:
OX-LDL, 9-hydroxy-10, 12-octadecadienoic acid
- ACTR:
Activator of thyroid and retinoic acid receptor
- AF:
Activation function
- AIB1:
Amplified in breast cancer 1
- AR:
Androgen receptor
- BAF:
Brg1/Brm-associated factor
- BAT:
Brown adipose tissue
- CBP:
CREB-binding protein
- ChIP:
Chromatin immunoprecipitation
- CoRNR:
Corepressor nuclear receptor box
- CPT-I:
Carnitine O-palmitoyl transferase I
- CREB:
cAMP-responsive element binding protein
- DBD:
DNA-binding domain
- DKO:
Double knockout
- DRIP:
Vitamin D-interacting protein
- EMSA:
Electrophoretic mobility shift assay
- ER:
Estrogen receptor
- GRIP:
Glucocorticoid receptor interacting protein
- GST:
Glutathione s-transferase
- GyK:
Glycerol kinase
- HAT:
Histone acetyltransferase
- HDAC:
Histone deacetylase
- HMT:
Histone methyltransferase
- LBD:
Ligand binding domain
- LTB4:
Leukotriene B4
- MEF:
Mouse embryonic fibroblast
- NAD:
Nicotinamide adenine dinucleotide
- NCoA:
Nuclear coactivator
- NCoEx:
Nuclear corepressor exchange factors
- NCoR:
Nuclear corepressor
- NR:
Nuclear receptor
- NRC:
Nuclear hormone receptor coregulator
- p/CIP:
p300/CBP interacting protein
- PBP:
PPAR binding protein
- PDIP:
PPAR DNA-binding domain interacting protein
- PGC:
PPAR-gamma coactivator
- PGJ2:
Prostaglandin J2
- PIC:
Preinitiation complex
- PIMT:
PRIP interacting protein with methyltransferase domain
- PPAR:
Peroxisome proliferator-associated receptor
- PPRE:
PPAR-response element
- PRIC:
PPARα-interacting cofactor
- PRIP:
PPAR interacting protein
- PRMT:
Protein arginine methyltransferase
- RAP:
Receptor-associated protein
- RIP140:
Receptor interacting protein 140
- RRM:
RNA-recognition motif
- RXR:
Retinoid X receptor
- Sirt1:
Sirtuin 1
- SMRT:
Silencing mediator of retinoid and thyroid receptors
- SRC:
Steroid receptor coactivator
- SWI/SNF:
Mating type switching/sucrose nonfermenting
- TBL1:
Transducin β-like 1
- TBLR1:
Transducin β-like related 1
- TIF:
Transcriptional intermediary factor
- TRAP:
Thyroid receptor-associated protein
- TRBP:
Thyroid receptor-binding protein
- TZD:
Thiazolidinedione
- UCP-1:
Uncoupling protein 1
- WAT:
White adipose tissue
- Wy-14643:
(4-Chloro-6-[(2, 3-dimethylphenyl)amino]-2-pyrimidinyl) thioacetic acid
References
- 1.Issemann I, Green S. Activation of a member of the steroid hormone receptor superfamily by peroxisome proliferators. Nature. 1990;347(6294):645–650. doi: 10.1038/347645a0. [DOI] [PubMed] [Google Scholar]
- 2.Berger J, Moller DE. The mechanisms of action of PPARs. Annual Review of Medicine. 2002;53(1):409–435. doi: 10.1146/annurev.med.53.082901.104018. [DOI] [PubMed] [Google Scholar]
- 3.Evans RM, Barish GD, Wang Y-X. PPARs and the complex journey to obesity. Nature Medicine. 2004;10(4):355–361. doi: 10.1038/nm1025. [DOI] [PubMed] [Google Scholar]
- 4.Feige JN, Gelman L, Michalik L, Desvergne B, Wahli W. From molecular action to physiological outputs: peroxisome proliferator-activated receptors are nuclear receptors at the crossroads of key cellular functions. Progress in Lipid Research. 2006;45(2):120–159. doi: 10.1016/j.plipres.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 5.Vidal-Puig AJ, Considine RV, Jimenez-Liñan M, et al. Peroxisome proliferator-activated receptor gene expression in human tissues: effects of obesity, weight loss, and regulation by insulin and glucocorticoids. Journal of Clinical Investigation. 1997;99(10):2416–2422. doi: 10.1172/JCI119424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ren D, Collingwood TN, Rebar EJ, Wolffe AP, Camp HS. PPARγ knockdown by engineered transcription factors: exogenous PPARγ2 but not PPARγ1 reactivates adipogenesis. Genes & Development. 2002;16(1):27–32. doi: 10.1101/gad.953802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Werman A, Hollenberg A, Solanes G, Bjørbæk C, Vidal-Puig AJ, Flier JS. Ligand-independent activation domain in the N terminus of peroxisome proliferator-activated receptor γ (PPARγ). Differential activity of PPARγ1 and -2 isoforms and influence of insulin. Journal of Biological Chemistry. 1997;272(32):20230–20235. doi: 10.1074/jbc.272.32.20230. [DOI] [PubMed] [Google Scholar]
- 8.Tontonoz P, Hu E, Spiegelman BM. Stimulation of adipogenesis in fibroblasts by PPARγ2, a lipid-activated transcription factor. Cell. 1994;79(7):1147–1156. doi: 10.1016/0092-8674(94)90006-x. [DOI] [PubMed] [Google Scholar]
- 9.Tontonoz P, Hu E, Graves RA, Budavari AI, Spiegelman BM. mPPARγ2: tissue-specific regulator of an adipocyte enhancer. Genes & Development. 1994;8(10):1224–1234. doi: 10.1101/gad.8.10.1224. [DOI] [PubMed] [Google Scholar]
- 10.Yu S, Viswakarma N, Batra SK, Sambasiva Rao M, Reddy JK. Identification of promethin and PGLP as two novel up-regulated genes in PPARγ1-induced adipogenic mouse liver. Biochimie. 2004;86(11):743–761. doi: 10.1016/j.biochi.2004.09.015. [DOI] [PubMed] [Google Scholar]
- 11.Chawla A, Schwarz EJ, Dimaculangan DD, Lazar MA. Peroxisome proliferator-activated receptor (PPAR) γ: adipose-predominant expression and induction early in adipocyte differentiation. Endocrinology. 1994;135(2):798–800. doi: 10.1210/endo.135.2.8033830. [DOI] [PubMed] [Google Scholar]
- 12.Gelman L, Fruchart J-C, Auwerx J. An update on the mechanisms of action of the peroxisome proliferator-activated receptors (PPARs) and their roles in inflammation and cancer. Cellular and Molecular Life Sciences. 1999;55(6-7):932–943. doi: 10.1007/s000180050345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lehrke M, Lazar MA. The many faces of PPARγ . Cell. 2005;123(6):993–999. doi: 10.1016/j.cell.2005.11.026. [DOI] [PubMed] [Google Scholar]
- 14.Torchia J, Glass C, Rosenfeld MG. Co-activators and co-repressors in the integration of transcriptional responses. Current Opinion in Cell Biology. 1998;10(3):373–383. doi: 10.1016/s0955-0674(98)80014-8. [DOI] [PubMed] [Google Scholar]
- 15.Kliewer SA, Umesono K, Noonan DJ, Heyman RA, Evans RM. Convergence of 9-cis retinoic acid and peroxisome proliferator signalling pathways through heterodimer formation of their receptors. Nature. 1992;358(6389):771–774. doi: 10.1038/358771a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rosenfeld MG, Lunyak VV, Glass CK. Sensors and signals: a coactivator/corepressor/epigenetic code for integrating signal-dependent programs of transcriptional response. Genes & Development. 2006;20(11):1405–1428. doi: 10.1101/gad.1424806. [DOI] [PubMed] [Google Scholar]
- 17.Hermanson O, Glass CK, Rosenfeld MG. Nuclear receptor coregulators: multiple modes of modification. Trends in Endocrinology and Metabolism. 2002;13(2):55–60. doi: 10.1016/s1043-2760(01)00527-6. [DOI] [PubMed] [Google Scholar]
- 18.Westin S, Rosenfeld MG, Glass CK. Nuclear receptor coactivators. Advances in Pharmacology. 2000;47:89–112. doi: 10.1016/s1054-3589(08)60110-6. [DOI] [PubMed] [Google Scholar]
- 19.Xu L, Glass CK, Rosenfeld MG. Coactivator and corepressor complexes in nuclear receptor function. Current Opinion in Genetics & Development. 1999;9(2):140–147. doi: 10.1016/S0959-437X(99)80021-5. [DOI] [PubMed] [Google Scholar]
- 20.Heery DM, Hoare S, Hussain S, Parker MG, Sheppard H. Core LXXLL motif sequences in CREB-binding protein, SRC1, and RIP140 define affinity and selectivity for steroid and retinoid receptors. Journal of Biological Chemistry. 2001;276(9):6695–6702. doi: 10.1074/jbc.M009404200. [DOI] [PubMed] [Google Scholar]
- 21.Heery DM, Kalkhoven E, Hoare S, Parker MG. A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature. 1997;387(6634):733–736. doi: 10.1038/42750. [DOI] [PubMed] [Google Scholar]
- 22.Leo C, Chen JD. The SRC family of nuclear receptor coactivators. Gene. 2000;245(1):1–11. doi: 10.1016/s0378-1119(00)00024-x. [DOI] [PubMed] [Google Scholar]
- 23.Xu W. Nuclear receptor coactivators: the key to unlock chromatin. Biochemistry and Cell Biology. 2005;83(4):418–428. doi: 10.1139/o05-057. [DOI] [PubMed] [Google Scholar]
- 24.Martens JA, Winston F. Recent advances in understanding chromatin remodeling by Swi/Snf complexes. Current Opinion in Genetics & Development. 2003;13(2):136–142. doi: 10.1016/s0959-437x(03)00022-4. [DOI] [PubMed] [Google Scholar]
- 25.Salma N, Xiao H, Mueller E, Imbalzano AN. Temporal recruitment of transcription factors and SWI/SNF chromatin-remodeling enzymes during adipogenic induction of the peroxisome proliferator-activated receptor γ nuclear hormone receptor. Molecular and Cellular Biology. 2004;24(11):4651–4663. doi: 10.1128/MCB.24.11.4651-4663.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Debril MB, Gelman L, Fayard E, Annicotte JS, Rocchi S, Auwerx J. Transcription factors and nuclear receptors interact with the SWI/SNF complex through the BAF60c subunit. Journal of Biological Chemistry. 2004;279(16):16677–16686. doi: 10.1074/jbc.M312288200. [DOI] [PubMed] [Google Scholar]
- 27.Fondell JD, Ge H, Roeder RG. Ligand induction of a transcriptionally active thyroid hormone receptor coactivator complex. Proceedings of the National Academy of Sciences of the United States of America. 1996;93(16):8329–8333. doi: 10.1073/pnas.93.16.8329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rachez C, Lemon BD, Suldan Z, et al. Ligand-dependent transcription activation by nuclear receptors requires the DRIP complex. Nature. 1999;398(6730):824–828. doi: 10.1038/19783. [DOI] [PubMed] [Google Scholar]
- 29.Roeder RG. Role of general and gene-specific cofactors in the regulation of eukaryotic transcription. Cold Spring Harbor Symposia on Quantitative Biology. 1998;63:201–218. doi: 10.1101/sqb.1998.63.201. [DOI] [PubMed] [Google Scholar]
- 30.Malik S, Roeder RG. Transcriptional regulation through Mediator-like coactivators in yeast and metazoan cells. Trends in Biochemical Sciences. 2000;25(6):277–283. doi: 10.1016/s0968-0004(00)01596-6. [DOI] [PubMed] [Google Scholar]
- 31.Ge K, Guermah M, Yuan CX, et al. Transcription coactivator TRAP220 is required for PPAR γ 2-stimulated adipogenesis. Nature. 2002;417(6888):563–567. doi: 10.1038/417563a. [DOI] [PubMed] [Google Scholar]
- 32.Puigserver P, Adelmant G, Wu Z, et al. Activation of PPARγ coactivator-1 through transcription factor docking. Science. 1999;286(5443):1368–1371. doi: 10.1126/science.286.5443.1368. [DOI] [PubMed] [Google Scholar]
- 33.Wallberg AE, Yamamura S, Malik S, Spiegelman BM, Roeder RG. Coordination of p300-mediated chromatin remodeling and TRAP/mediator function through coactivator PGC-1α . Molecular Cell. 2003;12(5):1137–1149. doi: 10.1016/s1097-2765(03)00391-5. [DOI] [PubMed] [Google Scholar]
- 34.Monsalve M, Wu Z, Adelmant G, Puigserver P, Fan M, Spiegelman BM. Direct coupling of transcription and mRNA processing through the thermogenic coactivator PGC-1. Molecular Cell. 2000;6(2):307–316. doi: 10.1016/s1097-2765(00)00031-9. [DOI] [PubMed] [Google Scholar]
- 35.Puigserver P, Spiegelman BM. Peroxisome proliferator-activated receptor-γ coactivator 1 α (PGC-1 α): transcriptional coactivator and metabolic regulator. Endocrine Reviews. 2003;24(1):78–90. doi: 10.1210/er.2002-0012. [DOI] [PubMed] [Google Scholar]
- 36.Lin J, Handschin C, Spiegelman BM. Metabolic control through the PGC-1 family of transcription coactivators. Cell Metabolism. 2005;1(6):361–370. doi: 10.1016/j.cmet.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 37.Horlein AJ, Naar AM, Heinzel T, et al. Ligand-independent repression by the thyroid hormone receptor mediated by a nuclear receptor co-repressor. Nature. 1995;377(6548):397–404. doi: 10.1038/377397a0. [DOI] [PubMed] [Google Scholar]
- 38.Chen JD, Evans RM. A transcriptional co-repressor that interacts with nuclear hormone receptors. Nature. 1995;377(6548):454–457. doi: 10.1038/377454a0. [DOI] [PubMed] [Google Scholar]
- 39.Dowell P, Ishmael JE, Avram D, Peterson VJ, Nevrivy DJ, Leid M. Identification of nuclear receptor corepressor as a peroxisome proliferator-activated receptor α interacting protein. Journal of Biological Chemistry. 1999;274(22):15901–15907. doi: 10.1074/jbc.274.22.15901. [DOI] [PubMed] [Google Scholar]
- 40.Stanley TB, Leesnitzer LM, Montana VG, et al. Subtype specific effects of peroxisome proliferator-activated receptor ligands on corepressor affinity. Biochemistry. 2003;42(31):9278–9287. doi: 10.1021/bi034472c. [DOI] [PubMed] [Google Scholar]
- 41.Krogsdam A-M, Nielsen CA, Neve S, et al. Nuclear receptor corepressor-dependent repression of peroxisome-proliferator-activated receptor δ-mediated transactivation. Biochemical Journal. 2002;363(pt 1):157–165. doi: 10.1042/0264-6021:3630157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yu C, Markan K, Temple KA, Deplewski D, Brady MJ, Cohen RN. The nuclear receptor corepressors NCoR and SMRT decrease peroxisome proliferator-activated receptor γ transcriptional activity and repress 3T3-L1 adipogenesis. Journal of Biological Chemistry. 2005;280(14):13600–13605. doi: 10.1074/jbc.M409468200. [DOI] [PubMed] [Google Scholar]
- 43.Picard F, Kurtev M, Chung N, et al. Sirt1 promotes fat mobilization in white adipocytes by repressing PPAR-γ . Nature. 2004;429(6993):771–776. doi: 10.1038/nature02583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Perissi V, Rosenfeld MG. Controlling nuclear receptors: the circular logic of cofactor cycles. Nature Reviews. Molecular Cell Biology. 2005;6(7):542–554. doi: 10.1038/nrm1680. [DOI] [PubMed] [Google Scholar]
- 45.Yoon HG, Chan DW, Huang ZQ, et al. Purification and functional characterization of the human N-CoR complex: the roles of HDAC3, TBL1 and TBLR1. EMBO Journal. 2003;22(6):1336–1346. doi: 10.1093/emboj/cdg120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Perissi V, Aggarwal A, Glass CK, Rose DW, Rosenfeld MG. A corepressor/coactivator exchange complex required for transcriptional activation by nuclear receptors and other regulated transcription factors. Cell. 2004;116(4):511–526. doi: 10.1016/s0092-8674(04)00133-3. [DOI] [PubMed] [Google Scholar]
- 47.Privalsky ML. The role of corepressors in transcriptional regulation by nuclear hormone receptors. Annual Review of Physiology. 2004;66:315–360. doi: 10.1146/annurev.physiol.66.032802.155556. [DOI] [PubMed] [Google Scholar]
- 48.McKenna NJ, O'Malley BW. From ligand to response: generating diversity in nuclear receptor coregulator function. Journal of Steroid Biochemistry and Molecular Biology. 2000;74(5):351–356. doi: 10.1016/s0960-0760(00)00112-6. [DOI] [PubMed] [Google Scholar]
- 49.Xu J, O'Malley BW. Molecular mechanisms and cellular biology of the steroid receptor coactivator (SRC) family in steroid receptor function. Reviews in Endocrine & Metabolic Disorders. 2002;3(3):185–192. doi: 10.1023/a:1020016208071. [DOI] [PubMed] [Google Scholar]
- 50.Picard F, Gehin M, Annicotte J, et al. SRC-1 and TIF2 control energy balance between white and brown adipose tissues. Cell. 2002;111(7):931–941. doi: 10.1016/s0092-8674(02)01169-8. [DOI] [PubMed] [Google Scholar]
- 51.Wang Z, Qi C, Krones A, et al. Critical roles of the p160 transcriptional coactivators p/CIP and SRC-1 in energy balance. Cell Metabolism. 2006;3(2):111–122. doi: 10.1016/j.cmet.2006.01.002. [DOI] [PubMed] [Google Scholar]
- 52.Uldry M, Yang W, St-Pierre J, Lin J, Seale P, Spiegelman BM. Complementary action of the PGC-1 coactivators in mitochondrial biogenesis and brown fat differentiation. Cell Metabolism. 2006;3(5):333–341. doi: 10.1016/j.cmet.2006.04.002. [DOI] [PubMed] [Google Scholar]
- 53.Lin J, Wu PH, Tarr PT, et al. Defects in adaptive energy metabolism with CNS-linked hyperactivity in PGC-1α null mice. Cell. 2004;119(1):121–135. doi: 10.1016/j.cell.2004.09.013. [DOI] [PubMed] [Google Scholar]
- 54.Barak Y, Nelson MC, Ong ES, et al. PPAR γ is required for placental, cardiac, and adipose tissue development. Molecular Cell. 1999;4(4):585–595. doi: 10.1016/s1097-2765(00)80209-9. [DOI] [PubMed] [Google Scholar]
- 55.Zhu Y, Kan L, Qi C, et al. Isolation and characterization of peroxisome proliferator-activated receptor (PPAR) interacting protein (PRIP) as a coactivator for PPAR. Journal of Biological Chemistry. 2000;275(18):13510–13516. doi: 10.1074/jbc.275.18.13510. [DOI] [PubMed] [Google Scholar]
- 56.Antonson P, Schuster GU, Wang L, et al. Inactivation of the nuclear receptor coactivator RAP250 in mice results in placental vascular dysfunction. Molecular and Cellular Biology. 2003;23(4):1260–1268. doi: 10.1128/MCB.23.4.1260-1268.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhu YJ, Crawford SE, Stellmach V, et al. Coactivator PRIP, the peroxisome proliferator-activated receptor-interacting protein, is a modulator of placental, cardiac, hepatic, and embryonic development. Journal of Biological Chemistry. 2003;278(3):1986–1990. doi: 10.1074/jbc.C200634200. [DOI] [PubMed] [Google Scholar]
- 58.Leonardsson G, Steel JH, Christian M, et al. Nuclear receptor corepressor RIP140 regulates fat accumulation. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(22):8437–8442. doi: 10.1073/pnas.0401013101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Puigserver P, Wu Z, Park CW, Graves R, Wright M, Spiegelman BM. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell. 1998;92(6):829–839. doi: 10.1016/s0092-8674(00)81410-5. [DOI] [PubMed] [Google Scholar]
- 60.Wu Y, Chin WW, Wang Y, Burris TP. Ligand and coactivator identity determines the requirement of the charge clamp for coactivation of the peroxisome proliferator-activated receptor γ . Journal of Biological Chemistry. 2003;278(10):8637–8644. doi: 10.1074/jbc.M210910200. [DOI] [PubMed] [Google Scholar]
- 61.Guan HP, Ishizuka T, Chui PC, Lehrke M, Lazar MA. Corepressors selectively control the transcriptional activity of PPARγ in adipocytes. Genes & Development. 2005;19(4):453–461. doi: 10.1101/gad.1263305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wu Z, Puigserver P, Andersson U. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell. 1999;98(1):115–124. doi: 10.1016/S0092-8674(00)80611-X. [DOI] [PubMed] [Google Scholar]
- 63.Tiraby C, Langin D. Conversion from white to brown adipocytes: a strategy for the control of fat mass? Trends in Endocrinology and Metabolism. 2003;14(10):439–441. doi: 10.1016/j.tem.2003.10.001. [DOI] [PubMed] [Google Scholar]
- 64.Tsukiyama-Kohara K, Poulin F, Kohara M, et al. Adipose tissue reduction in mice lacking the translational inhibitor 4E-BP1. Nature Medicine. 2001;7(10):1128–1132. doi: 10.1038/nm1001-1128. [DOI] [PubMed] [Google Scholar]
- 65.Cederberg A, Gronning LM, Ahren B, Tasken K, Carlsson P, Enerback S. FOXC2 is a winged helix gene that counteracts obesity, hypertriglyceridemia, and diet-induced insulin resistance. Cell. 2001;106(5):563–573. doi: 10.1016/s0092-8674(01)00474-3. [DOI] [PubMed] [Google Scholar]
- 66.Kopecky J, Clarke G, Enerback S, Spiegelman B, Kozak LP. Expression of the mitochondrial uncoupling protein gene from the aP2 gene promoter prevents genetic obesity. Journal of Clinical Investigation. 1995;96(6):2914–2923. doi: 10.1172/JCI118363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lin J, Puigserver P, Donovan J, Tarr P, Spiegelman BM. Peroxisome proliferator-activated receptor γ coactivator 1β (PGC-1β ), a novel PGC-1-related transcription coactivator associated with host cell factor. Journal of Biological Chemistry. 2002;277(3):1645–1648. doi: 10.1074/jbc.C100631200. [DOI] [PubMed] [Google Scholar]
- 68.Lin J, Tarr PT, Yang R, et al. PGC-1β in the regulation of hepatic glucose and energy metabolism. Journal of Biological Chemistry. 2003;278(33):30843–30848. doi: 10.1074/jbc.M303643200. [DOI] [PubMed] [Google Scholar]
- 69.St-Pierre J, Lin J, Krauss S, et al. Bioenergetic analysis of peroxisome proliferator-activated receptor γ coactivators 1α and 1β (PGC-1α and PGC-1β) in muscle cells. Journal of Biological Chemistry. 2003;278(29):26597–26603. doi: 10.1074/jbc.M301850200. [DOI] [PubMed] [Google Scholar]
- 70.Gelman L, Zhou G, Fajas L, Raspe E, Fruchart JC, Auwerx J. p300 interacts with the N- and C-terminal part of PPARγ2 in a ligand-independent and -dependent manner, respectively. Journal of Biological Chemistry. 1999;274(12):7681–7688. doi: 10.1074/jbc.274.12.7681. [DOI] [PubMed] [Google Scholar]
- 71.Takahashi N, Kawada T, Yamamoto T, et al. Overexpression and ribozyme-mediated targeting of transcriptional coactivators CREB-binding protein and p300 revealed their indispensable roles in adipocyte differentiation through the regulation of peroxisome proliferator-activated receptor γ . Journal of Biological Chemistry. 2002;277(19):16906–16912. doi: 10.1074/jbc.M200585200. [DOI] [PubMed] [Google Scholar]
- 72.Sif S, Saurin AJ, Imbalzano AN, Kingston RE. Purification and characterization of mSin3A-containing Brg1 and hBrm chromatin remodeling complexes. Genes & Development. 2001;15(5):603–618. doi: 10.1101/gad.872801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bultman S, Gebuhr T, Yee D, et al. A Brg1 null mutation in the mouse reveals functional differences among mammalian SWI/SNF complexes. Molecular Cell. 2000;6(6):1287–1295. doi: 10.1016/s1097-2765(00)00127-1. [DOI] [PubMed] [Google Scholar]
- 74.Reyes JC, Barra J, Muchardt C, Camus A, Babinet C, Yaniv M. Altered control of cellular proliferation in the absence of mammalian brahma (SNF2α) EMBO Journal. 1998;17(23):6979–6991. doi: 10.1093/emboj/17.23.6979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lemon B, Inouye C, King DS, Tjian R. Selectivity of chromatin-remodelling cofactors for ligand-activated transcription. Nature. 2001;414(6866):924–928. doi: 10.1038/414924a. [DOI] [PubMed] [Google Scholar]
- 76.Yuan CX, Ito M, Fondell JD, Fu ZY, Roeder RG. The TRAP220 component of a thyroid hormone receptor- associated protein (TRAP) coactivator complex interacts directly with nuclear receptors in a ligand-dependent fashion. Proceedings of the National Academy of Sciences of the United States of America. 1998;95(14):7939–7944. doi: 10.1073/pnas.95.14.7939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhu Y, Qi C, Jain S, Rao MS, Reddy JK. Isolation and characterization of PBP, a protein that interacts with peroxisome proliferator-activated receptor. Journal of Biological Chemistry. 1997;272(41):25500–25506. doi: 10.1074/jbc.272.41.25500. [DOI] [PubMed] [Google Scholar]
- 78.Zhu Y, Qi C, Jia Y, Nye JS, Rao MS, Reddy JK. Deletion of PBP/PPARBP, the gene for nuclear receptor coactivator peroxisome proliferator-activated receptor-binding protein, results in embryonic lethality. Journal of Biological Chemistry. 2000;275(20):14779–14782. doi: 10.1074/jbc.C000121200. [DOI] [PubMed] [Google Scholar]
- 79.Caira F, Antonson P, Pelto-Huikko M, Treuter E, Gustafsson JA. Cloning and characterization of RAP250, a novel nuclear receptor coactivator. Journal of Biological Chemistry. 2000;275(8):5308–5317. doi: 10.1074/jbc.275.8.5308. [DOI] [PubMed] [Google Scholar]
- 80.Zhu Y , Qi C, Cao W-Q, et al. Cloning and characterization of PIMT, a protein with a methyltransferase domain, which interacts with and enhances nuclear receptor coactivator PRIP function. Proceedings of the National Academy of Sciences of the United States of America. 2001;98(18):10380–10385. doi: 10.1073/pnas.181347498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Misra P, Qi C, Yu S, et al. Interaction of PIMT with transcriptional coactivators CBP, p300, and PBP differential role in transcriptional regulation. Journal of Biological Chemistry. 2002;277(22):20011–20019. doi: 10.1074/jbc.M201739200. [DOI] [PubMed] [Google Scholar]
- 82.Surapureddi S, Yu S, Bu H, et al. Identification of a transcriptionally active peroxisome proliferator-activated receptor α -interacting cofactor complex in rat liver and characterization of PRIC285 as a coactivator. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(18):11836–11841. doi: 10.1073/pnas.182426699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Cavailles V, Dauvois S, L'Horset F, et al. Nuclear factor RIP140 modulates transcriptional activation by the estrogen receptor. EMBO Journal. 1995;14(15):3741–3751. doi: 10.1002/j.1460-2075.1995.tb00044.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Treuter E, Albrektsen T, Johansson L, Leers J, Gustafsson JA. A regulatory role for RIP140 in nuclear receptor activation. Molecular Endocrinology. 1998;12(6):864–881. doi: 10.1210/mend.12.6.0123. [DOI] [PubMed] [Google Scholar]
- 85.Hu X, Lazar MA. The CoRNR motif controls the recruitment of corepressors by nuclear hormone receptors. Nature. 1999;402(6757):93–96. doi: 10.1038/47069. [DOI] [PubMed] [Google Scholar]
- 86.Nagy L, Kao HY, Love JD, et al. Mechanism of corepressor binding and release from nuclear hormone receptors. Genes & Development. 1999;13(24):3209–3216. doi: 10.1101/gad.13.24.3209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Perissi V, Staszewski LM, McInerney EM, et al. Molecular determinants of nuclear receptor-corepressor interaction. Genes & Development. 1999;13(24):3198–3208. doi: 10.1101/gad.13.24.3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Krogsdam A-M, Nielsen CA, Neve S, et al. Nuclear receptor corepressor-dependent repression of peroxisome-proliferator-activated receptor δ-mediated transactivation. Biochemical Journal. 2002;363(pt 1):157–165. doi: 10.1042/0264-6021:3630157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Guan HP, Ishizuka T, Chui PC, Lehrke M, Lazar MA. Corepressors selectively control the transcriptional activity of PPARγ in adipocytes. Genes & Development. 2005;19(4):453–461. doi: 10.1101/gad.1263305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Picard F, Kurtev M, Chung N, et al. Sirt1 promotes fat mobilization in white adipocytes by repressing PPAR-γ . Nature. 2004;429(6993):771–776. doi: 10.1038/nature02583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kodera Y, Takeyama K, Murayama A, Suzawa M, Masuhiro Y, Kato S. Ligand type-specific interactions of peroxisome proliferator-activated receptor γ with transcriptional coactivators. Journal of Biological Chemistry. 2000;275(43):33201–33204. doi: 10.1074/jbc.C000517200. [DOI] [PubMed] [Google Scholar]