Abstract
DNA vaccination can elicit both humoral and cellular immune responses and can confer protection against several pathogens. However, DNA vaccines expressing the envelope (Env) protein of human immunodeficiency virus (HIV) have been relatively ineffective at generating high titer, long-lasting, neutralizing antibodies in a variety of animal models. In this study, we report that fusion of Env and the complement component, C3d, in a DNA vaccine, enhances the titers of antibody to Env. Plasmids were generated that expressed a secreted form of Env (sgp120) from three isolates of HIV and these same forms fused to three tandem copies of the murine homologue of C3d (sgp120-3C3d). Analyses of titers and avidity maturation of the raised antibody indicated that immunizations with each of the sgp120-3C3d-expressing DNAs accelerated both the onset and the avidity maturation of antibody to Env.
INTRODUCTION
Human immunodeficiency virus (HIV), the etiologic agent of AIDS, has become the major cause of death in individuals 25 to 44 years of age in the United States.1 Nineteen million people have died worldwide since the first cases were identified in 1981 and the total number of people living with HIV/AIDS is currently estimated at 35 million.1 At the present rate, HIV is spreading faster in the human population than any infectious agent in the last 100 years. Despite the effectiveness of the current highly active antiretroviral therapy (HAART) drug cocktails in developed countries, a vaccine against HIV is the best method to combat the worldwide AIDS pandemic.
DNA vaccination has become the fastest growing field in vaccine technology (for reviews, see 2-4). These genetic vaccines consist of eukaryotic expression plasmids that are inoculated into target cells and translated into proteins.2 DNA vaccines are comparatively easy to develop and manufacture, and are likely to not require a cold chain for worldwide distribution. In animal models, DNA vaccination induces protective immunity against a variety of pathogens including influenza, herpes simplex, rabies, malaria, and measles.3,4 These studies have demonstrated that DNA vaccination effectively induces both humoral and cellular immune response to immunogens from diverse infectious agents. However, DNA immunizations have been less successful at generating neutralizing antibodies against HIV-1.5 Unlike most immunogens, multiple DNA immunizations are required to elicit even modest titers of neutralizing antibody to the HIV envelope (Env) glycoprotein.6-13 In addition, the antibody responses raised by DNA vaccination, like those to Env (gp120) subunit immunizations, are transient, rising and falling with each successive immunization.14-16 Studies using simian immunodeficiency virus (SIV) in a macaque animal model demonstrated that DNA immunization elicited neutralizing antibody that was only 10% that of SIV-infected monkeys.10 In natural infections, in HIV-infected patients, or in experimentally SIV-infected rhesus macaques, specific antibodies require 6 to 8 months to achieve avidity maturation. This maturation is associated with the appearance of neutralizing antibody.17 The time required for maturation of envelope-specific antibodies parallels the time necessary for the development of protective immunity to experimental challenge with SIV.17 Also, protection to virus challenge could be associated with a combination of antibody properties that includes high titer to Env, high avidity, and in vitro neutralization of the challenge virus. Therefore, we sought to increase the efficacy of DNA vaccines expressing HIV Env using a component of the innate immune system, C3d, to enhance both the levels of antibody and the avidity maturation of the elicited antibody.
In prior studies in mice, the fusion of two or three copies of C3d to a model antigen, hen egg lysozyme (HEL), increased the efficiency of immunizations by more than 1000-fold.18 In the human immune system, C3d is one of the final degradation products of the third complement protein, C3. One consequence of complement activation is the covalent attachment of the C3d to antigen. C3d in turn binds to CD21 on B lymphocytes, a molecule with B cell stimulatory functions that amplify B lymphocyte activation.18 Recently, we examined whether a DNA vaccine expressing a fusion of hemagglutinin (HA) from influenza virus and the C3d component of complement could achieve an earlier and more efficient protective immune response.19 Our results demonstrated that mice vaccinated with DNA expressing a secreted HA fused to three copies of C3d (sHA-3C3d) generated antibody that underwent more rapid avidity maturation than antibody generated by secreted or transmembrane forms of HA. This resulted in more rapid appearance of hemagglutination inhibition (HI) activity and protective immunity.19
In this study, we used a similar approach by fusing three copies of murine C3d to the carboxyl terminus of the HIV Env gp120 subunit. Using DNA vaccination, BALB/c mice were inoculated and assayed for enhanced immune responses. The fusion constructs induced higher antibody responses to Env and a faster onset of avidity maturation than did the respective wild-type gp120 sequences. These results suggest that the efficacy of DNA vaccines for raising antibody can be significantly improved by fusing proteins with C3d.
MATERIALS AND METHODS
Plasmid DNA
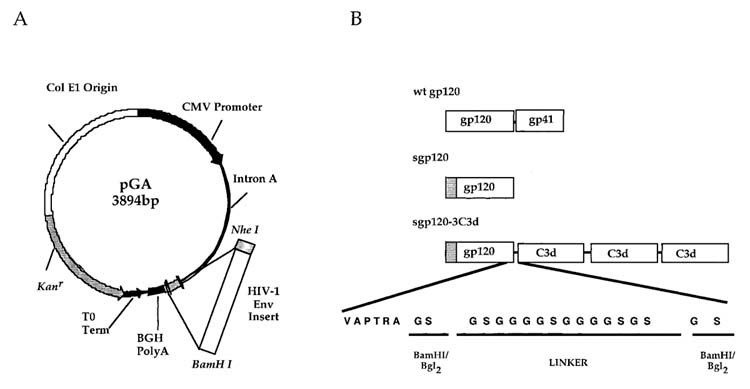
pGA, a eukaryotic expression vector, has been described previously.19 Briefly, the vector was constructed to contain the cytomegalovirus immediate-early promoter (CMV-IE) plus intron A (IA) for initiating transcription of eukaryotic inserts and the bovine growth hormone polyadenylation signal [BGH poly(A)] for termination of transcription (Fig. 1). The vector contains the Col E1 origin of replication for prokaryotic replication and the kanamycin resistance gene (Kanr) for selection in antibiotic media.
FIG. 1.
Schematic representation of vector DNA vaccine constructs. (A) The pGA vector contains the cytomegalovirus immediate-early promoter (CMV-IE) plus intron A (IA) for initiating transcription of eukaryotic inserts and the bovine growth hormone polyadenylation terminator [BGH poly(A)] for termination of transcription. The vector also contains the Col E1 origin of replication for prokaryotic replication as well as the kanamycin resistance (Kanr) gene for selection in antibiotic media. The lambda T0 terminator has been placed 3′ to the Kanr to increase the stability of eukaryotic inserts. Inserts were cloned into the vector using the HindIII and BamHI restriction endonuclease sites. (B) The top schematic represents the wild-type, transmembrane form of the Env protein. The middle schematic represents the secreted gp120 form of the Env. The bottom schematic represents the sgp120-3C3d construct used as a vaccine insert. Linkers composed of two repeats of four glycines and a serine {(G4S)2} were fused at the junctures of HA and C3d and between each C3d repeat.
HIV envelope sequences from the isolates, ADA, IIIB, and 89.6 encoding almost the entire gp120 region (Fig. 1A) and C3d sequences were cloned into the pGA vaccine vector using unique restriction endonuclease sites (Fig. 1B). The gp120 segment encodes a region from amino acid 32 to 465 and ended with the amino acid sequence VAPTRA. The first 32 amino acids were deleted from the N-terminus of each sgp120 and replaced with a leader sequenced from the trypsin plasminogen activator (tpA). The vectors expressing sgp120-C3d fusion proteins were generated by cloning three tandem repeats of the mouse homologue of C3d in frame with the sgp120-expressing DNA. The construct design was based upon Dempsey et al.18 Linkers composed of two repeats of four glycines and a serine {(G4S)2} were fused at the junctures of ENV and C3d and between each C3d repeat. Potential proteolytic cleavage sites between the junction of ENV and C3d and the junctions of 3C3d were mutated by ligating BamHI and BglII restriction endonuclease sites to mutate an Arg codon to a Gly codon.19
The plasmids were amplified in Escherichia coli strain DH5a, purified using anion-exchange resin columns (Qiagen, Valencia, CA) and stored at 220° in dH2O. Plasmids were verified by appropriate restriction enzyme digestion and gel electrophoresis. Purity of DNA preparations was determined by optical density reading at 260 and 280 nm.
Mice and DNA immunizations
Six- to 8-week-old BALB/c mice (Harlan Sprague Dawley, Indianapolis, IN) were used for inoculations. Mice, housed in microisolator units and allowed free access to food and water, were cared for under USDA guidelines for laboratory animals. Mice were anesthetized with 0.03-0.04 ml of a mixture of 5 ml ketamine-HCl (100 mg/ml) and 1 ml xylazine (20 mg/ml). Gene gun immunizations were performed on shaved abdominal skin using the hand-held Accell® gene delivery system as described previously.20-22 Mice were immunized with two gene gun doses containing 0.5 μg of DNA/0.5 mg of approximately 1-μm gold beads (DeGussa-Huls Corp., Ridgefield Park, NJ) at a helium pressure setting of 400 psi.
Transfections and expression analysis
The human embryonic kidney cell line, 293T (5 × 105 cells/transfection), was transfected with 2 μg of DNA using 12% lipofectamine according to the manufacturer’s (Life Technologies, Grand Island, NY) guidelines. Supernatants were collected and stored at 220°. Cell lysates were collected in 300 μl of RIPA lysis buffer [0.05 M Tris-HCl, pH 8.0, 0.1% sodium dodecyl sulfate (SDS), 1.0% Triton X-100, 2 mM phenylmethylsulfonyl fluoride (PMSF), 0.15 M NaCl] and stored at 220°. Quantitative antigen capture enzyme-linked immunosorbent assays (ELISAs) were conducted as previously described.23
For Western hybridization analysis, 15 μl of supernatant or cell lysate was diluted 1:2 in SDS sample buffer (Bio-Rad, Hercules, CA) and loaded onto a 10% polyacrylamide/SDS gel. The resolved proteins were transferred onto a nitrocellulose membrane (Bio-Rad, Hercules, CA) and incubated with a 1:1000 dilution of polyclonal human HIV-infected patient antisera in phosphate-buffered saline (PBS) containing 0.1% Tween 20 and 1% nonfat dry milk. After extensive washing, bound human antibodies were detected using a 1:2000 dilution of horseradish peroxidase-conjugated goat anti-human antiserum and enhanced chemiluminescence (Amersham, Buckinghamshire, UK).
ELISA and avidity assays
An end point ELISA was performed to assess the titers of anti-Env immunoglobulin G (IgG) in immune serum using purified HIV-1-IIIB gp120 Chinese hamster ovary (CHO)-expressed protein (Intracell) to coat plates as described.16 Alternatively, plates were coated with sheep anti-Env antibody (International Enzymes Inc., Fallbrook, CA) and used to capture sgp120 produced in 293T cells that were transiently transfected with sgp120 expression vectors. Mouse serum from vaccinated mice was allowed to bind and subsequently detected by anti-mouse IgG conjugated to horseradish peroxidase. End point titers were considered positive that were 2-fold higher than background. Avidity ELISAs were performed similarly to serum antibody determination ELISAs up to the addition of samples and standards. Samples were diluted to give similar concentrations of specific IgG by OD. Plates were washed three times with 0.05% PBS-Tween 20. Different concentrations of the chaotropic agent, sodium thiocyanate (NaSCN) in PBS, were then added (0, 1, 1.5, 2, 2.5, and 3 M NaSCN). Plates were allowed to stand at room temperature for 15 min and then washed six times with PBS-Tween 20. Subsequent steps were performed similarly to the serum antibody determination ELISA. Percent of initial IgG was calculated as a percent of the initial OD. All assays were done in triplicate.
Neutralizing antibody assays
Antibody-mediated neutralization of HIV-1 IIIB and 89.6 was measured in an MT-2 cell-killing assay as described previously.23 Briefly, cell-free virus (50 μl containing 108 TCID50 of virus) was added to multiple dilutions of serum samples in 100 μl of growth medium in triplicate wells of 96-well microtiter plates coated with poly-L-lysine and incubated at 37° for 1 h before MT-2 cells were added (105 cells in 100 μl added per well). Cell densities were reduced and the medium was replaced after 3 days of incubation when necessary. Neutralization was measured by staining viable cells with Finter’s neutral red when cytopathic effects in control wells were >70% but less than 100%. Percentage protection was determined by calculating the difference in absorption (A540) between test wells (cells + virus) and dividing this result by the difference in absorption between cell control wells (cells only) and virus control wells (virus only). Neutralizing titers are expressed as the reciprocal of the plasma dilution required to protect at least 50% of cells from virus-induced killing.
RESULTS
Expression of plasmids
The Env vaccine plasmids were constructed using the previously described pGA vector to express a secreted form of Env (sgp120) or a C3d fusion of the secreted form of Env (sgp120-3C3d) (Fig. 1A). The Env gene was cloned from one of three HIV isolates, ADA, IIIB, and 89.6. The molecularly cloned gp120 region represented the entire surface domain of Env, but excluded the oligomerization and transmembrane domains and the cytoplasmic regions (Fig. 1B). The sgp120-3C3d fusion protein was generated by cloning three tandem repeats of the mouse homologue of C3d18 in frame with the sgp120 gene (Fig. 1B). The proteolytic cleavage sites, found at the junction between each C3d molecule as well as the junction between the gp120 protein and the first C3d coding region, were destroyed by mutagenesis.
Env was expressed at overall similar levels by plasmids containing either the secreted form of the antigen, but at a 2- to 4-fold lower level by the sgp120-C3d-expressing plasmids (Table 1). Human 293T cells were transiently transfected with 2 μg of plasmid and both supernatants and cell lysates were assayed for gp120 using an antigen capture ELISA. The sgp120 constructs expressed from 450 to 800 ng/ml, whereas the 3C3d fusions expressed from 140 to 250 ng/ml. Approximately 90% of the Env protein was present in the supernatant for both sgp120 and sgp120-3C3d-DNA-transfected cells (data not shown). The approximately 2-fold differences in the levels of expression of the different sgp120s are likely a reflection in differences in the Env genes as well as differences in the efficiency that the capture and detection antibodies recognized the different Envs.
Table 1.
IN VITRO EXPRESSION LEVELS OF ENV VACCINE PLASMIDS
| Env | Coreceptor | sgp120 | sgp120-3C3d | Ratioa |
|---|---|---|---|---|
| IIIB | X4 | 800 | 175 | 4.5 |
| 89.6 | R5X4 | 600 | 250 | 2.4 |
| ADA | R5 | 450 | 140 | 3.2 |
Ratio is expressed as the amount of Env protein detected in the supernatant of 293T cells transfected with sgp120 expressing DNA over the amount of protein detected in supernatant of cells expressing the sgp120-3C3d-expressing constructs.
Western blot analyses revealed sgp120 and sgp120-3C3d proteins of the expected sizes (Fig. 2). Using human patient polyclonal antisera, Western blot analysis showed the expected broad band of 115-120 kDa corresponding to gp120 (Fig. 2). A higher molecular weight band at ∼240 kDa is consistent with the projected size of the sgp120-3C3d fusion protein (Fig. 2). Consistent with the antigen-capture assay (Table 1), intense protein bands were present in the supernatants of cells transfected with sgp120-DNA, whereas less intense bands were present in the supernatants of cells transfected with sgp120-3C3d-DNA (data not shown). No evidence for the proteolytic cleavage of the sgp120-C3d fusion protein was seen by Western analysis.
FIG. 2.
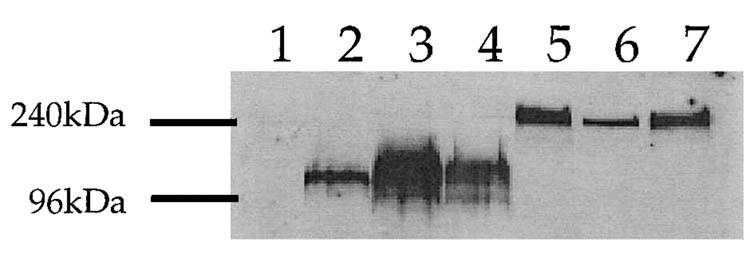
Expression of vaccine constructs in vitro. Human embryonic kidney cells, 293T, were transfected with 2 μg of each vaccine plasmid. Supernatant was collected and 1.5% of the total volume was electrophoresed on a 10% polyacrylamide gel. Lane 1: pGA vector; lane 2: sgp120 (ADA)-DNA; lane 3: sgp120-(IIIB)-DNA; lane 4; sgp120-(89.6)-DNA; lane 5: sgp120-(ADA)-3C3d-DNA; lane 6: sgp120-(IIIB)-3C3d-DNA; lane 7: sgp120-(89.6)-3C3d-DNA.
Antibody response to Env gp120-DNA immunizations
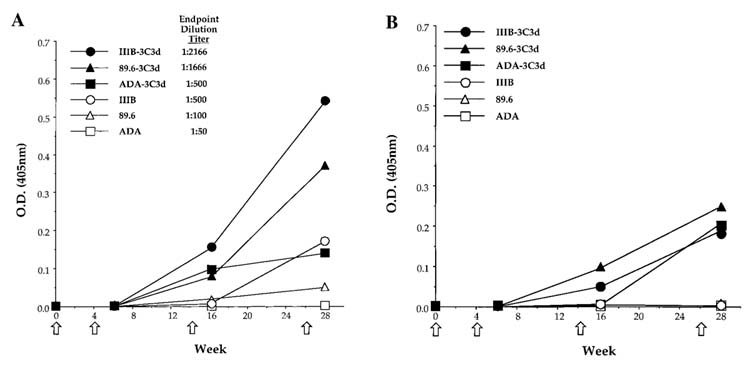
The sgp120-3C3d-expressing DNA plasmids raised higher titers of ELISA antibody than the sgp120 DNA (Fig. 3A). BALB/c mice were vaccinated by DNA-coated gold particles via gene gun with a 1 μg dose inoculum. Mice were vaccinated at Day 1 and then boosted at 4, 14, and 26 weeks with the same DNA given in the first immunization. When sera were assayed on gp120-IIIB-coated plates, mice vaccinated with the DNAs expressing the C3d fusion proteins had anti-Env antibodies three to seven times higher then the amount of antibody raised by the counterpart sgp120-expressing plasmids (Fig. 3A). Among the C3d constructs, mice vaccinated with sgp120-(IIIB)-3C3d had the highest levels of antibody and mice vaccinated with sgp120-(ADA)-3C3d expressing DNA had the lowest levels of anti-Env antibodies. The temporal pattern for the appearance of anti-Env antibody revealed titers being boosted at each of the inoculations for all constructs tested.
FIG. 3.
Anti-Env IgG raised by gene gun inoculation of DNAs expressing sgp120 proteins. Mice were primed at Day 0 and boosted at Weeks 4, 14, and 26. Sera were obtained from mice with vector (X), sgp120 (ADA)-DNA (□), sgp120-(IIIB)-DNA (○), sgp120-(89.6)-DNA (△), sgp120-(ADA)-3 C3d-DNA (■), sgp120-(IIIB)-3C3d-DNA (•), and sgp120-(89.6)-3C3d-DNA (▲). Sera collected at the indicated times from each group were pooled for determination of specific IgG levels by ELISA at a dilution of 1:100 via two different ELISA protocols. (A) 96-well plates were coated with recombinant gp120 protein-derived Chinese hamster ovary (CHO) cells expressing the HIV-1 isolate, IIIB. (B) 96-well plates were coated with supernatant containing sgp120s from 293T cells that were transiently transfected with sgp120 expression plasmids. Each ELISA plate was coated with the respective Env to detect antibodies from mice vaccinated with that particular sgp120, i.e., plates were coated with the ADA-Env in order to detect antibodies to Env in mice vaccinated with plasmids expressing sgp120-ADA proteins. Data are represented as the average of three individual assays. Preimmune sera from mice had no detectable specific IgG. End point dilution titers were conducted by diluting the sera until OD values reached background.
Differences in the levels of the antibody raised by the different Envs appeared to be determined by the specificity of the raised antibody (Fig. 3B). Using an alternative ELISA protocol, in which antibody was captured on the homologous Env, all of the C3d fusions appeared to raise similar levels of antibody. In this assay, sheep anti-Env antibody was used to capture transiently produced sgp120 proteins. This assay revealed low, but similar levels of antibody raised by each of the sgp120-3C3d constructs. The lower levels of antibody detected in this assay are likely to reflect the levels of transfection-produced Env used to capture antibody being lower than in the assays using commercially produced IIIB gp120 to coat plates. As expected using either ELISA method, booster immunizations were necessary to achieve even the most modest antibody response.
Avidity of mouse Env antiserum
Sodium thiocyanate (NaSCN) displacement ELISAs demonstrated that the avidity of the antibody generated with sgp120-3C3d-expressing DNA was consistently higher than that from sgp120-DNA-vaccinated mice (Fig. 4). Avidity assays were conducted on sera raised by sgp120-(IIIB) and sgp120-(IIIB-3C3d because of the type specificity of the raised antisera and the commercial availability of the IIIB protein, but not the other proteins, for use as capture antigen. The avidity of specific antibodies to Env was compared by using graded concentrations NaSCN, a chaotropic agent, to disrupt antigen-antibody interaction.24 The binding of antibodies with less avidity to the antigen is disrupted at lower concentrations of NaSCN than that of antibodies with greater avidity to the antigen. The effective concentration of NaSCN required to release 50% of antiserum (ED50) collected at 28 weeks, after four inoculations of the sgp120-(IIIB)-3C3d-DNA vaccine, was ∼2.0 M (Fig. 4). At this time, the avidity for the gp120-raised sera that could be tested was less than 1. These results indicate that the antibody from sgp120-(IIIB)-3C3d DNA-vaccinated mice had undergone more rapid affinity maturation than antibody from sgp120-DNA-vaccinated mice.
FIG. 4.
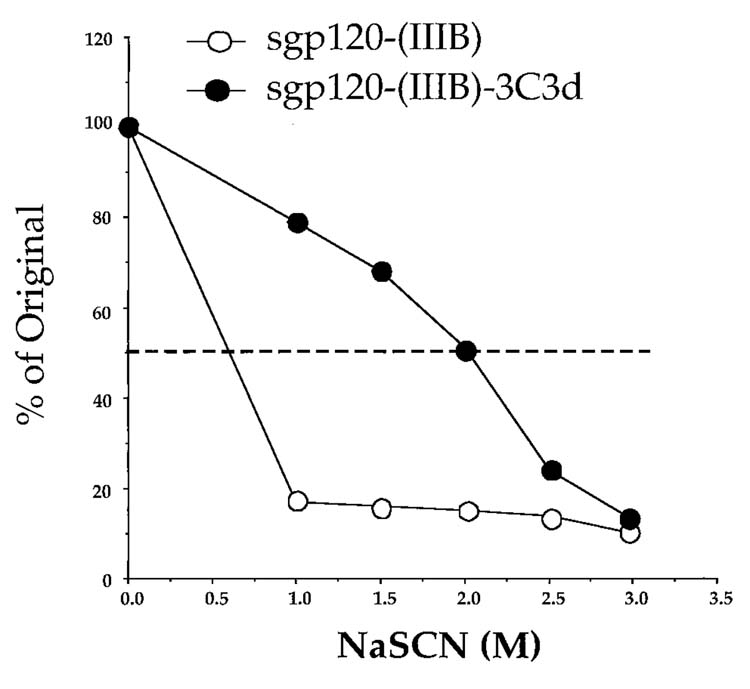
Avidity of the anti-Env IgG raised by the IIIB Env-DNA vaccines. Sera were analyzed from Week 28 in an Env-specific NaSCN-displacement ELISA. Plates were with recombinant gp120-(IIIB). Sera were obtained from mice immunized with sgp120-(IIIB)-DNA (○) and sgp120-(IIIB)-3C3d-DNA (•). Assays used pooled serum samples from each mouse group at a dilution of 1:50 for sgp120-(IIIB) and 1:200 for sgp120-(IIIB)-3C3d. Data are representative of two independent experiments.
Env-3C3d expressing plasmids elicit modest neutralizing antibody
Neutralizing antibody studies performed on MT-2 cells detected higher titers of neutralizing activity in the sera generated by the gp120-3C3d constructs than in the sera generated by the sgp120 constructs. (Table 2). Sera were tested against two syncytium-inducing viruses, IIIB (X4) and 89.6 (X4R5). Mice vaccinated with sgp120-3C3d-expressing plasmids had very modest levels of neutralizing antibody to the homologous strain of HIV tested by the protection of MT-2 cells from virus-induced killing as measured by neutral red uptake (Table 2). Titers of neutralizing antibody raised by the gp120-expressing DNAs were at the background of the assay.
TABLE 2.
NEUTRALIZATION ANTIBODY TITER
| Vaccine group | HIV-1 IIIBa | SHIV 89.6a |
|---|---|---|
| Week 16 | ||
| IIIB gp 120 | 1:87 | NTb |
| IIIB gp 120-3C3d | 1:178 | NT |
| 89.6 gp 120 | NT | 1:38 |
| 89.6 gp 120-3C3d | NT | 1:86 |
| Week 28 | ||
| IIIB gp 120 | 1:94 | NTb |
| IIIB gp 120-3C3d | 1:167 | NT |
| 89.6 gp 120 | NT | 1:48 |
| 89.6 gp 120-3C3d | NT | 1:84 |
| Vector | 1:61 | 1:58 |
Titers are the sample dilution at which 50% of MT-2 cells were protected from virus-induced killing as measured by neutral red uptake.
NT, not tested.
DISCUSSION
In this study, we showed that fusions of HIV-1 Env to three copies of murine C3d enhanced the antibody response to Env in vaccinated mice. Mice vaccinated with any of the three DNA plasmids expressing the sgp120 sequence had low or undetectable levels of antibody after four vaccinations (28 weeks postprime). In contrast, mice vaccinated with DNA expressing the fusion of sgp120 and 3C3d proteins elicited a faster onset of antibody (three vaccinations), as well as higher levels of antibodies (Fig. 3). C3d is likely to have supported the height of antibody responses by directly stimulating antibody-producing B cells through complement receptor 2 (CD21) and expanding the pool of anti-Env-specific B cells.25,26 CD21 is complexed with the costimulator CD19 on B cells.27,28 Differences in the titers of antibody elicited by different Envs appeared to reflect the type specificity of the raised antibody. This was shown by the sera having overall similar titers of antibody when assayed against the homologous Env (Fig. 3b).
Immunization with the C3d fusions also resulted in enhanced avidity maturation of anti-Env antibody. Env is a heavily glycosylated protein that may allow it to resemble a T cell-independent antigen.29 The formation of germinal centers, which are critical for antibody maturation, could be impeded by the glycosylation of Env.30,31 Previous studies have described the lengthy evolution of antibody responses to a variety of lentivirus infections.17,32 Antibodies gradually mature over a 6- to 8-month period from low avidity to reach a steady-state level of intermediate to high avidity.17,32 Avidity maturation occurs in germinal centers where the somatic hypermutation of immunoglobulin results in a large repertoire of Ag-specific B cells that undergo selection for high-affinity B cell receptors. The more rapid avidity maturation of antibodies elicited by the sgp120-3C3d fusions is consistent with C3d supporting the entry of B cells producing anti-Env antibody into germinal centers. This could reflect the C3d domain of the fusion proteins binding to CD21 on follicular dendritic cells and promoting the entry of B cells recognizing gp120 into a germinal center reaction. Prior studies on the avidity of DNA-raised anti-Env antibodies have also revealed low-avidity anti-Env antibodies.16
In contrast to the enhancement of antibody titers and avidity maturation of antibodies to Env, the amount of neutralizing antibody elicited in the vaccinated mice was low. Mice vaccinated with plasmids expressing sgp120 had low levels of neutralizing antibody that were only modestly increased in mice vaccinated with sp120-3C3d-expressing plasmids. However, the disappointing levels of neutralizing antibodies did seem to increase after the fourth immunization (Table 2). The poor titers of neutralizing antibody could have reflected an inherent poor ability of the sgp120-3C3d fusion protein to raise neutralizing antibody because of the failure to adequately expose neutralizing epitopes to responding B cells. The intrinsic high backgrounds for HIV-1 neutralization assays in mouse sera may have contributed to the poor neutralization titers. Also, the correlation between avidity maturation and neutralizing antibody production to gp120 has not always been clear. Some studies have indicated a good correlation,17,32,33 whereas others have not.29,34-37 Binley et al.29 demonstrated that avidity assays, based upon elution of antibody in an in vitro assay for monitoring the antibody response to HIV-1 gp120 during either vaccination with subunit proteins or natural infection, did not show a strong correlation between the avidity of the antibody response and the ability of these same antibodies to neutralize virus.
Much effort is being made to improve the efficacy of HIV vaccines. The approach we tried in this study was the use of C3d fusion proteins to increase the humoral immune response to the envelope protein of HIV. The results demonstrate the effectiveness of C3d fusions as a molecular adjuvant in enhancing antibody production and enhancing antibody maturation. However, the neutralizing antibody response to Env was poor and increased slightly in mice vaccinated with C3d fusion vaccines. Similar to results seen in our previous study using secreted versions of HA from influenza virus,19 C3d-enhanced antibody responses were achieved with plasmids expressing only half as much protein as plasmids expressing nonfused sgp120 (Table 1). In contrast to the present study, virus neutralization titers were dramatically increased in mice vaccinated with C3d fusion constructs and may have been a result of the low glycosylation state of HA. These results underscore the importance of increasing our understanding of the relationship between innate and acquired immunity. They also underscore the importance of identifying forms of Env that when coupled with C3d are capable of raising neutralizing antibody. If this can be accomplished, C3d fusions could have a major impact on the development of the neutralizing component for an AIDS vaccine.
ACKNOWLEDGMENTS
This research was supported by faculty grant award 265515 to T.M.R. from East Carolina University and grant awards R21 AI44325 to T.M.R. and P01 AI43045 to H.L.R. from the National Institute of Allergy and Infectious Diseases. The authors thank Douglas Fearon for supplying the murine C3d constructs, David Campbell and Sean Winburn for technical assistance with serum collection, and Stephanie Oberhaus for helpful discussions.
REFERENCES
- 1. http://www.us.unaids.org.epidemic_update/report/epicore/2001
- 2.Donnelly JJ, Ulmer JB, Liu MA. DNA vaccines. Dev Biol Stand. 1998;95:43–53. [PubMed] [Google Scholar]
- 3.Robinson HL, Pertmer TM. DNA vaccines for viral infections: Basic studies and applications. Adv Virus Res. 2000;55:1–74. doi: 10.1016/s0065-3527(00)55001-5. [DOI] [PubMed] [Google Scholar]
- 4.Liu MA, Fu TM, Donnelly JJ, Caulfield MJ, Ulmer JB. DNA vaccines: Mechanisms for generation of immune responses. Adv Exp Med Biol. 1998;452:187–191. [PubMed] [Google Scholar]
- 5.Robinson HL. DNA vaccines for immunodeficiency viruses. AIDS. 1997;11(Suppl A):S109–S119. [PubMed] [Google Scholar]
- 6.Barouch DH, Santra S, Schmitz JE, et al. Control of viremia and prevention of clinical AIDS in rhesus monkeys by cytokine-augmented DNA vaccination. Science. 2000;290:486–492. doi: 10.1126/science.290.5491.486. [DOI] [PubMed] [Google Scholar]
- 7.Barnett SW, Rajasekar S, Legg H, et al. Vaccination with HIV-1 gp120 DNA induces immune responses that are boosted by a recombinant gp120 protein subunit. Vaccine. 1997;15:869–873. doi: 10.1016/s0264-410x(96)00264-2. [DOI] [PubMed] [Google Scholar]
- 8.Boyer JD, Wang B, Ugen KE, et al. In vivo protective anti-HIV immune responses in non-human primates through DNA immunization. J Med Primatol. 1996;25:242–250. doi: 10.1111/j.1600-0684.1996.tb00022.x. [DOI] [PubMed] [Google Scholar]
- 9.Coney L, Wang B, Ugen KE, et al. Facilitated DNA inoculation induces anti-HIV-1 immunity in vivo. Vaccine. 1994;12:1545–1550. doi: 10.1016/0264-410x(94)90082-5. [DOI] [PubMed] [Google Scholar]
- 10.Lu S, Arthos J, Montefiori DC, et al. Simian immunodeficiency virus DNA vaccine trial in macaques. J Virol. 1996;70:3978–3991. doi: 10.1128/jvi.70.6.3978-3991.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lu S, Santoro JC, Fuller DH, Haynes JR, Robinson HL. Use of DNAs expressing HIV-1 env and noninfectious HIV-1 particles to raise antibody responses in mice. Virology. 1995;209:147–154. doi: 10.1006/viro.1995.1238. [DOI] [PubMed] [Google Scholar]
- 12.Okuda K, Bukawa H, Hamajima K, et al. Induction of potent humoral and cell-mediated immune responses following direct injection of DNA encoding the HIV type 1 env and rev gene products. AIDS Res Hum Retroviruses. 1995;11:933–943. doi: 10.1089/aid.1995.11.933. [DOI] [PubMed] [Google Scholar]
- 13.Shiver JW, Davies M-E, Yasutomi Y, et al. Anti-HIV env immunities elicited by nucleic acid vaccines. Vaccine. 1997;15:884–887. doi: 10.1016/s0264-410x(96)00251-4. [DOI] [PubMed] [Google Scholar]
- 14.Graham BS, Wright PF. Candidate AIDS vaccines. N Engl J Med. 1995;333:1331–1339. doi: 10.1056/NEJM199511163332007. [DOI] [PubMed] [Google Scholar]
- 15.Mustafa F, Richmond JFL, Fernandez-Larsson R, et al. HIV-1 Env glycoproteins from two series of primary isolates: Replication phenotype and immunogenicity. Virology. 1997;229:269–278. doi: 10.1006/viro.1997.8445. [DOI] [PubMed] [Google Scholar]
- 16.Richmond JFL, Lu S, Santoro C, et al. Studies of the neutralizing activity and avidity of anti-human immunodeficiency virus type 1 Env antibody elicited by DNA priming and protein boosting. J Virol. 1998;72:9092–9100. doi: 10.1128/jvi.72.11.9092-9100.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cole KS, Rowles JL, Hagerski BA, et al. Evolution of envelope-specific antibody responses in monkeys experimentally infected or immunized with simian immunodeficiency virus and its association with the development of protective immunity. J Virol. 1997;71:5069–5079. doi: 10.1128/jvi.71.7.5069-5079.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dempsey PW, Allison MED, Akkaraju S, Goodnow CC, Fearon DT. C3d of complement as a molecular adjuvant: Bridging innate and acquired immunity. Science. 1996;271:348–350. doi: 10.1126/science.271.5247.348. [DOI] [PubMed] [Google Scholar]
- 19.Ross TM, Xu Y, Bright RA, Robinson HL. C3d enhancement of antibodies to hemagglutinin accelerates protection against influenza virus challenge. Nat Immunol. 2000;1:127–131. doi: 10.1038/77802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haynes JR, Fuller DH, Eisenbraun MD, Ford MJ, Pertmer TM. Accell particle-mediated DNA immunization elicits humoral, cytotoxic, and protective immune responses. AIDS Res Hum Retro-viruses. 1994;10:S43–45. [PubMed] [Google Scholar]
- 21.Pertmer TM, Eisenbraun MD, McCabe D, Prayaga SK, Fuller DH, Haynes JR. Gene gun based nucleic acid immunization. Eliciting of humoral and cytotoxic T lymphocyte response following epidermal delivery of nanogram quantitation of DNA. Vaccine. 1995;13:1427–1430. doi: 10.1016/0264-410x(95)00069-d. [DOI] [PubMed] [Google Scholar]
- 22.Pertmer TM, Robinson HL. Studies on antibody responses following neonatal immunization with influenza hemagglutinin DNA or protein. Virology. 1999;257:406–414. doi: 10.1006/viro.1999.9666. [DOI] [PubMed] [Google Scholar]
- 23.Montefiori DC, Robinson WE, Schuffman SS, Mitchell WM. Evaluation of antiviral drugs and neutralizing antibodies to HIV by a rapid and sensitive microtiter infection assay. J Clin Microbiol. 1988;26:231–237. doi: 10.1128/jcm.26.2.231-235.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pullen GR, Fitzgerald MG, Hosking CS. Antibody avidity determination by ELISA using thiocyanate elution. J Immunol Methods. 1986;86:83–87. doi: 10.1016/0022-1759(86)90268-1. [DOI] [PubMed] [Google Scholar]
- 25.Berek C, Berger A, Apel M. Maturation of the immune response in germinal centers. Cell. 1991;67:1121–1129. doi: 10.1016/0092-8674(91)90289-b. [DOI] [PubMed] [Google Scholar]
- 26.Jacob J, Kelsoe G, Rajewsky K, Weiss U. Intraclonal generation of antibody mutants in germinal centers. Nature (London) 1991;354:389–392. doi: 10.1038/354389a0. [DOI] [PubMed] [Google Scholar]
- 27.Fearon DT, Carter RH. The CD19/CR2/TAPA-1 complex of B lymphocytes: Linking natural to acquired immunity. Annu Rev Immunol. 1995;13:127–149. doi: 10.1146/annurev.iy.13.040195.001015. [DOI] [PubMed] [Google Scholar]
- 28.Fearon DT. The complement system and adaptive immunity. Semin Immunol. 1998;10:355–361. doi: 10.1006/smim.1998.0137. [DOI] [PubMed] [Google Scholar]
- 29.Binley JM, Arashad H, Fouts RR, Moore JP. An investigation of the high avidity antibody response to glycoprotein 120 of human immunodeficiency virus type 1. AIDS Res Hum Retroviruses. 1997;13:1007–1015. doi: 10.1089/aid.1997.13.1007. [DOI] [PubMed] [Google Scholar]
- 30.MacLennan ICM. Germinal centers. Annu Rev Immunol. 1994;12:117–139. doi: 10.1146/annurev.iy.12.040194.001001. [DOI] [PubMed] [Google Scholar]
- 31.Tew JG, DiLosa RM, Burton GF. Germinal centers and antibody production in bone marrow. Immunol Rev. 1992;126:99–112. doi: 10.1111/j.1600-065x.1992.tb00633.x. [DOI] [PubMed] [Google Scholar]
- 32.Hammond SA, Cook SJ, Lichtenstein DL, Issel CJ, Monterlaro RC. Maturation of the cellular and humoral responses to persistent infection in horses by equine infectious anemia virus is a complex and lengthy process. J Virol. 1997;71:3840–3852. doi: 10.1128/jvi.71.5.3840-3852.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.MacDonald RA, Hosking CS, Jones CL. The measurement of relative antibody affinity by ELISA using thiocyanate elution. J Immunol Methods. 1988;106:191–194. doi: 10.1016/0022-1759(88)90196-2. [DOI] [PubMed] [Google Scholar]
- 34.Chargelegue D, Stanley CM, O’Toole CM, Colvin BT, Steward MW. The absence or loss of antibodies of high affinity to human immunodeficiency virus (HIV) is associated with disease progression in HIV-1 infected patients. J Infect Dis. 1995;172:897–898. doi: 10.1093/infdis/172.3.897. [DOI] [PubMed] [Google Scholar]
- 35.Hall JT, Heckel C. Thiocyanate elution estimation of relative antibody avidity. J Immunol Methods. 1988;115:153–154. doi: 10.1016/0022-1759(88)90324-9. [DOI] [PubMed] [Google Scholar]
- 36.VanCott TC, Bethke FR, Burke DS, Redfield RR, Birx DL. Lack of indication of antibodies specific for conserved, discontinuous epitopes of HIV-1 envelope glycoproteins by candidate AIDS vaccines. J Immunol. 1995;155:4100–4110. [PubMed] [Google Scholar]
- 37.Berman FW, Gregory TJ, Riddle L, et al. Protection of chimpanzees from infection by HIV-1 after vaccination with recombinant glycoprotein gp120 but not gp160. Nature (London) 1990;345:622–625. doi: 10.1038/345622a0. [DOI] [PubMed] [Google Scholar]