Abstract
Control of actin assembly nucleated by the Arp2/3 complex plays a crucial role during budding yeast endocytosis. The yeast Eps15-related Arp2/3 complex activator, Pan1p, is essential for endocytic internalization and proper actin organization. Pan1p activity is negatively regulated by Prk1 kinase phosphorylation after endocytic internalization. Phosphorylated Pan1p is probably then dephosphorylated in the cytosol. Pan1p is recruited to endocytic sites ∼25 s before initiation of actin polymerization, suggesting that its Arp2/3 complex activation activity is kept inactive during early stages of endocytosis by a yet-to-be-identified mechanism. However, how Pan1p is maintained in an inactive state is not clear. Using tandem affinity purification–tagged Pan1p, we identified End3p as a stoichiometric component of the Pan1p complex, and Sla2p, a yeast Hip1R-related protein, as a novel binding partner of Pan1p. Interestingly, Sla2p specifically inhibited Pan1p Arp2/3 complex activation activity in vitro. The coiled-coil region of Sla2p was important for Pan1p inhibition, and a pan1 partial loss-of-function mutant suppressed the temperature sensitivity, endocytic phenotypes, and actin phenotypes observed in sla2ΔCC mutant cells that lack the coiled-coil region. Overall, our results establish that Sla2p's regulation of Pan1p plays an important role in controlling Pan1p-stimulated actin polymerization during endocytosis.
INTRODUCTION
The actin cytoskeleton plays an important role in cellular processes including cell polarity development, cell migration, cytokinesis, and endocytosis. Endocytosis is critical for controlling nutrient uptake, plasma membrane homeostasis, synaptic machinery recycling, and signaling pathway regulation. In budding yeast, endocytosis requires dynamic interactions between the plasma membrane and the actin cytoskeleton (Engqvist-Goldstein and Drubin, 2003). In both yeast and mammals, a transient burst of actin polymerization at endocytic sites is essential for endocytic internalization (Merrifield et al., 2002; Kaksonen et al., 2003). Mutations in actin cytoskeletal genes, including the Arp2/3 complex, or treatment of yeast with the actin polymerization inhibitor, latrunculin A, block endocytic vesicle internalization (Ayscough et al., 1997; Lappalainen and Drubin, 1997; Belmont and Drubin, 1998; Martin et al., 2005). Therefore, actin polymerization is thought to provide force for plasma membrane invagination, vesicle scission, and/or movement into the cytoplasm (Munn, 2001; Engqvist-Goldstein and Drubin, 2003; Kaksonen et al., 2006).
Recent analyses using yeast genetics and real-time, live-cell fluorescence microscopy demonstrated that proteins are recruited to endocytic sites, or cortical patches, in a highly predictable order. In yeast, clathrin is recruited to cortical patches ∼1–2 min before vesicle internalization (Kaksonen et al., 2005; Newpher et al., 2005). Early endocytic components, including Pan1p (Eps15-related Arp2/3 complex activator), Las17p (yeast WASP), Sla1p (an adaptor for NPFx(1,2)D-mediated endocytosis) and Sla2p (yeast Hip1R), then appear at cortical patches and are later joined by actin, Abp1p and the Arp2/3 complex (Kaksonen et al., 2003). Abp1p, a marker for actin polymerization in cortical patches, has a patch lifetime of 10–15 s, whereas early endocytic components, including the Arp2/3 activators Pan1p and Las17p, have lifetimes ranging from 30 to 40 s (Kaksonen et al., 2003). Thus, the Arp2/3 activators Pan1p and Las17p arrive at endocytic sites ∼25 s before actin assembly is initiated. How activities of Pan1p and Las17p are regulated and how the Arp2/3 complex is recruited to endocytic sites are not well understood. One possibility is that the Arp2/3 complex binding regions of these activators are masked by interactions with other proteins, which dissociate in response to yet-to-be-identified stimuli, triggering activation of these activators. Las17p's activity is inhibited by Sla1p and Bbc1p (Rodal et al., 2003), and the combination of sla1Δ and bbc1Δ mutations leads to formation of dramatic actin protrusions that associate with endocytic sites (Kaksonen et al., 2005). Importantly, how Pan1p activity is inhibited at early stages of endocytosis is not known.
Pan1p is an essential protein that is involved in both the internalization step of endocytosis and the organization of the actin cytoskeleton (Tang and Cai, 1996; Wendland et al., 1996). Pan1p interacts with many endocytic proteins, suggesting that Pan1p forms the core of an endocytic complex. The two Eps15 homology (EH) domains located at the N-terminal region of Pan1p bind to the NPF motifs of Ent1/2p and Yap180A/Bp, which also interact with clathrin (Wendland and Emr, 1998; Wendland et al., 1999). The central coiled-coil region of Pan1p contains a WH2-like region, which binds to filamentous actin (Toshima et al., 2005), and a C-terminal acidic region, which binds to the Arp2/3 complex (Duncan et al., 2001). Pan1p also has two N-terminal LR (long repeat) regions, which contain 18 Prk1p consensus phosphorylation sequences ([L/I/V/M]xx[Q/N/T/S]xTG), and also interact with Sla1p and the EH domain protein End3p (Tang et al., 1997; Zeng and Cai, 1999; Tang et al., 2000; Huang et al., 2003). The Ark1p/Prk1p kinases have been suggested to negatively regulate the Pan1p-Sla1p-End3p interaction and Pan1p's Arp2/3 complex activation activity (Zeng et al., 2001; Toshima et al., 2005). Inhibition of both Ark1p and Prk1p activities caused a severe endocytic defect and abnormal cytoplasmic actin aggregates, and a mutant in which the Prk1p-targeted threonines in Pan1p were mutated to alanines exhibited a phenotype similar to the ark1Δ prk1Δ mutant, demonstrating that Pan1p is a key Prk1p target in vivo (Cope et al., 1999; Sekiya-Kawasaki et al., 2003; Toshima et al., 2005). Moreover, phosphorylation by Prk1p inhibits Pan1p's abilities to bind to F-actin and to activate the Arp2/3 complex (Toshima et al., 2005). Therefore, these observations suggest that phosphoregulation of Pan1p by Prk1p is an important mechanism to shut off Arp2/3-mediated actin polymerization on endocytic vesicles. Importantly, when Pan1p is dephosphorylated and whether dephosphorylation of Pan1p causes initiation of actin assembly during endocytosis are not known.
Among early endocytic components, Sla2p arrives at cortical patches slightly earlier than Sla1p and is important for clathrin organization and for progression of early endocytic patches to the late stages of endocytosis (Newpher and Lemmon, 2006). Sla2p is a modular protein that is composed of an N-terminal AP180 N-terminal homology (ANTH) domain that interacts with phosphatidylinositol-4,5-bisphosphate [PtdIns(4,5)P2; Sun et al., 2005], a coiled-coil region that interacts with clathrin (Henry et al., 2002; Newpher and Lemmon, 2006), and a C-terminal talin-like actin-binding domain (Yang et al., 1999). Sla2p appears to regulate actin polymerization because sla2Δ cells exhibit continuous actin assembly at nonmotile endocytic sites (Kaksonen et al., 2003). An identical phenotype was also observed when Hip1R (mammalian Sla2p homologue) expression was lowered by RNA interference (Engqvist-Goldstein et al., 2004), suggesting that Sla2p and Hip1R share a conserved function and that these proteins negatively regulate actin assembly associated with endocytic vesicle formation.
In this study, we identified Sla2p as a novel binding partner of Pan1p. Using biochemical and genetic analyses, we showed that Sla2p specifically inhibits Pan1p Arp2/3 complex activation activity and regulates actin assembly during endocytic vesicle internalization.
MATERIALS AND METHODS
Yeast Strains, Growth Conditions, and Plasmids
The yeast strains used in this study are listed in Table 1. Yeast strains were grown in standard rich media (YPD) or synthetic media (SM) supplemented with the appropriate amino acids. The carbon sources used were 2% glucose, 2% galactose, or 2% raffinose, depending on the experiment. C-terminal tandem affinity purification (TAP) or GFP tags were integrated at the endogenous loci of the gene using the method described in Longtine et al. (1998). The sla2ΔCC mutant was generated as follows: First, to create a sla2 integration plasmid, the EcoRI fragment of the SLA2 gene was cloned into pBluescript II SK, and the SmaI fragment of the URA3 gene was inserted into the NruI site 150 base pairs downstream of the SLA2 ORF (PBS-SLA2-URA3). sla2ΔCC (Δ1081-2187) was generated by ligating the NarI-digested PCR product amplified by primers 5′-CCGGCGCCGGCGCCCAGAAGTCCGGCTGCATTTGTGCC-3′ and 5′-CCGGCGCCGGCGCCGAACCGTTATTGAACATACAATCTG-3′ using PBS-SLA2-URA3 as a template. To integrate the sla2ΔCC mutant at the endogenous locus, the plasmid was digested with EcoRI and transformed into sla2Δ::LEU2/SLA2 diploid strains. Integrated sla2ΔCC mutants were selected on SC plates lacking uracil and sporulated to obtain sla2ΔCC mutants. The Yap180Ap and Ent2p expression plasmids were constructed by inserting their ORFs into BamHI- and NotI-digested pRS426 containing the GAL1-10 promoter, a TEV protease recognition site, and encoding the myc epitope (Rodal et al., 2003). The Sla1p and End3p expression plasmids were constructed by inserting their ORFs into the BamHI site of pBS1479 (Rigaut et al., 1999). The Prk1p and Pan1-TAP expression plasmids were constructed as described previously (Toshima et al., 2005). Sla2p truncation constructs were generated as described previously (Yang et al., 1999).
Table 1.
Yeast strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| DDY1810 | Mataleu2ura3-52 lys2-801trp1 prb1-1122 pep4-3pre1-451 | Drubin lab |
| DDY2887 | Mataleu2ura3-52 lys2-801trp1 prb1-1122 pep4-3pre1-451 PAN1-S TAP-TEV-ZZ::TRP1 | Drubin lab |
| DDY3326 | Matahis3-Δ200leu2-3,112ura3-52 SLA1-GFP::KanMX6 ABP1-RFP::HIS3 | This study |
| DDY3327 | Matα his3-Δ200leu2-3, 112ura3-52 pan1Δ::pan1Δ855-1480::LEU2 SLA1-GFP::KanMX6 ABP1-RFP::HIS3 | This study |
| DDY3328 | Matα his3-Δ200leu2-3, 112ura3-52 lys2-801 pan1Δ::pan1Δ855-1480::LEU2 las17Δ547-633-13myc::HIS3 SLA1-GFP::KanMX6 ABP1-RFP::HIS3 | This study |
| DDY3329 | Matahis3-Δ200leu2-3, 112ura3-52 lys2-801pan1Δ::pan1ΔWA::LEU2 | This study |
| DDY3330 | Matahis3-Δ200leu2-3, 112ura3-52lys2-801 sla2Δ::LEU2 | This study |
| DDY3331 | Matahis3-Δ200leu2-3, 112ura3-52 lys2-801 sla2Δ:LEU2 pan1Δ::pan1ΔWA::LEU | This study |
| DDY3332 | Matahis3-Δ200leu2-3, 112ura3-52 lys2-801 sla2Δ::sla2ΔCC::URA3 pan1Δ::PAN1::LEU | This study |
| DDY3333 | Matahis3-Δ200leu2-3, 112ura3-52 lys2-801 sla2Δ::sla2ΔCC::URA3 pan1Δ::pan1ΔWA::LEU | This study |
| DDY3334 | Matahis3-Δ200leu2-3, 112ura3-52 lys2-801 sla2Δ::sla2ΔCC::URA3 PAN1-GFP:KanMX6 ABP1-RFP::HIS3 | This study |
| DDY3335 | Matahis3-Δ200leu2-3, 112ura3-52 lys2-801 sla2Δ::sla2ΔCC::URA3 pan1Δ::pan1ΔWA-GFP:KanMX6 ABP1-RFP::HIS3 | This study |
All strains are derived from strain S288C.
Protein Expression and Purification
DDY1810 strains expressing the relevant plasmid were grown to saturation in 20 ml SD supplemented with all amino acids except uracil. This inoculum was added to 1.5 l of synthetic media with 2% wt/vol raffinose and dropout uracil and was grown to OD600 of 1.0. Protein expression was induced upon addition of 30 g bacto-peptone, 15 g yeast extract, and a final concentration of 2% wt/vol galactose for 8–12 h, as described (Toshima et al., 2005). Cells were harvested by centrifugation, washed with 500 ml water, resuspended at a 1/5 wt/vol ratio of yeast to water, and drop-frozen in liquid N2. Cells were lysed five times in a Waring blender with liquid N2, as described (Goode et al., 1999). TAP-tagged proteins (Pan1p, Sla1p, and End3p) were purified according to the TAP method as described elsewhere (Rigaut et al., 1999). The Pan1p-End3p complex was purified from the DDY1810 strain overexpressing both End3p and TAP-tagged Pan1p, using the same method (Rigaut et al., 1999). TEV-myc–tagged proteins (Yap180Ap and Ent2p) and GST proteins were purified as described (Duncan et al., 2001 and Rodal et al., 2003, respectively). Arp2/3 complex (Martin et al., 2005) and yeast actin (Goode et al., 1999) were purified as described previously. Pyrene-labeled rabbit muscle actin was prepared as described elsewhere (Cooper et al., 1983).
Actin Filament Cosedimentation and Actin Assembly Assay
Pan1p (0.5 μM) and 0, 1, or 2 μM GST-fused Sla2-CC were added to 2 μM of assembled yeast actin, incubated for 30 min at room temperature, and centrifuged for 30 min in a TLA100 rotor (Beckman, Fullerton, CA) at 90,000 rpm. Pellets and supernatants were separated on 9% SDS-PAGE gels that were subsequently stained with Coomassie blue. To measure actin assembly kinetics, pyrene fluorescence was monitored as described previously (Goode et al., 1999). Actin (2 μM yeast monomeric actin and 0.1 μM pyrene-labeled rabbit muscle actin) was centrifuged at 90,000 rpm in a TLA100 rotor (Beckman) for 45 min to remove nuclei before use. Reaction conditions were as follows: 53 μl actin in G-buffer (50 mM Tris-Cl, pH 7.5, 0.2 mM CaCl2, 0.2 mM ATP, and 0.2 mM DTT) was mixed rapidly with 7 μl initiation buffer (250 mM KCl, 20 mM MgCl2, and 5 mM ATP) and 10 μl HEKG5 (20 mM HEPES, pH 7.5, 1 mM EDTA, 50 mM KCl, and 5% glycerol) containing 10 nM Arp2/3, and/or Pan1p and then immediately transferred to a cuvette in the fluorometer. Fluorescence data were collected by using a Fluoromax 3 fluorometer (Jobin-Yvon Horiba, Edison, NJ).
Immunoblotting and In Vitro Pulldown Assay
Immunoblot analysis was performed as described previously (Toshima et al., 2005). Anti-Pan1p (Duncan et al., 2001), anti-Sla2p (Yang et al., 1999) and anti-Act1p antibodies (Goode et al., 1999) were used at a dilution of 1:1000. Immunoreactive protein bands were visualized, using a SuperSignal West Pico Chemiluminescent Substrate System (Pierce, Rockford, IL). For in vitro pulldown assays, GST-Sla2 was overexpressed in DDY1810 and purified as described above. GST-Sla2 pelleted with glutathione-Sepharose beads (GE Healthcare, Waukesha, WI) was washed three times with wash buffer (10 mM Tris-Cl, pH 7.4, 150 mM NaCl, 0.5% NP40) and incubated with purified Pan1p in 100 μl binding buffer (10 mM Tris-Cl, pH 7.4, 150 mM NaCl, 5 mM EGTA, 1 mM EDTA, 7.5% glycerol, 0.5% NP40, 10 mM β-mercaptoethanol, 1 mM PMSF, and 0.5 μg/ml leupeptin, antipain, pepstatin A, and aprotinin). After incubation overnight at 4°C, the precipitates were washed three times with wash buffer and used for immunoblotting. In vitro pulldown assays using other protein combinations were also performed in the same way.
Fluorescence Microscopy
Fluorescence microscopy was performed using an Olympus IX81 microscope equipped with a 100×/NA 1.40 (Olympus, Melville, NY) objective and Orca-ER cooled CCD camera (Hamamatsu, Bridgewater, NJ). Simultaneous imaging of red and green fluorescence was performed using an Olympus IX81 microscope, described above, and an image splitter (Dual-View; Optical Insights, Tucson, AZ) that divided the red and green components of the images with a 565-nm dichroic mirror and passed the red component through a 630/50-nm filter and the green component through a 530/30-nm filter.
RESULTS
Localization of Phosphorylated Pan1p In Vivo
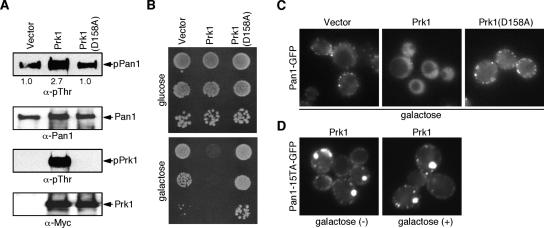
In yeast and mammals, several Arp2/3 complex activators associate with endocytic sites where they appear to promote actin assembly (Schafer, 2002; Kaksonen et al., 2006; Sun et al., 2006). Our group previously showed that Eps15-related Arp2/3 complex activator, Pan1p, functions redundantly with WASP-related protein, Las17p, and that these proteins are important for initiating actin assembly during yeast endocytosis (Duncan et al., 2001; Toshima et al., 2005; Sun et al., 2006). Pan1p is recruited to endocytic sites ∼25 s earlier than Abp1p (Kaksonen et al., 2003), a marker for actin assembly, suggesting that Pan1p's Arp2/3 complex activation activity is kept inactive during early stages of endocytosis. However, how Pan1p is maintained inactive before initiation of actin assembly has not been explored. One possibility is that phosphorylated/inactive Pan1p is recruited to cortical patches and that Pan1p dephosphorylation occurs immediately before the onset of actin polymerization (Toshima et al., 2005). To test this possibility, we examined the effects of Prk1p kinase overexpression in cells. Prk1p overexpression resulted in about a 2.7-fold increase in the level of Pan1p phosphorylation, and a severe growth defect that depended on Prk1p kinase activity (Figure 1, A and B). Interestingly, Prk1p overexpression caused Pan1p to be mislocalized in the cytosol, although Pan1p cortical localization was not significantly altered by overexpressing a kinase-dead mutant of Prk1p, Prk1(D158A) (Figure 1C). In contrast, the localization of Pan1-15TAp, in which 15 potential Prk1p phosphorylation sites in Pan1p were mutated to alanine (Toshima et al., 2005), at cortical patches was not affected by Prk1 overexpression (Figure 1D). These observations suggest that Prk1p-phosphorylated Pan1p localizes in the cytosol and that Pan1p dephosphorylation in the cytosol may be required for recruitment to endocytic sites. Therefore, Pan1p phosphorylation is not likely to maintain Pan1p in an inactive state during early stages of endocytosis.
Figure 1.
Localization of phosphorylated Pan1p in vivo. (A) In vivo phosphorylation of Pan1p by Prk1p. Pan1p were precipitated with TAP-tag from cells overexpressing vector alone, Prk1 or Prk1(D158A) and immunoblotted with anti-pThr or anti-Pan1p antibody. Relative phosphorylation levels are indicated under the top panel. Prk1 and Prk1(D158A) were immunoprecipitated with anti-Myc antibody and immunoblotted with anti-pThr or anti-Myc antibody. (B) Viability of cells overexpressing Prk1 or Prk1(D158A). Cells were grown in YPD media overnight, and serial dilutions were spotted on glucose and glactose plates. (C) Localization of Pan1-GFP in cells overexpressing Prk1 or Prk1(D158A). The same cells as in B were grown to an OD600 of ∼0.3 in selective synthetic medium with 2% raffinose, and then Prk1 or Prk1(D158A) was induced with 2% galactose for 120 min. (D) Localization of Pan1-15TA-GFP in cells overexpressing Prk1. Pan1-15TA cells were grown to an OD600 of ∼0.3 in selective synthetic medium with 2% raffinose (left), and then Prk1 was induced with 2% galactose for 120 min (right).
Purification of Pan1p-binding Proteins
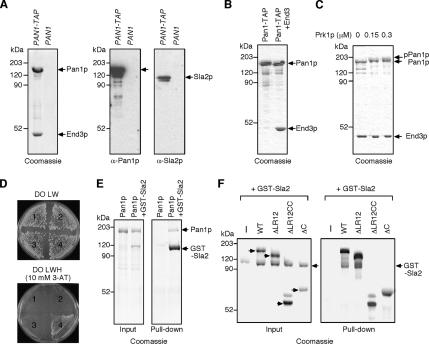
To determine how Pan1p might be inactivated at cortical patches, we next investigated Pan1p protein–protein interactions. Pan1p is known to interact with several endocytic proteins, such as Sla1p, End3p, Ent1/2p, and Yap180A/Bp (Wendland and Emr, 1998, 1999; Tang et al., 2000; Krogan et al., 2006). To purify the endogenous Pan1p complex, a TAP tag was fused to the C terminus of Pan1p and expressed from the endogenous PAN1 locus in a protease-deficient yeast strain. Interacting proteins were identified by mass spectrometry and immunoblotting, revealing that native End3p and Pan1p appear to form a stable complex (Figure 2A). To further examine the interaction between these proteins, we purified the Pan1p complex from yeast overexpressing TAP-tagged full-length Pan1p in strains overexpressing or not overexpressing End3p (Figure 2B). Similar to the native complex, Pan1p and End3p were purified as a stable complex in about a 1:1 binding stoichiometry (Figure 2B, right lane). It was previously reported that the interaction between Pan1p and End3p is negatively regulated by Prk1p phosphorylation (Zeng et al., 2001). However, in our experiments, phosphorylation of the Pan1p-End3p complex by Prk1p kinase did not affect the stability of the complex (Figure 2C). We previously showed that endogenous Pan1p purified from ark1Δ prk1Δ cells also makes a complex with End3p (Toshima et al., 2005). These observations, taken together, suggest that End3p can stably bind to Prk1p-phosphorylated Pan1p, as well as to the unphosphorylated form of Pan1p.
Figure 2.
Pan1p binds to End3p and Sla2p. (A) Coomassie-stained gel of TAP-tagged Pan1p and its binding proteins purified in two steps using immunoglobulin G (IgG)-coupled beads and S protein beads (left panel). The TAP-tag was expressed at the Pan1p C-terminus in DDY1810 cells via a chromosomal integration (PAN1-TAP). Immunoblots showing that Sla2p is coisolated with TAP-tagged Pan1p (center and right panels). The center blot was probed with an anti-Pan1p antibody, and the right blot was probed with anti-Sla2p antibody. DDY1810 (PAN1) was used as a control. (B) Coomassie-stained gel of the purified Pan1p or Pan1p-End3p complex. TAP-tagged Pan1p alone (left lane) or TAP-tagged Pan1p and End3p (right lane) were expressed in DDY1810 and purified in two steps using IgG-coupled beads and calmodulin-coupled agarose. (C) Effect of Prk1p phosphorylation on Pan1p-End3p complex. Purified Pan1p-End3p complex was incubated with 0–0.3 μM Prk1p at 30°C, separated by SDS-PAGE, and analyzed using Coomassie blue staining. (D) Interaction between Pan1p and Sla2p in the two-hybrid analysis. 1, pDB (bait vector) + pPC86 (prey vector); 2, pDB-PAN1 and pPC86; 3, pDB and pPC86-SLA2; 4, pDB-PAN1 and pPC86-SLA2. DO LW, drop-out Leu and Trp; DO LWH, drop-out Leu, Trp, and His; 3-AT, 3-amino-1,2,4-triazole. (E) In vitro binding of Pan1p to Sla2p. Purified Pan1p was pulled down with GST-Sla2. The pelleted proteins were separated by SDS-PAGE and analyzed using Coomassie blue staining. (F) Coomassie-stained gel for a cosedimentation assay using Pan1p truncation mutants. Purified GST-Sla2p was pulled down with indicated TAP-tagged Pan1 truncation mutants and analyzed as in E.
Because Sla2p appears at cortical patches at almost the same timing as Pan1p (Kaksonen et al., 2003), we also examined whether Sla2p copurifies with TAP-tagged Pan1p. Interestingly, we identified Sla2p as a novel Pan1p-interacting protein (Figure 2A). The interaction between Sla2p and Pan1p was much weaker, suggesting that this interaction might be more transient when compared with the Pan1p-End3p complex (Figure 2A). Both Pan1p and Sla2p are reported to bind to F-actin, but we did not find actin copurifying with TAP-tagged Pan1p, suggesting that this interaction is not mediated by F-actin (data not shown). We further confirmed the Pan1p-Sla2p interaction using the yeast two-hybrid system (Figure 2D). To examine the binding in vitro, we performed pulldown assays with purified Pan1p and GST-Sla2p and demonstrated that Pan1p binds to Sla2p in vitro as well as in vivo (Figure 2E). We next purified TAP-tagged Pan1p containing deletions of the LR regions (ΔLR12), the LR regions and the coiled-coil region (ΔLR12CC), or the C-terminal region including the coiled-coil region (ΔCC) (Toshima et al., 2005; Figure 2F) and attempted to pull down GST-Sla2 with these proteins. Pan1-ΔLR12 could bind to Sla2p, but neither ΔLR12CC nor ΔCC bound to Sla2p (Figure 2F). Therefore, Sla2p appears to bind to the central coiled-coil region of Pan1p, which contains the WH2-like region that binds to F-actin (Toshima et al., 2005).
Inhibition of Pan1p's Activity by Sla2p
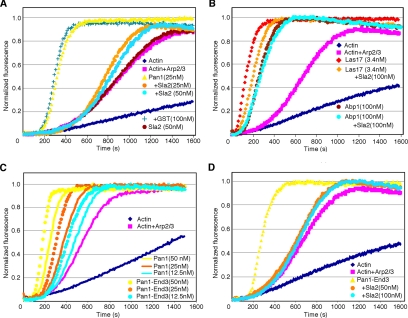
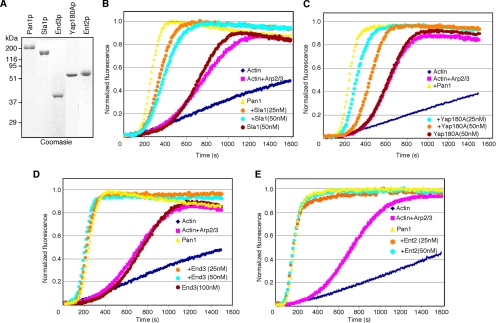
To examine the effect of Pan1p-interacting proteins on Pan1p's ability to activate the Arp2/3 complex, pyrene-actin assembly assays were performed. As reported previously (Toshima et al., 2005), 25 nM of full-length Pan1p enhances Arp2/3-mediated actin polymerization (Figure 3A). However, preincubation of Pan1p with 25 nM GST-Sla2p dramatically reduced its Arp2/3 activation activity, whereas GST alone, or Sla2p in the absence of Pan1p, did not affect actin assembly (Figure 3A). The effect of Sla2p was specific because neither Las17p, nor Abp1p activation of Arp2/3 was significantly inhibited by Sla2p, even when using 100 nM Sla2p with 3.4 nM Las17p (Figure 3B). The Pan1p-End3p complex exhibited slightly higher Arp2/3 activation activity than Pan1p alone (Figure 3C). The activity of the Pan1p-End3p complex was also inhibited by Sla2p (Figure 3D), suggesting that Sla2p regulates the native form of the Pan1p complex. To examine the effects of other previously identified Pan1p binding partners on Pan1p Arp2/3 complex activation activity, we purified Sla1p, Yap180Ap, End3p, and Ent2p (Figure 4A) and tested their effects on Pan1p using the pyrene-actin assembly assay. Sla1p and Yap180Ap slightly reduced Arp2/3 complex activation by Pan1p, but these effects were modest compared with those of Sla2p (Figure 4, B and C). Although the Pan1p-End3p complex exhibited a slightly higher activity compared with Pan1p alone (Figure 3C), End3p, when purified separately from Pan1p and added into the reaction, had no effect on Pan1p's activity (Figure 4D). This result might indicate that the N-terminal End3p-binding region of Pan1p was not correctly folded when purified separately from End3p and that it therefore could not form a stable complex with End3p. Ent2p had no effect on Pan1p's activity (Figure 4E). These results further support the conclusion that Sla2p specifically inhibits Pan1p's Arp2/3 complex activation activity.
Figure 3.
Effects of Sla2p on Pan1p-induced actin nucleation. (A) Pan1p-mediated Arp2/3 activation is inhibited by Sla2p. Graph shows assembly of 2 μM yeast actin and 0.1 μM pyrene-labeled rabbit muscle actin, with 10 nM yeast Arp2/3 complex and 25 nM Pan1p with or without indicated amounts of GST-Sla2p. GST or Sla2p alone was also added to the reaction as a control. (B) Effect of Sla2p on Las17p- or Abp1p-induced actin nucleation. Las17p with or without Sla2p was added to identical reactions of actin and Arp2/3 complex. (C) The Pan1p-End3p complex exhibits similar level of Arp2/3 activation activity to Pan1p. Indicated amounts of Pan1p or Pan1p-End3p complex was added to identical reactions of actin and Arp2/3 complex. (D) Pan1p-End3p complex mediated Arp2/3 activation is inhibited by Sla2p. Indicated amounts of GST-Sla2p were added to identical reactions of actin, Arp2/3 complex, and 50 nM Pan1p-End3p complex.
Figure 4.
Effects of Pan1p binding proteins on Pan1p's activity. (A) Coomassie-stained gels of purified proteins. (B–E) Indicated amounts of Sla1p (B), Yap180Ap (C), End3p (D), and Ent2p (E) were added to assembly of 2 μM yeast actin and 0.1 μM pyrene-labeled rabbit muscle actin, with 10 nM yeast Arp2/3 complex and 25 nM Pan1p.
The Coiled-Coil Region of Sla2p Binds to Pan1p and Inhibits Pan1p Activity
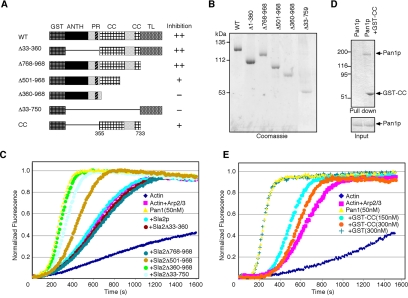
To identify the region of Sla2p that is important for the inhibition of Pan1p's activity, we purified GST-fused Sla2p containing deletions of the ANTH domain and the proline-rich region (Δ33-360), the C-terminal talin-like domain (Δ768-968), a part of the coiled-coil and talin-like domains (Δ501-968), the whole coiled-coil and talin-like domains (Δ360-968), or the N-terminal ANTH and the coiled-coil domains (33-750) (Figure 5, A and B). As shown in Figure 5C, truncation mutants that retained the coiled-coil region were able to fully or partially inhibit Pan1p's activity (also see summarized result in Figure 5A, right). In addition, deletions that lacked the coiled-coil region, Sla2Δ360-968 and Sla2Δ33-750, failed to inhibit Pan1p activity. We next purified the coiled-coil region (355-733aa) of Sla2p (GST-CC; Figure 5, A and D) and performed in vitro binding and actin assembly assays. GST-CC directly bound to Pan1p (Figure 5D) and inhibited Pan1p's Arp2/3 activation activity (Figure 5E), although a higher concentration of GST-CC was necessary to fully inhibit Pan1p's activity. This result suggests that either the 355-733aa region of Sla2p is not sufficient for forming correct 3D structure of the CC region or that a slightly wider region is required for complete inhibition.
Figure 5.
The coiled-coil region of Sla2p binds to Pan1p and inhibits Pan1p activity. (A) Diagram of GST-fused Sla2p constructs. The ANTH (AP180 N-terminal homology) domain, proline-rich (PR), predicted coiled-coil (CC), and talin-like (TL) regions are indicated. The results shown in C are summarized to the right of each diagram under “inhibition.” (B) Coomassie-stained gels of purified GST-fused Sla2 mutants. (C) Effects of Sla2 truncation mutants on Pan1p's activity. Full-length Sla2p, Δ33-360, Δ768-968, Δ501-968, Δ360-968, or Δ33-750, 100 nM, was added to assembly reactions containing 2 μM yeast actin and 0.1 μM pyrene-labeled rabbit muscle actin, with 10 nM yeast Arp2/3 complex and 50 nM Pan1p. (D) In vitro binding of Pan1p to GST-CC. Purified Pan1p was pulled down with (right) or without (left) GST-CC. The precipitates were separated by SDS-PAGE and analyzed using Coomassie blue staining. (E) Effect of GST-CC on Pan1p's activity. Indicated amounts of GST-CC were added to identical reactions of actin and Arp2/3 complex.
A Pan1 Partial Loss-of-Function Mutation Partially Suppresses the Temperature Sensitivity of the sla2Δ Mutant
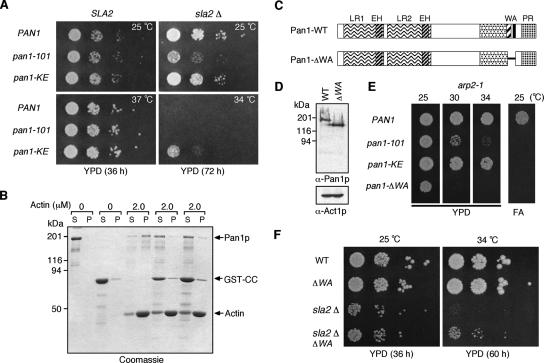
Cells lacking Sla2p are temperature sensitive for growth at 34°C and exhibit abnormal actin comet tail-like structures associated with endocytic complexes at the cell cortex (Yang et al., 1999; Kaksonen et al., 2003). We have shown that Sla2p directly binds to Pan1p and inhibits its Arp2/3 complex activation activity in vitro. Therefore, Pan1p may be constitutively active in sla2Δ cells, and a Pan1p loss-of-function mutant might suppress sla2Δ mutant phenotypes. To test this possibility, we crossed two Pan1 inactive mutants, the pan1-101 or pan1-KE, which impair Arp2/3 complex binding or F-actin binding, respectively (Duncan et al., 2001; Toshima et al., 2005), to the sla2Δ mutant. Interestingly, the pan1-KE mutation partially suppresses the growth defect of the sla2Δ mutant at 34°C, but the pan1-101 mutant does not (Figure 6A). This result suggests that Sla2p might inhibit Pan1p's activity by competing with F-actin. In support of this hypothesis, the addition of GST-CC markedly reduced the interaction between Pan1p and F-actin (Figure 6B), consistent with the observation that Sla2p binds to the Pan1p coiled-coil region, including the WH2-like region (Figure 2F). Because Pan1-KEp might retain some activity, and the pan1-KE mutation might therefore only suppress the phenotype of the sla2Δ mutant partially, we next used a pan1 mutant that lacks both F-actin binding and Arp2/3 binding regions (pan1-ΔWA; Figure 6C). Immunoblotting showed that Pan1-ΔWAp is expressed at levels similar to wild-type Pan1p (Figure 6D). As expected, the pan1-ΔWA mutant exhibited a more severe synthetic growth phenotype when combined with the arp2-1 mutant than either the pan1-101 or pan1-KE mutants (Figure 6E). However, although the pan1-ΔWA mutant suppressed the temperature sensitivity of the sla2Δ mutant a little more efficiently than the pan1-KE mutant, the suppression was still partial (Figure 6F). This result suggests that the sla2Δ mutant phenotype is caused in part by inappropriate activation of Pan1p and additionally by loss of other Sla2p functions, such as PIP2 binding via the ANTH domain (Sun et al., 2005) and/or actin binding via the talin-like domain (McCann and Craig, 1997).
Figure 6.
A Pan1 partial loss-of-function mutation partially suppresses the temperature sensitivity of the sla2Δ mutant. (A) The pan1-KE mutation partially suppresses the temperature sensitivity of the sla2Δ mutant at 34°C. A dilution series of cells was plated on YPD medium and incubated for the indicated times at 25, 34, or 37°C. (B) SDS-PAGE of a cosedimentation assay using full-length Pan1p, GST-CC, and F-actin. The gel was stained with Coomassie blue. P, pellets; S, supernatants. Indicated amounts of yeast actin were used with 100 nM Pan1p and 0, 250, or 500 nM GST-CC. (C) Diagram of wild-type Pan1p and Pan1ΔWA mutant. (D) Immunoblots showing that Pan1ΔWA is expressed at levels similar to wild-type Pan1p. Top panel was probed with anti-Pan1p antibody and bottom panel was probed with anti-Act1p antibody as a loading control. (E) Plates showing growth phenotypes of double-mutant combinations. Only pan1ΔWA results in a synthetic temperature sensitivity at 30°C when combined with arp2-1. All pan1 mutants result in synthetic lethality with arp2-1 on 3% formamide (FA) plates at 25°C. (F) The pan1ΔWA mutation partially suppresses the temperature sensitivity of the sla2Δ mutant at 34°C. A dilution series of cells was plated on YPD medium and incubated for indicated time at 25 or 34°C, respectively.
The pan1-ΔWA Mutant Suppresses the Temperature Sensitivity, Endocytic Defect, and Actin Phenotypes of sla2ΔCC Mutant
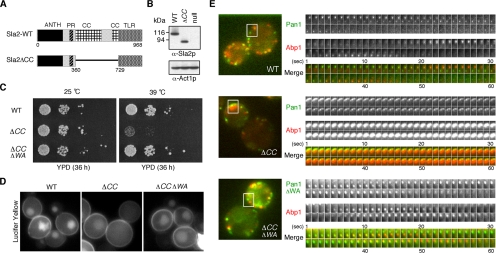
We showed that the coiled-coil region of Sla2p inhibits Pan1p's Arp2/3 complex activation activity (Figure 5). Therefore, a sla2 mutant lacking the coiled-coil region might result in a phenotype caused by uncontrolled Pan1p Arp2/3 complex activation. To test this possibility, we first made a sla2ΔCC mutant that lacks amino acids 360–729 of Sla2p and integrated this construct into the endogenous SLA2 locus (Figure 7A). Sla2ΔCCp was expressed at levels similar to the wild-type Sla2p protein (Figure 7B). Although the growth rate of the sla2ΔCC mutant was similar to that of the wild-type strain at 25°C, sla2ΔCC cells exhibited a severe defect in growth at 39°C (Figure 7C). Importantly, the pan1-ΔWA mutant suppressed the temperature sensitivity of the sla2ΔCC mutant at 39°C (Figure 7C), and it suppressed the endocytosis defect that was observed for the sla2ΔCC mutant at 39°C (Figure 7D). We also examined Pan1p and actin dynamics in the sla2ΔCC mutant using cells coexpressing Pan1-GFP and Abp1-RFP. In wild-type cells, Pan1p and Abp1p have lifetimes of ∼36 and 13.6 s, respectively (Figure 7E, top panels; Supplementary Movie 1). In contrast, the sla2ΔCC mutant exhibited abnormal actin tail structures (∼74.6%, n = 126 patches) and impaired Pan1p internalization into the cytosol at 37°C (Figure 7E, center panels; Supplementary Movie 1), much like the sla2Δ mutant. Similar to the growth phenotype and endocytosis defect, actin tail formation (∼5.7%, n = 132 patches) and the defect in Pan1p internalization were mostly suppressed by the pan1-ΔWA mutation (Figure 7E, bottom panels; Supplementary Movie 1), suggesting that the Sla2p coiled-coil region regulates Pan1p both in vitro and in vivo.
Figure 7.
The pan1ΔWA mutation suppresses the temperature sensitivity, endocytic defect, and actin phenotype of sla2ΔCC mutants. (A) Diagram of wild-type Sla2p and the Sla2ΔCC mutant. (B) Immunoblots showing that Sla2ΔCC is expressed at similar levels to wild-type Sla2p. Top panel was probed with an anti-Sla2p antibody and bottom panel was probed with anti-Act1p antibody as a loading control. (C) Plates showing growth phenotypes of sla2ΔCC mutant or sla2ΔCC pan1ΔWA double mutant. Dilution series of these mutants were plated on YPD medium and incubated for 36 h at 25 or 39°C, respectively. (D) The pan1ΔWA mutation suppresses the endocytosis defect observed in the sla2ΔCC mutant. Lucifer yellow uptake was performed for 60 min at 39°C and was assessed by fluorescence microscopy. (E) Left panels are single frames from a two-color movie showing Abp1-RFP (red) and Pan1-GFP (green) in wild-type (top), sla2ΔCC (middle), or sla2ΔCC pan1ΔWA (bottom) cells. Right panels are time series of boxed area in left panels. The time to acquire one image pair was 1 s.
DISCUSSION
Conserved Role for Hip1R/Sla2 Family Proteins in Actin Cytoskeleton Regulation
In both yeast and mammals, a transient burst of actin assembly occurs coincident with endocytic vesicle internalization (Merrifield et al., 2002; Kaksonen et al., 2003). Arp2/3 complex activators play a key role in actin assembly, but how activities of these proteins are regulated is poorly understood. Our group previously showed that cells lacking the SLA2 gene engage in continuous actin nucleation from nonmotile endocytic sites (Kaksonen et al., 2003). A similar phenotype is also observed in human cells depleted of a Sla2p-related protein, Hip1R, by RNA interference (Engqvist-Goldstein et al., 2004). These observations suggested that proteins of the Hip1R/Sla2 family might negatively regulate actin assembly during endocytosis. We showed here that Sla2p interacts with Pan1p and inhibits its Arp2/3 complex activation activity, possibly by preventing Pan1p's binding to F-actin. We also showed that a Pan1p loss-of-function mutant suppressed the continuous actin assembly phenotype observed in the sla2ΔCC mutant. Taken together, these findings clearly demonstrate that Sla2p negatively regulates Pan1p's activity during endocytosis. Interestingly, the formation of abnormal actin structures observed in Hip1R-depleted human cells was shown to be suppressed by knockdown of another Arp2/3 complex activator, cortactin, which localizes at clathrin-coated pits and functions in clathrin-mediated endocytosis (Cao et al., 2003; Engqvist-Goldstein et al., 2004). Thus, negative regulation of actin assembly by the Hip1R/Sla2 family of proteins at endocytic site is likely to be a conserved mechanism for regulating endocytic actin assembly in yeast and mammals.
Possible Regulatory Mechanism for Pan1p Activation
One possible factor that might regulate the interaction between Pan1p and Sla2p is the lipid PtdIns(4,5)P2. Our group previously showed that Sla2p directly binds to PtdIns(4,5)P2 through the ANTH domain (Sun et al., 2005). The ANTH domain of Sla2p is shown to be important for proper actin organization and endocytic internalization (Sun et al., 2005), but a physiological role of the interaction of Sla2p with PtdIns(4,5)P2 was not identified. Yeast expresses three synaptojanin-like proteins (Sjl1, Sjl2, and Sjl3), which are phosphoinositide phosphatases, and an sjl1Δ sjl2Δ double mutant exhibited severe defects in both actin organization and endocytosis (Singer-Kruger et al., 1998; Simonsen et al., 2001; Czech, 2003). Recently, it has been reported that Sjl2p recruitment to endocytic sites is dependent on F-actin. Therefore PtdIns(4,5)P2 may be converted to PI subsequent to actin assembly (Stefan et al., 2005). Interestingly, a strain lacking synaptojanin-like proteins, which probably overproduces PtdIns(4,5)P2, induces abnormal, enlarged plasma membrane structures in an actin-dependent manner, and the formation of the abnormal membrane structures is suppressed by Sla2p overexpression (Stefan et al., 2005). These observations suggest that the accumulation of PtdIns(4,5)P2 at endocytic sites could induce actin polymerization. Binding of PtdIns(4,5)P2 to Sla2p also suggests that accumulation of PtdIns(4,5)P2 might lead to dissociation of Sla2p from Pan1p and activation of Pan1p. Mutants of Pan1p show genetic interactions with mutants of Sjl1p (Wendland and Emr, 1998), further suggesting the importance of PtdIns(4,5)P2 regulation for Pan1p function.
The coiled-coil region of Sla2p was also reported to be important for interaction with Clc1p, the light chain subunit of yeast clathrin (Newpher and Lemmon, 2006). Although a sla2 mutant lacking the coiled-coil region (289-583aa) completely disrupts Clc1p binding in vitro, the physiological role of the clathrin-Sla2p interaction has yet to be established. Our group previously showed that Sla1p patch lifetime is reduced by 30% in clc1Δ or chc1Δ cells (Kaksonen et al., 2005), suggesting that the absence of mature clathrin structures could advance the timing of initiation of actin polymerization at endocytic sites. Therefore, clathrin may act as a negative regulator of the Pan1p-Sla2p interaction to control the timing of endocytic vesicle internalization.
End3p Stably Binds to Pan1p and Functions Together with Pan1p throughout Endocytosis
The association of Pan1p-Sla1p-End3p in a complex has been suggested to be negatively regulated by Prk1p (Zeng and Cai, 1999; Zeng et al., 2001). However, our results showed that Pan1p stably binds to End3p and that this interaction is not affected by Prk1p phosphorylation. In a previous study, Zeng et al. (2001) showed that the phosphorylated Pan1p LR2 (long repeat 2) fragment tagged with HA does not associate with GST-End3p. However, whether the pre-formed Pan1p-End3p complex can be dissociated by phosphorylation was not determined. In addition, Pan1p fragments containing both LR1 and LR2 regions bind to End3p about twofold more strongly than the LR2 fragment alone (Tang et al., 1997), suggesting that the LR1 region also contributes to the strength of this interaction. Prk1p appears to phosphorylate the Pan1p complex after endocytic internalization (Sekiya-Kawasaki et al., 2003). Therefore, it follows that our experimental conditions may better reflect the in vivo state of Pan1p-End3p. Earlier observations that Pan1p cortical patch localization decreases in end3Δ cells add further support to the importance of a stable interaction between Pan1p and End3p (Tang et al., 1997; Kaksonen et al., 2005). Besides Pan1p, the Pan1p binding proteins, Sla1p and Ent1/2p, are also Prk1p substrates (Watson et al., 2001; Zeng et al., 2001). Compared with the interaction between Pan1p and End3p, the interactions of these proteins with Pan1p appear much weaker because we did not find these proteins associated with TAP-tagged Pan1p. This finding suggests that these interactions might occur transiently, and potentially could be negatively regulated by Prk1p phosphorylation.
Phosphoregulation of Pan1p Localization during Endocytosis
Our results demonstrate that the phosphorylated form of Pan1p probably localizes in the cytosol and is dephosphorylated by a yet-to-be-identified phosphatase. One of the candidates to regulate the dephosphorylation of Prk1p-phosphorylated endocytic proteins is the Ser/Thr protein phosphase-1 (PP1/Glc7p). A glc7 mutant has been shown to affect cortical actin organization (Andrews and Stark, 2000). Scd5p, which is identified as a multicopy suppressor of a clathrin-deficient mutant (Nelson and Lemmon, 1993; Nelson et al., 1996), plays a crucial role in endocytosis and cortical actin organization (Henry et al., 2002), and Scd5p binding to the PP1/Glc7p is important for function (Chang et al., 2002; Henry et al., 2002). Furthermore, deletion of PRK1 suppresses the phenotypes of an scd5 mutant that has a mutation in the PP1-binding site (Henry et al., 2003; Huang et al., 2003), suggesting that Scd5p-PP1 could antagonize the function of Prk1p-dependent phosphorylation of endocytic proteins. A recent study showed that Scd5p recruitment to the cell surface is not essential for its function, and that Scd5p/PP1 can act on its targets in the cytosol (Chang et al., 2006). These results support our idea that phosphorylated Pan1p could be dephosphorylated in the cytosol.
Pan1p Activity Is Important for Initiating Actin Assembly during Yeast Endocytosis
Pan1p functions redundantly with Las17p, and these proteins are known to be important for endocytic internalization (Duncan et al., 2001; Toshima et al., 2005; Sun et al., 2006). We previously showed that a double mutant of pan1Δ855-1480, which lacks the C-terminal region containing the F-actin binding and Arp2/3 activating sites, and the las17ΔWCA mutant, has a severe endocytic defect (Toshima et al., 2005). However, we had not determined which step of endocytosis is impaired in the double mutant. In wild-type cells, Sla1p has a lifetime of ∼35.8 s, appearing at endocytic sites ∼24.2 s earlier than Abp1p, which has a shorter lifetime (∼13.6 s) and is internalized into the cytosol together with Abp1p (Supplementary Figure S1A; Kaksonen et al., 2003). The pan1Δ855-1480 or las17ΔWCA single mutant extended the Sla1-GFP lifetime (∼60.0 and ∼58.0 s, respectively) and delayed actin polymerization (∼48.0 s) at endocytic sites (Supplementary Figures S1, B and C). In addition, the double mutant of pan1Δ855-1480 and las17ΔWCA exhibited a significant delay in the initiation of actin assembly (∼156.1 s; Supplementary Figure S1D). Interestingly, the double mutant also exhibited a severe defect in Sla1p movement, which represents invagination of the plasma membrane into the cytosol (Supplementary Figure S1E; Kaksonen et al., 2003). These observations indicate that Pan1p and Las17p are important for initiating actin assembly and endocytic vesicle internalization. Our group previously showed that a double mutant of pan1ΔWA and las17ΔWCA exhibits a delay in the initiation of actin assembly (∼89.6 s), although the single mutant of pan1-ΔWA barely affected the timing of actin assembly (Sun et al., 2006). The pan1Δ855-1480 mutant likely affects a Pan1 activity in addition to the ability to activate the Arp2/3 complex. Compared with Las17p, Pan1p has much lower Arp2/3 activation activity in vitro (Sun et al., 2006). It remains possible that a yet-to-be-identified activator of Pan1p exists and strongly activates Pan1p in vivo, just as verprolin stimulates the Arp2/3-activating activity of the budding yeast type I myosin (Geli et al., 2000; Lechler et al., 2001; Sirotkin et al., 2005; Sun et al., 2006).
In summary, our data suggest that Pan1p's Arp2/3 activation activity is inhibited by binding to Sla2p during early stages of endocytosis. The coiled-coil region of Sla2p binds to the F-actin–binding region of Pan1p and inhibits Pan1p's activity. The sla2ΔCC mutant exhibited constitutive actin polymerization at endocytic sites, and the actin phenotype was suppressed by a pan1 loss-of-function mutation. Thus, these results suggest that Sla2p functions as a key regulator for initiating actin assembly during endocytic internalization. We propose that Pan1p is negatively regulated on the plasma membrane by Sla2p and on endocytic vesicles by Prk1p. It is now important to identify the positive regulators that counter the actions of Sla2p and Prk1p on Pan1p.
Supplementary Material
ACKNOWLEDGMENTS
We thank Beverly Wendland for the Ent2 and Yap1801 plasmid. We also thank Georjana Barnes and the members of the Drubin/Barnes lab for helpful discussions. This work was supported by Toyobo Biotechnology Foundation, Mochida Memorial Foundation for Medical and Pharmaceutical Research, and Yamanouchi Foundation for Research on Metabolic Disorders to J.T. and National Institutes of Health Grants GM50399 and GM42759 to D.G.D.
Abbreviations used:
- TAP
tandem affinity purification
- Arp
actin-related protein
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E06-09-0788) on December 6, 2006.

 The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
REFERENCES
- Andrews P. D., Stark M. J. Type 1 protein phosphatase is required for maintenance of cell wall integrity, morphogenesis and cell cycle progression in Saccharomyces cerevisiae. J. Cell Sci. 2000;113(Pt 3):507–520. doi: 10.1242/jcs.113.3.507. [DOI] [PubMed] [Google Scholar]
- Ayscough K. R., Stryker J., Pokala N., Sanders M., Crews P., Drubin D. G. High rates of actin filament turnover in budding yeast and roles for actin in establishment and maintenance of cell polarity revealed using the actin inhibitor latrunculin-A. J. Cell Biol. 1997;137:399–416. doi: 10.1083/jcb.137.2.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belmont L. D., Drubin D. G. The yeast V159N actin mutant reveals roles for actin dynamics in vivo. J. Cell Biol. 1998;142:1289–1299. doi: 10.1083/jcb.142.5.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H., Orth J. D., Chen J., Weller S. G., Heuser J. E., McNiven M. A. Cortactin is a component of clathrin-coated pits and participates in receptor-mediated endocytosis. Mol. Cell. Biol. 2003;23:2162–2170. doi: 10.1128/MCB.23.6.2162-2170.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang J. S., Henry K., Geli M. I., Lemmon S. K. Cortical recruitment and nuclear-cytoplasmic shuttling of Scd5p, a protein phosphatase-1-targeting protein involved in actin organization and endocytosis. Mol. Biol. Cell. 2006;17:251–262. doi: 10.1091/mbc.E05-10-0936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang J. S., Henry K., Wolf B. L., Geli M., Lemmon S. K. Protein phosphatase-1 binding to scd5p is important for regulation of actin organization and endocytosis in yeast. J. Biol. Chem. 2002;277:48002–48008. doi: 10.1074/jbc.M208471200. [DOI] [PubMed] [Google Scholar]
- Cooper J. A., Walker S. B., Pollard T. D. Pyrene actin: documentation of the validity of a sensitive assay for actin polymerization. J. Muscle Res. Cell Motil. 1983;4:253–262. doi: 10.1007/BF00712034. [DOI] [PubMed] [Google Scholar]
- Cope M. J., Yang S., Shang C., Drubin D. G. Novel protein kinases Ark1p and Prk1p associate with and regulate the cortical actin cytoskeleton in budding yeast. J. Cell Biol. 1999;144:1203–1218. doi: 10.1083/jcb.144.6.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czech M. P. Dynamics of phosphoinositides in membrane retrieval and insertion. Annu. Rev. Physiol. 2003;65:791–815. doi: 10.1146/annurev.physiol.65.092101.142522. [DOI] [PubMed] [Google Scholar]
- Duncan M. C., Cope M. J., Goode B. L., Wendland B., Drubin D. G. Yeast Eps15-like endocytic protein, Pan1p, activates the Arp2/3 complex. Nat. Cell Biol. 2001;3:687–690. doi: 10.1038/35083087. [DOI] [PubMed] [Google Scholar]
- Engqvist-Goldstein A. E., Drubin D. G. Actin assembly and endocytosis: from yeast to mammals. Annu. Rev. Cell Dev. Biol. 2003;19:287–332. doi: 10.1146/annurev.cellbio.19.111401.093127. [DOI] [PubMed] [Google Scholar]
- Engqvist-Goldstein A. E., Zhang C. X., Carreno S., Barroso C., Heuser J. E., Drubin D. G. RNAi-mediated Hip1R silencing results in stable association between the endocytic machinery and the actin assembly machinery. Mol. Biol. Cell. 2004;15:1666–1679. doi: 10.1091/mbc.E03-09-0639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geli M. I., Lombardi R., Schmelzl B., Riezman H. An intact SH3 domain is required for myosin I-induced actin polymerization. EMBO J. 2000;19:4281–4291. doi: 10.1093/emboj/19.16.4281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goode B. L., Wong J. J., Butty A. C., Peter M., McCormack A. L., Yates J. R., Drubin D. G., Barnes G. Coronin promotes the rapid assembly and cross-linking of actin filaments and may link the actin and microtubule cytoskeletons in yeast. J. Cell Biol. 1999;144:83–98. doi: 10.1083/jcb.144.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry K. R., D'Hondt K., Chang J., Newpher T., Huang K., Hudson R. T., Riezman H., Lemmon S. K. Scd5p and clathrin function are important for cortical actin organization, endocytosis, and localization of sla2p in yeast. Mol. Biol. Cell. 2002;13:2607–2625. doi: 10.1091/mbc.E02-01-0012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry K. R., D'Hondt K., Chang J. S., Nix D. A., Cope M. J., Chan C. S., Drubin D. G., Lemmon S. K. The actin-regulating kinase Prk1p negatively regulates Scd5p, a suppressor of clathrin deficiency, in actin organization and endocytosis. Curr. Biol. 2003;13:1564–1569. doi: 10.1016/s0960-9822(03)00579-7. [DOI] [PubMed] [Google Scholar]
- Huang B., Zeng G., Ng A. Y., Cai M. Identification of novel recognition motifs and regulatory targets for the yeast actin-regulating kinase Prk1p. Mol. Biol. Cell. 2003;14:4871–4884. doi: 10.1091/mbc.E03-06-0362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaksonen M., Sun Y., Drubin D. G. A pathway for association of receptors, adaptors, and actin during endocytic internalization. Cell. 2003;115:475–487. doi: 10.1016/s0092-8674(03)00883-3. [DOI] [PubMed] [Google Scholar]
- Kaksonen M., Toret C. P., Drubin D. G. A modular design for the clathrin- and actin-mediated endocytosis machinery. Cell. 2005;123:305–320. doi: 10.1016/j.cell.2005.09.024. [DOI] [PubMed] [Google Scholar]
- Kaksonen M., Toret C. P., Drubin D. G. Harnessing actin dynamics for clathrin-mediated endocytosis. Nat. Rev. Mol. Cell Biol. 2006;7:404–414. doi: 10.1038/nrm1940. [DOI] [PubMed] [Google Scholar]
- Krogan N. J., et al. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature. 2006;440:637–643. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- Lappalainen P., Drubin D. G. Cofilin promotes rapid actin filament turnover in vivo. Nature. 1997;388:78–82. doi: 10.1038/40418. [DOI] [PubMed] [Google Scholar]
- Lechler T., Jonsdottir G. A., Klee S. K., Pellman D., Li R. A two-tiered mechanism by which Cdc42 controls the localization and activation of an Arp2/3-activating motor complex in yeast. J. Cell Biol. 2001;155:261–270. doi: 10.1083/jcb.200104094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine M. S., McKenzie A., 3rd, Demarini D. J., Shah N. G., Wach A., Brachat A., Philippsen P., Pringle J. R. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Martin A. C., Xu X. P., Rouiller I., Kaksonen M., Sun Y., Belmont L., Volkmann N., Hanein D., Welch M., Drubin D. G. Effects of Arp2 and Arp3 nucleotide-binding pocket mutations on Arp2/3 complex function. J. Cell Biol. 2005;168:315–328. doi: 10.1083/jcb.200408177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCann R. O., Craig S. W. The I/LWEQ module: a conserved sequence that signifies F-actin binding in functionally diverse proteins from yeast to mammals. Proc. Natl. Acad. Sci. USA. 1997;94:5679–5684. doi: 10.1073/pnas.94.11.5679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrifield C. J., Feldman M. E., Wan L., Almers W. Imaging actin and dynamin recruitment during invagination of single clathrin-coated pits. Nat. Cell Biol. 2002;4:691–698. doi: 10.1038/ncb837. [DOI] [PubMed] [Google Scholar]
- Munn A. L. Molecular requirements for the internalisation step of endocytosis: insights from yeast. Biochim. Biophys. Acta. 2001;1535:236–257. doi: 10.1016/s0925-4439(01)00028-x. [DOI] [PubMed] [Google Scholar]
- Nelson K. K., Holmer M., Lemmon S. K. SCD5, a suppressor of clathrin deficiency, encodes a novel protein with a late secretory function in yeast. Mol. Biol. Cell. 1996;7:245–260. doi: 10.1091/mbc.7.2.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson K. K., Lemmon S. K. Suppressors of clathrin deficiency: overexpression of ubiquitin rescues lethal strains of clathrin-deficient Saccharomyces cerevisiae. Mol. Cell. Biol. 1993;13:521–532. doi: 10.1128/mcb.13.1.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newpher T. M., Lemmon S. K. Clathrin is important for normal actin dynamics and progression of Sla2p-containing patches during endocytosis in yeast. Traffic. 2006;7:574–588. doi: 10.1111/j.1600-0854.2006.00410.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newpher T. M., Smith R. P., Lemmon V., Lemmon S. K. In vivo dynamics of clathrin and its adaptor-dependent recruitment to the actin-based endocytic machinery in yeast. Dev. Cell. 2005;9:87–98. doi: 10.1016/j.devcel.2005.04.014. [DOI] [PubMed] [Google Scholar]
- Rigaut G., Shevchenko A., Rutz B., Wilm M., Mann M., Seraphin B. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 1999;17:1030–1032. doi: 10.1038/13732. [DOI] [PubMed] [Google Scholar]
- Rodal A. A., Manning A. L., Goode B. L., Drubin D. G. Negative regulation of yeast WASp by two SH3 domain-containing proteins. Curr. Biol. 2003;13:1000–1008. doi: 10.1016/s0960-9822(03)00383-x. [DOI] [PubMed] [Google Scholar]
- Schafer D. A. Coupling actin dynamics and membrane dynamics during endocytosis. Curr. Opin. Cell Biol. 2002;14:76–81. doi: 10.1016/s0955-0674(01)00297-6. [DOI] [PubMed] [Google Scholar]
- Sekiya-Kawasaki M., et al. Dynamic phosphoregulation of the cortical actin cytoskeleton and endocytic machinery revealed by real-time chemical genetic analysis. J. Cell Biol. 2003;162:765–772. doi: 10.1083/jcb.200305077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonsen A., Wurmser A. E., Emr S. D., Stenmark H. The role of phosphoinositides in membrane transport. Curr. Opin. Cell Biol. 2001;13:485–492. doi: 10.1016/s0955-0674(00)00240-4. [DOI] [PubMed] [Google Scholar]
- Singer-Kruger B., Nemoto Y., Daniell L., Ferro-Novick S., De Camilli P. Synaptojanin family members are implicated in endocytic membrane traffic in yeast. J. Cell Sci. 1998;111(Pt 22):3347–3356. doi: 10.1242/jcs.111.22.3347. [DOI] [PubMed] [Google Scholar]
- Sirotkin V., Beltzner C. C., Marchand J. B., Pollard T. D. Interactions of WASp, myosin-I, and verprolin with Arp2/3 complex during actin patch assembly in fission yeast. J. Cell Biol. 2005;170:637–648. doi: 10.1083/jcb.200502053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefan C. J., Padilla S. M., Audhya A., Emr S. D. The phosphoinositide phosphatase Sjl2 is recruited to cortical actin patches in the control of vesicle formation and fission during endocytosis. Mol. Cell. Biol. 2005;25:2910–2923. doi: 10.1128/MCB.25.8.2910-2923.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Kaksonen M., Madden D. T., Schekman R., Drubin D. G. Interaction of Sla2p's ANTH domain with PtdIns(4,5)P2 is important for actin-dependent endocytic internalization. Mol. Biol. Cell. 2005;16:717–730. doi: 10.1091/mbc.E04-08-0740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Martin A. C., Drubin D. G. Endocytic internalization in budding yeast requires coordinated actin nucleation and myosin motor activity. Dev. Cell. 2006;11:33–46. doi: 10.1016/j.devcel.2006.05.008. [DOI] [PubMed] [Google Scholar]
- Tang H. Y., Cai M. The EH-domain-containing protein Pan1 is required for normal organization of the actin cytoskeleton in Saccharomyces cerevisiae. Mol. Cell. Biol. 1996;16:4897–4914. doi: 10.1128/mcb.16.9.4897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H. Y., Munn A., Cai M. EH domain proteins Pan1p and End3p are components of a complex that plays a dual role in organization of the cortical actin cytoskeleton and endocytosis in Saccharomyces cerevisiae. Mol. Cell. Biol. 1997;17:4294–4304. doi: 10.1128/mcb.17.8.4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H. Y., Xu J., Cai M. Pan1p, End3p, and S1a1p, three yeast proteins required for normal cortical actin cytoskeleton organization, associate with each other and play essential roles in cell wall morphogenesis. Mol. Cell. Biol. 2000;20:12–25. doi: 10.1128/mcb.20.1.12-25.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toshima J., Toshima J. Y., Martin A. C., Drubin D. G. Phosphoregulation of Arp2/3-dependent actin assembly during receptor-mediated endocytosis. Nat. Cell Biol. 2005;7:246–254. doi: 10.1038/ncb1229. [DOI] [PubMed] [Google Scholar]
- Watson H. A., Cope M. J., Groen A. C., Drubin D. G., Wendland B. In vivo role for actin-regulating kinases in endocytosis and yeast epsin phosphorylation. Mol. Biol. Cell. 2001;12:3668–3679. doi: 10.1091/mbc.12.11.3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendland B., Emr S. D. Pan1p, yeast eps15, functions as a multivalent adaptor that coordinates protein-protein interactions essential for endocytosis. J. Cell Biol. 1998;141:71–84. doi: 10.1083/jcb.141.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendland B., McCaffery J. M., Xiao Q., Emr S. D. A novel fluorescence-activated cell sorter-based screen for yeast endocytosis mutants identifies a yeast homologue of mammalian eps15. J. Cell Biol. 1996;135:1485–1500. doi: 10.1083/jcb.135.6.1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendland B., Steece K. E., Emr S. D. Yeast epsins contain an essential N-terminal ENTH domain, bind clathrin and are required for endocytosis. EMBO J. 1999;18:4383–4393. doi: 10.1093/emboj/18.16.4383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S., Cope M. J., Drubin D. G. Sla2p is associated with the yeast cortical actin cytoskeleton via redundant localization signals. Mol. Biol. Cell. 1999;10:2265–2283. doi: 10.1091/mbc.10.7.2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng G., Cai M. Regulation of the actin cytoskeleton organization in yeast by a novel serine/threonine kinase Prk1p. J. Cell Biol. 1999;144:71–82. doi: 10.1083/jcb.144.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng G., Yu X., Cai M. Regulation of yeast actin cytoskeleton-regulatory complex Pan1p/Sla1p/End3p by serine/threonine kinase Prk1p. Mol. Biol. Cell. 2001;12:3759–3772. doi: 10.1091/mbc.12.12.3759. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.