Abstract
We have studied assembly of chromatin using Xenopus egg extracts and single DNA molecules held at constant tension by using magnetic tweezers. In the absence of ATP, interphase extracts were able to assemble chromatin against DNA tensions of up to 3.5 piconewtons (pN). We observed force-induced disassembly and opening–closing fluctuations, indicating our experiments were in mechanochemical equilibrium. Roughly 50-nm (150-base pair) lengthening events dominated force-driven disassembly, suggesting that the assembled fibers are chiefly composed of nucleosomes. The ATP-depleted reaction was able to do mechanical work of 27 kcal/mol per 50 nm step, which provides an estimate of the free energy difference between core histone octamers on and off DNA. Addition of ATP led to highly dynamic behavior with time courses exhibiting processive runs of assembly and disassembly not observed in the ATP-depleted case. With ATP present, application of forces of 2 pN led to nearly complete fiber disassembly. Our study suggests that ATP hydrolysis plays a major role in nucleosome rearrangement and removal and that chromatin in vivo may be subject to highly dynamic assembly and disassembly processes that are modulated by DNA tension.
INTRODUCTION
Transcription, replication, and other in vivo DNA processing in eukaryotes take place in the context of chromatin. The processive nature of these activities, and the necessity to disrupt histone–DNA contacts to accomplish them, suggests that chromatin must be dynamic in its structure, with actively transcribing genes perhaps in a continual state of structural rearrangement. The simplest example of chromatin rearrangement that would allow base pair access is displacement or dissociation of part or all of the histone octamer (Felsenfeld, 1996).
Chromosome visualization in vivo gives insight into chromatin dynamics at large length scales (Belmont, 2003; Levi et al., 2005) but is as yet unable to reveal events at the scale of individual nucleosome displacements. A complementary approach is to study individual chromatin fibers by using micromanipulation (Cui and Bustamante, 2000; Ladoux et al., 2000; Bennink et al., 2001; Brower-Toland et al., 2002; Leuba et al., 2003; Claudet et al., 2005; Gemmen et al., 2005; Bancaud et al., 2006). A major objective of such experiments has been the study of mechanically triggered changes in protein–DNA contacts, with an emphasis on force-driven opening of nucleosomes.
However, biophysical micromanipulation experiments offer possibilities beyond simply disassembling chromatin by force; experiments in “active” solutions containing chromatin-organizing or chromatin-processing enzymes permit direct observation of chromatin dynamics, and they can reveal details of structure and mechanism concerning compaction of DNA into chromatin, chromatin remodeling, gene expression in chromatin, mitotic chromosome condensation, and how such processes are affected by DNA tension. DNA tension is physiologically relevant because pulling of chromatin is likely to occur in vivo, given the large forces that can be produced by RNA polymerase (Yin et al., 1995; Wang et al., 1998), and DNA polymerase (Maier et al., 2000). Tension in chromatin has been suggested to play a role in regulation of various aspects of chromosome dynamics (Marko and Siggia, 1997a, b; Kleckner et al., 2004), via mechanisms including force-driven modification of chromatin structure.
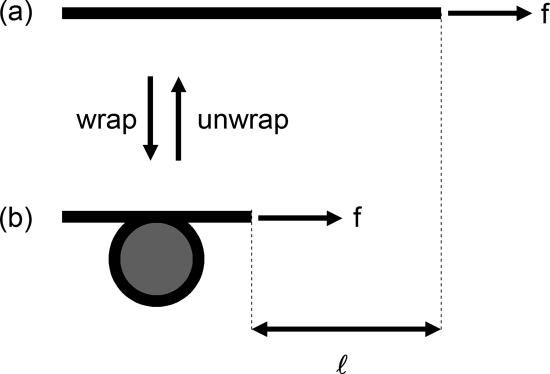
The concept underlying force–extension studies of chromatin assembly and disassembly is sketched roughly in Figure 1, applied to the example of the nucleosome core histone octamer. Force applied to the DNA favors nucleosome disassembly, because this liberates wrapped DNA of length l (expected to be ∼150 base pairs = 50 nm for a whole nucleosome), doing mechanical work equal to the product of force f and l (Marko and Siggia, 1997a). Acting against this, histone–DNA interactions stabilize the nucleosome. The free energy of interaction between the histones and DNA is thought to be roughly ΔG = 20 kcal/mol on the basis of chemical dissociation (Cotton and Hamkalo, 1981; Gottesfeld and Luger, 2001) and micromechanical (Brower-Toland et al., 2002) experiments. When the mechanical work done balances this interaction free energy, octamer “on” and “off” states will be in equilibrium. Thus, under conditions in which nucleosomes can assemble and disassemble, determination of the tension that “stalls” nucleosome assembly gives the free energy ΔG, which is problematic to measure otherwise (Thastrom et al., 2004). The same principle will apply for other proteins involved in folding of DNA into chromatin, although their disruption would likely yield a different DNA length per complex than the ∼50 nm expected for removal of one nucleosome.
Figure 1.
Crude sketch of force-controlled nucleosome assembly/disassembly. For naked DNA (a) under tension f, wrapping of a length l of DNA around histones is necessary to assemble a nucleosome (b), doing mechanical work against the applied force of fl. When wrapping and unwrapping occur at equal rates, no net assembly or disassembly will occur, corresponding to assembled nucleosomes and their diassembled components being in chemical equilibrium and therefore equal in free energy. The force at which assembly and disassembly are balanced can therefore be used to compute this free energy difference, via ΔG = fl.
The earliest force–extension experiments on chromatin used fibers isolated from cells (Cui and Bustamante, 2000), and found irreversible changes in fiber length to result from tensions of roughly 20 pN (1 pN = 10−12 newtons). The experimenters attributed this to force-driven disruption of histone–DNA interactions. Subsequent single chromatin fiber experiments have been based on in vitro reconstitution of chromatin. Using precisely defined biochemical conditions, Brower-Toland et al. (2002) and Brower-Toland and Wang (2004) assembled purified histones onto DNA molecules carrying tandem nucleosome-positioning sequences. After assembly, forces of 20–30 piconewtons (pN) led to abrupt length releases of roughly 25 nm each, which were interpreted as removals of DNA wrapped around nucleosomes.
One may assemble chromatin onto naked DNA by using reactions more closely approximating those occurring in vivo. Reactions using purified chromatin assembly factors (Leuba et al., 2003) showed that the ATP-independent chromatin assembly factor NAP-1 plus histones generates a chromatin assembly reaction that can overcome loads applied to the DNA template of up to 7.5 pN. Gemmen et al. (2005) examined nucleosomes assembled using NAP-1 and the ATP-dependent nucleosome assembly factor ACF and found that force-driven disassembly occurred over a wide range of forces (5–65 pN) and involved step events from 20 to 30 nm.
To even more closely approximate the complex biochemical environment of the nucleus, one may use Xenopus egg extracts (Murray, 1991; Smythe and Newport, 1991) to assemble chromatin onto DNA (Laskey et al., 1977; Glikin et al., 1984; Shimamura et al., 1988; Rodriguez-Campos et al., 1989; Hirano and Mitchison, 1991). These extracts contain a mixture of enzymes competent to assemble not only nucleosomes but also active chromatin. Despite the high complexity of extract reactions, drugs and antibodies have been used to identify and study the contribution of specific proteins to chromatin structure and function (Dilworth et al., 1987; Kleinschmidt et al., 1990; Hirano and Mitchison, 1994; MacCallum et al., 2002; Maresca et al., 2005). Extracts diluted from 2.5- to 400-fold have been used to assemble chromatin onto single DNAs against low piconewton forces (Ladoux et al., 2000; Wagner et al., 2005). After fiber assembly in diluted extracts, removal of nucleosomes by forces of 15–20 pN has been observed, via lengthening events predominantly of 50 nm, with some 25-nm events (Bennink et al., 2001; Pope et al., 2002, 2005).
In this article, we report the first single DNA measurements of chromatin assembly and disassembly in Xenopus extract solutions where ATP levels, DNA tension, and cell cycle conditions are all precisely controlled. We report experiments with extracts prepared from Xenopus eggs synchronized in interphase. Such “interphase” extracts are competent for a variety of physiological cellular reactions in vitro (Almouzni and Wolffe, 1993). All of our experiments are carried out in extract solutions competent to assemble nucleosomes onto naked DNA. To determine the role of ATP in these chromatin assembly reactions, we have carried out experiments in the absence of ATP (via enzymatic depletion, −ATP), in the presence of ATP plus an ATP regeneration system (+ATP), and finally in the presence of nonhydrolyzable ATP analogues [adenosine-5′-O-(3-thio)triphosphate (ATPγS) and adenyl-5′-yl imidodiphosphate (AMP-PNP)]. The main objectives of our experiments were twofold: to study the assembly and disassembly of chromatin under ATP conditions so as to measure the lengths, forces, and free energy associated with chromatin; and to determine the principal differences between the +ATP and −ATP reactions.
MATERIALS AND METHODS
Single-DNA Tethering
We end labeled 48.5-kilobase (kb) (referred to as 49 kb below) λ-DNA (Promega, Madison, WI) molecules using biotin- and digoxygenin-labeled oligonucleotides (oligos), as described in Smith et al. (1992). DNAs were then bound at one end to a 2.8-μm-diameter streptavidin-labeled paramagnetic particle (M-280; Dynal Biotech, Oslo, Norway), and at the other to either a 3-μm-diameter nonmagnetic polystyrene bead (Polybead-amino 3.0-μm microspheres; Polysciences, Warrington, PA) coated with anti-digoxygenin (Roche Diagnostics, Indianapolis, IN), or an anti-digoxygenin–coated coverglass.
Experiments were also carried out using pCWT25, a 15-kb plasmid with a λ-cos site. After linearization using λ-terminase (Epicenter, Madison WI), the resulting λ-cos ends were labeled using the same oligos used for λ-DNA. Ligations of pCWT25 and λ-DNA followed by end ligation of oligos were performed to construct longer end-labeled DNAs [97-kilobase pair λ-dimers, 64-kb λ + pCWT25, and 79-kb λ + 2(pCWT25)]. DNA contour lengths (l0) were computed from sequence lengths by using the factor 0.34 μm/kb.
Interphase Xenopus Egg Extracts
Crude cycling extracts were prepared as described in Murray (1991) with the following changes. Activated eggs were crushed at 10,200 rpm in a HB-6 (Sorvall, Newton, CT) rotor for 15 min. Cyclohexamide was added at 50 μg/ml to the crude extract to prevent entry into M phase; energy mix was not added. The crude extract was then centrifuged at 55,000 rpm (TLS-55 rotor; Beckman Coulter, Fullerton, CA) for 2 h at 4°C. The middle cytoplasmic layer was extracted using an 18-gauge needle and 1-ml syringe and was centrifuged again at 55,000 rpm for 30 min at 4°C to remove residual membranes. This high-speed interphase extract was immediately aliqouted, flash frozen in LN2, and stored at −80°C.
Experiments with Untreated Extracts
After calibration of a single-DNA tether, buffer was replaced with Xenopus interphase extract diluted 1:20 in HEPES buffer (50 mM HEPES-HCl, pH 7.5, and 50 mM KCl), whereas the tether is under a 5 pN force, which stops chromatin assembly and fully extends the molecule (Smith et al., 1992; Bustamante et al., 1994, 2000; Marko and Siggia, 1995). After extract was introduced, magnet position was adjusted to obtain the force desired.
Experiments with ATP-depleted Extracts
ATP was depleted by adding 0.02 U/μl apyrase (A6410; Sigma-Aldrich, St. Louis, MO) to the extracts diluted 1:20 with HEPES buffer, followed by 15-min incubation at 25°C. The ATP-depleted extracts were then injected into the sample cell, replacing the buffer.
Experiments with Extracts Containing ATP
Egg extracts were diluted 1:20 in HEPES buffer. For every 100 μl of diluted extract, 5 μl of “energy mix” (20 mM MgATP, 200 mM creatine phosphate, and 1 mg/ml creatine kinase) was added to obtain a final ATP concentration of 1 mM. Extract was incubated 15 min at 25°C before replacing the buffer in the sample cell.
Experiments with Extracts Containing Nonhydrolyzable ATP
Extract diluted 1:20 in HEPES buffer was depleted of ATP (see above) and then nonhydrolyzable ATP (either ATPγS or AMP-PNP; Sigma-Aldrich) was added to obtain a final 1 mM concentration. Extracts containing nonhydrolyzable ATP were then incubated for 15 min at 25°C before being put into the sample cell.
Micrococcal Nuclease (MNase) Gel Assay for Nucleosome Assembly by Extracts
Viability of extract to assemble chromatin was assayed using MNase gel analysis as described in Bonte and Becker (1999). Chromatin assembly reactions were run in extracts diluted 1:4 with HEPES buffer or in HEPES buffer with energy mix added (5 μl of energy mix was added per 100 μl of diluted extract). In either case, supercoiled pUC18 plasmid was added at a concentration of 2.5 ng/μl of diluted extract. Nucleosome (180 base pairs) and dinucleosome (360 base pairs) bands were observed in both cases, indicating the ability of the buffer-diluted extracts to assemble primarily nucleosomes onto DNA, in accord with previous studies (Ladoux et al., 2000; Bennink et al., 2001; Pope et al., 2002; Wagner et al., 2005).
RESULTS
Magnetic Tweezer (MT) Setups Allow Precise Observation and Control of Chromatin Assembly under Force Control
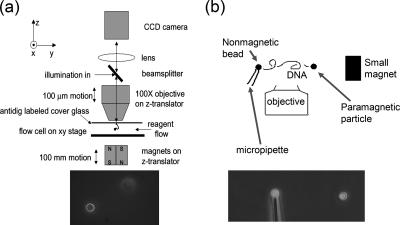
Our experiments were performed on single DNA molecules tethered at one end to a solid support, with the other end attached to a 2.8-μm-diameter paramagnetic bead (Figure 2). Controlled forces were applied to the bead by using permanent magnets (Smith et al., 1992; Strick et al., 1996, 1998, 2000), and bead positions were measured using digital videomicroscopy and bead-tracking computer software. We used two setups: in the first “vertical MT,” the bead is pulled perpendicular to the focal plane, so DNA extension is determined by autofocusing software (Figure 2a); details are described in Skoko et al. (2004). In the second “transverse MT” system the DNA is extended in the focal plane (Figure 2b); for details, see Yan et al. (2004). The vertical MT excels at low-force (<10 pN), low-noise measurements; the transverse MT has finer (20-nm) extension resolution and a higher acquisition rate.
Figure 2.
MT setups. (a) Vertical MT system. A DNA molecule is suspended from the top surface of a flow cell by one end, with its other end attached to a 3-μm-diameter paramagnetic particle that acts as a “handle” to which controlled forces can be applied, by using a permanent magnet below the flow cell. The bead is observed using a microscope objective; a piezoelectric focuser is used to locate the bead in the vertical (z) direction. Illumination of the bead is done through the objective and therefore a reflected-light (dark-field) image is obtained (bottom). Buffer exchanges are carried out using flow through the sample cell. (b) Transverse MT system. The principle is the same as for the vertical tweezer, but with the modifications that in addition to the paramagnetic particle at the right end of the molecule, a nonmagnetic particle is attached at its left end, held using suction on a thin glass capillary. By using a small magnet that can be positioned in the same plane or even below the end of the pipette, forces on the paramagnetic bead can be applied with direction in the plane of focus. The result is that both ends of the DNA molecule can be imaged simultaneously (bottom). An open sample cell is used allowing straightforward removal and replacement of buffer.
To examine chromatin assembly, first a tethered bead is identified, and forces measured at several magnet positions, by using bead thermal motion as described in Strick et al. (1996, 1998). This step ensures that a single DNA molecule is being studied. After this calibration in buffer, we adjusted the force on the bead to above 4 pN and introduced Xenopus extract. Use of force >4 pN keeps chromatin assembly from occurring until after flow of the extract into the sample cell is complete. After 5–10 min, force was reduced to below 4 pN, and chromatin assembly began. By adjusting the magnet position, different forces could be applied to the DNA template, and assembly/disassembly dynamics were recorded.
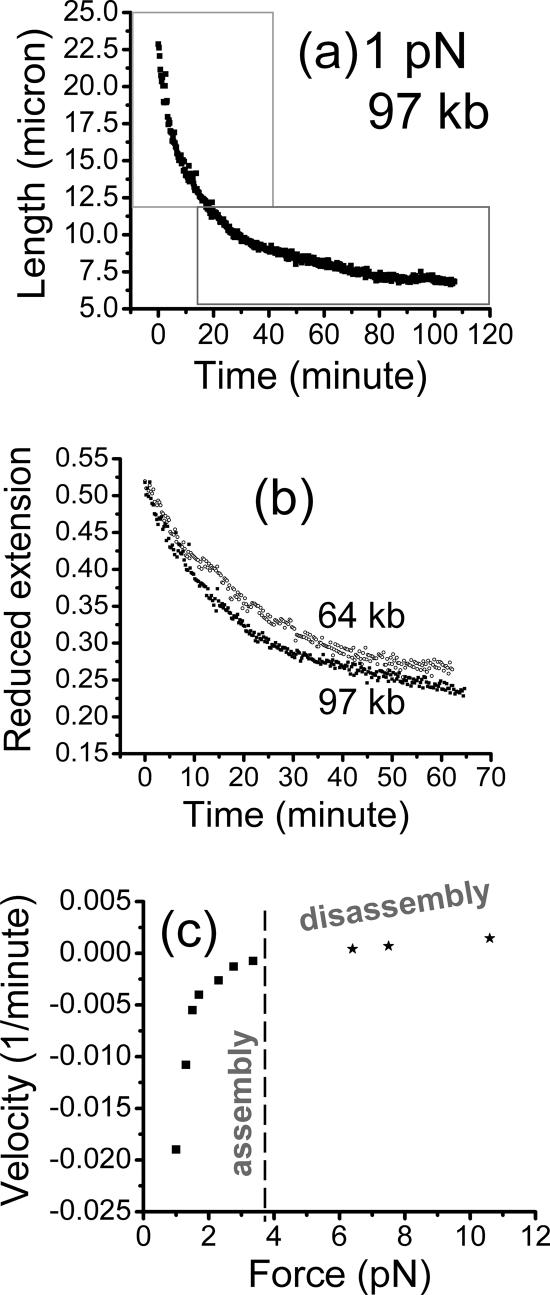
Figure 3a shows a time series recorded using the vertical MT during assembly of chromatin against a DNA tension of 1 pN on a 97-kb DNA (naked DNA contour length l0 = 32.8 μm), under −ATP conditions. At this force, the DNA is initially stretched out to ∼28-μm extension, which is then progressively shortened as chromatin assembles.
Figure 3.
Assembly of chromatin using diluted Xenopus egg extract, against constant forces, for −ATP (apyrase-treated) conditions. (a) Extension of a 97-kb DNA versus time measured by the vertical MT system, for most of the assembly reaction against a 1-pN force. The initial rapid contraction of the molecule during the early stage of assembly starting at the initial extension of 28 μm was not recorded. Boxes are drawn showing the two regions where exponential fits to the data can be made: the initial time course (top box) fits an exponential with a relaxation time of 10 min; later data in the bottom right box fit a slower relaxation time of 35 min. (b) Extension divided by naked DNA contour length l0 (reduced extension) for different-length assembly runs against forces of 1 pN (filled squares, 97 kb; open circles, 64 kb). Collapse of the curves onto one exponential demonstrates the simple scaling found for the −ATP assembly reaction. (c) Chromatin assembly and disassembly speeds for −ATP conditions show the force dependence expected in a simple force-controlled chemical reaction; figure shows “reduced velocities” (bead velocities divided by naked DNA contour length) of chromatin assembly (negative) and disassembly (positive). A smooth force–velocity relation is obtained, with a zero velocity or stall force of 3.5 pN (dashed line). At this point, assembly and disassembly processes compensate for one another, resulting in no net change in DNA compaction.
ATP-depleted Extracts Assemble Chromatin onto DNA in Two Phases below 1.2 pN
To determine the free energy balance and reaction kinetics of chromatin assembly, we performed a series of −ATP assembly experiments similar to Figure 3a at various forces. For forces <1.2 pN, two separate kinetic behaviors were observed during the assembly process; these are plainly visible in Figure 3, a and b, as an initial rapid decay, followed by a slower final decay. Fitting the two stages with separate exponential time courses, l/l0 = a∞ + ae−t/τ, the reaction times (τ) for the initial, fast and final, slow phases were 15 ± 5 and 35 ± 10 min, respectively (for an exponential time course, the reaction time τ indicates the time needed for the reaction to be approximately two thirds of the way to completion). The compaction parameter a∞ was found to be 0.35 ± 0.03 and 0.22 ± 0.02 for the initial fast phase and the final slow phase, respectively. Thus, at 1 pN the final fiber is compacted fivefold relative to naked DNA.
Similar −ATP experiments performed with different-length DNAs (48.5, 64, 79, and 97 kb) revealed the time courses to scale simply with DNA contour length l0. Figure 3b shows that assembly data for 64 and 97 kb against 1-pN tension collapse onto the same curve after extension is divided by l0. The −ATP initial fast phase reaction time constants were found to be independent of total DNA length (time constants were found to be within 20% for molecules between 15 and 97 kb with no systematic length dependence) in experiments on different-length DNAs.
For forces in the range 1.2–3 pN, the entire assembly time courses were fit by single exponentials corresponding to the “fast” phase discussed above. In experiments at constant forces between 1.2 and 3 pN, we found the fit value of a∞ to be 0.33 ± 0.03 with no force dependence. In this force range, the final compaction factor of the fiber was threefold relative to naked DNA.
ATP-depleted Extracts Can Assemble Chromatin against Loads of up to 3.5 pN
We observed the assembly rate (1/τ) to decrease with increasing force, approaching zero for forces near 3.5 pN. For larger forces, no assembly occurred. To determine whether forces beyond 3.5 pN could disassemble an assembled chromatin fiber, we carried out a series of experiments in which a fiber was first assembled against a 1-pN force, and then force was increased to above 3.5 pN. Fibers were observed to disassemble smoothly, allowing disassembly rates to be measured. The disassembly rate gradually increased with increasing force.
The rates can also be presented as bead velocities at the point of the reaction when extension was 0.66 times the extension of the initially naked DNA. These velocities, divided by contour length l0 of the naked DNA molecule used are shown in Figure 3c: negative velocities correspond to assembly, and positive velocities correspond to disassembly. The velocity versus force passes smoothly through zero at a force of 3.5 pN. Figure 3c is analogous to a velocity versus force curve for a molecular motor (Wang et al., 1998); the zero-velocity point at 3.5 pN is the “stall force” for the chromatin assembly reaction. Figure 3c shows data from a series of experiments on different-length DNAs; the smooth dependence of velocity divided by molecule length on force provides additional evidence for the length-scaling shown in Figure 3b (note the length-reduced velocities shown in Figure 3c can be converted to true velocities by multiplying by molecule contour length).
Observation of Stepwise Assembly and Disassembly Events
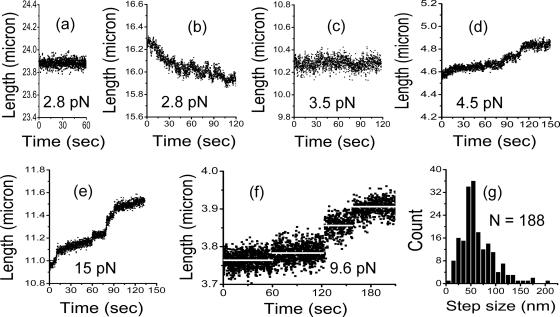
To determine whether chromatin folding and unfolding was reversible, i.e., whether opening and closing events could be observed at constant force, transverse MT experiments were performed. Figure 4a shows extension fluctuations for a 79-kb naked DNA under 2.8-pN tension, before adding extracts. For this force, the 26.9-μm-long DNA is extended to 23.9 μm, ∼0.9 times the molecule contour length as expected for naked DNA in physiological buffer under this tension (Bustamante et al., 2000). A random noise-like fluctuation of amplitude ∼100 nm was observed, due mainly to Brownian motion (Strick et al., 1996), because calibration experiments showed the mechanical noise in our horizontal MT setup generates an amplitude of 10–20 nm (Yan et al., 2004).
Figure 4.
Transverse MT results giving high-resolution DNA extension during chromatin assembly/disassembly, under −ATP (apyrase-treated) conditions. Each panel shows a time series of extension measurements at constant force as indicated. (a) Naked 79-kb DNA under a constant tension of 2.8 pN. The extension shows thermal fluctuations ∼100 nm around a fixed extension. (b) Same DNA as in a, following injection of −ATP extract. Assembly occurs as in Figure 3, but at the higher resolution of the transverse MT, step events of roughly 100-nm amplitude can be observed. Note that assembly and disassembly steps occur, but net assembly occurs due to a larger number of assembly steps. (c) Same DNA as a and b, but now at 3.5 pN, the stall force of the assembly reaction. Note the fluctuations of extension around its roughly constant value; by comparison with a, in addition to Brownian fluctuations, there are step-like fluctuations, of amplitude 50 and 100 nm. (d) Disassembly time series for 4.5 pN. After assembly of a fiber onto a 49-kb DNA at 1 pN, force was increased to 4.5 pN, leading to the disassembly trace shown here. The thermal fluctuations are slightly reduced by both the shorter DNA and the higher force. Step events of 50 and 100 nm are observed. (e) Same DNA as in d but now with force increased to 15 pN. Disassembly occurs more rapidly, and the steps are even more visible. Again, 50- and 100-nm steps are observed. (f) Disassembly time series of a chromatin fiber assembled onto a 15-kb DNA at 9.6 pN. Besides reducing the noise by the shorter molecule, plateau durations are increased due to the smaller number of nucleosomes along the DNA. Extension averages during the plateaus (white lines) allow step size measurement. A 25-nm step, a 50-nm step, and a 75-nm step can be seen. (g) Histogram of the step sizes for 188 steps collected from disassembly runs of 49-kb DNAs by using more than 9 pN. A well-defined peak occurs at ∼50 nm.
Figure 4b shows the same 79-kb molecule after addition of ATP-depleted egg extracts at the same force of 2.8 pN, during assembly. Compared with the longer, lower resolution traces of Figure 3, Figure 4b shows only a small portion of the assembly process where extension was decreased by a net amount of ∼500 nm; at this higher resolution, one observes folding and unfolding events. These step-like events were absent in the naked DNA trace (Figure 4a). Sharp extension changes of ∼50–100 nm are observed; thermal fluctuations of extension at this relatively low force make precise analysis of the steps problematic. However, it is clear that during assembly against 2.8-pN forces, disassembly events occur.
After assembly at 2.8 pN down to an extension of ∼10.3 μm, force was increased to 3.5 pN, the reaction stall force. The extension as a function of time is shown in Figure 4c after changing to this force (same 79-kb molecule as in Figure 4, a and b). Jump fluctuations of fiber length continued to be observed but with almost no net assembly or disassembly. Again, step-like fluctuations of amplitude of ∼100 nm can be observed against the roughly 100-nm amplitude thermal extension fluctuations, showing that near the stall point, stepwise opening and closing events occur at a rate of about one every 10 s; therefore, our experiments access a mechanically controlled chemical equilibrium.
Opening of Assembled Chromatin Fibers by Using Larger Forces
Above 3.5 pN, assembled fibers disassemble: as force is increased, thermal extension fluctuations are reduced (Strick et al., 1996) allowing chromatin-opening steps to be better resolved. Figure 4d shows an extension time series during disassembly of a chromatin fiber assembled onto a 49-kb DNA (different molecule from Figure 4, a–c). Before this 49-kb fiber was completely disassembled, the force was increased to 15 pN (Figure 4e), allowing small, roughly 50-nm steps to be resolved, consistent with the length change expected for release of the 150 base pairs of DNA wrapped around one nucleosome.
Using shorter 15-kb DNA, individual steps could be more clearly observed during the disassembly of a chromatin fiber. Figure 4f shows steps of roughly 30, 50, and 75 nm, estimated by the difference between the averages of successive plateaus. A histogram of 188 such steps collected from disassembly time series above 9 pN exhibits a well-defined peak near 50 nm, the length expected for individual nucleosome removal events (Figure 4g).
To determine the total number of 50-nm step events occurring during force-opening of assembled fibers, two experiments were performed with 49-kb DNAs compacted by ATP-depleted extracts against a force of 1 pN for 40 min. Subsequent opening by force led to a series of steps equivalent to 170 ± 20 50-nm events per λ-DNA. Under the hypothesis that the 50-nm events are due to nucleosome removals, we estimate that these fibers contained one nucleosome per 280 ± 35 base pairs.
ATP Stimulates Large-Amplitude Assembly/Disassembly (“V” and “Λ”) Events
We wanted to determine the effect of adding ATP and therefore activating ATP-dependent enzymes, on the −ATP reactions reported above. Our experimental approach allowed us to carry out a buffer replacement experiment, where we first assembled chromatin onto a single λ-DNA (48.5 kb, 16.4 μm) by using untreated diluted egg extracts (i.e., without addition of either ATP or apyrase). Assembly of chromatin was allowed to proceed and then was stalled at the −ATP reaction critical force of 3.5 pN (Figure 5, times before 180 s), and at a fiber length of ∼10 μm. Small, 50- to 100-nm step-like events were observed during this initial stage, as in our other −ATP experiments.
Figure 5.
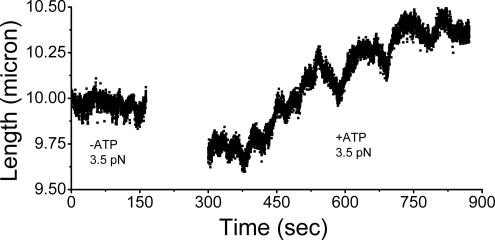
Effect of ATP addition on a chromatin fiber initially assembled with no ATP. Time series of extension measurements by using the transverse MT are shown, for a 49-kb DNA template. Before 180 s: After assembly of chromatin fiber at 1 pN by using untreated extracts, force was increased to 3.5 pN, the stall force for the −ATP reaction. As expected, the behavior of the fiber was similar to that observed under ATP-depleted conditions, with small step-like fluctuations plus thermal extension fluctuations. No net assembly or disassembly occurs. The behavior of the fiber is essentially the same as that observed for the ATP-depleted extract product (Figure 4C). After 300 s: After addition of +ATP extract solution into the experimental chamber, force still held at 3.5 pN. There are two main effects: the fiber tends to disassemble, and large-amplitude, apparently processive runs of extension are observed, in both assembly and disassembly directions.
Then, extract containing ATP plus energy mix was added to the reaction to obtain a final ATP concentration of 1 mM (see Materials and Methods) and incubated for ∼4 min (Figure 5, gap in data between 180 and 300 s). The times after 300 s in Figure 5 show chromatin extension (force was still at 3.5 pN) after the addition of +ATP extract. The +ATP extension began to undergo greatly increased length fluctuations of up to 400 nm, with sharp switching between assembly and disassembly (V and Λ shapes in Figure 5) on a time scale of ∼50–100 s. Both amplitude and time scale of the +ATP length fluctuations were much larger than in the absence of ATP. The +ATP extension exhibited a distinct type of assembly/disassembly dynamics, with a characteristic amplitude of ∼200–400 nm, instead of the randomly interspersed, roughly 50- to 100-nm chromatin assembly/disassembly step-like events observed in the −ATP case.
A second important difference between the +ATP and −ATP cases was that at the stall force for the −ATP case of 3.5 pN, addition of ATP greatly stimulated force-driven fiber disassembly. Figure 5 shows that the fiber greatly opens as a result of addition of ATP at 3.5 pN force. In numerous experiments with different extract preparations, addition of ATP was found to consistently result in a net opening (increase of extension) of an assembled fiber. In other experiments where the +ATP reaction was first carried out, and then the solution replaced with −ATP extracts, we observed a reduction in fiber extension with no V and Λ events (data not shown). Thus, although ATP is not required for assembly of the fiber, it stimulates fiber opening; we emphasize that the V and Λ events require ATP.
The 200- to 400-nm V and Λ events make the roughly 50-nm step on-off events difficult to observe in time series such as those of Figure 4. However, if, after +ATP assembly, force is abruptly increased to above 5 pN, step events similar to those observed in Figure 4, d and e, can be observed, with amplitudes of 50–100 nm indicating that nucleosomes are assembled onto DNA in the presence of ATP (data not shown).
Velocities of V and Λ Events during +ATP Assembly
To determine the nature of these large V and Λ events, we studied their shapes as a function of force (Figure 6). We were primarily concerned with whether they were large step-like events, or alternately whether the events were characterized by smooth (processive motor-like) motions. Figure 6a shows a +ATP assembly reaction at 0.7 pN, for a 32-μm-long 97-kb DNA. Gradual assembly occurred, but with a much slower rate than in the −ATP case, and also with large variations in velocity. Comparison of the +ATP assembly curve of Figure 6a with the −ATP assembly curve of Figure 3a indicates qualitative differences. The −ATP reaction shows a nearly exponential and smooth extension time course. However, the +ATP reaction shows a slow, nearly linear reduction of extension.
Figure 6.
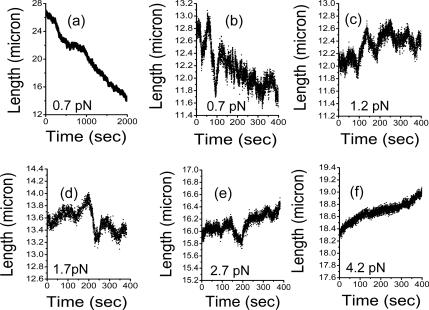
Extension dynamics during chromatin assembly/disassembly using extracts with added ATP. Panels show time series of extension for a 97-kb molecule measured using the transverse MT. (a) At a force of 0.7 pN, assembly gradually compacts the DNA. The initial DNA extension of roughly 28 μm is reduced by assembly. The shape of the assembly curve is rather linear apart from some large-scale variations, distinct from the nearly exponential assemblies observed for the −ATP case. (b) Higher resolution plot of extension variations for the fiber of a, for a time window 3 min after the end of a. Large extension Λ and V events are observed, characterized by a well-defined velocity which is the same for the opening and closing legs. Extension remains near 12 μm. (c and d) After increasing force on the fiber of b to 1.2 and 1.7 pN, extension gradually increases, with continuation of the Λ and V events. (e) After increase of force on the fiber of c to 2.7 pN, further extension occurs, again with large Λ and V events. (f) After increasing force to 4.2 pN, further extension occurs without Λ and V events.
The time series of Figure 6a shows almost all of the assembly reaction: after extension is reduced to ∼12 μm (∼0.4 times the extension of the initially naked DNA), it remains near that value, for force kept at 0.7 pN. Sharp switching events are apparent if the data are plotted at higher extension resolution; these are distinct from the small 50-and 100-nm steps observed in the −ATP case. Figure 6b shows a higher resolution plot of the extension, still at 0.7 pN, 3 min after the end of the time series of Figure 6a; the extension is undergoing runs of V and Λ events, causing the extension to vary over a range of ∼1 μm.
Subsequently, force was increased to 1.2 pN; Figure 6c shows the resulting extension behavior as the fiber opened, with V and Λ events (note that Figure 6, b–f, have the same time and length scales to allow comparison). Force was then increased to 1.7 pN, causing further opening with V and Λ events (Figure 6d). Figure 6e shows the extension after force was increased to 2.7 pN; faster overall disassembly occurred, but V and Λ events were still observed. Finally, larger forces of 4.2 pN (Figure 6f) eliminated large V or Λ events; only small <100-nm jumps were observed, although some small retraction events continued to occur.
The V and Λ events are not step-like, but instead they are rather smooth, with well-defined velocities. For low forces (<1 pN), this velocity was 42 nm/s; for medium forces (between 1 and 2 pN), the velocity was reduced to 12 nm/s. Finally, for forces of 3.5 pN, we found a velocity of 6 nm/s. Strikingly, the extension and retraction velocities were the same for each force in the range of forces examined. In all experiments, no V or Λ event was ever observed to have a pause at its trough or peak.
The V and Λ Events Require ATP Hydrolysis
To test whether the V and Λ events required ATP hydrolysis, or just ATP binding, we tested the effects of nonhydrolyzable ATP analogues added to extracts that had been treated with apyrase to remove ATP. We found addition of ATPγS or AMP-PNP to a final concentration 1 mM to reactions in the vertical MT flow cell resulted in assembly against a 2.6-pN force similar to that observed with −ATP (data not shown). On reduction of the force to 1.2 pN, fiber assembly proceeded to the fiber length observed in the −ATP experiments; reaction rates observed at 2.6 and 1.2 pN were comparable with those in the −ATP experiments of Figure 3. Similar results were obtained in the presence of higher concentrations (5 and 10 mM; data not shown) of nonhydrolyzable ATP analogues. We concluded that the V and Λ dynamics require ATP hydrolysis.
DISCUSSION
Chromatin Assembles onto Naked DNA in Cytoplasmic Extracts in the Absence of ATP
Although previous studies have observed chromatin assembly dynamics by using diluted egg extracts (Ladoux et al., 2000), and single-nucleosome removal events at higher forces (Bennink et al., 2001; Brower-Toland et al., 2002), our experiments are the first to document step opening and closing fluctuations in an active chromatin assembly reaction. In the absence of ATP, we observed compaction of naked DNA into chromatin, via an assembly reaction displaying first-order–like kinetics, for DNA tensions below 3.5 pN. Once the fiber assembled, a progressive disassembly reaction could be obtained by applying forces above 3.5 pN. At the stall force of 3.5 pN separating assembly and disassembly regimes, we could observe forward and reverse reaction “steps.” For large forces, the disassembly steps were dominated by events of 50 nm (Figure 4g).
These observations allow us to conclude that our −ATP experiments probe mechanical-chemical equilibrium. For forces ∼1 pN away from 3.5 pN, the process is either dominated by on-events (almost no off-events were observed for <2.5 pN) or by off-events (almost no on-events were observed) for >4.5 pN.
Are Nucleosomes Present in the Assembled Fibers?
In all single-chromatin fiber experiments, one may ask whether nucleosomes are present along the assembled fiber, and in experiments where cell extracts are used (Ladoux et al., 2000; Bennink et al., 2001; Pope et al., 2005), to what degree nonhistone proteins are present in the final assembled fiber. By conventional MNase digestions and DNA supercoiling assays, we found that both the +ATP and −ATP reaction conditions support assembly of nucleosomes onto DNA (data not shown). In single-molecule experiments, we have observed extract-assembled fibers to disassemble predominantly via 50-nm steps (Figure 4g). This is in accordance with other experiments where chromatin has been assembled using Xenopus egg extracts (Bennink et al., 2001; Pope et al., 2005), where disassembly proceeds predominantly via ∼50-nm steps.
Our count of one nucleosome per ∼280 base pairs based on step events is significantly lower than the one nucleosome per 160–180 base pairs found in MNase gel analyses of extract–assembly of chromatin on DNA plasmids (Rodriguez-Campos et al., 1989). There are a few possible reasons for this discrepancy. First, our count is based on −ATP reactions, which generate less precisely positioned nucleosome arrays than reactions where ATP is present. Second, our count is based on experiments where a 1-pN force was kept on the fiber during assembly, which may limit the degree to which nucleosomes can be packed together. Third, the single-DNA assembly reactions used for counting nucleosomes were carried out for 40-min periods, which even for +ATP reactions is too short a time for tightly spaced arrays to form. Our results (Figure 3, a and b) are consistent with those of Shimamura et al. (1988) who found that arrays with one nucleosome per 260 base pairs were assembled in 30-min reactions; formation of tight arrays with 160 base pairs per nucleosome required 6-h reactions.
The predominance of ∼50-nm jumps during disassembly of extract-assembled chromatin is at odds with results for single chromatin fibers assembled using purified components (Brower-Toland et al., 2002; Gemmen et al., 2005) where the main force-driven disassembly pathway is observed to be via 25-nm steps. This has been proposed to be due to a gradual removal of approximately one DNA turn around each nucleosome at low forces (<5 pN) followed by abrupt removal of the second turn in a single 25-nm step (Brower-Toland et al., 2002). However, Claudet et al. (2005) showed that for nucleosomes assembled using purified components, exposure to dilute solution conditions (below 5 nM nucleosome concentration) can result in dissociation of histones H2A/H2B, leading to observation of 25-nm steps when force is used to pull DNA away from the remaining H3/H4 proteins. Conversely, when assembled nucleosomes were present at levels in excess of 0.1 mg/ml, Claudet et al. (2005) found whole nucleosomes to be stable, and 50-nm steps in addition to 25-nm steps were observed during force-driven opening of chromatin. Therefore, a possible explanation for the preponderance of ∼25-nm steps during disassembly of chromatin reconstituted from purified components may be simply that half of the histones dissociated before the fiber was pulled.
It also should be noted that the extract-based reactions include linker histones, whereas all the purified component experiments to date do not. Although details of how linker histone binds to nucleosomes are still unknown, recent experiments indicate that it binds linker DNA to DNA near the dyad axis (Brown et al., 2006). Possibly linker histone stabilizes nucleosomes sufficiently so that 50-nm events corresponding to unwrapping of whole nucleosomes can be observed in pulling experiments. Linker histone-depleted extracts (Maresca et al., 2005) could be used to examine this possibility.
Given that we have used reagents that we have verified to be competent to assemble whole nucleosomes onto DNA, that we have taken care to carry out experiments in the 20-fold diluted extracts where free histones are present at ∼2 μg/ml concentrations, that we observe a dominance of 50-nm events during force opening, and finally that we observe a total number of opening events consistent with removal of 170 ± 30 nucleosomes along a 48.5-kb λ-DNA, corresponding to one nucleosome per ∼280 base pairs, we conclude that the 50-nm events are mainly due to removal of nucleosomes.
Initial Assembly of Nucleosomes onto DNA by ATP-depleted Reaction Is Not Processive
Two features of our data indicate that the −ATP reaction is not processive, i.e., nucleosomes are assembled in an uncoordinated manner onto naked DNA. First, experiments on DNA templates of different lengths showed that the assembly and disassembly velocity scaled linearly with DNA length (Figure 3b). This length-scaling property of our results indicates that nucleosome assembly proceeds at many places along the initial DNA in a parallel manner, and it rules out the possibility that the assembly proceeds processively from one initiation point, e.g., near an end, because in that alternative case, a linear dependence of extension on time would be observed. Second, the exponential time course during assembly suggests that nucleosomes are initially placed at different positions along the DNA independently. Given this interpretation of the data, the initial reaction rate (Figure 3a) indicates the characteristic time of nucleosome assembly. Note that the time to fill a given DNA with nucleosomes is also given by this rate.
For forces below 1.2 pN, the −ATP reaction displays two distinct phases. The initial rapid assembly phase likely corresponds to initially random placement of nucleosomes along our long DNAs. Further support for this conclusion comes from the fact that we find that the initial phase compacts the DNA to ∼35% of its initial, naked extension, close to the 30% expected from “random deposition” (Schaaf and Talbot, 1989) of nucleosomes. The second, slower phase possibly corresponds to a gradual reorganization of nucleosomes, which only slowly makes room for addition of new nucleosomes. A simple model for this would involve on- and off-rates for nucleosomes, plus a nucleosome diffusion constant describing the rate at which nucleosomes move (randomly slide or “1D-diffuse”) on the DNA. Given this hypothesis, the apparent absence of this slower phase at higher forces between 1.2 and 3 pN suggests either that tension in the DNA may suppress sliding of nucleosomes along DNA, which is reasonable, because force may well tend to suppress fluctuations of DNA off the histones. Alternately, it is possible that at higher forces, faster on- and off-rates at higher forces simply make sliding less important a process for reaching the assembled state.
Stall Force of Assembly Reaction Indicates Nucleosome Assembly Free Energy
The observation of a well-defined stall force, and on- and off-events near the stall force of 3.5 pN permits an estimate of the free energy associated with assembly of single nucleosomes. The mechanical work done assembling one nucleosome is the product of the force with the length change as measured in our experiment; at the stall force of 3.5 ± 0.2 pN, this equals the free energy per assembled nucleosome (Figure 4c). Per single-nucleosome length change of 50 nm, a free energy of 50 nm × 3.5 pN, or 175 pN.nm = 1.75 × 10−19 J (recall 1 N.m = 1 J). In chemical units, this is 27 ± 2 kcal/mol per assembled nucleosome.
Nucleosome–DNA interaction free energies have been measured previously in equilibrium dissociation experiments (Cotton and Hamkalo, 1981) to be roughly 20 kcal/mol. Recent reexamination of the dissociation approach by (Thastrom et al., 2004) criticized dissociation studies for not demonstrating thermodynamic reversibility. By contrast, we have demonstrated microscopic reversibility (on- and off-events in Figure 5). Our experiment, therefore, may be the first equilibrium measurement of the free energy difference between histone octamers on and off DNA. Our ability to do this is likely facilitated by the presence of ATP-independent nucleosome assembly factors in the egg extracts, which may greatly reduce barriers to assembly (and therefore to disassembly) of nucleosomes onto DNA. In Xenopus egg extracts, these factors are known: the protein nucleoplasmin is known to act primarily as a chaperone for histones H2A/B, proteins N1 and N2 are associated with H3/4 (Dilworth et al., 1987; Kleinschmidt et al., 1990; Laskey et al., 1993), whereas the protein NAP-1 is associated with the linker histone B4 the embryonic variant of H1 (Shintomi et al., 2005). Because they are ATP independent, these factors must mediate both assembly and disassembly processes: for example, yeast NAP-1 facilitates lateral “sliding” of nucleosomes along DNA through transient removal and replacement of H2A/B (Park et al., 2005), and it is known to play a role in formation of regularly spaced nucleosome arrays in more complex chromatin assembly reactions (Ito et al., 1996).
We note that although step-like events are clear in our data (Figure 4), we have not been able to definitively measure the statistical distribution of extension step events for forces near the 3.5 pN stall point. This is due mainly to thermal fluctuations of molecule extension that have an amplitude of ∼100 nm for 50- to 100-kb DNAs (Figure 4a). Because the width of these extension fluctuations is proportional to DNA length (Strick et al., 1996), we used 15-kb DNAs to more clearly observe single-nucleosome assembly and disassembly events (Figure 4f). However, even for the 15-kb template, we could not definitively measure the step size distribution near 3.5 pN.
Other experiments have observed extract-driven chromatin assembly onto stretched DNA. Notably, Ladoux et al. (2000) observed chromatin assembly even in the case where the original DNA molecule was stretched by up to 12-pN forces. Although at first glance this may seem inconsistent with our 3.5-pN force threshold, the molecules were in that case extended by hydrodynamic flow without an end-attached particle, resulting in inhomogeneous DNA stretching with tension varying from its maximum near the tether point to zero at the free end. Consequently, the tension in the DNA in such an experiment will always be below the threshold force for chromatin assembly near the free end, causing the assembly reaction to initiate there (Ladoux et al., 2000). Then, as assembly proceeds the complex shortens, and the hydrodynamic drag (roughly proportional to complex length; Berg, 1993) and therefore DNA tension (Marko and Siggia, 1995) drop, promoting further assembly. For DNA molecules extended in flow, one can therefore expect at least a partial assembly of chromatin starting from the free end for essentially any rate of flow.
To emphasize this point, we note that Bennink et al. (2001) observed that similar extract-based assembly reactions were stalled by flow-generated DNA tensions larger than ∼5 pN. In that case, an end-attached particle ensured that nonzero tension was maintained at all points along the DNA. Given the limited accuracy of the flow–force calibration of the study of (Bennink et al., 2001), we conclude that our 3.5-pN stall force measurement is consistent with previous experiments.
Nucleosome Removal by Force
Nucleosomes may be removed by forces just above the 3.5-pN stall point (e.g., 4 pN), significantly smaller than the ∼20-pN forces reported by Bennink et al. (2001) by using an extension-controlled laser tweezer setup. Similar, large 15-pN forces were reported to be necessary for unfolding of chromatin fibers isolated from cells by using laser tweezers (Cui and Bustamante, 2000).
A key point of our magnetic tweezer experiments is their use of constant force and long measurement times, in extract-rich solution. After assembly, complete removal of most of the ∼200 nucleosomes on a λ-DNA requires >100 min at a constant force of 14 pN (Figure 3). Laser tweezer experiments generally impose a time-varying extension and measure the resulting tension. In the experiments of Cui and Bustamante (2000) and Bennink et al. (2001), the fiber pulls were done in <1 min; relatively large nucleosome removal forces were likely observed simply because the pulling rates were far greater than that at which nucleosomes could be removed by near-to-equilibrium fluctuations.
Thus, the high nucleosome-removal forces observed by Cui and Bustamante (2000), Bennink et al. (2001), and Brower-Toland et al. (2002) may be in part a result of the high strain rates imposed. The effect of an increase in bond failure force with increasing strain rate has been well documented theoretically, mainly in connection with probing of receptor–ligand interactions (Evans, 2001). This theory has been shown to describe nucleosome removal experiments (Brower-Toland et al., 2002; Pope et al., 2005), and it has been discussed in terms of the barrier associated with removal of DNA wraps by (Kulic and Schiessel, 2004). Our force–velocity curve for nucleosome disassembly (Figure 3) shows that removal of nucleosomes on shorter time scales (i.e., at larger removal rates) requires larger forces, and explains why laser tweezer experiments give nucleosome disruption forces that are much larger than those expected from thermodynamic arguments (Marko and Siggia, 1997a). A further problem hampering laser tweezer experiments is the general necessity to replace cell extract solutions with optically clear buffers (Bennink et al., 2001); this removes nucleosome assembly factors that, as discussed above, are likely to facilitate nucleosome removal.
Experiments of (Leuba et al., 2003) using purified histones plus NAP-1 nucleosome assembly protein and fixed-force magnetic tweezer methods have observed forces of ∼10 pN during nucleosome disruption. Although this is lower than the laser tweezer results, it is still a surprisingly large force. This may be due to the different solution conditions, i.e., purified proteins in buffer in the experiment of (Leuba et al., 2003) versus diluted egg extracts, which contain many chromatin regulators and more closely approximate conditions found in vivo.
Chromatin Assembly Reaction in Presence of ATP
In the presence of ATP, the character of chromatin assembly is transformed. Most simply, ATP has the general effect of destabilizing nucleosomes. Forces below the 3.5-pN threshold no longer lead to gradual assembly as in the −ATP case, but instead they lead to a highly dynamic behavior with remarkably large (200- to 400-nm) Λ and V events with high local rates (up to 40 nm/s). Appreciable assembly only occurs when force is reduced to below 1 pN; forces in the range 1–3.5 pN result in large fluctuations in length and partially assembled fibers. Experiments with nonhydrolyzable ATP analogues indicate that this dynamic behavior is dependent on ATP hydrolysis. We also note that we have observed Λ and V events in reactions using crude interphase extracts, and for several different dilutions (data not shown), indicating that they are not extremely sensitive to details of extract preparation or protein concentration.
We suspect that the +ATP Λ and V events are due to processive ATPases, which remove or transport nucleosomes in chromatin. At present, we do not know the identity of these ATPases, but by selective depletion of enzymes from the extracts, it may be possible to identify what factors are responsible for these remarkable chromatin-reorganizing events. We have carried out experiments using +ATP assembly reactions in the presence of the DNA polymerase inhibitor aphidicolin as well as function-blocking antibodies to condensin subunits, and in both cases we observed Λ and V events, indicating that this activity is not due to either DNA polymerase or condensin function. In addition, we have included the topo II inhibitor ICRF-193 in +ATP reactions, and we have observed the Λ and V events, indicating that topo II is not the enzyme responsible for them.
Other ATPases that might be responsible for the Λ and V events include minichromosome maintenance complexes, which are loaded onto chromatin in Xenopus egg extracts when ATP is present, and chromatin-remodeling factors, which are again known to participate in assembly of chromatin in Xenopus egg extracts (MacCallum et al., 2002). The role played by these proteins in the Λ and V events could be probed by their immunodepletion from the extracts, by using methods such as those recently used to study the role of linker histone (Maresca et al., 2005).
An interesting feature of the +ATP assembly reaction (Figure 6a) is that, ignoring the Λ and V events, the overall behavior of the extension reduction is a slow linear-in-time reduction. This is markedly different from the −ATP case where initially exponential assembly time courses were observed (e.g., Figure 3). A possible explanation for the slow net assembly rate, compared with the same extract −ATP, and for the linear-in-time extension reduction may be that in the presence of ATP, the assembly reaction becomes highly processive, e.g., assembling nucleosomes serially at one or a few places along the fiber.
Physiological Relevance
Our results have implications for chromatin structure in vivo. Past studies where chromatin fiber is visualized using electron microscopy paint a static picture of chromatin. Recent single-chromatin fiber micromechanics experiments have enriched this view via direct observation of force-controlled chromatin assembly and disassembly. For example, the work of Bennink et al. (2001) observes assembly of chromatin onto naked DNA, followed by a progressive nucleosome-by-nucleosome disruption when high forces are applied. Our work on −ATP egg–extract assembly reaction builds on this preceding work: −ATP nucleosome assembly occurs in an uncoordinated manner along DNA and can be controlled and switched from assembly to disassembly, by using DNA tension. The physiological relevance of our −ATP results is mainly through their quantitation of mechanochemical aspects of nucleosome assembly, most notably in providing an estimate of the free energy of octamer–DNA interactions.
In vivo, +ATP conditions prevail: our experiments with ATP indicate that chromatin may be subject to dramatic disassembly and reassembly processes modulated by DNA tension. A number of experiments indicate that bulk rearrangements of core histones occur in vivo: fluorescence recovery after photobleaching experiments have revealed that 3% of chromatin-bound H2B is turned over with a 6-min half-life, whereas 40% is mobilized on a 2-hour time scale; in the same experiments, 20% of bound H3 was observed to exchange with a 2-h half-life (Kimura and Cook, 2001). Similar experiments show that 20% of H2A and nearly all of the variant H2ABbd exchanges in 1 h (Gautier et al., 2004) and that a fraction of core histones are turned over in a cell cycle-dependent manner (Chen et al., 2005). Our experiments indicate that rather moderate forces may play a role in these dynamics; we find that 1 to 3 pN in the DNA template stimulates partial disassembly of nucleosomes, resulting in a highly unfolded chromatin conformation relative to the highly compacted products of −ATP reactions on DNAs under such tensions. Although the effects of nucleosome chaperones and remodeling factors in our experiments could be exaggerated due to an excess of such factors relative to DNA, we feel our results more closely resemble in vivo conditions than previous single-DNA chromatin studies.
Our findings suggest how chromatin might respond in vivo to perturbation by transcription or replication, which involve polymerases that can generate >10-pN forces in their DNA templates (Wang et al., 1998; Maier et al., 2000). Our +ATP experiments indicate that forces of roughly 1 pN may be all that is required to drive chromatin reorganization, far less than the 10–20 pN necessary to rapidly displace nucleosomes under dilute solution conditions (Cui and Bustamante, 2000; Bennink et al., 2001; Brower-Toland et al., 2002; Gemmen et al., 2005). We note that if large 10- to 20-pN nucleosome-displacing forces were needed for polymerases to progress through chromatin in vivo, those enzymes would be significantly slowed: 20 pN is well in excess of the forces at which DNApol and RNApol are slowed to half their maximal velocities. Thus, active disassembly of chromatin in response to ∼1-pN tensions would greatly facilitate actions of polymerases and other DNA-processing enzymes. Finally, our result that ATP-dependent factors present in vivo enable DNA tensions as low as 1 pN to confer significant changes to chromatin structure indicates a mechanism through which weak forces applied to chromosomes could play a role in chromatin remodeling and gene regulation.
ACKNOWLEDGMENTS
We thank Benjamin Freedman for help with characterizing the Xenopus extracts. Work at Northwestern University and University of Illinois at Chicago was supported by the National Science Foundation Grants DMR-0203963, MCB-0240998, PHY-0445565, and DMR-0605895. R.H. was supported by the Cancer Research Coordinating Committee. M.O.C. was supported by the Deutsche Forschungsgemeinschaft Grants BO 910/3-2; SFB 503/B10, and GRK 1033.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E06-09-0800) on November 15, 2006.
REFERENCES
- Almouzni G., Wolffe A. P. Nuclear assembly, structure, and function: the use of Xenopus in vitro systems. Exp. Cell Res. 1993;205:1–15. doi: 10.1006/excr.1993.1051. [DOI] [PubMed] [Google Scholar]
- Bancaud A., et al. Structural plasticity of single chromatin fibers revealed by torsional manipulation. Nat. Struct. Mol. Biol. 2006;13:444–450. doi: 10.1038/nsmb1087. [DOI] [PubMed] [Google Scholar]
- Belmont A. Dynamics of chromatin, proteins, and bodies within the cell nucleus. Curr. Opin. Cell Biol. 2003;15:304–310. doi: 10.1016/s0955-0674(03)00045-0. [DOI] [PubMed] [Google Scholar]
- Bennink M. L., Leuba S. H., Leno G. H., Zlatanova J., de Grooth B. G., Greve J. Unfolding individual nucleosomes by stretching single chromatin fibers with optical tweezers. Nat. Struct. Biol. 2001;8:606–610. doi: 10.1038/89646. [DOI] [PubMed] [Google Scholar]
- Berg H. C. Random Walks in Biology. Princeton, NJ: Princeton University Press; 1993. [Google Scholar]
- Bonte E., Becker P. B. Preparation of chromatin assembly extracts from preblastoderm Drosophila embryos. Methods Mol. Biol. 1999;119:187–194. doi: 10.1385/1-59259-681-9:187. [DOI] [PubMed] [Google Scholar]
- Brower-Toland B., Wang M. D. Use of optical trapping techniques to study single-nucleosome dynamics. Methods Enzymol. 2004;376:62–72. doi: 10.1016/S0076-6879(03)76005-4. [DOI] [PubMed] [Google Scholar]
- Brower-Toland B. D., Smith C. L., Yeh R. C., Lis J. T., Peterson C. L., Wang M. D. Mechanical disruption of individual nucleosomes reveals a reversible multistage release of DNA. Proc. Natl. Acad. Sci. USA. 2002;99:1960–1965. doi: 10.1073/pnas.022638399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown D. T., Izard T., Misteli T. Mapping the interaction surface of linker histone H1(0) with the nucleosome of native chromatin in vivo. Nat. Struct. Mol. Biol. 2006;13:250–255. doi: 10.1038/nsmb1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustamante C., Marko J. F., Siggia E. D., Smith S. Entropic elasticity of lambda-phage DNA. Science. 1994;265:1599–1600. doi: 10.1126/science.8079175. [DOI] [PubMed] [Google Scholar]
- Bustamante C., Smith S. B., Liphardt J., Smith D. Single-molecule studies of DNA mechanics. Curr. Opin. Struct. Biol. 2000;10:279–285. doi: 10.1016/s0959-440x(00)00085-3. [DOI] [PubMed] [Google Scholar]
- Chen D., Dundr M., Wang C., Leung A., Lamond A., Misteli T., Huang S. Condensed mitotic chromatin is accessible to transcription factors and chromatin structural proteins. J. Cell Biol. 2005;168:41–54. doi: 10.1083/jcb.200407182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claudet C., Angelov D., Bouvet P., Dimitrov S., Bednar J. Histone octamer instability under single molecule experiment conditions. J. Biol. Chem. 2005;280:19958–19965. doi: 10.1074/jbc.M500121200. [DOI] [PubMed] [Google Scholar]
- Cotton R. W., Hamkalo B. A. Nucleosome dissociation at physiological ionic strengths. Nucleic Acids Res. 1981;9:445–457. doi: 10.1093/nar/9.2.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Y., Bustamante C. Pulling a single chromatin fiber reveals the forces that maintain its higher-order structure. Proc. Natl. Acad. Sci. USA. 2000;97:127–132. doi: 10.1073/pnas.97.1.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dilworth S. M., Black S. J., Laskey R. A. Two complexes that contain histones are required for nucleosome assembly in vitro: role of nucleoplasmin and N1 in Xenopus egg extracts. Cell. 1987;51:1009–1018. doi: 10.1016/0092-8674(87)90587-3. [DOI] [PubMed] [Google Scholar]
- Evans E. Probing the relation between force-lifetime- and chemistry in single molecular bonds. Annu. Rev. Biophys. Biomol. Struct. 2001;30:105–128. doi: 10.1146/annurev.biophys.30.1.105. [DOI] [PubMed] [Google Scholar]
- Felsenfeld G. Chromatin unfolds. Cell. 1996;86:13–19. doi: 10.1016/s0092-8674(00)80073-2. [DOI] [PubMed] [Google Scholar]
- Gautier T., Abbott D. W., Molla A., Verdel A., Ausio J., Dimitrov S. Histone variant H2ABbd confers lower stability to the nucleosome. EMBO Rep. 2004;5:715–720. doi: 10.1038/sj.embor.7400182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gemmen G. J., Sim R., Haushalter K. A., Ke P. C., Kadonaga J. T., Smith D. E. Forced unraveling of nucleosomes assembled on heterogeneous DNA using core histones, NAP-1, and ACF. J. Mol. Biol. 2005;351:89–99. doi: 10.1016/j.jmb.2005.05.058. [DOI] [PubMed] [Google Scholar]
- Glikin G. C., Ruberti I., Worcel A. Chromatin assembly in Xenopus oocytes: in vitro studies. Cell. 1984;37:33–41. doi: 10.1016/0092-8674(84)90298-8. [DOI] [PubMed] [Google Scholar]
- Gottesfeld J. M., Luger K. Energetics and affinity of the histone octamer for defined DNA sequences. Biochemistry. 2001;40:10927–10933. doi: 10.1021/bi0109966. [DOI] [PubMed] [Google Scholar]
- Hirano T., Mitchison T. J. Cell cycle control of higher-order chromatin assembly around naked DNA in vitro. J. Cell Biol. 1991;115:1479–1489. doi: 10.1083/jcb.115.6.1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano T., Mitchison T. J. A heterodimeric coiled-coil protein required for mitotic chromosome condensation in vitro. Cell. 1994;79:449–458. doi: 10.1016/0092-8674(94)90254-2. [DOI] [PubMed] [Google Scholar]
- Ito T., Bulger M., Kobayashi R., Kadonaga J. T. Drosophila NAP-1 is a core histone chaperone that functions in ATP-facilitated assembly of regularly spaced nucleosomal arrays. Mol. Cell. Biol. 1996;16:3112–3124. doi: 10.1128/mcb.16.6.3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura H., Cook P. R. Kinetics of core histones in living human cells: little exchange of H3 and H4 and some rapid exchange of H2B. J. Cell Biol. 2001;153:1341–1353. doi: 10.1083/jcb.153.7.1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleckner N., Zickler D., Jones G. H., Dekker J., Padmore R., Henle J., Hutchinson J. A mechanical basis for chromosome function. Proc. Natl. Acad. Sci. USA. 2004;101:12592–12597. doi: 10.1073/pnas.0402724101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinschmidt J. A., Seiter A., Zentgraf H. Nucleosome assembly in vitro: separate histone transfer and synergistic interaction of native histone complexes purified from nuclei of Xenopus laevis oocytes. EMBO J. 1990;9:1309–1318. doi: 10.1002/j.1460-2075.1990.tb08240.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulic I. M., Schiessel H. DNA spools under tension. Phys. Rev. Lett. 2004;92:228101. doi: 10.1103/PhysRevLett.92.228101. [DOI] [PubMed] [Google Scholar]
- Ladoux B., Quivy J. P., Doyle P., du Roure O., Almouzni G., Viovy J. L. Fast kinetics of chromatin assembly revealed by single-molecule videomicroscopy and scanning force microscopy. Proc. Natl. Acad. Sci. USA. 2000;97:14251–14256. doi: 10.1073/pnas.250471597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskey R. A., Mills A. D., Morris N. R. Assembly of SV40 chromatin in a cell-free system from Xenopus eggs. Cell. 1977;10:237–243. doi: 10.1016/0092-8674(77)90217-3. [DOI] [PubMed] [Google Scholar]
- Laskey R. A., Mills A. D., Philpott A., Leno G. H., Dilworth S. M., Dingwall C. The role of nucleoplasmin in chromatin assembly and disassembly. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 1993;339:263–269. doi: 10.1098/rstb.1993.0024. discussion, 268–269. [DOI] [PubMed] [Google Scholar]
- Leuba S. H., Karymov M. A., Tomschik M., Ramjit R., Smith P., Zlatanova J. Assembly of single chromatin fibers depends on the tension in the DNA molecule: magnetic tweezers study. Proc. Natl. Acad. Sci. USA. 2003;100:495–500. doi: 10.1073/pnas.0136890100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levi V., Ruan Q., Plutz M., Belmont A. S., Gratton E. Chromatin dynamics in interphase cells revealed by tracking in a two-photon excitation microscope. Biophys. J. 2005;89:4275–4285. doi: 10.1529/biophysj.105.066670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacCallum D. E., Losada A., Kobayashi R., Hirano T. ISWI remodeling complexes in Xenopus egg extracts: identification as major chromosomal components that are regulated by INCENP-aurora B. Mol. Biol. Cell. 2002;13:25–39. doi: 10.1091/mbc.01-09-0441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier B., Bensimon D., Croquette V. Replication by a single DNA polymerase of a stretched single-stranded DNA. Proc. Natl. Acad. Sci. USA. 2000;97:12002–12007. doi: 10.1073/pnas.97.22.12002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maresca T. J., Freedman B. S., Heald R. Histone H1 is essential for mitotic chromosome architecture and segregation in Xenopus laevis egg extracts. J. Cell Biol. 2005;169:859–869. doi: 10.1083/jcb.200503031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marko J. F., Siggia E. D. Stretching DNA. Macromolecules. 1995;28:8759–8770. [Google Scholar]
- Marko J. F., Siggia E. D. Driving proteins off DNA using applied tension. Biophys. J. 1997a;73:2173–2178. doi: 10.1016/S0006-3495(97)78248-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marko J. F., Siggia E. D. Polymer models of meiotic and mitotic chromosomes. Mol. Biol. Cell. 1997b;8:2217–2231. doi: 10.1091/mbc.8.11.2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray A. W. Cell cycle extracts. Methods Cell Biol. 1991;36:581–605. [PubMed] [Google Scholar]
- Park Y. J., Chodaparambil J. V., Bao Y., McBryant S. J., Luger K. Nucleosome assembly protein 1 exchanges histone H2A-H2B dimers and assists nucleosome sliding. J. Biol. Chem. 2005;280:1817–1825. doi: 10.1074/jbc.M411347200. [DOI] [PubMed] [Google Scholar]
- Pope L. H., Bennink M. L., Greve J. Optical tweezers stretching of chromatin. J. Muscle Res. Cell Motil. 2002;23:397–407. doi: 10.1023/a:1023450204528. [DOI] [PubMed] [Google Scholar]
- Pope L. H., Bennink M. L., van Leijenhorst-Groener K. A., Nikova D., Greve J., Marko J. F. Single chromatin fiber stretching reveals physically distinct populations of disassembly events. Biophys. J. 2005;88:3572–3583. doi: 10.1529/biophysj.104.053074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Campos A., Shimamura A., Worcel A. Assembly and properties of chromatin containing histone H1. J. Mol. Biol. 1989;209:135–150. doi: 10.1016/0022-2836(89)90177-0. [DOI] [PubMed] [Google Scholar]
- Schaaf P., Talbot J. Kinetics of random sequential adsorption. Phys. Rev. Lett. 1989;62:175–178. doi: 10.1103/PhysRevLett.62.175. [DOI] [PubMed] [Google Scholar]
- Shimamura A., Tremethick D., Worcel A. Characterization of the repressed 5S DNA minichromosomes assembled in vitro with a high-speed supernatant of Xenopus laevis oocytes. Mol. Cell. Biol. 1988;8:4257–4269. doi: 10.1128/mcb.8.10.4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shintomi K., Iwabuchi M., Saeki H., Ura K., Kishimoto T., Ohsumi K. Nucleosome assembly protein-1 is a linker histone chaperone in Xenopus eggs. Proc. Natl. Acad. Sci. USA. 2005;102:8210–8215. doi: 10.1073/pnas.0500822102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skoko D., Wong B., Johnson R. C., Marko J. F. Micromechanical analysis of the binding of DNA-bending proteins HMGB1, NHP6A and HU reveals their ability to cooperatively form highly stable DNA-protein complexes. Biochemistry. 2004;43:13867–13874. doi: 10.1021/bi048428o. [DOI] [PubMed] [Google Scholar]
- Smith S. B., Finzi L., Bustamante C. Direct mechanical measurements of the elasticity of single DNA molecules by using magnetic beads. Science. 1992;258:1122–1126. doi: 10.1126/science.1439819. [DOI] [PubMed] [Google Scholar]
- Smythe C., Newport J. W. Systems for the study of nuclear assembly, DNA replication, and nuclear breakdown in Xenopus laevis egg extracts. Methods Cell Biol. 1991;35:449–468. doi: 10.1016/s0091-679x(08)60583-x. [DOI] [PubMed] [Google Scholar]
- Strick T., Allemand J., Croquette V., Bensimon D. Twisting and stretching single DNA molecules. Prog. Biophys. Mol. Biol. 2000;74:115–140. doi: 10.1016/s0079-6107(00)00018-3. [DOI] [PubMed] [Google Scholar]
- Strick T. R., Allemand J. F., Bensimon D., Bensimon A., Croquette V. The elasticity of a single supercoiled DNA molecule. Science. 1996;271:1835–1837. doi: 10.1126/science.271.5257.1835. [DOI] [PubMed] [Google Scholar]
- Strick T. R., Allemand J. F., Bensimon D., Croquette V. Behavior of supercoiled DNA. Biophys. J. 1998;74:2016–2028. doi: 10.1016/S0006-3495(98)77908-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thastrom A., Gottesfeld J. M., Luger K., Widom J. Histone-DNA binding free energy cannot be measured in dilution-driven dissociation experiments. Biochemistry. 2004;43:736–741. doi: 10.1021/bi0302043. [DOI] [PubMed] [Google Scholar]
- Wagner G., Bancaud A., Quivy J. P., Clapier C., Almouzni G., Viovy J. L. Compaction kinetics on single DNAs: purified nucleosome reconstitution systems versus crude extract. Biophys. J. 2005;89:3647–3659. doi: 10.1529/biophysj.105.062786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M. D., Schnitzer M. J., Yin H., Landick R., Gelles J., Block S. M. Force and velocity measured for single molecules of RNA polymerase. Science. 1998;282:902–907. doi: 10.1126/science.282.5390.902. [DOI] [PubMed] [Google Scholar]
- Yan J., Skoko D., Marko J. F. Near-field-magnetic-tweezer manipulation of single DNA molecules. Phys. Rev. E. 2004;70 doi: 10.1103/PhysRevE.70.011905. 011905. [DOI] [PubMed] [Google Scholar]
- Yin H., Wang M. D., Svoboda K., Landick R., Block S. M., Gelles J. Transcription against an applied force. Science. 1995;270:1653–1657. doi: 10.1126/science.270.5242.1653. [DOI] [PubMed] [Google Scholar]