Abstract
Expression of yeast mitochondrial genes depends on specific translational activators acting on the 5′-untranslated region of their target mRNAs. Mss51p is a translational factor for cytochrome c oxidase subunit 1 (COX1) mRNA and a key player in down-regulating Cox1p expression when subunits with which it normally interacts are not available. Mss51p probably acts on the 5′-untranslated region of COX1 mRNA to initiate translation and on the coding sequence itself to facilitate elongation. Mss51p binds newly synthesized Cox1p, an interaction that could be necessary for translation. To gain insight into the different roles of Mss51p on Cox1p biogenesis, we have analyzed the properties of a new mitochondrial protein, mp15, which is synthesized in mss51 mutants and in cytochrome oxidase mutants in which Cox1p translation is suppressed. The mp15 polypeptide is not detected in cox14 mutants that express Cox1p normally. We show that mp15 is a truncated translation product of COX1 mRNA whose synthesis requires the COX1 mRNA-specific translational activator Pet309p. These results support a key role for Mss51p in translationally regulating Cox1p synthesis by the status of cytochrome oxidase assembly.
INTRODUCTION
Biogenesis of eukaryotic cytochrome c oxidase (COX), the terminal enzyme of the mitochondrial respiratory chain, involves the coordinated action of two genomes. Three proteins, encoded in the mitochondrial DNA (mtDNA), form the catalytic core with the heme A and copper prosthetic groups of the enzyme. The other nine to 10 subunits, encoded in nuclear DNA, interact with the core subunits to form a stable COX complex. In addition to the structural subunits, COX biogenesis requires the assistance of at least 20 other nuclear gene products some of which are essential for the expression of the mitochondrial encoded subunits (McEwen et al., 1986; Tzagoloff and Dieckmann, 1990; Barrientos et al., 2002a; Solans et al., 2004).
In Saccharomyces cerevisiae, translation of each mitochondrial COX mRNA depends on one or more translational activator (for review, see Towpik, 2005). These mRNA-specific translational factors are either integral or peripheral inner membrane proteins that recognize the 5′-untranslated region (UTR) of their target transcripts. Current speculations suggest that translational activators may couple translation to cotranslational insertion of the newly synthesized hydrophobic products into the membrane near or at the site of their assembly into multisubunit complexes (Michaelis et al., 1991; Naithani et al., 2003). Most activators, Mss51p being an exception, interact with each other (Brown et al., 1994; Naithani et al., 2003), suggesting some level of coregulation in the expression of the core-forming subunits of COX (Fiori et al., 2005). Interactions have also been noted between the transcription factor Nam1p and translational activators, including Pet111p, Pet309p, and Pet494p (Naithani et al., 2003; Krause et al., 2004) raising the possibility that mitochondrial transcription may be coupled to translation.
COX1 is the mitochondrial gene for subunit1/Cox1p, an important catalytic subunit of COX containing copper and heme A prosthetic groups. Translation of COX1 in S. cerevisiae is under the control of the MSS51 and PET309 gene products that are also involved in maturation of the COX1 mRNAs (Decoster et al., 1990; Manthey and McEwen, 1995). Mutations in MSS51 or overexpression of the wild type gene can suppress shy1 null mutants by enhancing synthesis of Cox1p (Barrientos et al., 2002b). The function of Shy1p in maturation and/or assembly of Cox1p (Barrientos et al., 2002b; Perez-Martinez et al., 2003; Smith et al., 2005) is of considerable interest because mutations in its human homologue, Surf1p, are responsible for most diagnosed cases of Leigh's syndrome presenting a COX deficiency (Tiranti et al., 1998; Zhu et al., 1998).
Cox1p synthesis is suppressed in most COX assembly mutants, including shy1 mutants, but it is restored to normal levels by mss51 suppressors of shy1 or by mutations in COX14 (Barrientos et al., 2004), which codes for a COX assembly factor (Glerum et al., 1995). Like other translational activators Mss51p acts on the 5′-UTR to initiate translation (Perez-Martinez et al., 2003); however, mss51 mutants, unlike pet309 mutants, cannot be suppressed by changes in the 5′-UTR of COX1 mRNA. (Perez-Martinez et al., 2003; Barrientos et al., 2004). In addition, Mss51p acts on a target in the protein coding sequence of COX1 mRNA to promote elongation (Perez-Martinez et al., 2003). Mss51p and Cox1p form a transient complex (Perez-Martinez et al., 2003; Barrientos et al., 2004) that is stabilized by Cox14p (Barrientos et al., 2004). These interactions have been postulated to down-regulate Cox1p synthesis when COX assembly is impaired (Barrientos et al., 2004). According to this model, the release of Mss51p from the ternary complex and its availability for Cox1p synthesis occur at a downstream step in the assembly pathway, most likely catalyzed by Shy1p (Barrientos et al., 2004). Further studies are necessary to clarify the functional significance of the interactions of Mss51p on the 5′-UTR and the coding sequence of COX1 mRNA.
In the present study, we report a novel mitochondrial translation product, named mp15, with an apparent mass of ∼15 kDa. This protein is detected in mss51 null mutants blocked in Cox1p synthesis and in strains carrying null alleles in nuclear genes coding for COX subunits or assembly factors in which the synthesis of Cox1p is reduced to 10% or less of wild type. We present evidence that mp15 is a truncated translation product of a partially unprocessed COX1 mRNA and that it is synthesized by an Mss51p- but not Pet309-independent mechanism. We also present evidence that binding of Mss51p to the 5′-UTR of COX1 mRNA is necessary for optimal initiation of translation by Pet309p, whereas the interaction of Mss51p with newly synthesized Cox1p may regulate elongation of the nascent polypeptide.
MATERIALS AND METHODS
Strains and Media
The genotypes and sources of the S. cerevisiae strains carrying null alleles of COX-related genes are listed in Table 1. Double mutants were constructed by crosses of the single mutants. The compositions of the growth media have been described previously (Myers et al., 1985).
Table 1.
Genotypes and sources of yeast strains
| Straina | Strains with mitochondrial DNA of different source and intron composition |
||
|---|---|---|---|
| Genotype | mtDNA | Source | |
| W303-1A | MATa ade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 | ρ+ bI+, aI1+, aI2+, aI3+, aI5γ+ | R. Rothstein (Department of Human Genetics, Columbia University, New York, NY) |
| W303-1B | MATα ade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 | ρ+ bI+, aI1+, aI2+, aI3+, aI5γ+ | R. Rothstein |
| W303/ρ0 | MATα ade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 | ρo | This study |
| D273-10B/A1 | MATa met6 | ρ+ bI+, aI1+, aI2+, aI3+, aI4+, aI5g+ | Tzagoloff et al. (1976) |
| JC3/ρo | MATa kar1-1 ade2 lys2 | ρo | ATCC 201577 |
| JC3/ρD273 | MATa kar1-1 ade2 lys2 | ρ+ bI+, aI1+, aI2+, aI3+, aI4+, aI5g+ | JC3 ρo × D273-10B/A21 |
| JC11/rCK5112 | MATα, kar1-1, his3 | ρ+ aI2+, aI3+, aI5γ+w+ | J. Lazowska (Centre de Génétique Moleculaire du Centre National de la Recherche Scientifique, Gif-sur-Yvette, France) |
| JC11/rWI04 | MATα, kar1-1, his3 | ρ+ bI+, aI1+, aI2+, aI3+, aI5 + | Labouesse (1990) |
| JC3/rGF134-6D | MATa kar1-1 ade2 lys2 | ρ+ bI+, aI4+ | Barros et al. (2006) |
| D273-10B/ρG1-224 | MATα met6 | ρ+ bI+, aI1+, aI2+, aI3+, aI4+ | Seraphin et al. (1987) |
| JC3/ρG1-224 | MATa kar1-1 ade2 lys2 | ρ+ bI+, aI1+, aI2+, aI3+, aI4+ | JC3ρo × D273-10B/G1-224 |
| D273-10B/ρG1-356-R5 | MATα met6 lys2 | ρ+ bI+, aI1+, aI4+, aI5g+ | Seraphin et al. (1987) |
| JC3/ρG1-356-R5 | MATa kar1-1 ade2 lys2 | ρ+ bI+, aI1+, aI4+, aI5g+ | JC3ρo × D273-10B/G1-356-R5 |
| aW303/ρIo | MATa ade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 | ρ+ Io | Barros et al. (2005) |
| JC3/ρW303 | MATa kar1-1 ade2 lys2 | ρ+ bI+, aI1+, aI2+, aI3+, aI5γ+ | JC3ρo × W303-1B |
| Δmss51/ρD273 | MATα ade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 Δmss51::HIS3 | ρ+ bI+, aI1+, aI2+, aI3+, aI4+, aI5g+ | W303Δmss51 × JC3/ρD273 |
| Δmss51/ρG1-224 | MATα ade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 Δmss51::HIS3 | ρ+ bI+, aI1+, aI2+, aI3+, aI4+ | W303Δmss51 × JC3/ρG1-224 |
| Δmss51/ρG1-356-R5 | MATα ade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 Δmss51::HIS3 | ρ+ bI+, aI1+, aI4+, aI5g+ | W303Δmss51 × JC3/ρG1-356-R5 |
| Δmss51/ρCK5112 | MATα ade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 Δmss51::HIS3 | ρ+ aI2+, aI3+, aI5γ+w+ | W303Δmss51 × JC3/ρCK5112 |
| Δmss51/ρGF134-6D | MATα ade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 Δmss51::HIS3 | ρ+ bI+, aI4+ | W303Δmss51 × JC3/ρGF134-6D |
| Δmss51/ρWI04 | MATα ade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 Δmss51::HIS3 | ρ+ bI+, aI1+, aI2+, aI3+, aI5γ+ | W303Δmss51 × JC11/ρWI04 |
| Δmss51/ρIo | MATα ade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 Δmss51::HIS3 | ρ+ Io | W303Δmss51 × W303/ρIo |
| Δshy1/ρG1-224 | MATα ade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 Δshy1::URA3 | ρ+ bI+, aI1+, aI2+, aI3+, aI4+ | W303Δmss51 × JC3/ρG1-224 |
| Δshy1/ρG1-356-R5 | MATα ade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 Δshy1::URA3 | ρ+ bI+, aI1+, aI4+, aI5g+ | W303Δmss51 × JC3/ρG1-356-R5 |
| Δshy1/ρCK5112 | MATα ade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 Δshy1::URA3 | ρ+ Ai2+, aI3+, aI5γ+w+ | W303Δmss51 × JC3/ρCK5112 |
| Δshy1/ρGF134-6D | MATα ade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 Δshy1::URA3 | ρ+ Bi+, aI4+ | W303Δ mss51 × JC3/ρGF134-6D |
| Δshy1/ρIo | MATα ade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 Δshy1::URA3 | ρ+ I° | W303Δ mss51 × JC3/ρIo |
| Structural subunitsb | |||
| W303Δcox4 | ade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 Δcox4::URA3 | W303 | Glerum and Tzagoloff (1997) |
| W303Δcox5a | ade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 Δcox5a::HIS3 | W303 | Glerum and Tzagoloff (1997) |
| W303Δcox6 | ade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 Δcox6::URA3 | W303 | Glerum and Tzagoloff (1997) |
| W303Δcox7 | ade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 Δcox7::URA3 | W303 | Barrientos et al. (2004) |
| W303Δcox9 | ade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 Δcox9::URA3 | W303 | Barrientos et al. (2004) |
| M5-16-A3 | ade1 cox1 | D273 | Tzagoloff et al. (1975) |
| COX1 expressionb | |||
| W303Δpet309 | ade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 Δpet309::HIS3 | W303 | Glerum and Tzagoloff (1997) |
| W303Δmss51 | ade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 Δmss51::HIS3 | W303 | Barrientos et al. (2002b) |
| C199 | met6mss51-199 | D273 | Tzagoloff, unpublished data |
| C283 | met6mss51-283 | D273 | Tzagoloff, unpublished data |
| Maturation of CuA or CuB centersb | |||
| W303Δcox17 | ade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 Δcox17::TRP1 | W303 | Glerum et al. (1996a) |
| W303Δsco1 | ade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 Δsco1::URA3 | W303 | Glerum et al. (1996b) |
| W303Δcox11 | ade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 Δcox11::HIS3 | W303 | Tzagoloff et al. (1990) |
| COX2 expressionb | |||
| W303Δoxa1 | ade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 Δoxa1::HIS3 | W303 | Hell et al. (2000) |
| W303Δcox18 | ade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 Δcox18::URA3 | W303 | Souza et al. (2000) |
| W303Δpet111 | ade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 Δpet111::HIS3 | W303 | Barros et al. (2002) |
| W303Δimp1 | ade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 Δimp1::HIS3 | W303 | Barrientos et al. (2004) |
| W303Δimp2 | ade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 Δimp2::URA3 | W303 | Barros et al. (2002) |
| Heme biosynthesisb | |||
| W303Δcox10 | ade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 Δcox10::HIS3 | W303 | Nobrega et al. (1990) |
| W303Δcox15 | ade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 Δcox15::HIS3 | W303 | Glerum et al. (1997) |
| Assembly/unknownb | |||
| W303Δpet117 | ade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 Δpet117::HIS3 | W303 | Barros et al. (2002) |
| W303Δshy1/U2 | ade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 Δshy1::URA3 | W303 | Barrientos et al. (2002b) |
| W303Δpet191 | ade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 Δpet191::HIS3 | W303 | Barrientos et al. (2004) |
| W303Δcox14 | ade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 Δcox14::TRP1 | W303 | Barrientos et al. (2004) |
| W303Δcox16 | ade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 Δcox14::URA3 | W303 | Carlson et al. (2003) |
| W303Δcox19 | ade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 Δcox19::URA3 | W303 | Nobrega et al. (2002) |
| Strains carrying mutant alleles of yta10 |
|||
| YHA103 | MATaade2-1 his3-11,15 trp1-1 leu2,112 YCplac111 (LEU2, CEN):ADH1-YTA10E559Qura3-52Δ yta10::URA3 | W303 | Arlt et al. (1996) |
| W303Δshy1Δyta10/yta10E559Q | MATaade2-1 his3-11,15 trp1-1 leu2,112 YCplac111 (LEU2, CEN):ADH1-YTA10E559Qura3-52Δ yta10Δshy1::URA3 | W303 | W303Δshy1 × YHA103 |
| W303Δcox11Δyta10/yta10E559Q | MATaade2-1 his3-11,15 trp1-1 leu2,112 YCplac111 (LEU2, CEN):ADH1-YTA10E559Qura3-52Δ yta10::URA3Δ cox11::HIS3 | W303 | W303Δcox11 × YHA103 |
a All null mutations have been created or are available in both a and α mating types.
b These headings indicate the functional category of the deleted gene products.
In Vivo Mitochondrial Protein Synthesis
Mitochondrial gene products were labeled with [35S]methionine (7 mCi/mmol; GE Healthcare, Little Chalfont, Buckinghamshire, United Kingdom) in whole cells at 30°C in the presence of cycloheximide (Barrientos et al., 2002b). Equivalent amounts of total cellular proteins were separated by SDS-PAGE on a 17.5% polyacrylamide gel, transferred to a nitrocellulose membrane, and exposed to a Kodak X-OMAT x-ray film. Deviations from this procedure are described in some figure legends.
Construction of COX Mutant Strains Carrying a Proteolytically Inactive Version of yta10
The haploid strain W303Δyta10/Yta10E559Q (Table 1) was obtained from Prof. T. Langer (Institut für Genetik und Zentrum für Molekulare Medizin, Universität zu Köln, Köln, Germany). This strain was crossed to a W303Δshy1 strain and to W303Δcox11. Diploid cells were sporulated and haploid progeny with the two parental mutant alleles (yta10 + shy1 or yta10 + cox11) and the yta10E559Q allele were isolated.
Protein Purification and Sequencing
Mss51p fused with the 26-kDa glutathione S-transferase (GST) was expressed from an integrative plasmid (pG96/ST13) in a strain carrying a null mutant allele of mss51 as reported previously (aW303Δmss51/ST13) (Barrientos et al., 2004). This strain was respiratory competent and grew on nonfermentable carbon sources with a doubling time similar to the parental wild type strain (Barrientos et al., 2004). Mitochondria were prepared from aW303Δmss51/ST13 strain by the method of Faye et al. (1974) except that zymolyase 20,000 (ICN Biochemicals, Aurora, OH) instead of glusulase was used to digest the cell wall. Mitochondrial proteins (5 mg) were solubilized with 1% lauryl maltoside in the presence of 0.5 M KCl and 1 mM phenylmethylsulfonyl fluoride. The extract was clarified by centrifugation at 50,000 × gav for 30 min and incubated with glutathione-Sepharose beads for 4 h at 4°C. After centrifugation at 1500 rpm for 5 min, the beads were washed three times with phosphate-buffered saline. The Mss51p–GST fusion protein was eluted with 10 mM glutathione, 50 mM Tris-base, pH 8.0, and concentrated using Vivaspin 500 columns. The concentrate was separated in a 12% SDS-PAGE system, transferred to a polyvinylidene fluoride membrane and stained with Coomassie blue. A prominent band of ∼68 kDa was excised from the membrane and used to sequence the N-terminal seven residues with a Procise Sequencer (Applied Biosystems, Foster City, CA) in the Proteomics facility of the University of Miami.
Yeast Three-Hybrid System (Y3H)
A Y3H system to map the interacting domain in Mss51p and its target in the 5′-UTR of COX1 mRNA was developed by using the commercially available RNA-Protein Hybrid Hunter kit (Invitrogen, Carlsbad, CA). Portions of the COX1 mRNA 5′-UTR were cloned into the plasmid pRH3′ and portions, of Mss51p were expressed from the plasmid pYESTrp2. The host strain L40-ura-ms2 was cotransformed with pairs of pRH3′/and pYESTrp2/constructs, and the transformants were selected on yeast synthetic medium missing the appropriate auxotrophic markers (Ura− and Trp−) and histidine. The His−-deficient medium selected for expression of the HIS3 reporter gene, e.g., for colonies in which the RNA–protein interaction was produced. Inclusion of 5 mM 3-minotriazole, a competitive inhibitor of the HIS3 reporter protein, suppressed background growth in the His− medium. Cotransformations of the host strain with pair-combinations of the parent plasmids and one of the constructs served as controls. Interactions were confirmed by assaying for β-galactoside, the second reporter using a filter lift assay as described previously (Breeden and Nasmyth, 1985).
Miscellaneous Procedures
Standard procedures were used for the preparation and ligation of DNA fragments and for transformation and recovery of plasmid DNA from Escherichia coli (Sambrook et al., 1989). Yeast were transformed by the method of Schiestl and Gietz (1989). The one-step gene insertion method (Rothstein, 1983) was used to integrate linear plasmids at the URA3 or LEU2 locus of yeast nuclear DNA. Mitochondrial RNA was prepared from isolated mitochondria by modified extraction with hot-acidic phenol (Ausubel et al., 1994).
RESULTS
Synthesis of a Novel Polypeptide from a COX1 Transcript in mss51 Null Mutants
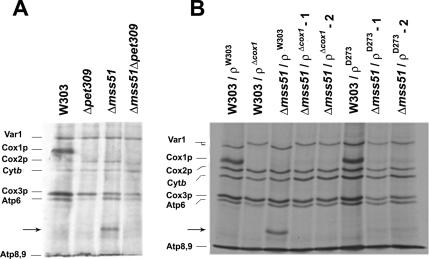
PET309 and MSS51 code for inner membrane proteins that are essential for translation of mitochondrial COX1 mRNA in S. cerevisiae (Decoster et al., 1990; Manthey and McEwen, 1995). In agreement with earlier studies, mutations in either of these genes abolish synthesis of Cox1p (Figure 1A). However, a new low-molecular-weight polypeptide with a migration intermediate between Atp6p and Atp8p in SDS-PAGE was detected among the mitochondrial translation products labeled with [35S]methionine in the mss51 null mutant. Expression of wild-type MSS51 from integrative and episomal plasmids in this mss51 null mutant strain completely restored its ability of synthesizing Cox1p and abolished the synthesis of the new polypeptide (data not shown). This protein was absent in the pet309 mutant and in a pet309/mss51 double mutant (Figure 1A). The absence of the novel polypeptide in the pet309 mutant suggested that it could be a truncated product of COX1 requiring Pet309p for initiation of translation but destined for premature termination in the absence of Mss51p. To further establish the origin of the new polypeptide, mtDNA of M5-16/A3, a mutant with a large deletion in COX1 (Tzagoloff et al., 1975) was transferred by cytoduction to an mss51 null mutant and its parental wild type, both lacking mtDNA (ρo strains). No synthesis of Cox1p or the novel protein was seen in the cox1 mutant either in the presence or absence of the mss51 null mutation (Figure 1B). These data constitute strong evidence that the novel protein, which we have named mp15, is encoded by a COX1 transcript.
Figure 1.
Pet309p and COX1-dependent synthesis of a novel protein in mss51 null mutants. (A) In vivo synthesis of mp15 by mss51 mutants requires the presence of Pet309p. Mitochondrial products of wild type (W303) and of pet309 and mss51 single or double mutants were labeled with [35S]methionine at 30°C for 10 min in the presence of cycloheximide (Barrientos et al., 2002b). (B) A partial deletion of COX1 blocks in vivo synthesis of mp15. Mitochondrial products were labeled as in A in the wild type W303-1A (W303) and mss51 null mutants (Δmss51) in different mitochondrial genetic backgrounds: W303-mtDNA (ρW303), D273-mtDNA (W303ρD273), Δmss51ρD273), or Δcox1-mtDNA (Tzagoloff et al., 1975), the later a D273 type of D273-mtDNA with a partial deletion of the COX1 gene obtained from strain M5.16-A3 (Table 1). Δmss51rDcox1 -1 and -2 and Δmss51ρD273 -1 and -2 are two different cytoductants of each type.
The mtDNA of D273-10B, the parent of the cox1 mutant M5-16/A3 was also transferred to the mss51 mutant and to the isogenic wild-type strain W303-1A. The introduction of the D273 mtDNA into the W303 nuclear background of the mss51 mutant resulted in significantly reduced synthesis and/or stability of mp15 (Figure 1B). No mp15 was detected in the parental strain with wild-type MSS51 and D273-10B mtDNA.
The mp15 detected in mss51 mutants could be a novel translation product of a COX1 transcript or a degradation product of Cox1p. The fact that no Cox1p is detected in mss51 mutants, even after pulses of up to 60 min (data not shown), suggested that there is no synthesis of full-length Cox1p. Detection of mp15 after a 2.5-min pulse, the shortest time needed to observe a clear pattern of labeled proteins in the in vivo experiments (data not shown), argues against degradation of Cox1p as the source of mp15.
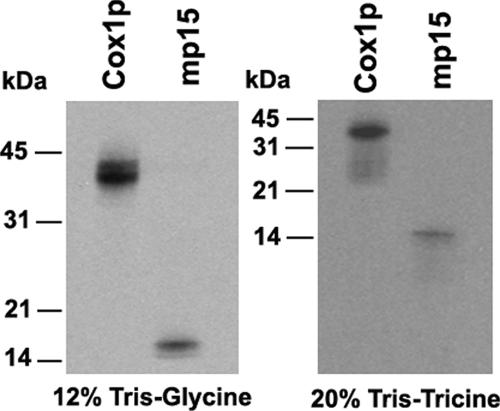
Radiolabeled mp15 separated by SDS-PAGE migrates slightly above 14-kDa marker (Figure 2) indicating a molecular weight of ∼15,000. Because hydrophobic proteins do not always separate true to size, the actual molecular weight could be larger.
Figure 2.
Electrophoretic mobility of mp15. Mitochondrial products were labeled in vivo in a wild type (W303-1A) and a mutant strain carrying a null allele of mss51 (Δmss51) as in Figure 1. Total cellular proteins were separated in a 17.5% PAGE (Barrientos et al., 2002b). Cox1p of the wild-type cells and mp15 from the mss51 mutant were excised from the SDS polyacrylamide gel and electroeluted using ElutaTube Protein Extraction kit (Fermentas, Ontario, Canada). The proteins were trichloroacetic acid (TCA) precipitated and resuspended in loading buffer. The radiochemical purity of the two proteins was tested by separation on a second 12% polyacrylamide gel (Laemmli, 1970) or a 20% polyacrylamide gel in Tris-Tricine buffer (Schagger and von Jagow, 1987).
Because mp15 is synthesized in low amounts, only detected because it is radiolabeled, and is highly labile (explained below), it is very difficult to purify enough protein to attempt mass spectrometry characterization. Attempts will be made in the future with this goal.
Pet309-dependent Synthesis of mp15 Polypeptide Occurs in COX Assembly-arrested Mutants Displaying Reduced Synthesis of Cox1p
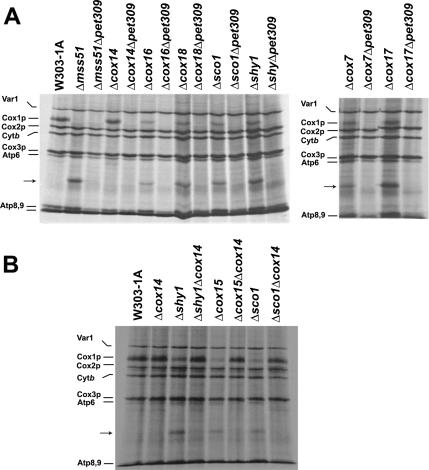
Mss51p was proposed to play a role in adjusting translation of Cox1p to the availability of its partners subunits during assembly of COX (Barrientos et al., 2002b, 2004). This was based on observations that synthesis of Cox1p is greatly reduced in most COX assembly mutants (Barrientos et al., 2004; Figure 3, A and B). The small amount of Cox1p detected in such mutants may reflect the fraction of this subunit that is associated with and stabilized by Mss51p and Cox14p (Perez-Martinez et al., 2003; Barrientos et al., 2004). Assembly-defective mutants in which Mss51p is tied up with unassembled Cox1p would be expected to have substantially reduced amounts of Mss51p available for new rounds of Cox1p translation, thereby allowing synthesis of mp15.
Figure 3.
Accumulates of mp15 in vivo in COX assembly-arrested mutants. (A) Wild type (W303-1A), mss51, cox14, cox16, cox18, sco1, cox7, and cox17 single null mutants or double mutants with a second null mutation in PET309 were labeled with [35S]methionine at 30°C for 15 min in the presence of cycloheximide as in Figure 1. (B) Wild type (W303-1A), cox14 single mutant or double mutants with a second mutation in shy1, cox15, and sco1 were labeled with [35S]methionine at 30°C for 15 min in the presence of cycloheximide as in Figure 1. The mitochondrial translation products are identified in the margin. The functions affected in the different strains are described in Table 1.
In vivo labeling of mitochondrial gene products indicated that with the exception of the cox14 mutant, all the other COX mutants tested synthesize mp15 (Figure 3A). As with the mss51 mutant, the presence of a second mutation in pet309 completely suppressed the synthesis of mp15 in all the COX mutants (Figure 3A). The efficiency of mp15 synthesis was variable but never as high as in the mss51 mutant. No mp15 was detected in the cox14 mutant, which as reported previously, synthesizes normal amounts of Cox1p (Barrientos et al., 2004; Figure 3, A and B). Synthesis of mp15 was also abolished or greatly reduced in double mutants in which a null allele in a COX assembly gene was combined with a deletion of COX14 (Figure 3B). Such strains were previously shown to synthesize Cox1p normally (Barrientos et al., 2004).
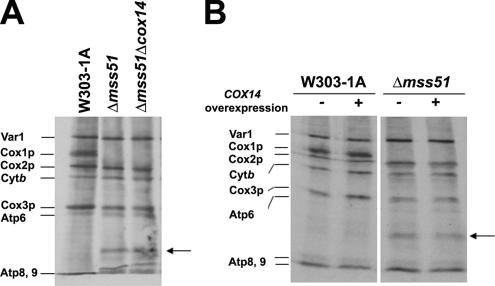
Synthesis of mp15 in the mss51 Mutant Is Not Affected by a cox14 Mutation
Cox14p plays an important role in regulating translation of COX1 mRNA. In other studies, we have shown that cox14 mutants synthesize normal amounts of Cox1p. Moreover, the cox14 mutation cures the Cox1p translational defect of most COX mutants (Barrientos et al., 2004). This was interpreted to indicate that Cox14p may be necessary to stabilize the interaction of Mss51p with Cox1p. Accordingly, mutants lacking Cox14p would have more Mss51p available for translation of Cox1p.
The synthesis of mp15 is independent of Cox14p, because incorporation of [35S]methionine into mp15 was the same in the mss51 single mutant and in a strain with mutation in both mss51 and cox14 (Figure 4A). Overexpression of COX14 from the episomal plasmid pG93/T1 (Glerum et al., 1995) in a mss51 null mutant strain also did not influence the synthesis of mp15. These results support a role of Cox14p in regulating the availability of Mss51p for translation by trapping the protein in a ternary complex with full-length Cox1p, rather than acting to inhibit translational elongation.
Figure 4.
COX14 does not affect synthesis and accumulation of mp15. (A) Wild type (W303-1A), an mss51 null mutant, and an mss5/cox14 double mutant were labeled with [35S]methionine at 30°C for 15 min in the presence of cycloheximide as described in Figure 1. (B) Wild type (W303-1A), and a null mutant of mss51 overexpressing or not COX14, were labeled in vivo as described in A. The mitochondrial translation products are identified in the margin.
Accumulation of mp15 Is Significantly Reduced In Vivo in COX Assembly Mutants Pretreated with Chloramphenicol
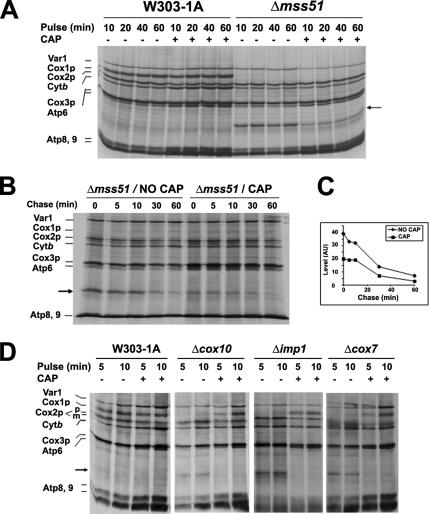
We have previously shown that, with the exception of the mss51, pet309, and oxa1 mutants, Cox1p labeling was increased in COX mutants when cells were preincubated in chloramphenicol (CAP) before the in vivo pulse (Barrientos et al., 2004), which was interpreted to be due to the accumulated larger pools of nuclear-encoded factors (i.e., Mss51p) required for mitochondrial gene expression. We wanted to test now if the enhanced synthesis of Cox1p in most COX assembly mutants had a modifying effect on the amount of newly synthesized mp15.
We first tested the effect of CAP preincubation on mp15 synthesis in mss51 mutants. Incorporation of [35S]methionine into mp15 in an mss51 null mutant strain was substantially lower in CAP-pretreated cells (Figure 5, A and B). Although at variable levels, in some experiments, the amount of newly synthesized mitochondrial ribosomal protein Var1 was also reduced in mss51 mutants pretreated with CAP, suggesting an effect of the antibiotic on ribosome metabolism that could be also related to the reduced synthesis of mp15. However, the amount of newly synthesized mp15 was consistently reduced in CAP-pretreated mss51 mutant cells even in experiments in which the amount of newly synthesized Var1 was not affected. The reduction in mp15 was unlikely to be due to faster turnover, because it was observed at the shortest times of labeling. Pulse-chase experiments also showed that mp15 synthesized in 10 min (Figure 5, B and C) or 15 min of pulse (data not shown) with or without the CAP pretreatment were degraded at similar rates.
Figure 5.
Synthesis and/or accumulation of mp15 are reduced in COX assembly mutants pretreated with chloramphenicol. (A) Effect of CAP pretreatment on in vivo labeling of mitochondrial gene products. Wild type (W303-1A) and the mss1 null mutant (Δmss51) were grown in YPGal. One-half of the cultures were further incubated at 30°C for 2 h in the presence of 2 mg/ml CAP. Cells were harvested and washed two times with a solution containing 40 mM potassium phosphate plus 2% galactose before labeling. Samples were removed after the indicated times of labeling and processed as in Figure 1. (B) Degradation of mp15 synthesized in cells pretreated or not with CAP. The mss1 null mutant (Δmss51) was incubated in the presence and absence of CAP as described in A. After harvesting and washing, cells were pulsed with [35S]methionine for 10 min. Labeling was terminated by addition of 80 μmol of cold methionine and 12 μg/ml puromycin (0 time). Samples of the cultures were collected after the indicated times of incubation at 30°C and processed as described in Figure 1. (C) Quantification of mp15 degradation in B. The radiolabeled bands were detected and quantified with a PhosphorImager (GE Healthcare). The values (arbitrary units) were plotted against the time of chase. (D) Effect of CAP pretreatment on the mitochondrial protein synthesis pattern of cox10, imp1, and cox7 null mutants. Cells were labeled as in A. Mitochondrial translation products are identified in the margin. Cox2p is not processed in Δimp1 mutant. The Cox2p precursor (pCox2p) in these strain migrates slower that the mature Cox2p (mCox2p). The functions affected in the different mutants are described in Table 1.
Pretreatment with CAP of different COX assembly-defective mutants reduced incorporation of [35S]methionine into mp15 to undetectable levels (Figure 5D). Synthesis of Cox1p in the CAP-treated mutants was comparable with that seen in wild type under normal pulse-labeling conditions. This confirms the presence of a fully functional translation apparatus in the COX mutants and supports the earlier conclusion that synthesis of Cox1p is down-regulated in such strains (Barrientos et al., 2004).
Mp15 Is a Membrane Protein and the Yta10p/Yta12p Protease Is Partially Responsible for Its Degradation
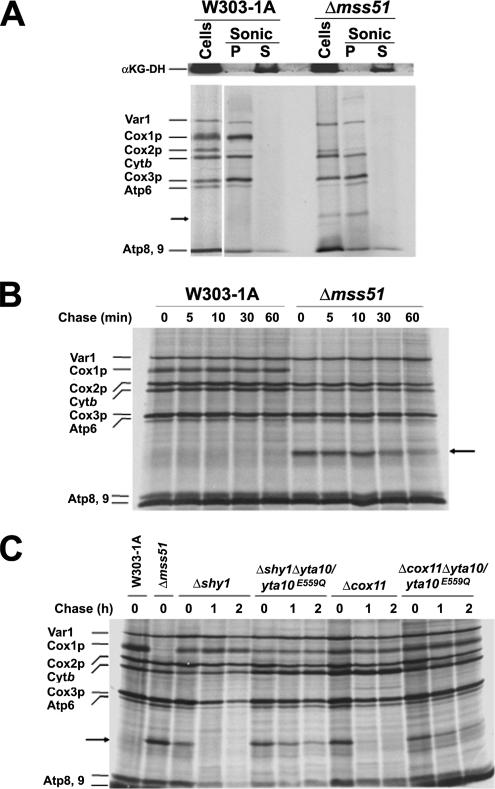
Cox1p is an integral protein, embedded in the mitochondrial inner membrane with 12 transmembrane α-helices (Tsukihara et al., 1996). To test whether mp15 is also a membrane-bound protein, we have determined its solubility properties. Sonic irradiation of Δmss51 spheroplasts containing newly synthesized 35S-labeled mitochondrial translation products solubilized α-ketoglutarate dehydrogenase, a soluble protein of the mitochondrial matrix, but not mp15 nor any of the other newly synthesized proteins that are known to be cotranslationally inserted in the membrane (Figure 6A). These results suggest that also mp15 is cotranslationally inserted in the mitochondrial inner membrane.
Figure 6.
mp15 is a membrane protein whose degradation depends partially on a proteolytically active YTA10/YTA12 complex. (A) mp15 is a membrane protein. Wild type (W303-1A) and a mutant carrying a null allele of mss51 were grown and labeled for 15 min at 30°C with [35S]methionine in the presence of cycloheximide. Cells were subsequently converted to spheroplasts by digestion of the cell wall with zymolyase and submitted to sonic radiation. Samples were centrifuged 25,000 rpm, and the pellet (P) and supernatant (S) fractions were collected, TCA precipitated, and resuspended in loading buffer. The proteins were separated into a 17.5% PAGE and transferred to a nitrocellulose membrane. The membrane was both, exposed to x-ray film (bottom) and used for a Western blot that was probed with a polyclonal antibody against the matrix soluble protein α-ketoglutarate dehydrogenase (top). (B) mp15 is rapidly degraded after synthesis. Wild type (W303-1A) and a mutant carrying a null allele of mss51 were grown and labeled for 15 min at 30°C with [35S]methionine. Labeling was terminated by addition of 80 μmol of cold methionine and 12 μg/ml puromycin (0 time). Samples of the cultures were collected after the indicated times of incubation at 30°C and processed as in Figure 1. (C) Degradation of mp15 is partially mediated by the YTA10/YTA12 complex. The wild type strain W303-1A, the shy1, and cox11 mutants, and the same mutants in which the endogenous wild-type YTA10 gene had been substituted by the catalytically inactive yta10E559Q mutant gene (Arlt et al., 1996) were labeled and chased for the indicated times as in A. Mitochondrial translation products are identified in the margin.
Our failure to detect mp15 with monoclonal antibodies against Cox1p (data not shown) suggested that either the Cox1p epitope is not present in mp15 or/and that most of this protein is degraded. Pulse-chase analysis of mp15 stability in an mss51 null mutant indicated ∼50% of the newly synthesized mp15 to be degraded after a 1-h chase (Figure 6B). The stability of unassembled Cox1p and of mp15 was also assessed in different COX mutants. In agreement with our previous findings, the small amount of labeled Cox1p present in the mutants was stable for at least 2 h of chase (Barrientos et al., 2004; Figure 6C). In contrast, mp15 was almost completely degraded after the 2-h chase.
Turnover of mitochondrially synthesized COX subunits that do not enter the assembly pathway is a function of different proteases, including the m-AAA protease complex Yta10p/Yta12p (Arlt et al., 1996), which also acts as a chaperone for assembly of the mitochondrial respiratory chain complexes (Arlt et al., 1996). The protease function can be inactivated by the E559Q mutation in the active site of Yta10p without affecting the chaperone function of the complex (Arlt et al., 1996). The role of the Yta10p/Yta12p in turnover of mp15 was studied in shy1/yta10 and a cox11/yta10 double mutant harboring the yta10E559Q allele. In vivo pulse-chase experiments of the double mutants revealed that mp15 is a substrate of Yta10p/Yta12p protease as a considerable amount of this novel protein was spared from degradation during the 1–2 h of chase in the background of the yta10E559Q allele (Figure 6C).
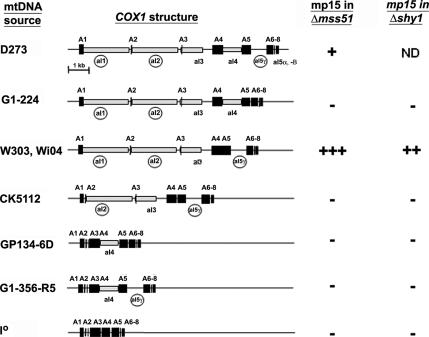
Synthesis of mp15 Is Affected by the Intron Composition of COX1
W303Δmss51, an mss51 null mutant containing W303 mtDNA, had significantly more mp15 than the same mutant with D273 mtDNA (Figure 1). The only significant difference in the COX1 genes of the two strains is the presence of additional introns in the gene of D273 mtDNA (Figure 7). In the absence of Mss51p, splicing of COX1 pre-mRNA containing a particular combination of introns may result in the accumulation of a particular splicing intermediate that serves as an mRNA for the aberrant mp15 translation product.
Figure 7.
Synthesis of mp15 depends on the intron composition of the COX1 gene. The figure represents the maps of COX1 in different strains of yeast. Group II introns are circled. mtDNA containing COX1 genes with different intron compositions (see details in Table 1) were transferred by cytoduction into to a kar1 mutant (Conde and Fink, 1976) devoid of mtDNA (ρo). The different mitochondrial genomes were transferred from the kar1 donor to ρo derivatives of mss51 and shy1 null mutants. To determine whether mp15 was synthesized in these strains, the mitochondrial gene products of the corresponding strains were labeled in vivo in the presence of cycloheximide and separated on a 17.5% polyacrylamide gel as in Figure 1. The last two columns on the right represent a summary of the results obtained.
To further ascertain the intron requirement for mp15 synthesis, mtDNA with different intron-containing COX1 genes (Figure 7) were transferred to ρo derivatives of mss51 and shy1 null mutants. In vivo labeling of the mitochondrial products in the resultant strains showed that mp15 was detected as a prominent band in strains containing the mitochondrial genome of W303 but not D273 (Figure 1B). To confirm that the efficient production of mp15 was related to its COX1 gene, mtDNA from Wi04, a strain with the same COX1 introns as W303, was also transferred to the mss51 and shy1 null mutants. Synthesis of mp15 in both mutants was comparable with that seen in the same strains with the W303 mtDNA (data not shown). Based on the in vivo labeling results obtained with other mss51 mutants harboring different COX1 genes, however, it was not possible to relate the synthesis of mp15 to the absence of a particular intron, although the presence of aI1 and 5γ seems to be an absolute requirement (Figure 7).
No synthesis of mp15 was seen in C199, a strain with D273 mtDNA and a point mutation in mss51 causing partial loss of function (data not shown). This was also true of five other mss51 point mutants in the same mitochondrial genetic background (data not shown). Substitution of D273 by W303 mtDNA in C199, however, allowed some limited synthesis of mp15. These results could suggest that expression of mp15 is at least partially dependent on the nuclear background or that the synthesis of mp15 is reduced in leaky mutants of mss51.
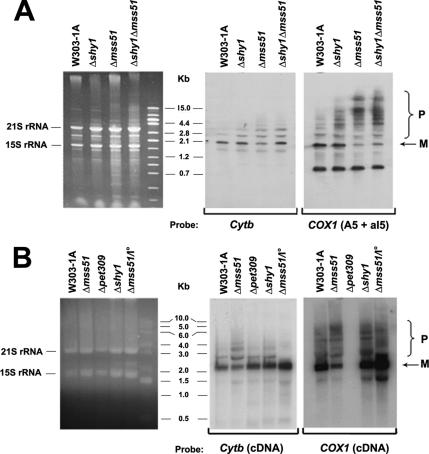
mp15 Is Translated from a Partially Processed COX1 Transcript
Synthesis of mp15 occurs in strains lacking Mss51p but not Pet309p, and additionally, it requires a COX1 gene with a particular set of introns. Because processing and translation of COX1 transcripts are interdependent events, splicing of some COX1 introns being dependent on translation of intron-encoded maturases (Carignani et al., 1983, 1986), mp15 is likely to be derived from an incompletely processed COX1 transcript. It was not excluded, however, that mp15 might be translated from an aberrantly spliced or partially degraded COX1 transcript. To rule out these possibilities, we examined the COX1 transcripts by Northern blot analysis of total mitochondrial RNAs of mss51, pet309, and shy1 mutants (Figure 8). Even though the amount of mature COX1 mRNA was reduced in the mss51 mutant, as reported previously (Simon and Faye, 1984), the exon A5/intron aI5 probe revealed a similar pattern of COX1 precursors in the mss51 and shy1 mutants (Figure 8A). No new small mRNA species were found in either strain (Figure 8A). The probe against the entire coding sequence of COX1, confirmed the presence both mature and partially processed transcripts in the mss51 mutant with a COX1 gene containing four or more introns (W303) (Figure 8B). In agreement with previous reports, neither mature nor precursor COX1 RNAs were present in the pet309 mutant because of their instability in this genetic background (Manthey and McEwen, 1995). Mature COX1 mRNA was also detected in two point mutants of mss51 in the D273 background, C199 and C283 (data not shown).
Figure 8.
Mature and unprocessed COX1 transcripts accumulate in mss51 and shy1 but not in pet309 null mutants. (A) Mitochondrial RNA was extracted from mitochondria purified of the wild-type strain W303-1A, a shy1 null mutant (Δshy1), an mss51 null mutant (Δmss51) and a double shy/1mss51 mutant (Δshy1 Δmss51). The RNA extracts were separated onto a 1% agarose gel, stained with ethidium bromide, photographed, and the RNAs blotted to a nylon membrane (Nytran, SuPerCharge; Whatman Schleicher and Schuell, Keene, NH). After cross-linking with UV light, the nylon membrane was prehybridized at 43°C with 125 μg of salmon sperm DNA in 5× SSC, 5× Denhardts, 0.5% SDS. The blotted RNAs were hybridized overnight at 43°C with probes containing exon 5 + intron 5 of COX1 and exon 1 of COB. Both probes were labeled with [α-32P]dATP by random priming (Feinberg and Vogelstein, 1983). (B) Mitochondrial RNA extracts were separated onto a denaturing 1.2% agarose gel, blotted to a nylon membrane and UV-light cross-linked. Northern blots of mitochondrial RNA of the wild-type W303-1A, the mss51, pet309, and shy1 null mutants with a W303 mtDNA background (Figure 7A) and the mss51 null mutant with an intronless mitochondrial genome were prehybridized at 42°C for 2 h with 1 μg of salmon sperm and hybridized overnight at 65°C in a solution containing 7% SDS, 1 μM EDTA, and 0.5 M Na2PO4 with probes containing the entire coding sequence of COX1 or COB. The mature (M) and unprocessed (P) COX1 transcripts are identified in the margins. The left panels in A–C shows the ethidium bromide-stained gels used for the Northern blots. The positions of the 15S and 21S mitochondrial rRNAs are indicated.
The presence of mature COX1 mRNA in the mss51 mutant suggests that there is sufficient translation of intron-encoded maturases for splicing of introns aI1, -2, and -3, although the possibility of enhanced self-splicing in the absence of Mss51p cannot be eliminated. As expected, normal levels of COX1 mRNA are present in the mss51 mutant with intronless mtDNA. The absence in this strain of mp15 also supports the notion that it is likely to be a translation product of a partially spliced COX1 transcript.
Mss51p Binds to the 5′-UTR of COX1 mRNA
Mss51p has been conjectured to act on the COXI mRNA 5′-UTR to initiate translation and on the coding sequence to promote elongation (Decoster et al., 1990; Perez-Martinez et al., 2003). Because mp15 is only detected in the W303 and D273 mtDNA backgrounds, it is conceivable that the presence of a particular sequence(s) in the 5′-UTR of COX1 of these strains could by-pass the requirement of Mss51p for translation initiation but not elongation. Under these circumstances, synthesis of Cox1p could be prematurely terminated.
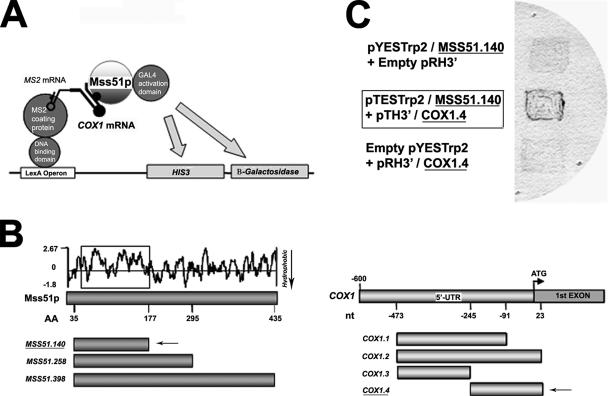
The putative interaction/s of Mss51p with the 5′-UTR of COX1 mRNA has not been characterized. We have used the Y3H depicted in Figure 9A to define both the RNA target(s) sequence and the Mss51p domain involved in the binding to the RNA. The constructs used as baits consisted of 460 base pairs 5′-UTR sequence (Manthey and McEwen, 1995) as well as different regions of the 5′-UTR with part of the first exon of the COX1 gene (Figure 9B). To ensure targeting of the Mss51p/Gal4 fusion proteins to the nucleus, a requirement in the Y3H system, the mitochondrial leader peptide of Mss51p was deleted. The mitochondrial import sequence of Mss51p determined by protein sequencing of Mss51p-GST purified from mitochondria consists of the N-terminal 35 residues encoded by the gene.
Figure 9.
Mss51p binds to the 5′-UTR of COX1 mRNA. (A) Y3H strategy used to explore the interaction of Mss51p with the 5′-UTR of COX1 mRNA. (B) Diagram showing the regions of Mss51p (including a hydrophobicity map) and the COX1 mRNA that were used to make the Y3H constructs. (C) A domain in the N′-terminus of Mss51p (Mss51.140; marked with a square in the hydrophobicity map in B) interacts with a domain in the 5′-UTR of COX1 mRNA (Cox1.4; marked with an arrow in B). Activation of the β-galactosidase reporter gene detected as a blue color occurred only when both the protein prey and RNA bait were expressed.
The results obtained with different COX1 and MSS51 constructs indicated an interaction between the MSS51-140 domain expressing the first 177 amino acids of the mature protein and a sequence located between nucleotides −245 and +23 of COX1 mRNA (Figure 9C). This domain did not interact with the MSS51-398 construct, which should produce the entire Mss51p (data not shown), perhaps because folding of the membrane protein interferes with recognition of the bait. No interaction was detected between MSS51-140 and COX1.1 (data not shown), suggesting that the interacting mRNA domain is located between nucleotides −97 and +23, although we cannot exclude the possibility that the longer COX1.1, as in COX1.2, made the mRNA bait unsuitable for the assay. The MSS51–140 sequence encodes a hydrophilic domain, which probably protrudes into the mitochondrial matrix where it interacts with the 5′-UTR of COX1 mRNA.
The region of the 5′-UTR of COX1 that was ascertained to interact with Mss51p did not display any nucleotide variation in the mtDNAs of D273, W303, and the strain with the intronless gene.
DISCUSSION
Mss51p is a mitochondrial inner membrane protein that faces the matrix (Siep et al., 2000). Together with Pet309p, it interacts with the 5′-UTR of the COX1 mRNA to initiate translation (Perez-Martinez et al., 2003). The yeast three-hybrid experiments reported here suggest that the target in the 5′-UTR of COX1 mRNA could be within 245 nucleotides upstream of the initiation codon. The sequence in Mss51p that could interact with this activation site has been mapped to a hydrophilic region located in the N-terminal 177 residues of the mature protein.
As noted previously, mss51 mutants lack Cox1p; unexpectedly, however, we found that they synthesize a novel 15-kDa protein. Several lines of evidence suggest that mp15 is translated from a COX1 transcript. This protein is not present in pet309 mutants in which initiation of COX1 translation is blocked and which do not accumulate mature or precursor COX1 mRNAs when this mitochondrial gene contains more than four introns (Manthey and McEwen, 1995). In addition, no mp15 is detected in an mss51/pet309 double mutant or in a mutant with a large deletion in COX1.
Translation of a Cox1p-related protein in mss51 mutants is at variance with previous observations that the interaction of Mss51p with the 5′-UTR of COX1 is essential for translation of Cox1p. This was evident by the failure of an mss51 mutant to express ARG8m when the recoded gene was substituted for COX1 and was fused to the normal 5′-UTR of COX1 (Perez-Martinez et al., 2003). The possibility that the requirement for Mss51p for translation is by-passed in mp15-producing strains as a result of a polymorphism in the COX1 5′-UTRs has been excluded as no sequence differences were found in their Mss51p-interacting domain. These observations suggest that translation of mp15 is initiated from an ATG other than the normal COX1 initiation codon. The dependence of mp15 expression on Pet309p, a protein implicated in translation initiation of COX1 mRNA (Manthey and McEwen, 1995), may be related to the already mentioned absence of COX1 transcripts in pet309 mutants and therefore does not exclude the use of an alternate start codon. Whether translation of mp15 starts in an exon or intron is unclear, although an exonic ATG is more likely for several reasons. The partially overlapping peptide patterns of Cox1p and mp15 indicate the presence in the latter of some exon-encoded sequences. Because the reading frames of COX1 introns are in register with the reading frames of their upstream but not downstream exons, a 15-kDa product could be expressed exclusively from an mRNA containing intron sequences but not from a reading frame initiated in an intron and encompassing the downstream exon.
Alternatively, mp15 could result from premature termination of translation of a COX1 transcript. Recent evidence has shown that Mss51p acts on target/s within the coding sequence of the COX1 mRNA or to the protein itself, to promote translational elongation and that in the absence of Mss51p, nascent Cox1p may block its own synthesis (Perez-Martinez et al., 2003). Translational regulation by nascent chains has been reported previously (for review, see Tenson and Ehrenberg, 2002).
The mp15 polypeptide is most evident in mss51 mutant containing mtDNA of W303, to a lesser extent in mss51 mutants with D273 mtDNA, and it is not detected in strains with an intronless COX1 or with combinations of introns other than those of W303 and D273. The appearance of novel mitochondrial translation products has been reported previously. For example, a cox1 mutant expressing both Pet309p and Mss51p but defective in intron aI1 splicing was shown to produce a 90-kDa protein translated from the continuous reading frame encoded by the first exon and part of the first intron of COX1. This protein is post-translationally cleaved into two polypeptides of 20 and 68 kDa, the latter constituting the maturase encoded by intron aI1 (Carignani et al., 1983). In contrast, synthesis of mp15 depends on the complete absence of Mss51p or presence of only limiting amounts of the translationally competent protein. That mp15 is not detected in an mss51 null mutant with intronless mtDNA supports the notion that this protein is translated from a COX1 precursor transcript with one or more introns. The COX1 genes of W303 and D273 contain all three group II introns (aI1, -2, and -5γ) and the group I intron aI3. The intron composition of COX1 in D273 is similar to W303 except for the presence of group I intron aI4, which seems to suppress mp15 synthesis. It is also noteworthy that the only difference between the COX1 genes of D273 and G1–224, which does not express mp15, is the absence in the latter of intron aI5γ. The lack of a clear correlation between the presence or absence of a particular intron and mp15 synthesis suggests that the overall intronic composition in the context of the mss51 mutation may be crucial for the production or stability of the transcript corresponding to the mp15 mRNA.
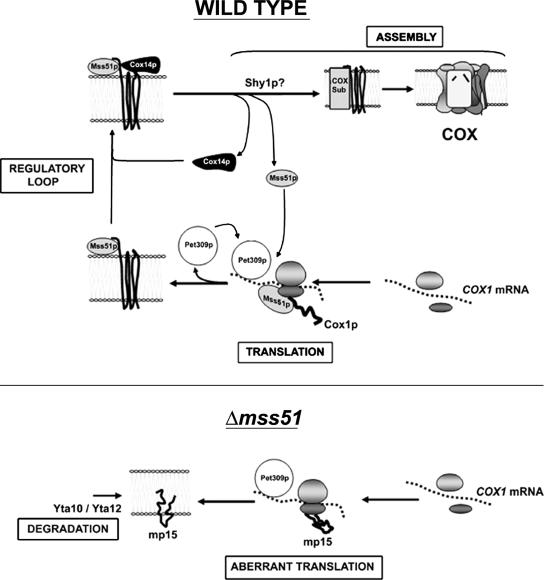
Cox1p synthesis is regulated by downstream events occurring during assembly of the COX holoenzyme (Perez-Martinez et al., 2003; Barrientos et al., 2004). In our model, entry of newly translated Cox1p into the COX assembly pathway depends on its interaction with Mss51p and Cox14p (Figure 10). When engaged in this ternary complex, Mss51p is forestalled from initiating new rounds of Cox1p synthesis. Dissociation of Mss51p from the complex is envisioned to occur when Cox1p acquires its prosthetic groups or interacts with other COX subunits. This step may be catalyzed by Shy1p (Perez-Martinez et al., 2003; Barrientos et al., 2004). This is supported by the severe repression of Cox1p translation in mutants that are blocked in COX assembly (Barrientos et al., 2004), in which mp15 is also synthesized because in these strains the amount of Mss51p available for normal COX1 mRNA translation is limited. The single exception are cox14 mutants, in which normal amounts of Cox1p are synthesized and Mss51p is available for translation even in the absence of COX assembly (Barrientos et al., 2004). For simplicity, our model depicted in Figure 10. does not include the role of Mss51p on the coding sequence of COX1 mRNA. As mentioned above, the interaction of Mss51p with Cox1p itself could be necessary for elongation of the nascent polypeptide and it could in addition regulate Cox1p synthesis (Perez-Martinez et al., 2003).
Figure 10.
Model depicting the hypothetical roles of Mss51p in Cox1p translation and coupling to Shy1p-dependent COX assembly. Mss51p is required for translation of Cox1p by acting, together with Pet309p on the 5′-UTR of COX1 mRNA. As shown before (Barrientos et al., 2004), Mss51p binds newly synthesized Cox1p forming a transient complex that is stabilized by Cox14p. A downstream event, maybe catalyzed by Shy1p, causing Cox1p to dissociate from the ternary complex makes Mss51p available for new rounds of translation. To simplify the model we have not included the action of Mss51p on the coding sequence of COX1, probably on Cox1p itself, interpreted as necessary for elongation of the nascent polypeptide (Perez-Martinez et al., 2003). In mss51 mutants (Δmss51) Mss51p-independent translation from alternate initiation sites in a COX1 mRNA precursor occur generating a new polypeptide, mp15, which is proteolytically degraded.
The regulatory model portrayed in Figure 10 is also supported by the results presented here. Synthesis of mp15 is Pet309p-dependent and Mss51p-independent, whereas synthesis of Cox1p requires both translational factors (Figure 10). Cox1p and mp15 synthesis are inversely related, suggesting that synthesis of Cox1p may suppress Mss51p-independent translation from alternate initiation sites by competing for Pet309p. Translation of mp15 in COX assembly mutants (not depicted in Figure 10 for simplicity) also points to the existence of a Cox14p-stabilized Mss51p-Cox1p complex. When trapped in this complex Mss51p is unavailable for translation of Cox1p. Mutations in COX14, either alone or in combination with mutations in other COX assembly factors (except MSS51), inhibit synthesis of mp15. This is consistent with the model as the instability of the Mss51p-Cox1p complex in the absence of Cox14p would be expected to make more Mss51p available for translation of Cox1p. The lack of an effect on mp15 synthesis of overexpression or deletion of COX14 in an mss51 null mutant, indicates that Cox14p does not directly repress Cox1p synthesis but rather does so indirectly by limiting Mss51p available for translation. Finally, incubation of cells in the presence of CAP before the [35S]methionine pulse enhances synthesis of Cox1p in most COX assembly mutants. This may be explained by the synthesis of extra Mss51p during the CAP preincubation leading to enhanced translation of Cox1p, whereas having the opposite effect on mp15.
ACKNOWLEDGMENTS
We thank Vladimir Malinovskii (Proteomics Facility, Department of Biochemistry, University of Miami, Miami, FL) for technical assistance with the protein sequencing experiments. We are indebted to Dr. Mario Barros (Department of Genetics, Instituto de Biociências de Botucatu, Universidade Estadual Paulista, Botucatu, Sãn Paulo, Brazil) for providing several strains with different mitochondrial DNA intron composition. This research was supported by National Institutes of Health Research Grants GM-071775A (to A.B.) and GM-50187 (to A.T.), a research grant from the Muscular Dystrophy Association (to A.B.), and Telethon-Italy fellowship GFP05008 (to F.F.).
Abbreviations used:
- CAP
chloramphenicol
- COX
cytochrome c oxidase
- GST
glutathione S-transferase
- UTR
untranslated region
- Y3H
yeast three-hybrid system.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E06-09-0803) on November 29, 2006.
REFERENCES
- Arlt H., Tauer R., Feldmann H., Neupert W., Langer T. The YTA10-12 complex, an AAA protease with chaperone-like activity in the inner membrane of mitochondria. Cell. 1996;85:875–885. doi: 10.1016/s0092-8674(00)81271-4. [DOI] [PubMed] [Google Scholar]
- Ausubel F. M., Brent R., Kingston R. E., Moore D. D., Seidman J. G., Smith J. A., Struhl K. Current Protocols in Molecular Biology. Vol. 2. New York: Wiley; 1994. Saccharomyces cerevisiae; p. 13. [Google Scholar]
- Barrientos A., Barros M. H., Valnot I., Rotig A., Rustin P., Tzagoloff A. Cytochrome oxidase in health and disease. Gene. 2002a;286:53–63. doi: 10.1016/s0378-1119(01)00803-4. [DOI] [PubMed] [Google Scholar]
- Barrientos A., Korr D., Tzagoloff A. Shy1p is necessary for full expression of mitochondrial COX1 in the yeast model of Leigh's syndrome. EMBO J. 2002b;21:43–52. doi: 10.1093/emboj/21.1.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrientos A., Zambrano A., Tzagoloff A. Mss51p and Cox14p jointly regulate mitochondrial Cox1p expression in Saccharomyces cerevisiae. EMBO J. 2004;23:3472–3482. doi: 10.1038/sj.emboj.7600358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barros M. H., Myers A. M., Van Driesche S., Tzagoloff A. COX24 codes for a mitochondrial protein required for processing of the COX1 transcript. J. Biol. Chem. 2006;281:3743–3751. doi: 10.1074/jbc.M510778200. [DOI] [PubMed] [Google Scholar]
- Barros M. H., Nobrega F. G., Tzagoloff A. Mitochondrial ferredoxin is required for heme A synthesis in Saccharomyces cerevisiae. J. Biol. Chem. 2002;277:9997–10002. doi: 10.1074/jbc.M112025200. [DOI] [PubMed] [Google Scholar]
- Breeden L., Nasmyth K. Regulation of the yeast HO gene. Cold Spring Harb. Symp. Quant. Biol. 1985;50:643–650. doi: 10.1101/sqb.1985.050.01.078. [DOI] [PubMed] [Google Scholar]
- Brown N. G., Costanzo M. C., Fox T. D. Interactions among three proteins that specifically activate translation of the mitochondrial COX3 mRNA in Saccharomyces cerevisiae. Mol. Cell. Biol. 1994;14:1045–1053. doi: 10.1128/mcb.14.2.1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carignani G., Groudinsky O., Frezza D., Schiavon E., Bergantino E., Slonimski P. P. An mRNA maturase is encoded by the first intron of the mitochondrial gene for the subunit I of cytochrome oxidase in S. cerevisiae. Cell. 1983;35:733–742. doi: 10.1016/0092-8674(83)90106-x. [DOI] [PubMed] [Google Scholar]
- Carignani G., Netter P., Bergantino E., Robineau S. Expression of the mitochondrial split gene coding for cytochrome oxidase subunit I in S. cerevisiae: RNA splicing pathway. Curr. Genet. 1986;11:55–63. doi: 10.1007/BF00389426. [DOI] [PubMed] [Google Scholar]
- Carlson C. G., Barrientos A., Tzagoloff A., Glerum D. M. COX16 encodes a novel protein required for the assembly of cytochrome oxidase in Saccharomyces cerevisiae. J. Biol. Chem. 2003;278:3770–3775. doi: 10.1074/jbc.M209893200. [DOI] [PubMed] [Google Scholar]
- Conde J., Fink G. R. A mutant of Saccharomyces cerevisiae defective for nuclear fusion. Proc. Natl. Acad. Sci. USA. 1976;73:3651–3655. doi: 10.1073/pnas.73.10.3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decoster E., Simon M., Hatat D., Faye G. The MSS51 gene product is required for the translation of the COX1 mRNA in yeast mitochondria. Mol. Gen. Genet. 1990;224:111–118. doi: 10.1007/BF00259457. [DOI] [PubMed] [Google Scholar]
- Faye G., Kujawa C., Fukuhara H. Physical and genetic organization of petite and grande yeast mitochondrial DNA. IV. In vivo transcription products of mitochondrial DNA and localization of 23 S. ribosomal RNA in petite mutants of Saccharomyces cerevisiae. J. Mol. Biol. 1974;88:185–203. doi: 10.1016/0022-2836(74)90304-0. [DOI] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal. Biochem. 1983;132:6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Fiori A., Perez-Martinez X., Fox T. D. Overexpression of the COX2 translational activator, Pet111p, prevents translation of COX1 mRNA and cytochrome c oxidase assembly in mitochondria of Saccharomyces cerevisiae. Mol. Microbiol. 2005;56:1689–1704. doi: 10.1111/j.1365-2958.2005.04658.x. [DOI] [PubMed] [Google Scholar]
- Glerum D. M., Koerner T. J., Tzagoloff A. Cloning and characterization of COX14, whose product is required for assembly of yeast cytochrome oxidase. J. Biol. Chem. 1995;270:15585–15590. doi: 10.1074/jbc.270.26.15585. [DOI] [PubMed] [Google Scholar]
- Glerum D. M., Muroff I., Jin C., Tzagoloff A. COX15 codes for a mitochondrial protein essential for the assembly of yeast cytochrome oxidase. J. Biol. Chem. 1997;272:19088–19094. doi: 10.1074/jbc.272.30.19088. [DOI] [PubMed] [Google Scholar]
- Glerum D. M., Shtanko A., Tzagoloff A. Characterization of COX17, a yeast gene involved in copper metabolism and assembly of cytochrome oxidase. J. Biol. Chem. 1996a;271:14504–14509. doi: 10.1074/jbc.271.24.14504. [DOI] [PubMed] [Google Scholar]
- Glerum D. M., Shtanko A., Tzagoloff A. SCO1 and SCO2 act as high copy suppressors of a mitochondrial copper recruitment defect in Saccharomyces cerevisiae. J. Biol. Chem. 1996b;271:20531–20535. doi: 10.1074/jbc.271.34.20531. [DOI] [PubMed] [Google Scholar]
- Glerum D. M., Tzagoloff A. Submitochondrial distributions and stabilities of subunits 4, 5, and 6 of yeast cytochrome oxidase in assembly defective mutants. FEBS Lett. 1997;412:410–414. doi: 10.1016/s0014-5793(97)00799-0. [DOI] [PubMed] [Google Scholar]
- Hell K., Tzagoloff A., Neupert W., Stuart R. A. Identification of Cox20p, a novel protein involved in the maturation and assembly of cytochrome oxidase subunit 2. J. Biol. Chem. 2000;275:4571–4578. doi: 10.1074/jbc.275.7.4571. [DOI] [PubMed] [Google Scholar]
- Krause K., Lopes de Souza R., Roberts D. G., Dieckmann C. L. The mitochondrial message-specific mRNA protectors Cbp1 and Pet309 are associated in a high-molecular weight complex. Mol. Biol. Cell. 2004;15:2674–2683. doi: 10.1091/mbc.E04-02-0126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labouesse M. The yeast mitochondrial leucyl-tRNA synthetase is a splicing factor for the excision of several group I introns. Mol. Gen. Genet. 1990;224:209–221. doi: 10.1007/BF00271554. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Manthey G. M., McEwen J. E. The product of the nuclear gene PET309 is required for translation of mature mRNA and stability or production of intron-containing RNAs derived from the mitochondrial COX1 locus of Saccharomyces cerevisiae. EMBO J. 1995;14:4031–4043. doi: 10.1002/j.1460-2075.1995.tb00074.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwen J. E., Ko C., Kloeckner-Gruissem B., Poyton R. O. Nuclear functions required for cytochrome c oxidase biogenesis in Saccharomyces cerevisiae. Characterization of mutants in 34 complementation groups. J. Biol. Chem. 1986;261:11872–11879. [PubMed] [Google Scholar]
- Michaelis U., Korte A., Rodel G. Association of cytochrome b translational activator proteins with the mitochondrial membrane: implications for cytochrome b expression in yeast. Mol. Gen. Genet. 1991;230:177–185. doi: 10.1007/BF00290666. [DOI] [PubMed] [Google Scholar]
- Myers A. M., Pape L. K., Tzagoloff A. Mitochondrial protein synthesis is required for maintenance of intact mitochondrial genomes in Saccharomyces cerevisiae. EMBO J. 1985;4:2087–2092. doi: 10.1002/j.1460-2075.1985.tb03896.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naithani S., Saracco S. A., Butler C. A., Fox T. D. Interactions among COX1, COX2, and COX3 mRNA-specific translational activator proteins on the inner surface of the mitochondrial inner membrane of Saccharomyces cerevisiae. Mol. Biol. Cell. 2003;14:324–333. doi: 10.1091/mbc.E02-08-0490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobrega M. P., Bandeira S. C., Beers J., Tzagoloff A. Characterization of COX19, a widely distributed gene required for expression of mitochondrial cytochrome oxidase. J. Biol. Chem. 2002;277:40206–40211. doi: 10.1074/jbc.M207348200. [DOI] [PubMed] [Google Scholar]
- Nobrega M. P., Nobrega F. G., Tzagoloff A. COX10 codes for a protein homologous to the ORF1 product of Paracoccus denitrificans and is required for the synthesis of yeast cytochrome oxidase. J. Biol. Chem. 1990;265:14220–14226. [PubMed] [Google Scholar]
- Perez-Martinez X., Broadley S. A., Fox T. D. Mss51p promotes mitochondrial Cox1p synthesis and interacts with newly synthesized Cox1p. EMBO J. 2003;22:5951–5961. doi: 10.1093/emboj/cdg566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothstein R. J. One-step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- Sambrook J., Fritsch E. F., Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Schagger H., von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- Schiestl R. H., Gietz R. D. High efficiency transformation of intact yeast cells using single stranded nucleic acids as a carrier. Curr. Genet. 1989;16:339–346. doi: 10.1007/BF00340712. [DOI] [PubMed] [Google Scholar]
- Seraphin B., Boulet A., Simon M., Faye G. Construction of a yeast strain devoid of mitochondrial introns and its use to screen nuclear genes involved in mitochondrial splicing. Proc. Natl. Acad. Sci. USA. 1987;84:6810–6814. doi: 10.1073/pnas.84.19.6810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siep M., van Oosterum K., Neufeglise H., van der Spek H., Grivell L. A. Mss51p, a putative translational activator of cytochrome c oxidase subunit-1 (COX1) mRNA, is required for synthesis of Cox1p in Saccharomyces cerevisiae. Curr. Genet. 2000;37:213–220. doi: 10.1007/s002940050522. [DOI] [PubMed] [Google Scholar]
- Simon M., Faye G. Steps in processing of the mitochondrial cytochrome oxidase subunit I pre-mRNA affected by a nuclear mutation in yeast. Proc. Natl. Acad. Sci. USA. 1984;81:8–12. doi: 10.1073/pnas.81.1.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith D., Gray J., Mitchell L., Antholine W. E., Hosler J. P. Assembly of cytochrome-c oxidase in the absence of assembly protein Surf1p leads to loss of the active site heme. J. Biol. Chem. 2005;280:17652–17656. doi: 10.1074/jbc.C500061200. [DOI] [PubMed] [Google Scholar]
- Solans A., Zambrano A., Barrientos A. Cytochrome c oxidase deficiency: from yeast to human. Preclinica. 2004;2:336–348. [Google Scholar]
- Souza R. L., Green-Willms N. S., Fox T. D., Tzagoloff A., Nobrega F. G. Cloning and characterization of COX18, a Saccharomyces cerevisiae PET gene required for the assembly of cytochrome oxidase. J. Biol. Chem. 2000;275:14898–14902. doi: 10.1074/jbc.275.20.14898. [DOI] [PubMed] [Google Scholar]
- Tenson T., Ehrenberg M. Regulatory nascent peptides in the ribosomal tunnel. Cell. 2002;108:591–594. doi: 10.1016/s0092-8674(02)00669-4. [DOI] [PubMed] [Google Scholar]
- Tiranti V., et al. Mutations of SURF-1 in Leigh disease associated with cytochrome c oxidase deficiency. Am. J. Hum. Genet. 1998;63:1609–1621. doi: 10.1086/302150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towpik J. Regulation of mitochondrial translation in yeast. Cell. Mol. Biol. Lett. 2005;10:571–594. [PubMed] [Google Scholar]
- Tsukihara T., Aoyama H., Yamashita E., Tomizaki T., Yamaguchi H., Shinzawa-Itoh K., Nakashima R., Yaono R., Yoshikawa S. The whole structure of the 13-subunit oxidized cytochrome c oxidase at 2.8 A. Science. 1996;272:1136–1144. doi: 10.1126/science.272.5265.1136. [DOI] [PubMed] [Google Scholar]
- Tzagoloff A., Akai A., Foury F. Assembly of the mitochondrial membrane system XVI. Modified form of the ATPase proteolipid in oligomycin-resistant mutants of Saccharomyces cerevisiae. FEBS Lett. 1976;65:391–395. doi: 10.1016/0014-5793(76)80154-8. [DOI] [PubMed] [Google Scholar]
- Tzagoloff A., Akai A., Needleman R. B., Zulch G. Assembly of the mitochondrial membrane system. Cytoplasmic mutants of Saccharomyces cerevisiae with lesions in enzymes of the respiratory chain and in the mitochondrial ATPase. J. Biol. Chem. 1975;250:8236–8242. [PubMed] [Google Scholar]
- Tzagoloff A., Capitanio N., Nobrega M. P., Gatti D. Cytochrome oxidase assembly in yeast requires the product of COX11, a homolog of the P. denitrificans protein encoded by ORF3. EMBO J. 1990;9:2759–2764. doi: 10.1002/j.1460-2075.1990.tb07463.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzagoloff A., Dieckmann C. L. PET genes of Saccharomyces cerevisiae. Microbiol. Rev. 1990;54:211–225. doi: 10.1128/mr.54.3.211-225.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z., et al. SURF1, encoding a factor involved in the biogenesis of cytochrome c oxidase, is mutated in Leigh syndrome. Nat. Genet. 1998;20:337–343. doi: 10.1038/3804. [DOI] [PubMed] [Google Scholar]