Abstract
The coherence of mitochondrial biogenesis relies on spatiotemporally coordinated associations of 800–1000 proteins mostly encoded in the nuclear genome. We report the development of new quantitative analyses to assess the role of local protein translation in the construction of molecular complexes. We used real-time PCR to determine the cellular location of 112 mRNAs involved in seven mitochondrial complexes. Five typical cases were examined by an improved FISH protocol. The proteins produced in the vicinity of mitochondria (MLR proteins) were, almost exclusively, of prokaryotic origin and are key elements of the core construction of the molecular complexes; the accessory proteins were translated on free cytoplasmic polysomes. These two classes of proteins correspond, at least as far as intermembrane space (IMS) proteins are concerned, to two different import pathways. Import of MLR proteins involves both TOM and TIM23 complexes whereas non-MLR proteins only interact with the TOM complex. Site-specific translation loci, both outside and inside mitochondria, may coordinate the construction of molecular complexes composed of both nuclearly and mitochondrially encoded subunits.
INTRODUCTION
Most mitochondrial proteins of eukaryotic cells are encoded by nuclear genes and synthesized by cytoplasmic ribosomes. However, a few proteins are encoded by the mitochondrial DNA and are translated inside mitochondria by bacterial-type ribosomes. The major mitochondrial import pathway of cytoplasmically translated proteins is now well understood (Koehler, 2004; Rehling et al., 2004). Although a posttranslational mechanism for import is widely accepted, it may concern only a limited class of proteins. Indeed, it was demonstrated 30 years ago that a subclass of cytoplasmic polysomes is bound to the surface of mitochondria (Kellems et al., 1975). Subsequently, experiments in yeast and human cells have suggested a cotranslational import process for some mitochondrial proteins (Fujiki and Verner, 1993; Mukhopadhyay et al., 2004). The evolutionary history of mitochondria might elucidate this apparent discrepancy between post- and cotranslational import processes. Phylogenetic studies of the yeast mitochondrial proteome have demonstrated a composite origin. It is estimated that half or more of the genes that code for the modern mitochondrial proteome originated directly from the host nuclear genome, and the other half are of bacterial origin, a consequence of a massive transfer of genes from the endosymbiont to the host nuclear genome (Marcotte et al., 2000; Karlberg et al., 2000). This symbiotic situation required the development of protein translocation machineries (translocases) in the mitochondrial membranes (Herrmann, 2003). Possibly, the two classes of nuclear genes coding for mitochondrial products may have used the same mitochondrial import pathway. However, a recent genome-wide analysis (Marc et al., 2002) suggested a strong correlation between the bacterial origin of the genes and their locus specific translation on mitochondria-linked polysomes. An index called MLR (mitochondrial localization of mRNA) was derived from microarray analyses to classify (from 0 to 100) the mRNAs according to their cellular localization. A recent analysis of the outer membrane proteome (Zahedi et al., 2006) presented strong support for the MLR concept: it included both bona fide outer membrane proteins and precursors for many MLR proteins. This strongly suggests that the MLR proteins are translated next to mitochondria, although the reason for this remains obscure. In some cases, this phenomenon may favor the formation of nucleo-proteocomplexes in the vicinity of mitochondria, as for Msk1, the mitochondrial lysyl-tRNA synthetase, which guides the import of tRNALys toward mitochondria (Entelis et al., 2006).
To describe the MLR phenomenon in more detail, we developed two highly sensitive methods to: 1) quantify, in vitro, the amount of mitochondrial-associated mRNA and 2) to visualize, in situ, the localization of some mRNA species. These approaches were applied to 112 nuclear mRNAs to establish a spatial map of the sites of translation of proteins involved in the assembly of seven mitochondrial complexes. We report a close link between core proteins and mitochondrially associated translation sites. The prokaryotic origin of most of these MLR proteins emphasizes the evolutionary stability of this phenomenon.
MATERIALS AND METHODS
Strains and Growth Conditions
The Saccharomyces cerevisiae strain CW252 has an intronless mitochondrial genome and is nuclearly isogenic to W303 (Saint-Georges et al., 2002). Cells were grown at 30°C in galactose medium (1% bactopeptone, 1% yeast extract, 2% galactose) for overnight precultures and FISH experiments; for purification of mitochondria, cells were grown in the same medium plus 0.1% KH2PO4 and 0.12% (NH4)2SO4.
Quantitative PCR analysis of asymmetric mRNA localization. Schematically, mitochondrion-bound polysomes were purified as in Kellems et al. (1975), and RNA was extracted using the hot-phenol method. Aliquots of 50 ng of either total or mitochondria-associated mRNA were used for reverse transcription (RT), and the cDNA was purified using the Macherey-Nagel PCR extract kit. Serial dilutions (1/10, 1/50, 1/100) of cDNAs were subjected to real-time quantitative PCR with oligonucleotides specific for each ORF (Supplementary Table S1) and the Sybr Green PCR kit (QuantiTech).
For a reliable evaluation of the mRNA associated with mitochondria, various correction factors were used to account for the difficulties of the purification procedure. COX1 and COX2 mRNAs were used to normalize the data, assuming they have a 100% mitochondrial localization. ACT1 mRNA was used to assess the extent of contamination by nonmitochondrial fractions. The SD error by this approach varies from 0 to 5%, whereas previous microarray-based methods gave much less reproducible results (Marc et al., 2002). A detailed description of this approach (PCR programs and calculations) is available in Garcia et al. (2006) and in supplemental document S1.
FISH Analysis and In Situ mRNA Localization
The combined hybridization of three to five antisense oligonucleotides was used to enhance the visualization of each mRNA. Typically, each oligonucleotide was designed to contain 50–55 nucleotides with five aminoallyl thymidines and a coherent Tm value (±2°C for the set of oligonucleotide). After synthesis (Eurogentec, San Diego, CA) the oligonucleotides were directly labeled with CY3 or CY5 fluorochromes as previously described (Garcia et al., 2006). The probes used in this study are presented in Supplementary Table S2.
For in situ hybridization, yeast cells were grown to midlog phase and immediately fixed with a final concentration of 4% paraformaldehyde, spheroplasted with lyticase, adhered to poly-l-lysine–treated coverslips, and stored in 70% ethanol at −20°C. After overnight hybridization at 37°C, washing, and DAPI staining, coverslips were mounted with anti-fade solution and imaged on an Olympus BX61 upright microscope (Melville, NY) using a 100× 1.35 NA objective. For 3D sampling 41 images were acquired at a spacing of 200 nm in the Z-axis. ImageJ was used for image processing; several stacks can be combined using a maximum projection algorithm. We developed an extension ImageJ plug-in for calculating the distance from mRNA to mitochondrion objects (Jourdren and Garcia, unpublished results).
RESULTS
Quantitative Analysis of Asymmetric mRNA Localization
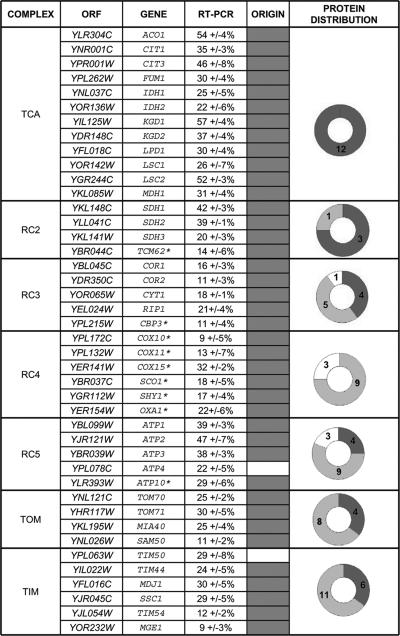
Most experimental evidence that mRNA molecules are unevenly distributed within the cell is from in situ fluorescent analyses (Darzacq et al., 2003). A few studies (Diehn et al., 2000; Marc et al., 2002; de Jong et al., 2006) used DNA microarrays to establish a list of genes whose mRNA is preferentially translated on membrane-bound polysomes. Though providing an abundance of data, these studies were limited by substantial heterogeneity of the biochemical preparations and variations in the yield due to the purification procedure. For instance, the general conclusions of the DNA microarray analyses are probably sound concerning mitochondria-bound polysomes (Marc et al., 2002), but gene-specific data may be absent or inaccurate. Because it is possible to enrich mitochondrial fractions carrying associated cytoplasmic polysomes (Kellems et al., 1975), we developed a protocol with better discrimination. We used real-time quantitative RT-PCR with normalization and correction of the results to take into account the purification yield and contaminating factors (see Materials and Methods). Mitochondrial mRNAs were analyzed to assess the exact quantity of mitochondrial products in each extract as an internal control. The technical reproducibility of the procedure was verified by conducting three independent polysomal fractionations from different yeast cultures. mRNA from 112 genes was studied, and the percentage of each mRNA associated with mitochondria-bound polysomes was between 0 and 57% (Supplementary Table S1 and Supplementary Figure S1). Three classes of gene were distinguished. The first class includes the 56 genes that have between 0 and 3% of their mRNA linked to mitochondria; these mRNAs were considered to be translated on free cytoplasmic polysomes. The second class includes 50 genes for which 9–57% of their mRNA was linked to mitochondria: the corresponding proteins are probably produced in the vicinity of mitochondria, and they were considered to be MLR proteins (Figure 1). The third class, the intermediate group (4–6%), includes six genes that cannot be unambiguously classified to either class 1 or class 2. Classes 1 and 2 contain approximately the same number of genes, 56 and 50 respectively, as previously observed in a more global analysis (Marc et al., 2002). In contrast, data from the Saccharomyces Genome Database (Christie et al., 2004) and PSI-BLAST analysis (Altschul et al., 1997) indicate that 96% of the MLR genes, but only 8% of genes with nonlocalized mRNAs, are prokaryotic homologues (Figure 1 and Supplementary Table S1).
Figure 1.
MLR proteins of seven mitochondrial complexes. The association between mitochondrial-bound polysomes and 107 mRNAs coding for all the subunits of mitochondrial complexes: respiratory complexes 2, 3, 4, and 5; import complexes TOM and TIM; and TCA cycle complexes, and for assembly factors (starred) was quantified by RT-PCR analyses with several correction factors (see Materials and Methods and Garcia et al., 2006). Only the genes that are translated in the vicinity of mitochondria (MLR proteins) are represented (all data are available in Supplementary Material). Most of these MLR genes have prokaryotic homologues (gray shaded box), whereas most of the other genes have no known prokaryotic homologue (Supplementary Material). The right column gives a schematic composition of the assembled complexes according to gene origin: mitochondrially encoded (white), nuclearly encoded either of prokaryotic (dark gray), or eukaryotic (light gray) origin.
FISH Analysis of the In Situ mRNA Localization
Single-cell FISH experiments are complementary to the biochemical analyses described. New developments in probe design and in fluorophore chemistry allow detection of single molecules of mRNA in their cellular environment (Femino et al., 1998, 2003). The protocol we used here is adapted from Long et al. (1995, 1997) and can be used to compare, simultaneously, the spatial distribution of different mRNAs relative to each other or to the mitochondrion (see Materials and Methods). Mitochondria were unambiguously detected using probes recognizing mitochondrial ribosomal RNAs; this allowed simultaneous detection of mRNAs and mitochondria in a single step.
Both haploid and diploid strains were examined, and because of their size, findings with diploid strains were easier to interpret. However, it was evident that in the results in the two genetic backgrounds were equivalent.
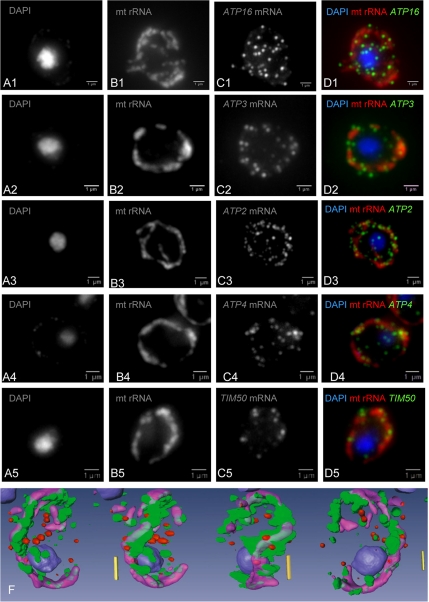
The findings of FISH experiments were entirely consistent with the results of the RT-PCR analyses. For all genes classified as MLR (>6% asymmetrical localization of the mRNA), as exemplified by ATP1, ATP2, and ATP3 (Figure 2), the FISH signal localized to punctuate structures that aligned with mitochondria. Conversely ATP16 (0% by RT-PCR) had its mRNA isotropically dispersed in the cell with no apparent association with mitochondria (Figure 2 and Supplementary Figure 2S for 3D quantification analysis). Almost all MLR genes seem to be of prokaryotic origin (Figure 1), although a few, including ATP4 and TIM50, have no prokaryotic homolog (Saccharomyces Genome Database; Christie et al., 2004); FISH analyses fully confirmed that the corresponding mRNAs of these two atypical cases were translated adjacent to mitochondria (Figure 2). This is also consistent with the detection of precursor forms of these proteins in the outer mitochondrial membrane proteome (Zahedi et al., 2006).
Figure 2.
Live-cell imaging of nuclear-encoded mRNAs for mitochondrial proteins. Five fluorescent oligonucleotides specific for mitochondrial rRNA (lane B) and for specific mRNAs (ATP16-C1, ATP3-C2, ATP2-C3, ATP4-C4, and TIM50-C5) were used to probe wild-type yeast cells for hybridization. The merged figures (lane D) indicate the relative localizations of mt rRNA (red) and the various mRNAs (green). 3D quantitative analysis was used to compare ATP16 mRNA, which is isotropically distributed, with the specifically localized ATP3 mRNA (Supplementary Figure 2S). A 3D reconstitution of these relative localizations is represented on the bottom line for ATP2 mRNA (green), mt rRNA (violet), and the negative control YRA1 mRNA (red). These 3D analyses allow a quantitative assessment of the distance between a particular mRNA molecule and the edge of the mitochondrion (see Supplementary Data).
MLR Proteins and Mitochondrial Complexes
All the mitochondrial complexes examined contained MLR proteins. We therefore examined whether these proteins have a particular role in these complexes.
The Krebs TCA cycle enzymes are believed to be highly organized in supramolecular complexes (Velot et al., 1997) connected to the other supermacromolecular complex, succinate:quinone oxidoreductase (SQR, Complex II; Horsefield et al., 2004). All the 16 proteins comprising these two complexes are unambiguously MLR proteins. This applies even to Tcm62, which is considered to be a putative protein chaperone required for complex II assembly.
The ATP synthase complex (RCC5) is composed of proteins of diverse origins, and its structure and biosynthesis are well documented (Ackerman and Tzagoloff, 2005). Three MLR proteins, ATP1 (subunit α), ATP2 (subunit β), and ATP3 (subunit γ) are components of the F1 subunit, and three other MLR proteins, ATP4 (subunit b), ATP5 (OSCP), and ATP10 (essential chaperone for F0) are essential for F0 subunit construction. Genetic and biochemical experiments have demonstrated that the α, β, γ, b, and OSCP subunits are the basic elements of the F1 and F0 complexes (see Ackerman and Tzagoloff, 2005 for a review). The case of ATP10 is less straightforward. There is genetic interaction between ATP10 and ATP6, the mitochondrial gene for subunit a, suggesting that Atp10 may act as a subunit a-chaperone at early stages of F0 assembly (Tzagoloff et al., 2004). Interestingly, the five basic subunits of the ATP synthase (α, β, γ, b, and OSCP) are all translated in the vicinity of mitochondria. Conversely, none of the 13 accessory genes required for yeast ATP synthase biogenesis (Ackerman and Tzagoloff, 2005) are MLR proteins. Thus, translation near the mitochondrion appears to be a characteristic of the early nucleation process of the core enzyme.
The case of the COX complex (RC4) deserves a special mention: none of the mRNAs of nuclear-encoded subunits were translated in the vicinity of mitochondria. However, Shy1, Sco1, Oxa1, Cox10, Cox11, and Cox15 (Figure 1), important for the assembly of this complex (Nijtmans et al., 2001; Szyrach et al., 2003), are undoubtedly MLR proteins.
The bc1 complex (RC3) is a complex of tripartite origin: the apocytochrome b is always encoded in the mitochondrial genome, and some of its nuclear-encoded subunits are MLR proteins, whereas others are translated on free cytoplasmic polysomes. The core proteins of the complex—Cor1, Cor2, Rip1 and Cyt1—clearly belong to the MLR protein class and they are unambiguously of a prokaryotic origin. Cbp3, like Atp10 for complex V, is a chaperone essential for complex III biosynthesis (Kronekova and Rodel, 2005), suggesting that there is in all cases at least one chaperone associated with the assembly of these core proteins. Thus, the bc1 complex shares various similarities with the ATP synthase complex.
The TOM and TIM complexes differ from the previous examples from an evolutionary point of view. This import machinery emerged as a consequence of the endosymbiosis. The synthesis of mitochondrial proteins in the cytosol required the development of protein translocation systems to ensure appropriate localization. Consequently, few of these translocase components have obvious bacterial homologues and most are of eukaryotic origin. Systematic analysis of the MLR subunits composing these complexes provided an interesting observation. At least one of the components of each subcomplex of the translocase machinery (Koehler, 2004) is unambiguously an MLR protein (SAM = SAM50, TOM = TOM70, TIM22 = TIM54, Export = OXA1). This is consistent with the suggestion that the ability of mitochondria to bind mRNAs for the genes coding for targeted proteins would alleviate the problem of targeting the prokaryotic proteins (Herrmann, 2003). As in the respiratory complexes, most of the MLR proteins are critical to the biogenesis of the complex. SAM50, for instance, is required for viability and probably forms the core of the SAM complex (Paschen et al., 2003; Kozjak et al., 2003; Pfanner et al., 2004). The TOM translocase is composed of seven proteins and the MLR proteins Tom70 and Tom40 are crucial to the properties of the complex. Tim50, an essential protein of the complex TIM23, has no prokaryotic homolog but its MLR localization and the FISH analyses identify it as an MLR protein. This might be an example of a protein becoming MLR through acquisition of specific targeting signals. Finally, Mia40, a recently discovered protein (Chacinska et al., 2004), is a central component of the protein import and assembly machinery. Mia40 is required for the assembly of small proteins of the intramembrane space (Rissler et al., 2005). The identification of Mia40 as an MLR protein (of prokaryotic origin) is entirely consistent with its importance in the construction of this mitochondrial domain.
DISCUSSION
The Functional Implications of Site-specific Translation of MLR Genes
Forty-eight of the 50 MLR proteins we identified are of prokaryotic origin, according to the phylogenic data available from SGD (Christie et al., 2004), whereas 53 of the 62 non-MLR proteins translated on free cytoplasmic polysomes are of eukaryotic origin (no homology found in the available prokaryotic sequences). This suggests that a strong selective pressure has maintained a mitochondria-linked translation mechanism for MLR proteins. This apparently ancestral process raises the following various interesting considerations.
MLR Proteins and Protein-Translocation Pathways
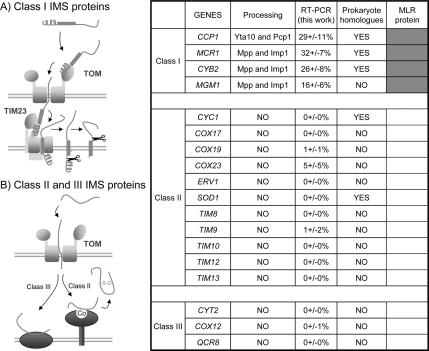
A recent survey (Herrmann and Hell, 2005) of the mechanisms by which proteins of the mitochondrial intermembrane space (IMS) are transported from the cytosol has revealed three main classes. These classes differ in the energy sources used to drive the translocation reactions. We examined the MLR values of most of these proteins (Figure 3) and found that class I proteins are exclusively MLR proteins, whereas the class II and III proteins are non-MLR proteins. The well-characterized class I proteins have bipartite presequences composed of matrix-targeting N-terminal sequences followed by hydrophobic sorting domains. Unlike class II or III proteins, class I protein importation into IMS is TIM23 complex dependent, and consequently class I (MLR) protein importation relies on a physical chain of interactions between TOM and TIM23 complexes. Thus, class I protein import may require a tight connection between cytoplasmically localized translation and sites of outer–inner membrane association.
Figure 3.
Class I intermembrane space proteins are MLR proteins. Protein-translocation pathways into the intermembrane space (IMS) of mitochondria have been classified into three classes (Herrmann and Hell, 2005). Class I proteins have typical mitochondrial presequences followed by hydrophobic sorting domains and are directed to the TIM23 complex. Class II and III proteins are imported independently of the inner-membrane TIM23 complex. Polysome localization of mRNA coding for the IMS proteins described was examined by RT-PCR analysis. Class I proteins were clearly produced from mRNA translated in the vicinity of mitochondria (MLR proteins-RT-PCR value above 6%, indicated in dark gray in the right column), whereas none of the mRNAs for the class II or III proteins were significantly linked to mitochondria (non-MLR proteins, white; RT-PCR value <6%).
MLR Proteins and Mitochondrial Complexes
We describe several examples of essential core elements of the early steps of the complex biogenesis being MLR proteins. This is especially clear for the ATP synthase complex for which the assembly of the subunits during the biogenesis of the complex is well described (Ackerman and Tzagoloff, 2005). Subunits α (ATP1), β (ATP2), and γ (ATP3) of the F1 subunit and subunit b (ATP4), OSCP (ATP5), and the essential chaperone Atp10 (ATP10) of the F0 subunit are all essential for the early steps of ATP synthase biogenesis, and are all MLR proteins. MLR proteins appear to be critical early during biogenesis of most of the complexes described above. This strongly suggests that the MLR-specific translation sites (the punctate structures in Figure 2) might also be the sites of initiation of the early steps of complex construction. Systematic relative localization analysis of these translation sites may further elucidate the space and time-controlled biogenesis of mitochondrial complexes.
MLR Protein Translation and Membrane Contact Sites
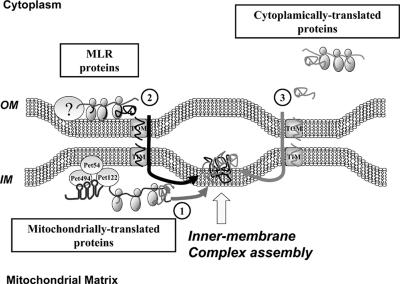
We can suggest a model (Figure 4) in which MLR proteins are translated at specific sites near the outer mitochondrial membrane. This is reminiscent of the mitochondrial mRNA translation process: for some mRNAs, including those of the COX1, COX2, and CYTb genes, specific translation sites at the inner mitochondrial membrane have been demonstrated. This site-specific translation depends on the mRNA associating with identified protein complexes (Naithani et al., 2003; Krause et al., 2004). The protein Oxa1 establishes, in some cases, a link with ribosomes to ensure cotranslational insertion of the nascent mitochondrial-encoded polypeptides into the membrane (Szyrach et al., 2003). Possibly, mitochondrial genes translocated to the nucleus after the symbiotic event have continued to use the same translation process. This membrane-controlled production might have facilitated the early steps of symbiosis when primitive mitochondrial genes became nuclear at a time when the mitochondrial import machineries did not exist.
Figure 4.
Proposed model of inner-membrane mitochondrial complex biogenesis. Three types of proteins compose the inner-membrane complexes: (1) Mitochondrial-encoded proteins translated on membrane-bound mRNA, depending on specific activator proteins. (2) MLR proteins, identified in this work, are nuclear-encoded and translated in the vicinity of the outer mitochondrial membrane. (3) Nuclear-encoded proteins translated on free cytoplasmic polysomes and that are posttranslationally imported. The model assumes that pathways 1 and 2 are spatially connected, thus facilitating the formation of the core protein complex with which accessory proteins (pathway 3) will combine.
There is no evidence for a mechanism coordinating the output of the mitochondrial and nuclear gene products in yeast. We propose that MLR protein translation is a key event in the early step of the biogenesis of these mitochondrial complexes. The corresponding prediction is that each mitochondrial complex is constructed at particular sites where cotranslationally imported nuclear-encoded proteins congregate with proteins of the same complex translated within the mitochondria. This implies that outer and inner mitochondrial membranes contact at sites allowing a process of cotranslation on both sides of the membranes. Cytoplasmic ribosomes have been observed bound to regions of the outer membrane that are in intimate association or contact with the inner membrane (Kellems et al., 1975). Also the fact that, in the intermembrane space, MLR proteins are imported via a chain of physical contacts between the two membranes gives further credence to this idea (see above and Figure 3). The improved FISH methodology presented in this work should allow us to address this issue experimentally.
MLR Proteins and Cellular Compartments
There may be damaging effects of site-specific translation of MLR proteins. The location of many mRNAs close to mitochondria may be deleterious if reactive oxygen species (ROS) production alters these molecules. For example, in Alzheimer's disease, it was observed that ROS production in the mitochondrial environment may cause excessive mRNA oxidation (Shan et al., 2003). Peri-mitochondrial translation might thus promote exposure of the mRNAs concerned to free radicals, with subsequent aggregation of the translation products (Swerdlow and Khan, 2004). However, a healthy wild-type cell is likely to minimize this possibility by a time-dependent compartmentalization process that disconnects mitochondria biogenesis and ROS production (Tu et al., 2005). It would be interesting to study how MLR protein production is regulated temporally.
Supplementary Material
ACKNOWLEDGMENTS
We thank Anne Lombès, Shailesh M. Shenoy, Geneviève Dujardin, Frédéric Devaux, and Yann Saint Georges for precious advice and comments. This work was supported by The French Ministry of Research Grants ANR-BLAN06-1-135513 (C.J.) and ANR-JCJC06-136140 (X.D.)
Abbreviations used:
- MLR
mitochondria-localized mRNA.
Footnotes

 The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E06-09-0827) on November 15, 2006.
REFERENCES
- Ackerman S. H., Tzagoloff A. Function, structure, and biogenesis of mitochondrial ATP synthase. Prog. Nucleic Acid Res. Mol. Biol. 2005;80:95–133. doi: 10.1016/S0079-6603(05)80003-0. [DOI] [PubMed] [Google Scholar]
- Altschul S. F., Madden T. L., Schaffer A. A., Zhang J., Zhang Z., Miller W., Lipman D. J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chacinska A., Pfannschmidt S., Wiedemann N., Kozjak V., Sanjuan Szklarz L. K., Schulze-Specking A., Truscott K. N., Guiard B., Meisinger C., Pfanner N. Essential role of Mia40 in import and assembly of mitochondrial intermembrane space proteins. EMBO J. 2004;23:3735–3746. doi: 10.1038/sj.emboj.7600389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie K. R., et al. Saccharomyces Genome Database (SGD) provides tools to identify and analyze sequences from Saccharomyces cerevisiae and related sequences from other organisms. Nucleic Acids Res. 2004;32:D311–D314. doi: 10.1093/nar/gkh033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darzacq X., Powrie E., Gu W., Singer R. H., Zenklusen D. RNA asymmetric distribution and daughter/mother differentiation in yeast. Curr. Opin. Microbiol. 2003;6:614–620. doi: 10.1016/j.mib.2003.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jong M., van Breukelen B., Wittink F. R., Menke F. L., Weisbeek P. J., Van den Ackerveken G. Membrane-associated transcripts in Arabidopsis; their isolation and characterization by DNA microarray analysis and bioinformatics. Plant J. 2006;46:708–721. doi: 10.1111/j.1365-313X.2006.02724.x. [DOI] [PubMed] [Google Scholar]
- Diehn M., Eisen M. B., Botstein D., Brown P. O. Large-scale identification of secreted and membrane-associated gene products using DNA microarrays. Nat. Genet. 2000;25:58–62. doi: 10.1038/75603. [DOI] [PubMed] [Google Scholar]
- Entelis N., Brandina I., Kamenski P., Krasheninnikov I. A., Martin R. P., Tarassov I. A glycolytic enzyme, enolase, is recruited as a cofactor of tRNA targeting toward mitochondria in Saccharomyces cerevisiae. Genes Dev. 2006;20:1609–1620. doi: 10.1101/gad.385706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Femino A. M., Fay F. S., Fogarty K., Singer R. H. Visualization of single RNA transcripts in situ. Science. 1998;280:585–590. doi: 10.1126/science.280.5363.585. [DOI] [PubMed] [Google Scholar]
- Femino A. M., Fogarty K., Lifshitz L. M., Carrington W., Singer R. H. Visualization of single molecules of mRNA in situ. Methods Enzymol. 2003;361:245–304. doi: 10.1016/s0076-6879(03)61015-3. [DOI] [PubMed] [Google Scholar]
- Fujiki M., Verner K. Coupling of cytosolic protein synthesis and mitochondrial protein import in yeast. Evidence for cotranslational import in vivo. J. Biol. Chem. 1993;268:1914–1920. [PubMed] [Google Scholar]
- Garcia M., Darzacq X., Devaux F., Singer R. H., Jacq C. In: Methods in Molecular Biology: Mitochondria. Leister D., Herrmann J. M., editors. Vol. 372. Totowa, NJ: Humana Press; 2006. pp. 505–527. [Google Scholar]
- Herrmann J. M. Converting bacteria to organelles: evolution of mitochondrial protein sorting. Trends Microbiol. 2003;11:74–79. doi: 10.1016/s0966-842x(02)00033-1. [DOI] [PubMed] [Google Scholar]
- Herrmann J. M., Hell K. Chopped, trapped or tacked—protein translocation into the IMS of mitochondria. Trends Biochem. Sci. 2005;30:205–211. doi: 10.1016/j.tibs.2005.02.005. [DOI] [PubMed] [Google Scholar]
- Horsefield R., Iwata S., Byrne B. Complex II from a structural perspective. Curr. Protein Pept. Sci. 2004;5:107–118. doi: 10.2174/1389203043486847. [DOI] [PubMed] [Google Scholar]
- Karlberg O., Canback B., Kurland C. G., Andersson S. G. The dual origin of the yeast mitochondrial proteome. Yeast. 2000;17:170–187. doi: 10.1002/1097-0061(20000930)17:3<170::AID-YEA25>3.0.CO;2-V. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellems R. E., Allison V. F., Butow R. A. Cytoplasmic type 80S ribosomes associated with yeast mitochondria. IV. Attachment of ribosomes to the outer membrane of isolated mitochondria. J. Cell Biol. 1975;65:1–14. doi: 10.1083/jcb.65.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koehler C. M. New developments in mitochondrial assembly. Annu. Rev. Cell Dev. Biol. 2004;20:309–335. doi: 10.1146/annurev.cellbio.20.010403.105057. [DOI] [PubMed] [Google Scholar]
- Kozjak V., Wiedemann N., Milenkovic D., Lohaus C., Meyer H. E., Guiard B., Meisinger C., Pfanner N. An essential role of Sam50 in the protein sorting and assembly machinery of the mitochondrial outer membrane. J. Biol. Chem. 2003;278:48520–48523. doi: 10.1074/jbc.C300442200. [DOI] [PubMed] [Google Scholar]
- Krause K., Lopes de Souza R., Roberts D. G., Dieckmann C. L. The mitochondrial message-specific mRNA protectors Cbp1 and Pet309 are associated in a high-molecular weight complex. Mol. Biol. Cell. 2004;15:2674–2683. doi: 10.1091/mbc.E04-02-0126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronekova Z., Rodel G. Organization of assembly factors Cbp3p and Cbp4p and their effect on bc(1) complex assembly in Saccharomyces cerevisiae. Curr. Genet. 2005;47:203–212. doi: 10.1007/s00294-005-0561-9. [DOI] [PubMed] [Google Scholar]
- Long R. M., Elliott D. J., Stutz F., Rosbash M., Singer R. H. Spatial consequences of defective processing of specific yeast mRNAs revealed by fluorescent in situ hybridization. RNA. 1995;1:1071–1078. [PMC free article] [PubMed] [Google Scholar]
- Long R. M., Singer R. H., Meng X., Gonzalez I., Nasmyth K., Jansen R. P. Mating type switching in yeast controlled by asymmetric localization of ASH1 mRNA. Science. 1997;277:383–387. doi: 10.1126/science.277.5324.383. [DOI] [PubMed] [Google Scholar]
- Marc P., Margeot A., Devaux F., Blugeon C., Corral-Debrinski M., Jacq C. Genome-wide analysis of mRNAs targeted to yeast mitochondria. EMBO Rep. 2002;3:159–164. doi: 10.1093/embo-reports/kvf025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcotte E. M., Xenarios I., van Der Bliek A. M., Eisenberg D. Localizing proteins in the cell from their phylogenetic profiles. Proc. Natl. Acad. Sci. USA. 2000;97:12115–12120. doi: 10.1073/pnas.220399497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay A., Ni L., Weiner H. A co-translational model to explain the in vivo import of proteins into HeLa cell mitochondria. Biochem. J. 2004;382:385–392. doi: 10.1042/BJ20040065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naithani S., Saracco S. A., Butler C. A., Fox T. D. Interactions among COX1, COX2, and COX3 mRNA-specific translational activator proteins on the inner surface of the mitochondrial inner membrane of Saccharomyces cerevisiae. Mol. Biol. Cell. 2003;14:324–333. doi: 10.1091/mbc.E02-08-0490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nijtmans L. G., Artal Sanz M., Bucko M., Farhoud M. H., Feenstra M., Hakkaart G. A., Zeviani M., Grivell L. A. Shy1p occurs in a high molecular weight complex and is required for efficient assembly of cytochrome c oxidase in yeast. FEBS Lett. 2001;498:46–51. doi: 10.1016/s0014-5793(01)02447-4. [DOI] [PubMed] [Google Scholar]
- Paschen S. A., Waizenegger T., Stan T., Preuss M., Cyrklaff M., Hell K., Rapaport D., Neupert W. Evolutionary conservation of biogenesis of beta-barrel membrane proteins. Nature. 2003;426:862–866. doi: 10.1038/nature02208. [DOI] [PubMed] [Google Scholar]
- Pfanner N., Wiedemann N., Meisinger C., Lithgow T. Assembling the mitochondrial outer membrane. Nat. Struct. Mol. Biol. 2004;11:1044–1048. doi: 10.1038/nsmb852. [DOI] [PubMed] [Google Scholar]
- Rehling P., Brandner K., Pfanner N. Mitochondrial import and the twin-pore translocase. Nat. Rev. Mol. Cell Biol. 2004;5:519–530. doi: 10.1038/nrm1426. [DOI] [PubMed] [Google Scholar]
- Rissler M., Wiedemann N., Pfannschmidt S., Gabriel K., Guiard B., Pfanner N., Chacinska A. The essential mitochondrial protein Erv1 cooperates with Mia40 in biogenesis of intermembrane space proteins. J. Mol. Biol. 2005;353:485–492. doi: 10.1016/j.jmb.2005.08.051. [DOI] [PubMed] [Google Scholar]
- Saint-Georges Y., Bonnefoy N., di Rago J. P., Chiron S., Dujardin G. A pathogenic cytochrome b mutation reveals new interactions between subunits of the mitochondrial bc1 complex. J. Biol. Chem. 2002;277:49397–49402. doi: 10.1074/jbc.M207219200. [DOI] [PubMed] [Google Scholar]
- Shan X., Tashiro H., Lin C. L. The identification and characterization of oxidized RNAs in Alzheimer's disease. J. Neurosci. 2003;23:4913–4921. doi: 10.1523/JNEUROSCI.23-12-04913.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swerdlow R. H., Khan S. M. A “mitochondrial cascade hypothesis” for sporadic Alzheimer's disease. Med. Hypotheses. 2004;63:8–20. doi: 10.1016/j.mehy.2003.12.045. [DOI] [PubMed] [Google Scholar]
- Szyrach G., Ott M., Bonnefoy N., Neupert W., Herrmann J. M. Ribosome binding to the Oxa1 complex facilitates co-translational protein insertion in mitochondria. EMBO J. 2003;22:6448–6457. doi: 10.1093/emboj/cdg623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu B. P., Kudlicki A., Rowicka M., McKnight S. L. Logic of the yeast metabolic cycle: temporal compartmentalization of cellular processes. Science. 2005;310:1152–1158. doi: 10.1126/science.1120499. [DOI] [PubMed] [Google Scholar]
- Tzagoloff A., Barrientos A., Neupert W., Herrmann J. M. Atp10p assists assembly of Atp6p into the F0 unit of the yeast mitochondrial ATPase. J. Biol. Chem. 2004;279:19775–19780. doi: 10.1074/jbc.M401506200. [DOI] [PubMed] [Google Scholar]
- Velot C., Mixon M. B., Teige M., Srere P. A. Model of a quinary structure between Krebs TCA cycle enzymes: a model for the metabolon. Biochemistry. 1997;36:14271–14276. doi: 10.1021/bi972011j. [DOI] [PubMed] [Google Scholar]
- Zahedi R. P., Sickmann A., Boehm A. M., Winkler C., Zufall N., Schonfisch B., Guiard B., Pfanner N., Meisinger C. Proteomic analysis of the yeast mitochondrial outer membrane reveals accumulation of a subclass of preproteins. Mol. Biol. Cell. 2006;17:1436–1450. doi: 10.1091/mbc.E05-08-0740. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.