Abstract
Drs2p family P-type ATPases (P4-ATPases) are required in multiple vesicle-mediated protein transport steps and are proposed to be phospholipid translocases (flippases). The P4-ATPases Drs2p and Dnf1p cycle between the exocytic and endocytic pathways, and here we define endocytosis signals required by these proteins to maintain a steady-state localization to internal organelles. Internalization of Dnf1p from the plasma membrane uses an NPFXD endocytosis signal and its recognition by Sla1p, part of an endocytic coat/adaptor complex with clathrin, Pan1p, Sla2p/End4p, and End3p. Drs2p has multiple endocytosis signals, including two NPFXDs near the C terminus and PEST-like sequences near the N terminus that may mediate ubiquitin (Ub)-dependent endocytosis. Drs2p localizes to the trans-Golgi network in wild-type cells and accumulates on the plasma membrane when both the Ub- and NPFXD-dependent endocytic mechanisms are inactivated. Surprisingly, the pan1-20 temperature-sensitive mutant is constitutively defective for Ub-dependent endocytosis but is not defective for NPFXD-dependent endocytosis at the permissive growth temperature. To sustain viability of pan1-20, Drs2p must be endocytosed through the NPFXD/Sla1p pathway. Thus, Drs2p is an essential endocytic cargo in cells compromised for Ub-dependent endocytosis. These results demonstrate an essential role for endocytosis in retrieving proteins back to the Golgi, and they define critical cargos of the NPFXD/Sla1p system.
INTRODUCTION
Drs2p is a resident P-type ATPase of the yeast trans-Golgi network (TGN) that is required for vesicle-mediated protein transport from this organelle. Most well-characterized P-type ATPases are cation pumps that control the concentration of ions in both intracellular and extracellular spaces (for example, the Na+/K+ ATPase, Ca++ ATPase, and H+/K+ ATPase) (Kuhlbrandt, 2004). Drs2p, in contrast, is the founding member of a large P-type ATPase subfamily, called P4-ATPases (Catty et al., 1997), that are proposed to translocate phospholipid rather than ions. This flippase activity is responsible for translocating specific phospholipid molecules from the exoplasmic leaflet to the cytosolic leaflet to establish asymmetry of the membrane bilayer (Graham, 2004; Pomorski et al., 2004; Holthuis and Levine, 2005; Paulusma and Oude Elferink, 2005; Devaux et al., 2006). For Drs2p, ATPase activity and presumably phospholipid translocation are essential, because mutation of the aspartic acid that forms an aspartyl–phosphate intermediate during catalysis (D560N) renders Drs2p nonfunctional in vivo (Chen et al., 1999). In addition, after shifting to the nonpermissive temperature, a drs2 temperature-sensitive (ts) allele causes a rapid loss of exocytic vesicle formation in vivo (Gall et al., 2002) and the loss of an ATP-dependent phosphatidylserine (PS) flippase activity in purified Golgi membranes containing Drs2-ts (Natarajan et al., 2004). Mammalian homologues of Drs2p include the chromaffin granule ATPase II (now called ATP8A1) (Tang et al., 1996), which is likely responsible for a PS translocase activity observed with these exocytic vesicles (Zachowski et al., 1989), and FIC1 (ATP8B1), for which mutations in humans cause an impairment of bile flow through the liver (cholestasis) (Bull et al., 1998; Klomp et al., 2004). In addition, deletions removing the mouse Atp10c gene cause diet-induced obesity and type 2 diabetes phenotypes (Dhar et al., 2004). P4-ATPases are also agriculturally important, because they are required for pathogenesis of the rice blast fungus Magneporthe griseus (Balhadere and Talbot, 2001; Gilbert et al., 2006) and growth of plants at cold temperatures (Gomes et al., 2000).
The yeast Drs2p family of P4-ATPases, including Neo1p, Dnf1p, Dnf2p, and Dnf3p, are all involved in protein transport in the secretory and endocytic pathways, but at different stages (Graham, 2004). Drs2p and the Dnf proteins form an essential group, and at least one of these proteins must be present to sustain yeast viability. The Drs2/Dnf P4-ATPases have both overlapping and nonoverlapping functions in protein transport (Hua et al., 2002). Strains carrying a deletion of DRS2 (drs2Δ) are viable but strongly cold sensitive for growth, and they exhibit defects in forming one of the two classes of exocytic vesicles targeted to the plasma membrane (Gall et al., 2002). The drs2Δ mutant also exhibits defects in protein trafficking between the TGN and early endosome that are comparable with clathrin mutant phenotypes (Chen et al., 1999; Hua et al., 2002). Thus, the Dnf ATPases cannot compensate for loss of Drs2p in these pathways; moreover, deletion of all three DNF genes does not perturb these Drs2p-dependent pathways. Dnf1p and Dnf2p are 69% identical in amino acid sequence, localize to the plasma membrane and internal membranes (TGN, early endosomes, and transport vesicles), and have redundant functions in the internalization step of endocytosis (at cold temperatures) and an early endosome to TGN transport pathway traveled by the Snc1p soluble N-ethylmaleimide-sensitive factor attachment protein receptor (SNARE) (Hua et al., 2002; Pomorski et al., 2003). Drs2p is also required for Snc1p recycling, suggesting that Drs2p and Dnf1,2p are partially redundant in the this pathway. In addition, Drs2p and Dnf1p have redundant functions in the adaptor protein (AP)-3–dependent transport of alkaline phosphatase from the TGN directly to the vacuole. The individual drs2Δ or dnf1Δ mutants show little to no defect in the AP-3 pathway, whereas this pathway is blocked in the drs2Δ dnf1Δ double mutant (Hua et al., 2002). The mechanism for coupling a specific P4-ATPase to a specific protein transport pathway is unclear, but it likely involves translocation substrate specificity, unique protein interactions, and appropriate localization.
Localization of Drs2p to the Golgi requires interaction with the Cdc50p chaperone subunit for the Drs2p/Cdc50p complex to exit the ER (Saito et al., 2004). In cdc50Δ, Drs2p is retained in the ER, and these cells show protein transport defects at the TGN comparable with drs2Δ (Chen et al., 2006). Similarly, the Cdc50p homologue Lem3p (also called Ros3p) is required for transport of Dnf1p and presumably Dnf2p to the plasma membrane and so lem3Δ phenocopies dnf1Δ dnf2Δ (Saito et al., 2004). The Drs2p carboxyl-terminal cytosolic tail (C-tail) makes an essential contribution to its function, apparently by mediating protein interactions and/or TGN localization. Drs2p is linked to the vesicle budding machinery by a direct interaction between the ARF-GEF Gea2p and a short motif in the C-tail (called GIM, for Gea2p interaction Motif) (Chantalat et al., 2004). Adjacent to GIM, there is a region highly conserved among all, including mammalian, Drs2p homologues. Function of this conserved motif (CM) is still unknown, although a mutational analysis suggested that the CM is primarily responsible for the essential function of the C-tail (Chantalat et al., 2004). At the membrane distal end of the C-tail, there are two NPFX(1,2)D motifs (hereafter referred to as NPFXD), which are potential endocytosis signals (Tan et al., 1996; Howard et al., 2002).
In yeast, two types of endocytosis signals have been characterized that recruit membrane proteins into a clathrin/actin-based endocytic pathway for internalization from the plasma membrane: sequences that mediate phosphorylation and ubiquitination of cargo, such as PEST-like sequences, and the NPFXD motif (Tan et al., 1996; Roth et al., 1998; Rotin et al., 2000; Howard et al., 2002; Hicke and Dunn, 2003). The NPFXD signal is recognized by the Sla1p subunit of an endocytic coat complex consisting of clathrin, Pan1p, End3p, Sla2p/End4p (related to mammalian Hip1R), and Sla1p (related to mammalian CIN85 and intersectin) (Tang et al., 1997, 2000; Howard et al., 2002; Newpher et al., 2005; Kaksonen et al., 2006). Pan1p, a member of the Eps15 family of modular scaffolding proteins, interacts with the clathrin binding proteins AP180 and epsin, and it also binds to and stimulates the ARP2/3 complex (Wendland and Emr, 1998; Duncan et al., 2001; Aguilar et al., 2003). Therefore, Pan1p has the capacity to link adaptor-bound cargo proteins to clathrin-coated pits and sites of actin assembly. Pan1p, End3p, and actin assembly are required for both ubiquitin (Ub)-dependent and NPFXD-dependent endocytosis, although Sla1p is only required for endocytosis of cargo bearing the NPFXD signal (Howard et al., 2002; Miliaras et al., 2004). Drs2p not only has the potential to physically interact with the Sla1p/Pan1p/End3p complex but also it is functionally linked to this complex as drs2Δ is synthetically lethal with the temperature-conditional pan1–20 allele (Chen et al., 1999). However, the nature of these relationships between Drs2p and this endocytic complex is unclear.
Drs2p exhibits a steady-state localization to the TGN, although recent reports showed accumulation of Drs2p on the plasma membrane of a verprolin (vrp1) mutant and the presence of Drs2p in exocytic vesicles, suggesting that Drs2p transits the plasma membrane as part of its trafficking itinerary (Saito et al., 2004; Alder-Baerens et al., 2006). Dnf1p also seems to cycle between the exocytic and endocytic pathways (Hua et al., 2002; Saito et al., 2004). In this work, we further examined the trafficking itinerary of Drs2p and Dnf1p and tested whether the NPFXD motifs contribute to the function and localization of these P4-ATPases.
MATERIALS AND METHODS
Media and Strains
Yeast were grown in standard rich medium (YPD) or synthetic defined (SD) minimal media containing the required nutritional supplements (Sherman, 1991). Yeast transformations were performed using the lithium acetate method. Escherichia coli strains DH5α and XL1-Blue were used for plasmid construction and amplification.
Yeast strains used in this study are summarized in Table 1. The yeast knockout strain collection was originally purchased from Research Genetics (Huntsville, AL), which is now Resgen, Invitrogen (Carlsbad, CA). Strains carrying multiple disruptions were generated by standard genetic crosses and tetrad dissection. The genotype of each spore was determined by a PCR method as described by the Saccharomyces genome deletion project (http://sequence-www.stanford.edu/group/yeast_deletion_project/deletions3.html). Strains expressing Myc and HA tagged Dnf1p were generated by PCR-based targeting into BY4741 and BY4741 sla1Δ by using pPF6a-13Myc-HisMX6 or pPF6a-3HA-HisMX6 as the PCR template (Longtine et al., 1998). Transformants were selected on SD plates without histidine and the integrated tags were confirmed by PCR.
Table 1.
Yeast strains used in this study
| Strain | Genotype | Reference or source |
|---|---|---|
| BY4741 | MATa his3Δ1 leu2Δ0 ura3Δ0 met15Δ0 | Research Genetics |
| BY4742 | MATα his3Δ1 leu2Δ0 ura3Δ0 lys2Δ0 | Research Genetics |
| BY4741 YBL007C | BY4741 sla1Δ::KanMX6 | Research Genetics |
| KLY011 | MATα his3Δ1 leu2Δ0 ura3Δ0 lys2Δ0 sla1Δ::KanMX6 | This study |
| BY4742 YNL084C | BY4742 end3Δ::KanMX6a | Research Genetics |
| KLY201 | MATα his3Δ1 leu2Δ0 ura3Δ0 lys2Δ0 end3Δ::KanMX6 | This study |
| BY4742 YLR337C | BY4742 vrp1Δ::KanMX6 | Research Genetics |
| BY4741 YPR173C | BY4741 vps4Δ::KanMX6 | Research Genetics |
| BY4741 YNR006W | BY4741 vps27Δ::KanMX6 | Research Genetics |
| ZHY615D1C | MATa his3Δ1 leu2Δ0 ura3Δ0 lys2Δ0 drs2Δ::KanMX6 | Hua et al. (2002) |
| ZHY615M2D | MATα his3Δ1 leu2Δ0 ura3Δ0 lys2Δ0 drs2Δ::KanMX6 | Hua et al. (2002) |
| ZHY2149D | MATα his3Δ1 leu2Δ0 ura3Δ0 lys2Δ0 drs2Δ::KanMX6 dnf1Δ::KanMX6 | Hua et al. (2002) |
| KLY041 | BY4742 DNF1::13×MYC | This study |
| KLY054 | KLY011 DNF1::13×MYC | This study |
| KLY035 | MATα his3Δ1 leu2Δ0 ura3Δ0 drs2Δ::KanMX6 sla1Δ::KanMX6 | This study |
| SEY6210 | MATα leu2-3,112 ura3-52 his3-Δ200 trp1-Δ901 lys2-801 suc2-Δ9 | Robinson et al. (1988) |
| SEY6211 | MATΔ leu2-3,112 ura3-52 his3-Δ200 trp1-Δ901 ade suc2-Δ9 | Robinson et al. (1988) |
| SEY6210 drs2Δ | SEY6210 drs2Δ::TRP1 | Chen et al. (1999) |
| TGY1907 | MATΔ leu2-3,112 ura3-52 his3-Δ200 trp1-Δ901 suc2-Δ9 pan1-20 | Chen et al. (1999) |
| TGY1906 | MATa leu2-3,112 ura3-52 his3-Δ200 trp1-Δ901 suc2-Δ9 pan1-20 | Chen et al. (1999) |
| TGY1912 | MATa leu2-3,112 ura3-52 his3-Δ200 trp1-Δ901 lys2-801 suc2-Δ9 end4-1 | Chen et al. (1999) |
| ZHY823 | MATα leu2-3,112 ura3-52 his3-Δ200 trp1-Δ901 suc2-Δ9 pan1-20 drs2Δ::TRP1 (pRS416-DRS2) | This study |
| CCY2808 | MATa leu2 ura3-52 his3 trp1-Δ901 ade2 ade3 arf1Δ::HIS3 drs2-2 (pRS416-DRS2) | Chen and Graham (1998) |
| YJF1165 | MATa trp1-901 leu2-3,112 ura3-52 his3-200 gal4Δ gal80Δ LYS2::GAL1-HIS3 GAL2-ADE2 met2::GAL7-lacZ | James et al. (1996) |
aAn extragenic suppressor mutation was found in this strain.
Plasmid Construction
Plasmids used in this study are listed in Table 2. To generate pGBT9-Drs2CT used in the two-hybrid test, a BamHI fragment from pDHS279 (Chantalat et al., 2004) containing the Drs2p C-tail (amino acids 1230–1355) was cloned into the pGBT9 BamHI site, and the orientation was confirmed by PCR. PCR products used to generate DRS2 C-tail truncation plasmids pRS315-Drs2-ΔCT, pRS315-Drs2-ΔEnd, pRS315-Drs2-ΔNPF2, pRS315-Drs2-ΔNPF, and pRS315-Drs2-Δ1274 were produced using a forward primer (CAGCTGATATAGCTCTTGG) that anneals 5′ of an endogenous NcoI site in DRS2 and reverse primers with a stop codon and MluI site added to the 3′ end. PCR products were then used to replace the NcoI/MluI region of pRS315-DRS2. The DRS2 internal deletion mutant pRS315-Drs2-ΔCM was generated from the truncation plasmid pRS315-Drs2-Δ1274 with the C-terminal sequence added as a MluI/SalI PCR fragment. A megaprimer PCR method (Barik and Galinski, 1991) was used to introduce point mutations into the MscI/SalI fragment of the DRS2 gene to produce plasmids pRS315-Drs2-NPW1 and pRS315-Drs2-NPW2. Using similar methods, pRS315-Drs2-ΔGIM-NPW1,2 was generated from pSC33 (pRS315-Drs2-ΔGIM). Sequencing of the resulting plasmids indicated that the specific mutations were introduced with no additional mutations. All other clones generated from the PCR fragments described below were also sequenced for confirmation.
Table 2.
Plasmids used in this study
| Plasmid | Description | Reference or source |
|---|---|---|
| pRS416 GFP-SNC1 | GFP-SNC1 URA3 CEN | Lewis et al. (2000) |
| pGO41 | GFP-ALPURA32μ | Cowles et al. (1997) |
| pRS426-KEX2-GFP | KEX2-GFP URA3 2μ | Gift from Tom Vida |
| pSM1493 | STE6-GFPURA32μ | Kelm et al. (2004) |
| pRS315-DRS2 | DRS2 LEU2 CEN | Chen et al. (1999) |
| pDRS2(D560N) | drs2(D560N) LEU2 CEN | Chen et al. (1999) |
| pRS315-Drs2-ΔCT | drs2-Δ1259-1355 LEU2 CEN | Chantalat et al. (2003) |
| pRS315-Drs2-ΔEnd | drs2-Δ1337-1355 LEU2 CEN | Chantalat et al. (2003) |
| pRS315-Drs2-ΔNPF2 | drs2-Δ1315-1355 LEU2 CEN | Chantalat et al. (2003) |
| pRS315-Drs2-ΔNPF | drs2-Δ1310-1355 LEU2 CEN | Chantalat et al. (2003) |
| pRS315-Drs2-ΔCM | drs2-Δ1274-1283 LEU2 CEN | Chantalat et al. (2003) |
| pSC33 [pRS315-Drs2-ΔGIM] | drs2-Δ1250-1270 LEU2 CEN | Chantalat et al. (2003) |
| pRS315-Drs2-ΔGIM-ΔNPF | drs2-Δ1250-1263, Δ1310-1355 LEU2 CEN | This study |
| pRS315-Drs2-NPW1 | drs2(F1313W) LEU2 CEN | This study |
| pRS315-Drs2-NPW2 | drs2(F1335W) LEU2 CEN | This study |
| pRS315-Drs2-NPW1,2 | drs2(F1313W,F1335W) LEU2 CEN | This study |
| pRS315-Drs2-ΔGIM-NPW1,2 | drs2-Δ1250-1270, F1313W, F1335W LEU2 CEN | This study |
| pGBT9-Drs2-CT | DRS2(aa 1230-1355) TRP1 2μ | This study |
| pZH429 [pGBT9-Drs2-CTΔNPF] | DRS2(aa 1230-1310) TRP1 2μ | This study |
| pGBT9-Drs2-CT-NPW1,2 | DRS2(aa 1221-1355, F1313W,F1335W) TRP1 2μ | This study |
| pGBT9-Drs2-CTΔGIM | DRS2(aa 1221-1355, Δ1250-1263) TRP1 2μ | This study |
| pPAN1.1 [Pan1-EH] | PAN1(aa 96-713) TRP1 2μ | Wendland and Emr (1998) |
| pGAD-SLA1-555 [Sla1-SHD1] | SLA1(aa 471-555) LEU2 2μ | Gift from Gregory Payne lab |
| pGAD-SLA1charged [Sla1-Charged] | SLA1(aa 511-855) LEU2 2μ | Gift from Gregory Payne lab |
| pRS313-DNF1 | DNF1 HIS3 CEN | This study |
| pRS313-Dnf1-NAI | DNF1(PF147,148AI) HIS3 CEN | This study |
| PGOGFP | CPY5′UTR-GFP URA3 2μ | Cowles et al. (1997) |
| pRS416-GFP | CPY5′UTR-GFP URA3 CEN | This study |
| pGFP-DRS2 | CPY5′UTR-GFP-DRS2 URA3 CEN | This study |
| pGFP-Drs2-ΔNPF2 | CPY5′UTR-GFP-drs2Δ1333-1355 URA3 CEN | This study |
| pGFP-Drs2-ΔNPF | CPY5′UTR-GFP-drs2Δ1310-1355 URA3 CEN | This study |
| pGFP-Drs2-NPW1,2 | CPY5′UTR-GFP-drs2(F1313W,F1335W) URA3 CEN | This study |
| pGFP-Drs2-ΔN2 | CPY5′UTR-GFP-drs2Δ1-72 URA3 CEN | This study |
| pGFP-Drs2-ΔN3 | CPY5′UTR-GFP-drs2Δ1-103 URA3 CEN | This study |
| pGFP-Drs2-ΔN3-NPW1,2 | CPY5′UTR-GFP-drs2Δ1-103 (F1313W,F1335W) URA3 CEN | This study |
| pRS425-CDC50 | CDC50 LEU2 2μ | Chen et al. (2006) |
| pGFP-CDC50 | CPY5′UTR-GFP-CDC50 URA3 CEN | This study |
The full-length DNF1 gene was cloned by PCR amplification using primers JN01F (CTATGTAATCACCTACTTCCC) and GR02R (CTGGAGTGCTACATGAGCC) and subcloned into pRS416 after treating both the vector and PCR product with SpeI and HindIII. The SpeI/XhoI fragment of pRS416-DNF1 was inserted into SpeI/XhoI site of pRS313 to produce pRS313-DNF1. Site-directed mutagenesis of DNF1 to produce pRS313-Dnf1-NAI was carried out in plasmid pRS313-DNF1 by using the QuickChange protocol (Stratagene, La Jolla, CA).
For construction of GFP-DRS2, a 1.3-kilobase (kb) SpeI/ClaI fragment from pGOGFP (Cowles et al., 1997) consisting of the PRC1 promoter and GFP(S65T) was inserted into pRS416 to generate pRS416-GFP. The plasmid pRS315-DRS2 was used as a PCR template with primers SalI-Drs2-F (ACGTAGTCGACAATGACGACAGAGAAACCCCC) and Drs2-CT-R (CCCCTCGAGGTCGACGGTA) to generate a 3.7-kb fragment that placed SalI sites at both the start and end of the DRS2 coding region. This fragment was subcloned into SalI site of pRS416-GFP, creating the plasmid pGFP-DRS2. To eliminate mutations produced by PCR, most of the DRS2 coding sequence in pGFP-DRS2 was further replaced by an AgeI/ClaI fragment from pRS315-DRS2. This form of GFP-DRS2 fully complemented the cell growth defect of drs2Δ at 20°C. To generate C-terminal tail mutated GFP-DRS2 (pGFP-Drs2-ΔNPF2, pGFP-Drs2-ΔNPF, or pGFP-Drs2-NPW1,2), PCR amplifications of different regions of DRS2 C terminus were used to replace the NheI/ClaI region of pGFP-DRS2. To generate N-terminal–truncated GFP-DRS2 (pGFP-Drs2-ΔN2 or pGFP-Drs2-ΔN3), primers Drs2ΔN2F (GATGAGATCTCATGAAAATCTATTTATGAGCAAT) or Drs2ΔN3F (GACTGAGATCTCGAGCAGTCAAGCCTCCC) were used with Drs2NR (GAACCACAGTTGGGGTATCAG) to produce fragments to replace the BglII region of pGFP-DRS2. The 1.4-kb NheI/ClaI fragment of pGFP-Drs2-NPW1,2 was used to replace the corresponding sequence in pGFP-Drs2-ΔN3 to generate pGFP-Drs2-ΔN3-NPW1,2. The PRC1 promoter is stronger than the DRS2 promoter, and so to avoid accumulation of GFP-Drs2p in the ER (Saito et al., 2004), we cotransformed yeast strains with a multicopy vector carrying CDC50 (pRS425-CDC50).
To generate pGFP-CDC50, pRS315-CDC50 was used as template with primers CDC50KpnIF (CGGTACCGTTTCATTGTTCAAAAGAGGTA) and CDC50KpnIR (CGGTACCCACAAATACCTACAGGCACTA) to produce a 1.2-kb fragment with KpnI sites at both ends of the CDC50 coding region. The fragment was subcloned into the KpnI site of pRS416-GFP.
Microscopy
Cells were observed using an Axioplan microscope (Carl Zeiss, Thornwood, NY). Fluorescent images were captured with a charge-coupled device camera and processed with MetaMorph 4.5 software (Molecular Devices, Sunnyvale, CA). To visualize green fluorescent protein (GFP)-tagged proteins, cells were grown to early mid-logarithmic phase, harvested, and resuspended in imaging buffer (10 mM Tris-HCl, pH 7.4, and 2% glucose). Cells were mounted on glass slides and observed immediately using a GFP (green) bandpass filter set.
To study the kinetics of GFP-Drs2p transport to the plasma membrane, mid-log phase cells were collected and resuspended in SD medium containing 200 μM latrunculin A. Samples of cells were harvested at different time points and imaged. To label endosomes, we incubated cells in ice-cold SD medium containing 10 μg/ml FM4-64 (Invitrogen) for 20 min. Cells were washed twice with ice-cold medium without FM4-64 and then incubated for 30 min at 30°C before microscopic examination.
Subcellular Fractionation and Immunological Methods
For subcellular fractionation experiments, ∼25 OD600 units of each strain were grown to an OD600 of 0.5–1.0. The cells were harvested and converted to spheroplasts in HB buffer (1.4 M sorbitol, 50 mM KPi, pH 7.5, 10 mM NaN3, 10 mM NaF, and 40 mM β-mercaotpethanol), by using 200 μg/ml Zymolyase 100T (MP Biomedicals, Irvine, CA) at 30°C for 30 min. The spheroplasts were washed twice with HB buffer and lysed by resuspension in triethanolamine (TEA) lysis buffer (0.5 M sorbitol, 25 mM TEA, pH 8.0, and 1 mM EDTA) containing 1X Complete protease inhibitor cocktail (PIC) lacking EDTA (Roche Diagnostics, Basel, Switzerland). The extract was centrifuged at 400 × g for 5 min, and the resulting supernatant was centrifuged at 13,000 × g for 15 min in a refrigerated microcentrifuge. After each centrifugation step, the supernatant was transferred to a separate tube, and the pellet was resuspended in an equal volume of TEA lysis buffer supplemented with PIC. SDS/urea buffer was added to 1X (20 mM Tris-HCl, pH 6.8, 4 M urea, 0.05 mM EDTA, 0.5% β-mercaptoethanol, 2.5% SDS, and 0.125% bromphenol blue), and the samples were heated at 65°C for 10 min before electrophoresis.
Immunoblotting and immunofluorescence experiments were performed as described previously (Chen et al., 1999). The 9E10 mouse monoclonal c-Myc antibody (Oncogene Research Products, Darmstadt, Germany) was used at 1:2000 for Western blot and 1:100 for immunofluorescence. Polyclonal rabbit anti-Pma1p antibody was a gift from Amy Chang (University of Michigan, Ann Arbor, MI), and was used at 1:1000 to detect Pma1p by Western blot. Polyclonal rabbit anti-G6PDH antibody (Sigma-Aldrich, St. Louis, MO) was used at 1:10,000 dilution. Alexa-594 goat anti-mouse IgG (Invitrogen) was used at 1:200 as secondary antibodies for immunofluorescence.
RESULTS
The C-tail Is Essential for Drs2p Function in Protein Transport
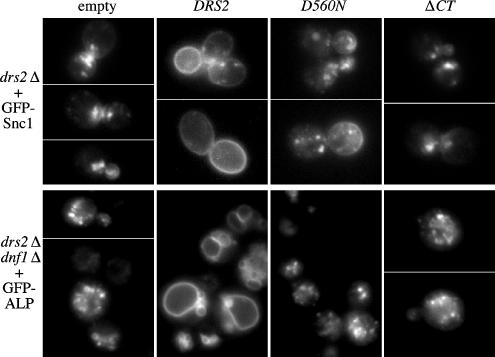
Previously, we had shown that the ATPase dead drs2-D560N allele and a drs2 allele bearing a truncation of the last 96 amino acids of the C-tail (drs2-ΔCT) could not complement the cold-sensitive growth defect of drs2Δ (Chen et al., 1999; Chantalat et al., 2004). To test whether these alleles could complement drs2Δ trafficking defects, we examined the localization of the exocytic vesicle SNARE Snc1p, which cycles between the plasma membrane, early endosomes, and the TGN (Lewis et al., 2000). In wild-type cells, although a small fraction of GFP-Snc1p localizes to punctate structures within the cell, GFP-Snc1p primarily localizes to the plasma membrane, concentrating in the bud or the regions of polarized growth. In contrast, drs2Δ cells carrying an empty plasmid exhibited very little GFP-Snc1p at the plasma membrane, and most of this fusion protein was found in internal structures, which may be either early endosomes or the TGN (Figure 1, empty; Hua et al., 2002). Introduction of a plasmid bearing wild-type DRS2 restored normal plasma membrane localization of GFP-Snc1p, but neither the drs2-D560N mutant nor the drs2-ΔCT allele was able to restore normal localization of GFP-Snc1p (Figure 1; D560N, ΔCT).
Figure 1.
ATPase activity and the C-tail region are essential for Drs2p function in protein trafficking. (A) Requirement for Drs2p ATPase activity and the C-tail region in Snc1p-GFP recycling. Plasmids pRS315 (empty), pRS315-DRS2 (DRS2), pDRS2-D560N (D560N, ATPase dead), and pRS315-Drs2-ΔCT (ΔCT) were introduced into strain ZHY615M2D (drs2Δ) along with pRS416-GFP-SNC1(GFP-Snc1). Transformants were grown at 30°C to mid-log phase and examined by fluorescence microscopy. Snc1-GFP is at the plasma membrane of DRS2 (wild-type) cells and is trapped in internal membranes in the drs2 mutants. (B) Requirement for Drs2p ATPase activity and the C-tail region in ALP transport to the vacuole. The same DRS2 plasmids as in A were cotransformed into strain ZHY2149D (drs2Δdnf1Δ) with pGO41 (GFP-ALP). Cells were grown at 30°C to mid-log phase, shifted to 15°C for 2 h, and then imaged. Fluorescent rings in the DRS2 (wild-type) cells are vacuoles, whereas GFP-ALP was mislocalized to extravacuolar puncta in drs2dnf1 mutants.
We also tested whether these two drs2 mutant alleles could complement the alkaline phosphatase (ALP) trafficking defect shown by drs2Δ dnf1Δ cells. Drs2p and Dnf1p have redundant functions in the transport of GFP-ALP from the TGN to the vacuole (Hua et al., 2002), a pathway mediated by AP-3–coated vesicles (Cowles et al., 1997). In wild-type cells, GFP-ALP primarily localizes to the vacuole membrane, and one to three punctate structures outside of the vacuole in a small percentage of cells. In drs2Δ dnf1Δ cells, however, most of the GFP-ALP localizes to extravacuolar puncta (Figure 1, empty; Hua et al., 2002). Wild-type DRS2 complemented this phenotype, but drs2-ΔCT and drs2-D560N failed to restore the normal vacuolar GFP-ALP localization pattern (Figure 1; D560N, ΔCT). These results indicate that both the ATPase activity and the C-tail are critical for Drs2p function in protein trafficking from the TGN.
Functional Requirement for the Drs2p NPFXD Motifs
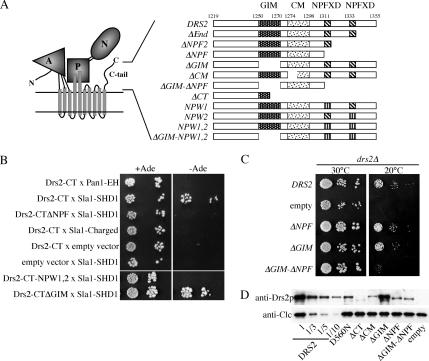
Three motifs have been mapped within the Drs2 C-tail thus far (Figure 2A): the Gea2p interaction motif (GIM), a highly conserved motif (CM), and the two NPFXD motifs, which could potentially interact with the Sla1p homology domain 1 (SHD1) of Sla1p (Howard et al., 2002). A two-hybrid analysis was done to test for an interaction between the Drs2p C-tail and the Sla1p SHD1 domain. The Eps15 homology (EH) domain of Pan1p interacts with NPF motifs (Wendland and Emr, 1998), and so the Pan1-EH domain was also tested for interaction with the Drs2p C-tail. The Drs2p C-tail gave a positive two-hybrid interaction with the Sla1p SHD1 domain but did not interact with the Pan1p EH domain (Figure 2B). The Drs2p C-tail also failed to interact with a fragment of Sla1p containing the SHD2 domain and a number of charged amino acids (Figure 2B, Sla1-charged). Deletion of the C-terminal region containing the two NPFXD motifs abolished the two-hybrid interaction as did mutating both NPFXDs to NPWXD (Drs2-CTΔNPF and Drs2-CT-NPW1,2). The F-to-W mutation was previously shown to disrupt the two-hybrid interaction of Sla1p with the NPFXD motif of Kex2p and to disrupt the ability of this motif to serve as an endocytosis signal (Howard et al., 2002). In contrast, deletion of the C-tail GIM sequence had no effect on the interaction (Drs2-CTΔGIM). These data indicate that the interaction between Drs2p and Sla1p is mediated by the NPFXD motifs.
Figure 2.
C-tail sequences containing the NPFXD motifs bind Sla1p and contribute to Drs2p function. (A) Predicted topology and domain structure of Drs2p based on the crystal structure of the related sarcoplasmic reticulum Ca++ ATPase (A, actuator; P, phosphorylation; N, nucleotide binding) (Toyoshima and Inesi, 2004). Schematic diagram of motifs in the Drs2p C-tail and constructs used in this study (GIM and CM). (B) Two-hybrid test for interaction between Drs2p C-tail and Sla1p SHD1 domain. Bait and prey plasmids used are described in Table 2. Serial dilutions of the cells were spotted on minimal medium with or without adenine (Ade). Growth in the absence of adenine indicates a two-hybrid interaction. (C) Synergistic defect caused by deleting both the GIM and C-terminal 44 amino acids bearing the two NPFXD motifs. Serial dilutions of drs2Δ strains (ZHY615M2D) expressing the indicated constructs were tested for their ability to grow at 20°C. DRS2 is the wild-type gene and “empty” received the vector without an insert, thus showing the drs2Δ growth phenotype. (D) Western blot of cell lysates probed for Drs2p and clathrin light chain (Clc1p). Lysates were normalized for cell equivalents and compared with a dilution series from cells expressing wild-type DRS2.
As shown previously, deletion of the conserved motif (ΔCM) partially abrogated the ability of this drs2 allele to complement the cold-sensitive growth defect of drs2Δ, while deletion of both GIM and CM abolished Drs2p function. In contrast, deletion of C-terminal sequences containing the NPFXD motifs (ΔNPF) did not appear to perturb Drs2p function or exacerbate the defect caused by ΔCM (Chantalat et al., 2004). These data initially suggested that the NPFXD motifs did not contribute to the essential function of the Drs2p C-tail. However, while neither deletion of the NPFXD motifs (ΔNPF) nor deletion of GIM (ΔGIM) perturbed complementation of the drs2Δ cold-sensitive growth defect, the ΔGIM-ΔNPF double mutant failed to complement (Figure 2C). These results indicate that the interaction with ARF-GEF and the C-terminal 44 residues bearing the two NPFXD motifs make important contributions to Drs2p function in vivo.
To determine how mutations of the C-tail affect expression of Drs2p, we performed a Western blot with whole cell lysates from the strains indicated in Figure 2C. Most of the mutants were expressed at lower levels than wild-type Drs2p (DRS2), with ΔCT being most affected at significantly <10% expression (Figure 2D). However, we have previously shown that coexpression of ΔCT and the ATPase dead D560N allele from two separate plasmids complements the cold-sensitive (cs) growth defect of drs2Δ, which indicates that this small amount of ΔCT provides sufficient ATPase activity to support Drs2p function reasonably well (Chantalat et al., 2004). Importantly, all other C-tail mutant proteins were more stable than ΔCT. Therefore, whereas protein stability might be a factor that influences the ability of C-tail mutants to complement drs2Δ, each mutant should supply sufficient Drs2p ATPase activity for in vivo function, and loss of specific sequence within the Drs2p C-tail is primarily responsible for reduced Drs2p function. Particularly relevant is the observation that ΔNPF and ΔGIM-ΔNPF are expressed at a similar level, but the ΔGIM-ΔNPF double mutant is much more defective than either single mutant (Figures 2, C and D).
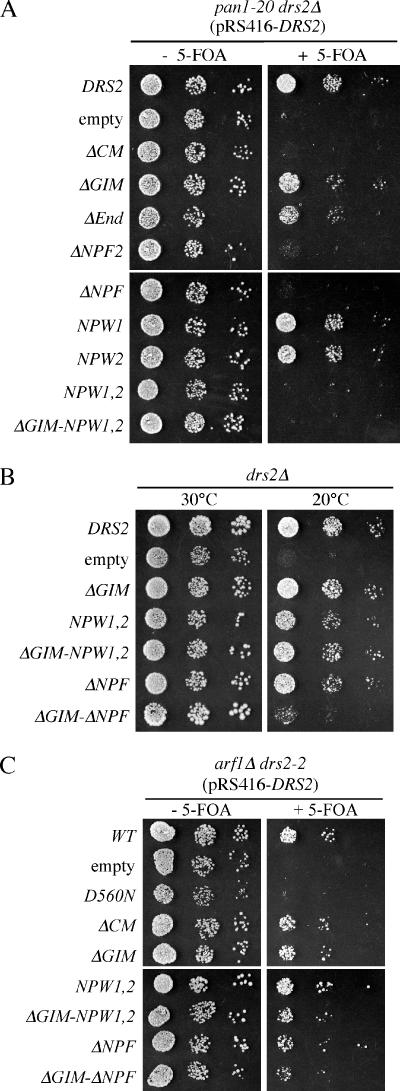
Moreover, we were surprised to find that deletion of even one NPFXD motif was sufficient to cause synthetic lethality with pan1-20 (Figure 3A). For this experiment, viability of a drs2Δ pan1-20 strain was maintained by the presence of wild-type DRS2 on a URA3-based plasmid (pRS416-DRS2). Various mutant drs2 alleles were introduced into this strain on LEU2 plasmids and cells capable of losing the wild-type DRS2-URA3 plasmid were selected on medium containing 5-fluoroorotic acid (5-FOA). Growth in the absence of 5-FOA shows the phenotype of the pan1-20 single mutant, whereas the 5-FOA plate shows the drs2 pan1-20 double mutant phenotype. The drs2Δ (empty) and ΔCM alleles are synthetically lethal with pan1-20, although the ΔGIM allele is not. Deletion of the C-terminal 22 residues (ΔEnd) had little effect on Drs2p function by this assay, but constructs bearing additional deletions removing one (ΔNPF2) or both NPFXDs (ΔNPF) failed to support growth of a pan1-20 drs2Δ strain. To better define the role of the NPFXDs in this genetic interaction, we mutated them to NPWXD. In this case, each individual drs2-NPW allele supported growth of the drs2Δ pan1-20 strain (NPW1 and NPW2), but the drs2-NPW1,2 double mutant (NPW1,2) failed to complement the drs2Δ pan1-20 synthetic lethality (Figure 3A). This allele-specific genetic interaction indicates that a yeast strain compromised for Pan1p activity relies on an NPFXD-dependent function of Drs2p to sustain life.
Figure 3.
Mutation of Drs2p NPFXD motifs causes synthetic lethality with pan1-20. (A) Serial dilutions of ZHY823 (drs2Δ pan1-20 pRS416-DRS2) expressing the indicated constructs from LEU2-based plasmids were tested for growth at 30°C on medium with or without 5-FOA. Failure to grow on the +5-FOA medium indicates synthetic lethality between pan1-20 and the drs2 allele expressed from the LEU2 plasmid. (B) The experiment shown in Figure 2C was repeated to include ΔGIM combined with the NPW point mutations. (C) The drs2-npw1,2 allele is not synthetically lethal with arf1Δ. Serial dilutions of CCY2808 (arf1Δ drs2-2 pRS416-DRS2) expressing the indicated constructs from LEU2-based plasmids were tested for growth at 30°C on medium with or without 5-FOA.
Because the ΔNPF2 C-terminal truncation showed a stronger phenotype in the pan1-20 synthetic lethality test than the NPW2 point mutation, we considered the possibility that other sequences in the C-terminal 44 residues contribute to Drs2p function independently of the NPFXDs. This possibility was tested by combining the ΔGIM and NPW1,2 mutations (ΔGIM-NPW1,2) and comparing the ability of this new allele to complement drs2Δ relative to the alleles used in Figure 2C. As shown in Figure 3B, the ΔGIM-NPW1,2 allele complemented the cs growth defect of drs2Δ, whereas ΔGIM-ΔNPF again failed to complement. These data suggest that C-terminal 44 amino acids have an NPFXD-independent function that acts redundantly with the GIM. Alternatively, it is possible that the mutant NPW motif retains some function.
To further test whether the genetic interaction is specific to pan1-20, we also examined the synthetic lethality between drs2 mutants and arf1Δ (Figure 3C). As expected, the ATPase dead drs2-D560N allele failed to support growth of a drs2–2 arf1Δ strain. The drs2-ΔCT allele causes slow growth when combined with arf1Δ (data not shown), but drs2 alleles carrying deletions of CM, GIM, or NPFXD motifs do not substantially perturb growth of arf1Δ cells. Therefore, the synthetic lethal interaction between drs2-npf alleles and pan1-20 is specific.
Functional Requirement for the Dnf1p NPFXD Motif
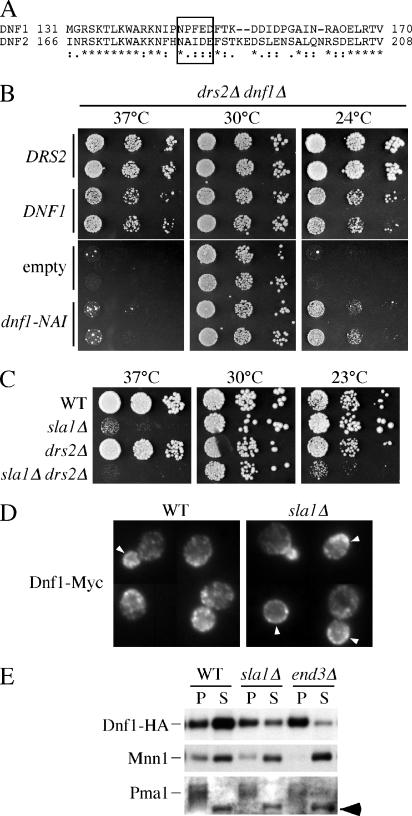
Interestingly, Dnf1p contains an NPFXD within its N-terminal cytosolic tail (N-tail) that is not present in the closely related Dnf2p. In addition, Dnf1p, but not Dnf2p, has redundant functions with Drs2p at the TGN, suggesting that the NPFXD-dependent endocytosis of Dnf1p may be required for its Golgi function. To test whether the NPFXD is important for Dnf1p function, we mutated NPF to NAI, the sequence found in Dnf2p (Figure 4A). Because there is no significant phenotype associated with deleting DNF1 alone, we tested the dnf1-NAI allele for complementation of growth defects associated with the drs2Δ dnf1Δ double mutant. On minimal medium, drs2Δ dnf1Δ grows well at 30°C but is strongly cs and ts for growth (Figure 4B, empty). Transformation with wild-type DRS2 completely complements these growth defects (Figure 4B, DRS2), showing the robust growth of a dnf1 single mutant. Transformation with wild-type DNF1 allowed growth at 24 and 37°C, although these drs2Δ DNF1 cells grew more slowly than the DRS2 dnf1Δ cells. In contrast, dnf1-NAI weakly complemented the cs growth defect and failed to complement the ts growth defect. Therefore, the NPFXD motif plays an important role in the ability of Dnf1p to compensate for the loss of Drs2p.
Figure 4.
An NPFXD/Sla1p interaction contributes to Dnf1p function and endocytosis. (A) Sequence alignment of Dnf1p and Dnf2p in the region surrounding the Dnf1p NPFXD motif. (B) Plasmids bearing wild-type DRS2 (pRS315-DRS2), DNF1 (pRS313-DNF1), empty vector, and NPF to NAI mutated DNF1 (pRS313-Dnf1-NAI) were introduced into drs2Δdnf1Δ (ZHY2149D). Cell growth on minimal medium at 37, 30, and 24°C was examined. (C) Growth of wild-type (BY4742), sla1Δ (KLY011), drs2Δ (ZHY615M2D), and sla1Δ drs2Δ (KLY035) was examined at 37, 30, and 23°C. (D) Fluorescence microscopy of wild-type and sla1Δ cells expressing Dnf1-Myc and stained with a mouse monoclonal anti-Myc antibody. Arrowheads indicate regions of labeled plasma membrane. (E) Distribution of Dnf1p-HA between the plasma membrane and internal membranes. Wild-type, sla1Δ, and end3Δ cells expressing HA-tagged Dnf1p were osmotically lysed and centrifuged at 400 × g to clear the cell debris. The supernatants were subsequently centrifuged at 13,000 × g for 15 min to generate P and S fractions. Samples from each fraction were immunoblotted for Dnf1p-HA, the plasma membrane H+-ATPase (Pma1p) and the Golgi protein Mnn1p. The arrow indicates a background protein that distributes in the S fraction.
If the important role of the Dnf1p NPFXD is for Sla1p-dependent endocytosis, then a drs2Δ sla1Δ mutant should show a more severe growth defect than drs2Δ, similar to what was observed for drs2Δ dnf1-NAI. Deletion of SLA1 alone causes a ts growth defect; however, we could detect a slightly more severe growth defect of sla1Δ drs2Δ at 37°C (Figure 4C). However, the sla1Δ single mutant grows well at low temperatures, and we tested whether sla1Δ would exacerbate the cs growth of drs2Δ by using a slightly lower, more restrictive temperature than used in Figure 4B. Indeed, drs2Δ sla1Δ grew much more slowly at 23°C than drs2Δ or sla1Δ. Even though sla1Δ may have pleiotropic effects on trafficking of several proteins, the uniquely strong cs growth defect of drs2Δ dnf1Δ suggests the effect of sla1Δ we are scoring in this assay is reduced Dnf1p function.
NPFXD-dependent Endocytosis of Dnf1p and Drs2p
To directly determine whether the endocytosis of Dnf1p was dependent on Sla1p, the localization of Dnf1p-Myc was examined in both wild-type and sla1Δ cells by indirect immunofluorescence (Figure 4D). In wild-type cells, Dnf1p-Myc is localized to both the plasma membrane and internal membranes with a polarized distribution, which could be transient endocytic and/or exocytic vesicles. Dnf1p is concentrated at the emerging bud site, small buds, and the mother-daughter neck of dividing cells (Hua et al., 2002; Pomorski et al., 2003; Figure 4D). Relative to wild-type cells, sla1Δ cells showed an accumulation of Dnf1p-Myc on the plasma membrane, although the change in the localization pattern was subtle by this method. To more quantitatively address the distribution of Dnf1p, a fractionation approach was used (Figure 4E). Cells expressing HA-tagged Dnf1p were converted to spheroplasts, osmotically lysed, centrifuged at 400 × g to pellet unlysed cells and large membranes, and then centrifuged at 13,000 × g to produce pellet (P) and supernatant (S) fractions. In wild-type cells, most Dnf1p-HA was found in the S fraction, which was relatively devoid of plasma membrane as judged by the distribution of the plasma membrane H+-ATPase Pma1p. However, with sla1Δ and end3Δ, the amount of Dnf1p-HA in the S fraction was diminished with a concomitant increase in the plasma membrane P fraction. As a control, the distribution of an integral membrane glycoprotein of Golgi complex Mnn1p was examined, and no difference was observed between wild-type, sla1Δ, and end3Δ cells. Moreover, the HA-tagged Dnf1-NAI mutant showed a 1.4-fold increase in the pellet fraction relative to wild-type HA tagged Dnf1p (our unpublished data). The end3Δ strain showed a more substantial redistribution of Dnf1p to the P fraction than sla1Δ or wild-type cells expressing Dnf1-NAI. In total, these experiments indicate that the NPFXD/Sla1p interaction significantly contributes to endocytosis of Dnf1p, but other endocytosis signals likely exist in this protein.
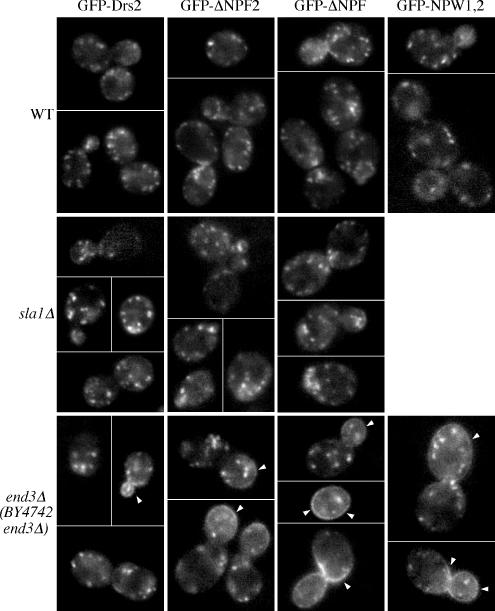
Even though Drs2p localizes to the TGN with Kex2p (Chen et al., 1999), the fact that Drs2p contains two NPFXD motifs suggests that it might travel to the plasma membrane and get rapidly endocytosed by the NPFXD/Sla1p pathway to maintain a steady-state TGN localization. If this is the case, we would expect to see accumulation of Drs2p on the cell surface when either the NPFXD motifs are mutated or SLA1 is deleted. To test this hypothesis, wild-type or NPFXD mutated GFP-Drs2 fusion proteins were expressed in either wild-type or sla1Δ cells (Figure 5). The GFP-DRS2 construct used here fully complements the drs2Δ cs growth defect and localizes appropriately to the TGN based on its colocalization with Sec7-RFP (Chen et al., 2006). We fused GFP to the N terminus of Drs2p to avoid interference with the potential trafficking signals near the C terminus. Surprisingly, neither truncation of the C-terminal region containing the two NPFXD motifs (GFP-ΔNPF) nor mutation of both NPFXD motifs to NPWXD (GFP-NPW1,2) caused accumulation of Drs2p on the plasma membrane of wild-type cells (Figure 5, WT). Similar results were obtained for sla1Δ cells, with neither the wild-type nor the ΔNPFXD GFP-Drs2p being mislocalized (Figure 5, sla1Δ).
Figure 5.
Drs2p does not rely on its NPFXDs and Sla1p for endocytosis unless the endocytic machinery is compromised. A series of plasmids harboring GFP-tagged wild-type (pGFP-DRS2) or NPFXD mutated drs2 alleles (pGFP-ΔNPF2, pGFP-ΔNPF, pGFP-NPW1,2) were cotransformed with pRS425-CDC50 into wild-type (BY4742), sla1Δ (KLY011), or end3Δ (BY4742 YNL084C) cells. Transformants were grown to early log phase at 30°C and imaged by fluorescence microscopy at room temperature. Arrowheads indicate plasma membrane fluorescence.
To test whether Drs2p trafficked to the plasma membrane and was retrieved by endocytosis signals other than NPFXD motifs, we expressed GFP-Drs2p in cells carrying a disruption of END3 (end3Δ), which should elicit an efficient block in endocytosis of all proteins that transit the cell surface. Only a modest amount of wild-type GFP-Drs2p was trapped on the end3Δ plasma membrane, most noticeably in small buds of a small percentage of cells (arrowheads in Figure 4, end3Δ and GFP-Drs2). Surprisingly, we found that deletion of one NPFXD motif (GFP-ΔNPF1) caused substantial accumulation of Drs2p on the end3Δ cell surface. Further truncation to remove the second NPFXD motif (GFP-ΔNPF) exacerbated this phenotype. Mutation of both NPFXDs to NPWXDs also resulted in accumulation of GFP-Drs2p on the end3Δ plasma membrane (Figure 5).
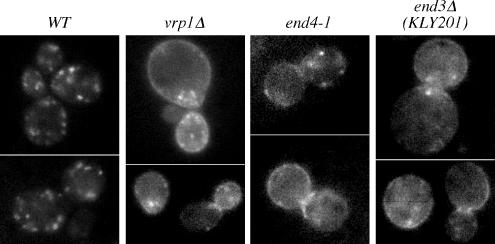
These results were unexpected and suggested that Drs2p did not normally travel to the plasma membrane, but deletion of the NPFXD motifs caused mislocalization of Drs2p to the plasma membrane where it could be trapped behind the end3 block. This was, however, contradictory to a published report showing Drs2p-GFP accumulates on the plasma membrane upon deletion of verprolin (vrp1Δ), a protein required for proper organization of cortical actin patches and the internalization step of endocytosis (Saito et al., 2004). In addition, NPFXD-mediated endocytosis reportedly requires End3p function (Tan et al., 1996). To resolve these discrepancies, we examined localization of GFP-Drs2 in vrp1Δ as well as end4-1 (also known as sla2), another endocytosis mutant. GFP-Drs2 accumulated at the plasma membrane of both vrp1Δ and end4-1 cells (Figure 6), in agreement with previously published vrp1Δ data (Saito et al., 2004). This result led us to suspect that the end3Δ strain from the yeast knockout collection contained an extragenic suppressor that specifically restored function of the NPFXD/Sla1p pathway. By backcrossing the original end3Δ strain to wild-type cells, new end3Δ strains were isolated that exhibited a tighter temperature-sensitive growth phenotype. Using a backcrossed end3Δ strain (KLY201), wild-type GFP-Drs2 was readily detected on the plasma membrane (Figure 6).
Figure 6.
Localization of GFP-tagged Drs2p to the plasma membrane of endocytosis mutants. pGFP-DRS2 was cotransformed with pRS425-CDC50 into wild-type (BY4742), vrp1Δ (BY4742 YLR337C), end3Δ (KLY201), and end4-1 (TGY1912) cells. Transformants were grown to early log phase at 27°C and examined by fluorescence microscopy.
The observation that GFP-Drs2p localizes to the TGN in wild-type cells but accumulates on the plasma membrane of end3, vrp1, and end4 suggests that Drs2p cycles between the exocytic and endocytic pathways. Neither deletion of SLA1 nor deletion of the Drs2-NPFXD motifs led to accumulation at the plasma membrane. Therefore, Drs2p must contain a second endocytosis signal that acts independently of Sla1p, but requires the actin-based endocytic machinery. Because GFP-Drs2p was only trapped on the plasma membrane of the original end3Δ strain when the NPFXD motifs were mutated, this strain must contain an extragenic mutation that suppresses Sla1p/NPFXD-mediated endocytosis but does not suppress end3Δ defects in endocytosis mediated by other signals. Thus, the original end3Δ strain (BY4742 end3Δ) was useful because it demonstrated an active role for the Drs2p NPFXD motifs in endocytosis.
Cargo-selective Endocytosis Defect of pan1-20
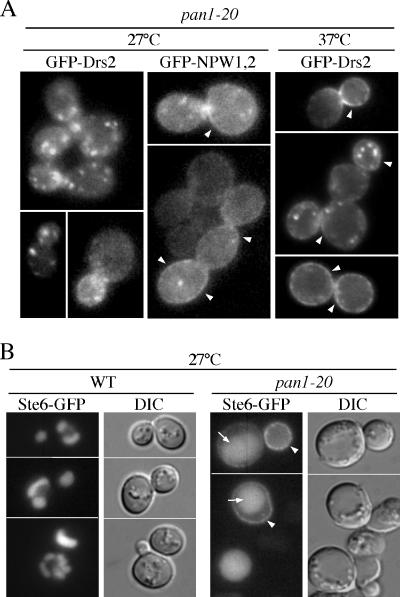
Mutation of both NPFXD motifs in Drs2p results in synthetic lethality with pan1-20 (Figure 3), suggesting that pan1-20 must maintain an active Sla1p/NPFXD-dependent endocytosis pathway at the permissive growth temperature. Otherwise, it is difficult to understand how mutation of an NPFXD endocytosis signal would further exacerbate growth of pan1-20. To test the Pan1p requirement for endocytosis of Drs2p, we examined the localization of GFP-Drs2p and GFP-Drs2-NPW1,2p in pan1-20 at the permissive (27°C) and nonpermissive (37°C) growth temperatures. GFP-Drs2p localized appropriately to the TGN at 27°C, but in stark contrast, GFP-Drs2-NPW1,2p was primarily localized to the plasma membrane. After a 1-h shift to 37°C, both GFP-Drs2p and GFP-Drs2-NPW1,2p accumulated on the plasma membrane. Kex2-GFP, another TGN resident, did not accumulate on the plasma membrane of pan1-20 at either temperature (our unpublished data). These results indicate that at the permissive growth temperature, pan1-20 cells are defective in the Sla1p/NPFXD-independent endocytosis pathway but retain a functional Sla1p/NPFXD-dependent pathway. Both pathways are blocked at the nonpermissive temperature, causing accumulation of wild-type GFP-Drs2p at the plasma membrane.
The Sla1p/NPFXD-independent endocytosis pathway blocked in pan1-20 at all temperatures is mostly likely dependent on Ub-dependent endocytosis signals. To test this possibility, we examined the localization of Ste6p-GFP in pan1-20 cells. Ste6p is an ATP-binding cassette transporter that uses a Ub-dependent signal for endocytosis (Kolling and Hollenberg, 1994; Kelm et al., 2004; Krsmanovic et al., 2005). In wild-type cells, Ste6-GFP accumulates in the vacuole over time, and so the cells show primarily vacuolar patterns of fluorescence. However, Ste6-GFP accumulated at the plasma membrane of pan1-20 cells at 27°C (Figure 7B). Thus, Ub-dependent endocytosis is abrogated in pan1-20 at permissive growth temperatures.
Figure 7.
The pan1-20 mutant exhibits a constitutive defect in Ub-dependent endocytosis and a temperature conditional defect in NPFXD-dependent endocytosis. (A) Localization of GFP-Drs2p and GFP-Drs2-NPW1,2p in pan1-20 (TGY1906) cells. This pan1-20 strain expresses wild-type Drs2p from its endogenous locus to support viability. Cells were grown to mid-log phase at 27°C with or without shifting to 37°C for 1 h before imaging. Arrowheads indicate plasma membrane fluorescence. (B) Ste6p-GFP localization in wild-type and pan1-20 cells at 27°C. Wild-type (SEY6211) and pan1-20 (TGY1906) cells transformed with a plasmid harboring Ste6p-GFP (pSM1493) were grown at 27°C to mid-log phase and examined by fluorescence microscopy. Arrows indicate vacuoles. Arrowheads indicate plasma membrane fluorescence.
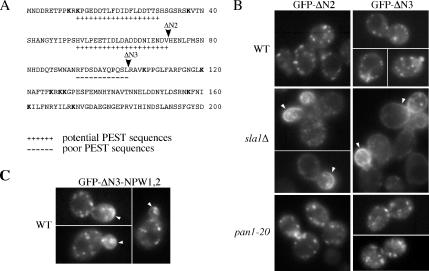
Additional Endocytosis Signals in the Drs2p N-Tail
The dependence of Drs2p on its NPFXD signals for TGN localization in pan1-20 and end3Δ (suppressor) cells but not wild-type cells strongly suggested that Drs2p must contain an additional endocytosis signal that is Ub dependent. A search for this signal using the PESTfind algorithm identified two “potential” and one “poor” PEST sequence along with 11 lysines in the N-tail of Drs2p (Figure 8A). Several other “poor” PEST sequences were found throughout Drs2p although none were in the C-tail. Deletions removing two (ΔN2, amino acids 1-72) or all three possible PEST sequences (ΔN3, amino acids 1-103) in the N-tail were constructed and the localization of GFP-Drs2-ΔN proteins was examined in wild-type, sla1Δ, and pan1-20 cells. As predicted, GFP-Drs2-ΔN2 and GFP-Drs2-ΔN3 were localized to the TGN in wild-type and pan1-20 cells at 30°C and mislocalized to the plasma membrane in sla1Δ cells (Figure 8B). Both GFP-Drs2-ΔN2 and GFP-Drs2-ΔN3 complemented the cs growth defect of drs2Δ, indicating that this deletion did not perturb Drs2p ATPase activity or function in protein transport. These data indicate that Drs2p has an endocytosis signal(s) in the N-tail that is recognized by components of the endocytic apparatus specifically disrupted by the pan1-20 mutation. Combining the ΔN3 and NPW mutations caused accumulation of a small amount of GFP-Drs2p on the plasma membrane of wild-type cells (Figure 8C). The localization of most Drs2-ΔN3-NPW1,2 to intracellular compartments suggests that these mutations have not eliminated all of the endocytosis signals in Drs2p. In addition, the observation that Drs2-ΔN3-NPW1,2 does not accumulate on the plasma membrane as much as Drs2-ΔN3 expressed in sla1Δ cells suggests that Sla1p contributes more to endocytosis than just the recruitment of NPFXD cargo.
Figure 8.
Sequences in the Drs2p N-tail bearing PEST-like motifs mediate endocytosis redundantly with the NPFXD motifs. (A) The N-tail of Drs2p contains two potential PEST sequences and one poor PEST sequence. Arrowheads indicate the N-terminal truncation boundaries for the GFP-DRS2-ΔN2 and -ΔN3 alleles. (B) Localization of the GFP-Drs2p with N-terminal truncations in wild-type, sla1Δ, and pan1-20 cells. pGFP-DRS2, pGFP-Drs2-ΔN2, and pGFP-Drs2-ΔN3 were cotransformed with pRS425-CDC50 into wild-type (BY4742), sla1Δ (KLY011), or pan1-20 (TGY1907) cells. Transformants were grown to early log phase at 30°C and imaged by fluorescence microscopy at room temperature. Arrowheads indicate plasma membrane fluorescence. (C) Localization of the GFP-Drs2p with both the N-terminal truncation and the NPW1,2 mutations in wild-type cells. pGFP-Drs2-ΔN3-NPW1,2 was cotransformed with pRS425-CDC50 into wild-type cells. Transformants were grown to early log phase at 30°C and imaged by fluorescence microscopy at room temperature. Arrowheads indicate plasma membrane fluorescence.
Slow Exit of Drs2p from the TGN
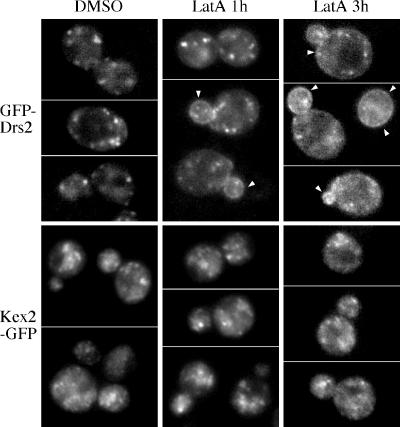
The vrp1 and end mutants described above are constitutively defective for endocytosis, and so these studies do not indicate how frequently GFP-Drs2p travels to the plasma membrane. To determine the kinetics of GFP-Drs2p transport to the plasma membrane, wild-type cells expressing GFP-Drs2 were treated with latrunculin A (lat-A), an inhibitor of actin assembly and endocytosis, and imaged over time (Figure 9). After 1 h of treatment, GFP-Drs2p was still primarily retained intracellularly in small puncta, although it could be detected on the plasma membrane (Figure 9, lat-A 1 h). After 3 h of treatment, the cell surface GFP-Drs2p further increased, concomitant with a reduction in the intensity of GFP-Drs2p intracellular fluorescence, and it approached the distribution observed in the end mutants (Figure 9, lat-A 3 h). Actin assembles on Golgi membranes, and so we considered the possibility that perturbation of actin caused mislocalization of all late Golgi proteins, comparable with clathrin mutants. Therefore, Kex2p-GFP was also examined in this experiment. Even when overexpressed, Kex2p-GFP did not show any plasma membrane staining after 3 h of lat-A treatment (Figure 9, Kex2-GFP). These data demonstrate that unlike Kex2p, another TGN-localizing, NPFXD-containing protein, Drs2p slowly cycles between the TGN and the plasma membrane.
Figure 9.
Drs2p accumulates slowly on the plasma membrane after disrupting endocytosis. Wild-type cells expressing either GFP-Drs2p or Kex2p-GFP were grown to early log phase at 30°C and then incubated in SD medium containing 200 μM lat-A or dimethyl sulfoxide at 30°C for the times shown. Arrowheads indicate plasma membrane fluorescence.
Drs2p and Cdc50p Do Not Significantly Transit the Late Endosome
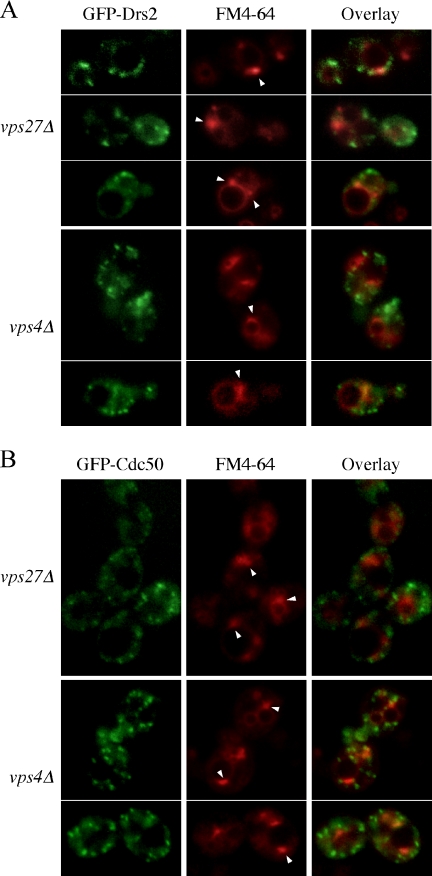
Several different TGN proteins cycle through the late endosome as part of their normal trafficking itinerary (Wilcox et al., 1992; Nothwehr et al., 1993; Cooper and Stevens, 1996; Brickner and Fuller, 1997; Foote and Nothwehr, 2006). We tested whether Drs2p transited through the late endosome/prevacuolar compartment (PVC) by examining its localization in class E vps mutants, which block protein and lipid transport out of the PVC and result in the formation of an enlarged endosomal compartment adjacent to the vacuole called the class E compartment (Conibear and Stevens, 1998). Proteins that traffic through the PVC accumulate in the class E compartment of vps4 or vps27 cells. We expressed GFP-Drs2p in vps4Δ or vps27Δ and labeled the class E compartments with the endocytic tracer FM4-64 (Vida and Emr, 1995) before analysis by fluorescence microscopy. Although a small amount of GFP-Drs2 was detected in class E compartments at steady state, the large majority of this protein did not colocalize with the FM4-64 and remained in small puncta (Figure 10A). In addition, we could not detect Drs2-Myc (C-terminal tag) in the class E compartment of vps27Δ by immunofluorescence (data not shown).
Figure 10.
GFP-Drs2p and GFP-Cdc50p do not accumulate in the prevacuolar compartment of class E vps mutants. vps27Δ and vps4Δ cells carrying pGFP-DRS2 and pRS425-CDC50 (A) or pGFP-CDC50 and pRS425-DRS2 (B) were grown to early mid-log phase at 30°C, labeled with 10 μg/ml FM4-64 on ice for 20 min, washed with fresh medium, and then chased at 30°C for 30 min before analysis. GFP and FM4-64 images were acquired separately and merged to show the coincidence of the two patterns. Arrowheads indicate the prevacuolar compartment.
The Drs2p chaperone Cdc50p has been described as a late endosomal protein because Cdc50-GFP (GFP fused to C terminus of Cdc50p) accumulated in the class E compartment (Misu et al., 2003). However, this phenotype does not discriminate between a TGN or endosomal protein, and it is inconsistent with our observations with GFP-Drs2p. Therefore, we examined the localization of GFP-Cdc50p (GFP fused to the N terminus of Cdc50p) in the vps mutants. This fusion protein is functional based on complementation of the cs growth defect of cdc50Δ. However, we were unable to detect a significant amount of colocalization between FM4-64 and GFP-Cdc50 in the class E compartment of vps27Δ or vps4Δ cells (Figure 10B). Because neither GFP-Drs2p nor GFP-Cdc50p fluorescence collapsed into the class E compartments of either mutant, the PVC does not seem to be a significant destination in the trafficking itinerary of these proteins. We also considered the possibility that NPFXD motifs play a role in retrieval of Drs2p from early endosomes back to the TGN. If so, deletion of the NPFXD motifs may force Drs2p to transit through the PVC more frequently. To test this possibility, we examined localization of GFP-Drs2p-ΔNPF and GFP-Drs2p-NPW1,2 in the vps cells. Neither GFP-Drs2p-ΔNPF nor GFP-Drs2p-NPW1,2 accumulated in the class E compartments (our unpublished data), indicating that potential sorting signals that prevent Drs2p trafficking to the PVC do not lie in the last 44 amino acids.
DISCUSSION
Trafficking of Drs2p and Dnf1p
TGN-dwelling proteins have evolved different mechanisms to maintain their steady-state localization in this organelle, although transport to the endosomal system and retrieval back to the TGN seems to be a common theme. Recycling mechanisms are critical for localization of Golgi proteins because a growing body of evidence indicates that Golgi cisternae are not stable structures but instead mature from cis to trans by changing the content of resident enzymes (Losev et al., 2006; Matsuura-Tokita et al., 2006). Cisternae mature with a half-time of 1 to 2 min in the yeast system and the TGN is apparently consumed into transport carriers with multiple destinations. Therefore, TGN resident proteins must recycle back to younger cisternae every few minutes to maintain Golgi residence. Some mammalian TGN proteins, such as the mannose-6-phosphate receptor, TGN38, and furin, cycle to the plasma membrane and endosomes before returning to the TGN. These proteins require endocytic signals for removal from the plasma membrane and retrograde sorting signals to mediate endosome to TGN transport (Thomas, 2002; Ghosh et al., 2003; Traub, 2005). In yeast, the vacuolar hydrolase sorting receptors Vps10p and Mrl1p rapidly cycle between the TGN and the late endosome/PVC (Cereghino et al., 1995; Cooper and Stevens, 1996; Whyte and Munro, 2001). The TGN markers Kex2p and Ste13p are thought to primarily cycle between the TGN and early endosomes, occasionally transiting the PVC but not the plasma membrane (Abazeed et al., 2005; Foote and Nothwehr, 2006).
None of the yeast TGN proteins mentioned above accumulate on the plasma membrane when endocytosis is blocked (Cooper and Bussey, 1992; Roberts et al., 1992; Wilcox et al., 1992; Bryant and Stevens, 1997). In contrast, most (but not all) Drs2p accumulates on the plasma membrane of mutants constitutively defective for endocytosis. However, acute inactivation of endocytosis by latrunculin A treatment of cells causes a rather slow accumulation of Drs2p on the plasma membrane over the course of ∼3 h. These findings suggest that Drs2p is inefficiently incorporated into exocytic vesicles, and is rapidly endocytosed upon arrival at the plasma membrane. The slow delivery to the plasma membrane and rapid endocytosis leads to an undetectable amount of Drs2p on the plasma membrane of wild-type cells unless endocytosis is inhibited. It is formally possible that disruption of endocytosis causes an aberrant incorporation of Drs2p into exocytic vesicles leading to plasma membrane accumulation. However, other TGN proteins do not share this fate in endocytosis mutants arguing against a reduced fidelity of sorting TGN residents from exocytic cargo. Moreover, Drs2p can be found in exocytic vesicles that accumulate in the sec6 mutant, providing a method independent of disrupting actin or blocking endocytosis to show targeting of some Drs2p to the plasma membrane (Alder-Baerens et al., 2006). Thus, it is much more likely that disrupting endocytosis simply traps Drs2p on the plasma membrane as it undergoes its normal trafficking itinerary.
After endocytosis, Drs2p must also be efficiently removed from the endocytic pathway and transported from the early endosome to the TGN, because we fail to see significant accumulation of Drs2p or its Cdc50p subunit in the late endosome of class E vps mutants. This is similar to the recycling pathway described for the SNARE protein Snc1p and chitin synthase (Chs3p). In contrast, class E vps mutants accumulate most of Vps10p, Mrl1p, Kex2p, and Ste13p in the PVC (Cereghino et al., 1995; Brickner and Fuller, 1997; Bryant and Stevens, 1997; Whyte and Munro, 2001). Moreover, we never detect GFP-Drs2p in the vacuole, which is a common destination for many proteins that use Ub for an endocytosis signal because this modification also directs proteins into the multivesicular body pathway at the late endosome for delivery to the vacuole lumen (Katzmann et al., 2002; Hicke and Dunn, 2003). If Drs2p is ubiquitinated, it must either avoid the late endosome to escape vacuolar delivery, or the Ub must be removed to allow retrieval of Drs2p to the TGN/early endosomal system. A previous report suggested that Drs2p is a late endosomal protein based on a relatively minor localization of Drs2p-GFP to the prevacuolar compartment of a class E vps mutant (Saito et al., 2004). We disagree with this interpretation because at the rate of recycling suggested by the cisternal maturation model, the majority of Drs2p would relocate to the PVC in class E vps mutants if even a small percentage of molecules transited the PVC in each round of recycling (as observed for Ste13p and Kex2p). In contrast, a majority of Cdc50-GFP (C-terminally tagged) was reported to localize to the PVC (Misu et al., 2003), whereas we found that GFP-Cdc50p (N-terminally tagged) was primarily excluded from the PVC. We suggest that the GFP-Cdc50p fusion protein more faithfully represents the localization of the endogenous Drs2p–Cdc50p complex as these data are more consistent with the localization data for Drs2p. In summary, it is likely that Drs2p rapidly cycles between the TGN and early endosome as suggested for several other TGN proteins, and slowly cycles in the TGN → plasma membrane → early endosome → TGN loop.
Dnf1p is localized to both the plasma membrane and cytoplasmic punctate structures typically found near the plasma membrane. The steady-state localization pattern of Dnf1p and functional studies suggest that this protein cycles in a plasma membrane → endosome → TGN → plasma membrane pathway (Hua et al., 2002). This hypothesis was confirmed by studies of the Tanaka group (Saito et al., 2004) and those reported here, which extend these studies to demonstrate that endocytosis of Dnf1p involves the NPFXD motif and Sla1p. Interestingly, the majority of Dnf1p was reported to fractionate in membranes with the same density as the plasma membrane (Pomorski et al., 2003; Alder-Baerens et al., 2006), which we have confirmed (our unpublished data), but most Dnf1p seems to be inside the cell by immunofluorescence localization and is separable from the plasma membrane by differential centrifugation techniques. We suggest that these data could be explained by efficient incorporation of Dnf1p into exocytic vesicles at the TGN (and/or endosome) and efficient endocytosis at the plasma membrane, thus giving a primary steady-state localization to vesicles trafficking to and from the plasma membrane.
Synthetic Lethality between drs2 and pan1
Endocytosis plays an important role in remodeling the plasma membrane, internalizing extracellular nutrients, down-regulating signal transduction pathways by internalizing receptors, and retrieving proteins required for Golgi/endosome function that have escaped to the plasma membrane. The synthetic lethality between pan1-20 and drs2-npw1,2 provides the best example we are aware of for the essential role of endocytosis in retrieving Golgi proteins. The synthetic lethality between drs2Δ and pan1-20 was initially surprising because Drs2p is required for Golgi/early endosome function, whereas Pan1p acts at the internalization step of endocytosis. However, the relationship between Drs2p and Pan1p is now clearer. The Ub-dependent endocytosis defect of pan1-20 likely depletes the TGN/endosomal system of several proteins required for the essential function of these organelles in protein transport. In spite of this deficit, Golgi/early endosome function can be maintained as long as Drs2p is present in these organelles. In pan1-20, Drs2p localization to the TGN/early endosome requires its endocytosis by the NPFXD/Sla1p pathway, thus explaining the inviability of pan1-20 drs2-npw1,2 (shown here) and pan1 sla1Δ (Tang et al., 2000).
We have also considered the possibility that the pan1-20 drs2-npw1,2 synthetic lethality could be explained by a requirement for Drs2p in the internalization step of endocytosis. However, Dnf1p and Dnf2p seem to provide a much more important contribution to endocytosis than Drs2p (Hua et al., 2002; Pomorski et al., 2003). In addition, Dnf1p should be present in the plasma membrane of pan1-20 cells and capable of engaging Sla1p through its NPFXD motif, thus satisfying a potential requirement to couple a phospholipid translocase to the endocytic machinery. Surprisingly, Drs2p does not seem to be active on the plasma membrane because end4 cells accumulate Drs2p on the plasma membrane, but there does not seem to be a loss of phosphatidylserine translocation across the plasma membrane when DRS2 is deleted in end4 cells (Marx et al., 1999). Therefore, it is much more likely that TGN/early endosome residence of Drs2p is required to sustain viability of pan1-20 cells. Moreover, endocytosis of Dnf1p is required for this protein to compensate for loss of Drs2p, further arguing for critical Golgi/endosomal functions for these proteins. Class I myosins (Myo3p and Myo5p) are required for endocytosis in yeast and CDC50 was recovered in a screen for multicopy suppressors of a myo3Δ myo5-360 double mutant growth defect (Misu et al., 2003). This suppression could also be explained by an increased Drs2p/Cdc50p concentration in the Golgi/early endosome that overcomes the loss caused by the endocytosis defect.
It is also possible that Pan1p has some unrecognized function in the TGN/early endosome system because end3Δ, end4Δ, and vrp1Δ mutants have a defect in both the Ub- and NPFXD-dependent endocytic pathways, accumulate Drs2p on the plasma membrane, and yet these mutants are viable, whereas pan1Δ is not. In addition, drs2Δ end4-1 and drs2Δ sla1Δ double mutants are viable but the drs2Δ pan1-20 double mutant is dead (Chen et al., 1999; this study). One potential explanation for this discrepancy is that Pan1p contributes to the clathrin-dependent localization of TGN proteins independently of End4p, End3p, and Sla1p, perhaps through its interaction with the clathrin assembly protein AP180. In this case, loss of Pan1p function would accelerate transport of TGN proteins to the plasma membrane while trapping them there behind the endocytosis block. Moreover, it seems that the redistribution of GFP-Drs2p from the TGN to the plasma membrane occurs faster and more completely in pan1-20 after shift to the nonpermissive temperature relative to latrunculin A-treated cells. This potential influence of Pan1p on Golgi localization seems to be specific to Drs2p because Kex2p is not mislocalized to the plasma membrane of pan1-ts cells. However, further work is required to determine whether these differences are significant and whether Pan1p has any functions beyond the internalization step of endocytosis.
Interactions between Cargo and the Endocytic Machinery
The nature of the pan1-20 mutation is also of interest because it causes cargo-selective defects in endocytosis at the permissive growth temperature, and the mechanism for recognition of Ub-dependent endocytic cargo is incompletely understood. Pan1p, End3p, End4p, and Sla1p are all recruited to the same endocytic sites and are considered a coat complex for clathrin/actin-based endocytosis. The Ub-dependent signal typically requires phosphorylation before ubiquitination by the Rsp5p ubiquitin ligase (Rotin et al., 2000; Hicke and Dunn, 2003), then recognition of the ubiquitinated cargo by epsins (Ent1p, Ent2p) and Ede1p, another Eps15-related protein, for inclusion into forming vesicles (Shih et al., 2002). Any of these steps could be defective in the pan1-20 strain at the permissive temperature, and so it will be informative to test whether Drs2-npw1,2p accumulates on the pan1-20 plasma membrane in a phosphorylated and/or ubiquitinated form. Interestingly, the pan1-20 mutation is a frameshift very near the carboxy terminus within the proline-rich domain of Pan1p (Wendland, personal communication), suggesting this mutation abrogates interaction with a Src homology (SH)3-containing protein. One possibility is that this region could potentially interact with the SH3 domain of Rvs167p (amphiphysin/endophilin). A point mutation in the SH3 domain of Rvs167p causes a synthetic lethal interaction with sla1Δ (Friesen et al., 2006), as does pan1 allele (Tang et al., 2000). Moreover, Sla1p forms a complex with Rvs167 that interacts with Rsp5p, comparable with the Cin85–edophilin interaction with the Cbl ubiquitin ligase in mammalian cells (Stamenova et al., 2004). This network of interactions may play a critical role in coupling cargo recognition (by Sla1p, Rsp5p, and epsins) to packaging of cargo into forming vesicles.
In summary, the observations reported here are novel in several respects. To our knowledge, the inviability of pan1-20 drs2-npw2 represents the first example where mutation of an endocytosis signal in a single protein is lethal to a cell. This result highlights the importance of endocytosis in retrieving proteins whose primary function is in the Golgi/endosomal system. In addition, the selective defect of pan1-20 in Ub-dependent endocytosis should facilitate a better understanding of how cargo is recognized and packaged into endocytic vesicles through characterization of effectors of the Pan1p proline-rich domain.
ACKNOWLEDGMENTS
We thank Scott Emr, Hugh Pelham, Jeff Flick, Tom Vida, Catherine Jackson, Gregory Payne, Susan Michaelis, and Beverly Wendland for providing plasmids, strains, and antibodies and for providing unpublished data. We also thank Sophie Chen and Guoxiang Ruan for assistance with plasmid construction. This work was supported by Grant GM-62367 from the National Institutes of Health (to T.R.G.).
Abbreviations used:
- 5-FOA
5 fluoroorotic acid
- ALP
alkaline phosphatase
- CM
conserved motif
- cs
cold sensitive
- EH
Eps15 homology
- GIM
Gea2p interaction motif
- PVC
prevacuolar compartment
- SHD
Sla1p homology domain
- TGN
trans-Golgi network.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E06-07-0592) on November 22, 2006.
REFERENCES
- Abazeed M. E., Blanchette J. M., Fuller R. S. Cell-free transport from the trans-Golgi network to late endosome requires factors involved in formation and consumption of clathrin-coated vesicles. J. Biol. Chem. 2005;280:4442–4450. doi: 10.1074/jbc.M412553200. [DOI] [PubMed] [Google Scholar]
- Aguilar R. C., Watson H. A., Wendland B. The yeast Epsin Ent1 is recruited to membranes through multiple independent interactions. J. Biol. Chem. 2003;278:10737–10743. doi: 10.1074/jbc.M211622200. [DOI] [PubMed] [Google Scholar]
- Alder-Baerens N., Lisman Q., Luong L., Pomorski T., Holthuis J. C. Loss of P4 ATPases Drs2p and Dnf3p disrupts aminophospholipid transport and asymmetry in yeast post-Golgi secretory vesicles. Mol. Biol. Cell. 2006;17:1632–1642. doi: 10.1091/mbc.E05-10-0912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balhadere P. V., Talbot N. J. PDE1 encodes a P-type ATPase involved in appressorium-mediated plant infection by the rice blast fungus Magnaporthe grisea. Plant Cell. 2001;13:1987–2004. doi: 10.1105/TPC.010056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barik S., Galinski M. S. “Megaprimer” method of PCR: increased template concentration improves yield. Biotechniques. 1991;10:489–490. [PubMed] [Google Scholar]
- Brickner J. H., Fuller R. S. SOI1 encodes a novel, conserved protein that promotes TGN-endosomal cycling of Kex2p and other membrane proteins by modulating the function of two TGN localization signals. J. Cell Biol. 1997;139:23–36. doi: 10.1083/jcb.139.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant N. J., Stevens T. H. Two separate signals act independently to localize a yeast late Golgi membrane protein through a combination of retrieval and retention. J. Cell Biol. 1997;136:287–297. doi: 10.1083/jcb.136.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bull L. N., et al. A gene encoding a P-type ATPase mutated in two forms of hereditary cholestasis. Nat. Genet. 1998;18:219–224. doi: 10.1038/ng0398-219. [DOI] [PubMed] [Google Scholar]
- Catty P., de Kerchove d'Exaerde A., Goffeau A. The complete inventory of the yeast Saccharomyces cerevisiae P-type transport ATPases. FEBS Lett. 1997;409:325–332. doi: 10.1016/s0014-5793(97)00446-8. [DOI] [PubMed] [Google Scholar]
- Cereghino J. L., Marcusson E. G., Emr S. D. The cytoplasmic tail domain of the vacuolar protein sorting receptor Vps10p and a subset of VPS gene products regulate receptor stability, function, and localization. Mol. Biol. Cell. 1995;6:1089–1102. doi: 10.1091/mbc.6.9.1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chantalat S., Park S. K., Hua Z., Liu K., Gobin R., Peyroche A., Rambourg A., Graham T. R., Jackson C. L. The Arf activator Gea2p and the P-type ATPase Drs2p interact at the Golgi in Saccharomyces cerevisiae. J. Cell Sci. 2004;117:711–722. doi: 10.1242/jcs.00896. [DOI] [PubMed] [Google Scholar]
- Chen C. Y., Ingram M. F., Rosal P. H., Graham T. R. Role for Drs2p, a P-type ATPase and potential aminophospholipid translocase, in yeast late Golgi function. J. Cell Biol. 1999;147:1223–1236. doi: 10.1083/jcb.147.6.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S., Wang J., Muthusamy B. P., Liu K., Zare S., Andersen R. J., Graham T. R. Roles for the Drs2p-Cdc50p complex in protein transport and phosphatidylserine asymmetry of the yeast plasma membrane. Traffic. 2006;7:1503–1517. doi: 10.1111/j.1600-0854.2006.00485.x. [DOI] [PubMed] [Google Scholar]
- Conibear E., Stevens T. H. Multiple sorting pathways between the late Golgi and the vacuole in yeast. Biochim. Biophys. Acta. 1998;1404:211–230. doi: 10.1016/s0167-4889(98)00058-5. [DOI] [PubMed] [Google Scholar]
- Cooper A., Bussey H. Yeast Kex1p is a Golgi-associated membrane protein: deletions in a cytoplasmic targeting domain result in mislocalization to the vacuolar membrane. J. Cell Biol. 1992;119:1459–1468. doi: 10.1083/jcb.119.6.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper A. A., Stevens T. H. Vps10p cycles between the late-Golgi and prevacuolar compartments in its function as the sorting receptor for multiple yeast vacuolar hydrolases. J. Cell Biol. 1996;133:529–541. doi: 10.1083/jcb.133.3.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowles C. R., Odorizzi G., Payne G. S., Emr S. D. The AP-3 adaptor complex is essential for cargo-selective transport to the yeast vacuole. Cell. 1997;91:109–118. doi: 10.1016/s0092-8674(01)80013-1. [DOI] [PubMed] [Google Scholar]
- Devaux P. F., Lopez-Montero I., Bryde S. Proteins involved in lipid translocation in eukaryotic cells. Chem. Phys. Lipids. 2006;141:119–132. doi: 10.1016/j.chemphyslip.2006.02.007. [DOI] [PubMed] [Google Scholar]
- Dhar M. S., Sommardahl C. S., Kirkland T., Nelson S., Donnell R., Johnson D. K., Castellani L. W. Mice heterozygous for Atp10c, a putative amphipath, represent a novel model of obesity and type 2 diabetes. J. Nutr. 2004;134:799–805. doi: 10.1093/jn/134.4.799. [DOI] [PubMed] [Google Scholar]
- Duncan M. C., Cope M. J., Goode B. L., Wendland B., Drubin D. G. Yeast Eps15-like endocytic protein, Pan1p, activates the Arp2/3 complex. Nat. Cell Biol. 2001;3:687–690. doi: 10.1038/35083087. [DOI] [PubMed] [Google Scholar]
- Foote C., Nothwehr S. F. The clathrin adaptor complex 1 directly binds to a sorting signal in Ste13p to reduce the rate of its trafficking to the late endosome of yeast. J. Cell Biol. 2006;173:615–626. doi: 10.1083/jcb.200510161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friesen H., Humphries C., Ho Y., Schub O., Colwill K., Andrews B. Characterization of the yeast amphiphysins Rvs161p and Rvs167p reveals roles for the Rvs heterodimer in vivo. Mol. Biol. Cell. 2006;17:1306–1321. doi: 10.1091/mbc.E05-06-0476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gall W. E., Geething N. C., Hua Z., Ingram M. F., Liu K., Chen S. I., Graham T. R. Drs2p-dependent formation of exocytic clathrin-coated vesicles in vivo. Curr. Biol. 2002;12:1623–1627. doi: 10.1016/s0960-9822(02)01148-x. [DOI] [PubMed] [Google Scholar]
- Ghosh P., Dahms N. M., Kornfeld S. Mannose 6-phosphate receptors: new twists in the tale. Nat. Rev. Mol. Cell Biol. 2003;4:202–212. doi: 10.1038/nrm1050. [DOI] [PubMed] [Google Scholar]
- Gilbert M. J., Thornton C. R., Wakley G. E., Talbot N. J. A P-type ATPase required for rice blast disease and induction of host resistance. Nature. 2006;440:535–539. doi: 10.1038/nature04567. [DOI] [PubMed] [Google Scholar]
- Gomes E., Jakobsen M. K., Axelsen K. B., Geisler M., Palmgren M. G. Chilling tolerance in Arabidopsis involves ALA1, a member of a new family of putative aminophospholipid translocases. Plant Cell. 2000;12:2441–2454. [PMC free article] [PubMed] [Google Scholar]
- Graham T. R. Flippases and vesicle-mediated protein transport. Trends Cell Biol. 2004;14:670–677. doi: 10.1016/j.tcb.2004.10.008. [DOI] [PubMed] [Google Scholar]
- Hicke L., Dunn R. Regulation of membrane protein transport by ubiquitin and ubiquitin-binding proteins. Annu. Rev. Cell Dev. Biol. 2003;19:141–172. doi: 10.1146/annurev.cellbio.19.110701.154617. [DOI] [PubMed] [Google Scholar]
- Holthuis J. C., Levine T. P. Lipid traffic: floppy drives and a superhighway. Nat. Rev. Mol. Cell Biol. 2005;6:209–220. doi: 10.1038/nrm1591. [DOI] [PubMed] [Google Scholar]
- Howard J. P., Hutton J. L., Olson J. M., Payne G. S. Sla1p serves as the targeting signal recognition factor for NPFX(1,2)D-mediated endocytosis. J. Cell Biol. 2002;157:315–326. doi: 10.1083/jcb.200110027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua Z., Fatheddin P., Graham T. R. An essential subfamily of Drs2p-related P-type ATPases is required for protein trafficking between Golgi complex and endosomal/vacuolar system. Mol. Biol. Cell. 2002;13:3162–3177. doi: 10.1091/mbc.E02-03-0172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaksonen M., Toret C. P., Drubin D. G. Harnessing actin dynamics for clathrin-mediated endocytosis. Nat. Rev. Mol. Cell Biol. 2006;7:404–414. doi: 10.1038/nrm1940. [DOI] [PubMed] [Google Scholar]
- Katzmann D. J., Odorizzi G., Emr S. D. Receptor downregulation and multivesicular-body sorting. Nat. Rev. Mol. Cell Biol. 2002;3:893–905. doi: 10.1038/nrm973. [DOI] [PubMed] [Google Scholar]
- Kelm K. B., Huyer G., Huang J. C., Michaelis S. The internalization of yeast Ste6p follows an ordered series of events involving phosphorylation, ubiquitination, recognition and endocytosis. Traffic. 2004;5:165–180. doi: 10.1111/j.1600-0854.2004.00168.x. [DOI] [PubMed] [Google Scholar]
- Klomp L. W., et al. Characterization of mutations in ATP8B1 associated with hereditary cholestasis. Hepatology. 2004;40:27–38. doi: 10.1002/hep.20285. [DOI] [PubMed] [Google Scholar]
- Kolling R., Hollenberg C. P. The ABC-transporter Ste6 accumulates in the plasma membrane in a ubiquitinated form in endocytosis mutants. EMBO J. 1994;13:3261–3271. doi: 10.1002/j.1460-2075.1994.tb06627.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krsmanovic T., Pawelec A., Sydor T., Kolling R. Control of Ste6 recycling by ubiquitination in the early endocytic pathway in yeast. Mol. Biol. Cell. 2005;16:2809–2821. doi: 10.1091/mbc.E04-10-0941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhlbrandt W. Biology, structure and mechanism of P-type ATPases. Nat. Rev. Mol. Cell Biol. 2004;5:282–295. doi: 10.1038/nrm1354. [DOI] [PubMed] [Google Scholar]
- Lewis M. J., Nichols B. J., Prescianotto-Baschong C., Riezman H., Pelham H. R. Specific retrieval of the exocytic SNARE Snc1p from early yeast endosomes. Mol. Biol. Cell. 2000;11:23–38. doi: 10.1091/mbc.11.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine M. S., McKenzie A., 3rd, Demarini D. J., Shah N. G., Wach A., Brachat A., Philippsen P., Pringle J. R. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Losev E., Reinke C. A., Jellen J., Strongin D. E., Bevis B. J., Glick B. S. Golgi maturation visualized in living yeast. Nature. 2006;441:1002–1006. doi: 10.1038/nature04717. [DOI] [PubMed] [Google Scholar]
- Marx U., Polakowski T., Pomorski T., Lang C., Nelson H., Nelson N., Herrmann A. Rapid transbilayer movement of fluorescent phospholipid analogues in the plasma membrane of endocytosis-deficient yeast cells does not require the Drs2 protein. Eur. J. Biochem. 1999;263:254–263. doi: 10.1046/j.1432-1327.1999.00497.x. [DOI] [PubMed] [Google Scholar]
- Matsuura-Tokita K., Takeuchi M., Ichihara A., Mikuriya K., Nakano A. Live imaging of yeast Golgi cisternal maturation. Nature. 2006;441:1007–1010. doi: 10.1038/nature04737. [DOI] [PubMed] [Google Scholar]
- Miliaras N. B., Park J. H., Wendland B. The function of the endocytic scaffold protein Pan1p depends on multiple domains. Traffic. 2004;5:963–978. doi: 10.1111/j.1600-0854.2004.00238.x. [DOI] [PubMed] [Google Scholar]
- Misu K., Fujimura-Kamada K., Ueda T., Nakano A., Katoh H., Tanaka K. Cdc50p, a conserved endosomal membrane protein, controls polarized growth in Saccharomyces cerevisiae. Mol. Biol. Cell. 2003;14:730–747. doi: 10.1091/mbc.E02-06-0314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natarajan P., Wang J., Hua Z., Graham T. R. Drs2p-coupled aminophospholipid translocase activity in yeast Golgi membranes and relationship to in vivo function. Proc. Natl. Acad. Sci. USA. 2004;101:10614–10619. doi: 10.1073/pnas.0404146101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newpher T. M., Smith R. P., Lemmon V., Lemmon S. K. In vivo dynamics of clathrin and its adaptor-dependent recruitment to the actin-based endocytic machinery in yeast. Dev. Cell. 2005;9:87–98. doi: 10.1016/j.devcel.2005.04.014. [DOI] [PubMed] [Google Scholar]
- Nothwehr S. F., Roberts C. J., Stevens T. H. Membrane protein retention in the yeast Golgi apparatus: dipeptidyl aminopeptidase A is retained by a cytoplasmic signal containing aromatic residues. J. Cell Biol. 1993;121:1197–1209. doi: 10.1083/jcb.121.6.1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulusma C. C., Oude Elferink R. P. The type 4 subfamily of P-type ATPases, putative aminophospholipid translocases with a role in human disease. Biochim. Biophys. Acta. 2005;1741:11–24. doi: 10.1016/j.bbadis.2005.04.006. [DOI] [PubMed] [Google Scholar]
- Pomorski T., Holthuis J. C., Herrmann A., van Meer G. Tracking down lipid flippases and their biological functions. J. Cell Sci. 2004;117:805–813. doi: 10.1242/jcs.01055. [DOI] [PubMed] [Google Scholar]
- Pomorski T., Lombardi R., Riezman H., Devaux P. F., van Meer G., Holthuis J. C. Drs2p-related P-type ATPases Dnf1p and Dnf2p are required for phospholipid translocation across the yeast plasma membrane and serve a role in endocytosis. Mol. Biol. Cell. 2003;14:1240–1254. doi: 10.1091/mbc.E02-08-0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts C. J., Nothwehr S. F., Stevens T. H. Membrane protein sorting in the yeast secretory pathway: evidence that the vacuole may be the default compartment. J. Cell Biol. 1992;119:69–83. doi: 10.1083/jcb.119.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth A. F., Sullivan D. M., Davis N. G. A large PEST-like sequence directs the ubiquitination, endocytosis, and vacuolar degradation of the yeast a-factor receptor. J. Cell Biol. 1998;142:949–961. doi: 10.1083/jcb.142.4.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotin D., Staub O., Haguenauer-Tsapis R. Ubiquitination and endocytosis of plasma membrane proteins: role of Nedd4/Rsp5p family of ubiquitin-protein ligases. J. Membr. Biol. 2000;176:1–17. doi: 10.1007/s00232001079. [DOI] [PubMed] [Google Scholar]
- Saito K., Fujimura-Kamada K., Furuta N., Kato U., Umeda M., Tanaka K. Cdc50p, a protein required for polarized growth, associates with the Drs2p P-type ATPase implicated in phospholipid translocation in Saccharomyces cerevisiae. Mol. Biol. Cell. 2004;15:3418–3432. doi: 10.1091/mbc.E03-11-0829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F. Getting started with yeast. Methods Enzymol. 1991;194:3–21. doi: 10.1016/0076-6879(91)94004-v. [DOI] [PubMed] [Google Scholar]
- Shih S. C., Katzmann D. J., Schnell J. D., Sutanto M., Emr S. D., Hicke L. Epsins and Vps27p/Hrs contain ubiquitin-binding domains that function in receptor endocytosis. Nat. Cell Biol. 2002;4:389–393. doi: 10.1038/ncb790. [DOI] [PubMed] [Google Scholar]
- Stamenova S. D., Dunn R., Adler A. S., Hicke L. The Rsp5 ubiquitin ligase binds to and ubiquitinates members of the yeast CIN85-endophilin complex, Sla1-Rvs167. J. Biol. Chem. 2004;279:16017–16025. doi: 10.1074/jbc.M313479200. [DOI] [PubMed] [Google Scholar]
- Tan P. K., Howard J. P., Payne G. S. The sequence NPFXD defines a new class of endocytosis signal in Saccharomyces cerevisiae. J. Cell Biol. 1996;135:1789–1800. doi: 10.1083/jcb.135.6.1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H. Y., Munn A., Cai M. EH domain proteins Pan1p and End3p are components of a complex that plays a dual role in organization of the cortical actin cytoskeleton and endocytosis in Saccharomyces cerevisiae. Mol. Cell Biol. 1997;17:4294–4304. doi: 10.1128/mcb.17.8.4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H. Y., Xu J., Cai M. Pan1p, End3p, and S1a1p, three yeast proteins required for normal cortical actin cytoskeleton organization, associate with each other and play essential roles in cell wall morphogenesis. Mol. Cell Biol. 2000;20:12–25. doi: 10.1128/mcb.20.1.12-25.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X., Halleck M. S., Schlegel R. A., Williamson P. A subfamily of P-type ATPases with aminophospholipid transporting activity. Science. 1996;272:1495–1497. doi: 10.1126/science.272.5267.1495. [DOI] [PubMed] [Google Scholar]
- Thomas G. Furin at the cutting edge: from protein traffic to embryogenesis and disease. Nat. Rev. Mol. Cell Biol. 2002;3:753–766. doi: 10.1038/nrm934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyoshima C., Inesi G. Structural basis of ion pumping by Ca2+-ATPase of the sarcoplasmic reticulum. Annu. Rev. Biochem. 2004;73:269–292. doi: 10.1146/annurev.biochem.73.011303.073700. [DOI] [PubMed] [Google Scholar]
- Traub L. M. Common principles in clathrin-mediated sorting at the Golgi and the plasma membrane. Biochim. Biophys. Acta. 2005;1744:415–437. doi: 10.1016/j.bbamcr.2005.04.005. [DOI] [PubMed] [Google Scholar]
- Vida T. A., Emr S. D. A new vital stain for visualizing vacuolar membrane dynamics and endocytosis in yeast. J. Cell Biol. 1995;128:779–792. doi: 10.1083/jcb.128.5.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendland B., Emr S. D. Pan1p, yeast eps15, functions as a multivalent adaptor that coordinates protein-protein interactions essential for endocytosis. J. Cell Biol. 1998;141:71–84. doi: 10.1083/jcb.141.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whyte J. R., Munro S. A yeast homolog of the mammalian mannose 6-phosphate receptors contributes to the sorting of vacuolar hydrolases. Curr. Biol. 2001;11:1074–1078. doi: 10.1016/s0960-9822(01)00273-1. [DOI] [PubMed] [Google Scholar]
- Wilcox C. A., Redding K., Wright R., Fuller R. S. Mutation of a tyrosine localization signal in the cytosolic tail of yeast Kex2 protease disrupts Golgi retention and results in default transport to the vacuole. Mol. Biol. Cell. 1992;3:1353–1371. doi: 10.1091/mbc.3.12.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zachowski A., Henry J. P., Devaux P. F. Control of transmembrane lipid asymmetry in chromaffin granules by an ATP-dependent protein. Nature. 1989;340:75–76. doi: 10.1038/340075a0. [DOI] [PubMed] [Google Scholar]