Abstract
Most misfolded secretory proteins remain in the endoplasmic reticulum (ER) and are degraded by ER-associated degradation (ERAD). However, some misfolded proteins exit the ER and traffic to the Golgi before degradation. Using model misfolded substrates, with or without defined ER exit signals, we found misfolded proteins can depart the ER by continuing to exhibit the functional export signals present in the corresponding correctly folded proteins. Anterograde transport of misfolded proteins utilizes the same machinery responsible for exporting correctly folded proteins. Passive ER retention, in which misfolded proteins fail to exit the ER due to the absence of exit signals or the inability to functionally present them, likely contributes to the retention of nonnative proteins in the ER. Intriguingly, compromising ERAD resulted in increased anterograde trafficking of a misfolded protein with an ER exit signal, suggesting that ERAD and ER exit machinery can compete for binding of misfolded proteins. Disabling ERAD did not result in transport of an ERAD substrate lacking an export signal. This is an important distinction for those seeking possible therapeutic approaches involving inactivating ERAD in anticipation of exporting a partially active protein.
INTRODUCTION
Secretory proteins enter the secretory pathway by translocation into the endoplasmic reticulum (ER; Deshaies et al., 1991; Johnson and van Waes, 1999). Once in the ER, proteins encounter an abundance of folding machinery, which aid in the achievement of native protein conformation, complex assembly, and addition of posttranslational modifications including carbohydrate addition and disulfide bond formation. Correctly folded proteins destined for distal compartments exit the ER via vesicular or tubular structures (Lee et al., 2004; Watanabe and Riezman, 2004). Vesicular transport of correctly folded proteins involves concentration of proteins in COPII vesicles by virtue of ER export signals within cargo proteins interacting with either COPII components or with ER export cargo receptors (Barlowe, 2003; Lee et al., 2004). Such ER export cargo receptors typically span the membrane with the luminal domain binding soluble cargo in the ER lumen, whereas the cytosolic domain of the receptor interacts with the COPII coat. Erv29p is an example of a yeast ER export cargo receptor responsible for the packaging of soluble cargo proteins such as correctly folded carboxypeptidase Y (wtCPY), proteinase A (wtPrA), and glycosylated pro-α-factor (gpαf; Belden and Barlowe, 2001; Caldwell et al., 2001), and a specific sequence within gpαf responsible for binding to Erv29p was recently identified (Otte and Barlowe, 2004). Many membrane-spanning proteins contain specific cytosolic exit signals, including the di-acidic (D/E-x-D/E) motif (Nishimura and Balch, 1997; Votsmeier and Gallwitz, 2001; Malkus et al., 2002; Otte and Barlowe, 2002; Wang et al., 2004), that interact directly with components of the COPII coat (Aridor et al., 1998; Kuehn et al., 1998). Recent studies demonstrated the direct binding of the cytosolic DxE exit signal of Sys1p to Sec24p, a component of the COPII coat involved in cargo selection (Votsmeier and Gallwitz, 2001; Miller et al., 2003).
A multifaceted quality control (ERQC) exists to monitor folding and assembly of proteins within the ER, whereas an associated process termed ERAD (ER-associated degradation) executes the degradation of defective and misfolded proteins remaining in the ER (Brodsky and McCracken, 1999; Ellgaard and Helenius, 2003). The need for ERQC and ERAD is evident by 1) the prediction that as much as a third of mammalian genes encode proteins associated with the secretory pathway and 2) the estimation that as much as 30% of nascent proteins are aberrantly synthesized (Schubert et al., 2000). ERAD involves many components that recognize aberrant proteins and implements their retrotranslocation to the cytosol before proteasomal degradation (Ellgaard and Helenius, 2003; Kostova and Wolf, 2003; McCracken and Brodsky, 2003). The Cdc48-Ufd1-Npl4 complex aids in dislocation of misfolded proteins from the ER and likely unfolding preceding degradation (Ye et al., 2001; Jarosch et al., 2002; Rabinovich et al., 2002). Misfolded proteins are ubiquitinated by E3 ubiquitin ligases (Hrd1p, Doa10p, and Rsp5p) before proteasomal degradation (Sommer and Jentsch, 1993; Hampton et al., 1996; Bordallo et al., 1998; Friedlander et al., 2000; Wilhovsky et al., 2000; Bays et al., 2001; Swanson et al., 2001; Haynes et al., 2002). Several additional components of ERAD have been identified, including Der1p and a related mammalian protein Derlin-1, whose function remains unknown, yet has been implicated in retrotranslocation (Knop et al., 1996; Lilley and Ploegh, 2004; Ye et al., 2004).
Although an active mechanism for retaining misfolded proteins in the ER has been proposed (Ellgaard and Helenius, 2003), a number of misfolded proteins exit the ER and traffic to the Golgi. For example, as much as 15% of the misfolded common mutant Z form of A1PiZ is secreted (Le et al., 1990). In addition, five mutations in the G protein–coupled V2 vasopressin receptor responsible for nephrogenic diabetes insipidus also exit the ER (Hermosilla et al., 2004). Furthermore, three mutations within the secreted cochlin protein are responsible for the autosomal dominant hearing loss and vestibular dysfunction disorder, DFNA9. These missense mutations result in protein misfolding yet the malfolded cochlin is secreted (Robertson et al., 2003). Finally, extensive work on ERAD substrates in Saccharomyces cerevisiae has found that some misfolded proteins, including CPY*, PrA*, and KHN, exit the ER as evidenced by incorporation into COPII vesicles, delivery to the Golgi and/or vacuole, and that the efficient degradation of these proteins is dependent on ER-Golgi trafficking (Hong et al., 1996; Caldwell et al., 2001; Vashist et al., 2001; Taxis et al., 2002; Coughlan et al., 2004; Vashist and Ng, 2004).
These observations led us to investigate in S. cerevisiae why some misfolded proteins are not retained in the ER and instead traffic to the Golgi. Our initial experiments with Erv29p suggest that this category of ERAD substrates, although misfolded, still present functional ER exit signals and consequently are incorporated into vesicles departing the ER (Caldwell et al., 2001). To test this hypothesis, we developed new model ERAD substrates, with or without defined ER exit signals. We found that misfolded proteins can depart the ER by presenting functional exit signals as do the corresponding correctly folded proteins. Conversely, missing or perturbed exit signals on many misfolded proteins likely contribute to these ERAD substrates remaining in the ER.
In addition, we found that compromising ERAD, through deletion of known ERAD components, resulted in increased forward trafficking of misfolded proteins containing ER exit signals. This data suggests that ERAD and ER exit machinery can compete for binding of misfolded proteins. Notably, a misfolded protein lacking an export signal did not traffic from the ER when ERAD was compromised.
MATERIALS AND METHODS
Materials
Restriction endonucleases and Endoglycosidase H were purchased from New England Biolabs (Beverly, MA). Oligonucleotides were purchased from IDT (Coralville, IA). Chemicals were purchased from Sigma-Aldrich (St. Louis, MO), Calbiochem (San Diego, CA), Seikagaku America (East Falmouth, MA), Fisher Scientific (St. Louis, MO), or Pierce (Rockford, IL).
Media, Strains, and Plasmid Construction
Media were prepared as described previously (Hill and Stevens, 1994). Yeast strains used in this work are described in Table 1. KHY30, KHY127, KHY163, KHY171, KHY265, KHY270, KHY271, KHY279, KHY298, KHY299, KHY306, KHY318, YPH499, CMY765, LHY434, and LHY433 were described previously (Thomas and Rothstein, 1989; Ghislain et al., 1993; Hill and Cooper, 2000; Caldwell et al., 2001; Dunn and Hicke, 2001; Haynes et al., 2002, 2004). KHY169 was created by repairing the vma22Δ locus of KHY127 (Hill and Cooper, 2000). The pep4Δ::TRP1 allele from SacI-XhoI–digested pLS1-10 (kindly provided by Dr. Steven Nothwehr) was introduced into KHY163 and KHY169 to create KHY252 and KHY264, respectively. This disruption and all others were confirmed by prototrophy and PCR analysis with oligonucleotides flanking the region originally amplified as described (Hill and Stevens, 1994). The pep4Δ::HIS3 allele was amplified by PCR and transformed into KHY163 and KHY171 to create KHY583 and KHY662, respectively. The prc1Δ::HIS3 allele from pAC556 (Haynes et al., 2002) was amplified by PCR and transformed into KHY306, KHY252, YPH499, and CMY765 to create KHY388, KHY401, KHY516, and KHY517, respectively. The SalI-StuI TRP1 fragment from pJJ246 was inserted into pAC550 (Haynes et al., 2002) digested SalI-NruI to create pAC754. The prc1Δ::TRP allele from pAC754 was amplified by PCR and transformed into LHY433 and LHY434 to create KHY747 and KHY741, respectively. pAC505, pAC530, pAC519, pMM322, and pAC460 were previously described (Manolson et al., 1992; Caldwell et al., 2001; Haynes et al., 2002). Briefly, pAC505 is the ERV29 allele inserted into pRS313 (Sikorski and Hieter, 1989), pAC530 is Erv29p-HA inserted into pRS313 (Sikorski and Hieter, 1989), pAC519 is the prc1-1 allele in YEp352 (Hill et al., 1986), pMM322 is Vph1p in pRS316 (Sikorski and Hieter, 1989), and pAC460 is Sec61-2p-HA in pTV3 (Rose and Broach, 1991). pAC812 and pAC815 were generated using modifications of the methodology previously described (Longtine et al., 1998). pAC723 was generated by PCR amplification of the Fus1p transmembrane and cytosolic domains and insertion of the fragment into pCR 2.1-TOPO (Invitrogen, Carlsbad, CA). The EcoRV-SacI Fus1p fragment from pAC723 was inserted into SnaBI-SacI–digested pBG15 (provided by Dr. Scott Moye-Rowley, University of Iowa, IA) to generate pAC724, CPY* luminal domain fused to the Fus1p transmembrane and cytosolic domains. SacI-digested pAC724 was cotransformed with the 3xHA-HIS3 PCR fragment from pFA6a-3HA-HIS3 (Longtine et al., 1998) into SEY6211a (Hill and Stevens, 1994) and plated on media lacking uracil and histidine to select for the new plasmid pAC757. SmaI-digested pAC757 was cotransformed into SEY6211a with either the PCR-amplified wild-type Sys1p cytosolic domain from CV1 or the mutant sys (AxA200) cytosolic domain from CV3 (Votsmeier and Gallwitz, 2001; provided by Dr. Dieter Gallwitz, Max-Planck-Institute of Biophysical Chemistry, Goettingen, Germany) and plated on media lacking uracil and histidine to select for the new plasmids pAC812 (CFS) and pAC815 (CFs′).
Table 1.
Yeast strains used in this work
| Strain | Genotype | Source |
|---|---|---|
| KHY30 | MATa vph1Δ::LEU2 trp1-Δ901 leu2-3,112 ura3-52 ade2-101 his3-Δ200 suc2-Δ9 | Caldwell et al. (2001) |
| KHY127 | MATa prc1-1 der1Δ::KANr vph1Δ::LEU2 vma22Δ-5::STOP trp1-Δ1901 leu2-3,112 ura3-52 ade2-101 his3-Δ200 suc2-Δ9 | Hill et al. (2000) |
| KHY163 | MATa prc1-1 vph1Δ::LEU2 trp1-Δ901 leu2-3,112 ura3-52 ade2-101 his3-Δ200 suc2-Δ9 | Caldwell et al. (2001) |
| KHY169 | vma22Δ locus repaired in KHY127 | This study |
| KHY171 | MATa prc1-1 hrd1Δ::LEU2 vph1Δ::KANr trp1-Δ901 leu2-3,112 ura3-52 ade2-101 his3-Δ200 suc2-Δ9 | Haynes et al. (2002) |
| KHY252 | pep4Δ::TRP1 in KHY163 | This study |
| KHY264 | pep4Δ::TRP1 in KHY169 | This study |
| KHY265 | pep4Δ::TRP1 in KHY171 | Haynes et al. (2002) |
| KHY270 | erv29Δ::KANr in KHY163 | Caldwell et al. (2001) |
| KHY271 | erv29Δ::KANr in KHY30 | Caldwell et al. (2001) |
| KHY279 | hrd1Δ::TRP1 in KHY270 | Haynes et al. (2002) |
| KHY298 | prc1Δ::HIS3 in KHY163 | Haynes et al. (2002) |
| KHY299 | prc1Δ::HIS3 in KHY171 | Haynes et al. (2002) |
| KHY306 | sec12-4 in KHY163 | Haynes et al. (2002) |
| KHY318 | prc1Δ::HIS3 in KHY270 | Haynes et al. (2004) |
| KHY388 | prc1Δ::HIS3 in KHY306 | This study |
| KHY401 | prc1Δ::HIS3 in KHY252 | This study |
| YPH499 | MATa ura3-52 leu2-Δ1 his3-Δ200 trp1-Δ63 lys2-801 ade2-101 | Ghislain et al. (1993) |
| KHY516 | prc1Δ::HIS3 in YPH499 | This study |
| CMY765 | MATα cim5-1 ura3-52 leu2-Δ1 his3-Δ200 | Ghislain et al. (1993) |
| KHY517 | prc1Δ::HIS3 in CMY765 | This study |
| KHY583 | pep4Δ::HIS3 in KHY163 | This study |
| KHY662 | pep4Δ::HIS3 in KHY171 | This study |
| LHY434 | MATa rsp5Δ::HIS3 leu2::RSP5::LEU2 bar1Δ::HIS3 his3 lys2 trp1 ura3 | Dunn et al. (2001) |
| KHY741 | prc1Δ::TRP1 in LHY434 | This study |
| LHY433 | MATa rsp5Δ::HIS3 leu2::rsp5-2::LEU2 bar1Δ::HIS3 his3 lys2 trp1 ura3 | Dunn et al. (2001) |
| KHY747 | prc1Δ::TRP1 in LHY433 | This study |
Radiolabeling, Immunoprecipitation, Cross-Linking, and Antibodies
Radiolabeling and immunoprecipitation were performed as described previously (Hill and Cooper, 2000; Haynes et al., 2002) with the following modification. Immunoprecipitated proteins were digested with endoglycosidase H (Endo H, New England Biolabs, Beverly, MA), according to manufacturers instructions, to “collapse” hyperglycosylated forms of substrates to a quantifiable form on SDS-PAGE. The hyperglycosylation of trafficked CPY* originally obscured the vacuolar delivery of CPY* (Haynes et al., 2002), which has now been revealed through degylcosylation by Endo H. Individual experiments were performed multiple times, and representative gels and quantification are shown. Sequential immunoprecipitation experiments were performed as described with some modifications (Graham et al., 1998). Briefly, 1 OD (600 nm) of cells was harvested and radiolabeled, and a primary immunoprecipitation was performed. Samples from the first immunoprecipitation were washed and resuspended in 50 μl of 8 M urea, 1% SDS, 2.5% β-mercaptoethanol, heated at 100°C for 5 min, and diluted, and a second immunoprecipitation was performed using anti-CPY or anti-α-1,6-mannose antibodies (provided by Dr. Howard Riezman, Biozentrum-University of Basel, Switzerland). Cross-linking experiments were as described previously with some modifications (Graham et al., 1998). Briefly, 2 ODs (600 nm) of cells were harvested and radiolabeled as described. Cells were pretreated with 50 mM Tris, pH 9.5, 10 mM DTT for 5 min at 30°C and converted to spheroplasts in 1.2 M sorbitol, 50 mM KPi, pH 7.5, 2 mM MgCl2, 10 mM sodium azide, and 32.5 μl 1 mg/ml zymolyase 100T (Seikagaku America, East Falmouth, MA) for 45 min at 30°C. Cells were washed in 1.2 M sorbitol before the addition of the reversible cross-linking reagent dithio-bis-succinimidylpropionate (DSP, Pierce) to a final concentration of 0.8 mM and then incubated for 40 min at room temperature. The cross-linker was quenched with the addition 50 mM Tris, pH 8, and placed on ice for 10 min. Cells were lysed with the addition of SDS to 1% and heated at 50°C for 10 min. Lysates were precleared, and primary immunoprecipitations were performed under nonreducing denaturing conditions using polyclonal antibodies against hemagglutinin or CPY. Samples from the first immunoprecipitation were washed and resuspended in 50 μl of 8 M urea, 1% SDS, 1.5% β-mercaptoethanol, heated at 100°C for 5 min, diluted, and a second immunoprecipitation performed using anti-CPY antibodies. Immunoprecipitated proteins were denatured and digested with Endo H, and samples were resolved by SDS-PAGE. The gels were fixed, dried, and exposed to a phosphor screen, and data were collected using a Phosphorimager system (Molecular Dynamics, Sunnyvale, CA) and quantified as previously described (Hill and Cooper, 2000). Rabbit polyclonal antiserum against CPY was raised against antigen expressed in Escherichia coli by Strategic Biosolutions (Newark, DE) as described previously (Hahm and Chung, 2001).
RESULTS
ER Exit of Both wtCPY and Misfolded CPY* Is Dependent on the ER Export Cargo Receptor Erv29p
Misfolded proteins typically remain in the ER where the ERAD machinery recognizes and translocates the misfolded proteins to the cytosol before ubiquitination and proteasomal degradation (Ellgaard and Helenius, 2003; McCracken and Brodsky, 2003). Surprisingly however, a number of ERAD substrates exit the ER and traffic to the Golgi (Caldwell et al., 2001; Vashist et al., 2001; Taxis et al., 2002; Vashist and Ng, 2004). An example of a misfolded protein capable of “escaping” the ER is CPY*, a luminal substrate that exits the ER and reaches the cis Golgi, as evidenced by Golgi-specific α-1,6-mannosylation (Franzusoff and Schekman, 1989; Haynes et al., 2002).
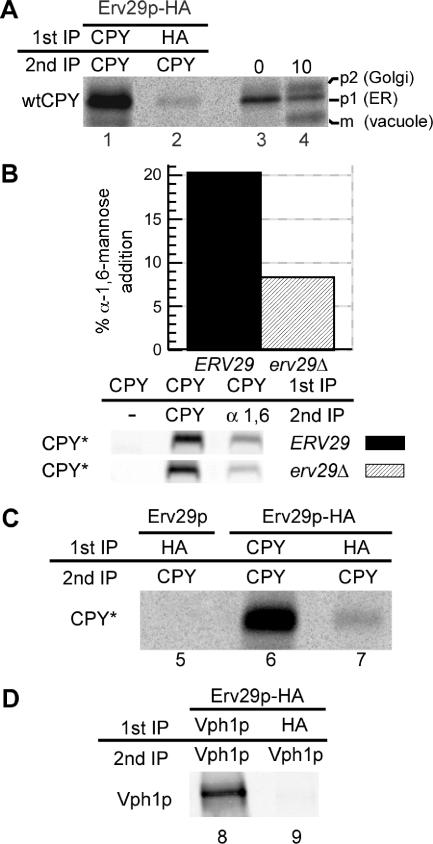
Why do some misfolded proteins remain in the ER, whereas others traffic to the Golgi? Is there a common pathway by which nonnative and correctly folded proteins exit the ER? Correctly folded proteins can exit the ER via incorporation into COPII vesicles by virtue of cis ER exit signals within cargo proteins interacting directly with COPII components or with ER export cargo receptors such as Erv29p (Belden and Barlowe, 2001; Barlowe, 2003; Lee et al., 2004; Otte and Barlowe, 2004). Erv29p is required for forward transport of correctly folded luminal cargo from the ER, such as wtCPY, and in erv29Δ cells the forward trafficking of wtCPY is significantly delayed but not abolished (Belden and Barlowe, 2001; Caldwell et al., 2001). The ER form of wtCPY (p1) is delivered to the Golgi and subsequently receives Golgi-specific glycosylation to produce the p2 form of wtCPY that upon delivery to the vacuole is proteolytically converted to the mature (m) form. Binding between Erv29p and wtCPY has not been observed, nor has binding between Erv29p and any of its cargoes been demonstrated in vivo. We expected Erv29p to bind the ER (p1) form of wtCPY, incorporate into COPII vesicles, and then dissociate from the wtCPY before or upon fusion with the cis Golgi. To observe this likely highly dynamic interaction in vivo, we used cross-linking and immunoprecipitation and found an association between Erv29p and wtCPY (Figure 1A). Approximately 2% of the radiolabeled wtCPY was cross-linked to Erv29p, comparable to the ∼1% of gpαf found cross-linked to Erv29p (Belden and Barlowe, 2001).
Figure 1.
ER exit of both wtCPY and misfolded CPY* is dependent on the ER export cargo receptor Erv29p. (A) erv29Δ cells (KHY271) expressing wtCPY and Erv29p-HA (pAC530) were grown at 30°C and radiolabeled with 35S methionine/cysteine for 6 min, and spheroplasts were treated with the cross-linking agent, DSP. Cell lysates were immunoprecipitated with anti-CPY (lane 1) or anti-HA antibodies (lane 2), the cross-linker was cleaved, and proteins reimmunoprecipitated with anti-CPY antibodies. Immunoprecipitated proteins were separated by SDS-PAGE and exposed to a phosphor screen. As markers of p1, p2, and mature wtCPY, KHY271 cells expressing Erv29 (pAC505) were grown at 30°C, radiolabeled for 5 min, and chased with unlabeled methionine/cysteine. Samples were removed at indicated times, and cell extracts were prepared, and wtCPY was immunoprecipitated with anti-CPY antibodies (lanes 3 and 4). Proteins were resolved by SDS-PAGE. (B) ERV29 (KHY171) and erv29Δ cells (KHY279) expressing CPY* (pAC519) were radiolabeled for 20 min and chased for 5 min, and CPY* was immunoprecipitated. The samples were solubilized, reimmunoprecipitated with either anti-CPY or anti-α-1,6-mannose antibodies, and treated with Endo H followed by SDS-PAGE and quantification. Graphic representation of the α-1,6-mannose modification of CPY* in ERV29 and erv29Δ cells is shown. (C) erv29Δ (KHY270) cells expressing Erv29p (pAC505, lane 5) or Erv29p-HA (pAC530, lane 6 and 7) were radiolabeled for 10 min before cross-linking and immunoprecipitation with anti-HA or anti-CPY antibodies and then reimmunoprecipitated with anti-CPY antibodies. Samples were treated with Endo H before SDS-PAGE. (D) erv29Δ (KHY270) cells expressing Vph1p (pMM322) and Erv29p-HA (pAC530) were radiolabeled for 6 min before cross-linking and immunoprecipitation with anti-HA or anti-Vph1 antibodies and then reimmunoprecipitated with anti-Vph1 antibodies.
In addition to mediating ER exit of the correctly folded protein wtCPY, Erv29p likely traffics the misfolded counterpart CPY* from the ER, as CPY* degradation is impaired in erv29Δ cells (Caldwell et al., 2001; Haynes et al., 2002). These data suggest that the subset of misfolded proteins capable of trafficking from the ER incorporate into vesicles in the same manner as correctly folded proteins. To determine if the trafficking of misfolded CPY* is mediated by Erv29p, we analyzed the extent of cis Golgi-specific α-1,6-mannosylation of CPY* in the presence or absence of Erv29p. CPY* α-1,6-mannosylation was decreased nearly 2.5-fold in erv29Δ cells relative to wild-type cells (Figure 1B), consistent with the trafficking defect of wtCPY in erv29Δ cells (Caldwell et al., 2001). Cross-linking provided direct evidence that Erv29p binds misfolded CPY* in vivo, as it does wtCPY (Figure 1C). The specificity of Erv29p binding of wtCPY and CPY* was demonstrated by the absence of cross-linking between Erv29p and newly synthesized Vph1p in the ER (Figure 1D). Together these data suggest that a nonnative protein may still contain a correctly folded domain that includes the functional ER export signal possessed by the corresponding correctly folded protein and therefore can traffic from the ER. In this model, the presence or absence of functional ER export signals distinguishes misfolded proteins that exit the ER from those that remain in the ER. Although termed misfolded, CPY* maintains the ability to bind Erv29p and exits the ER via the same mechanisms as correctly folded wtCPY. Disabling this interaction results in decreased delivery of CPY* to the Golgi as was observed in erv29Δ cells (Figure 1B).
ER Exit of Misfolded Proteins Is Dependent on Functional ER Exit Signals
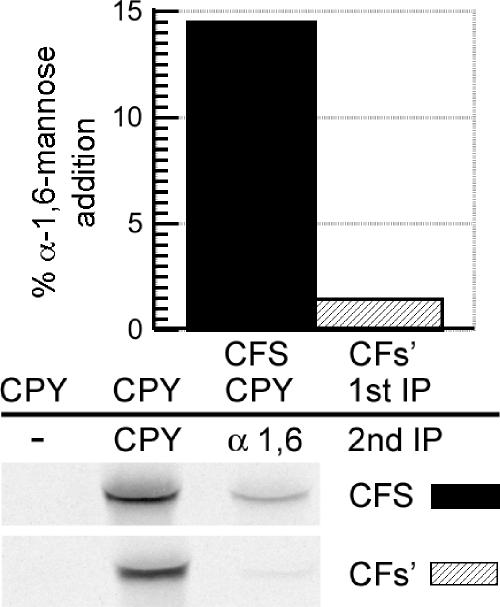
The involvement of Erv29p in the ER exit of CPY* suggests that, although an aberrant protein may not achieve the native conformation, it may still present a correctly folded ER exit signal. To further investigate the possibility that intact ER exit signals mediate trafficking of misfolded proteins, we utilized a misfolded protein to which a defined ER export signal was appended. The di-acidic motif DxE200 is a well-characterized ER exit signal in the cytosolic domain of the yeast membrane-spanning protein Sys1p. DxE200 is critical for ER exit because the AxA200 mutation significantly retarded exit, whereas addition of the DxE200 signal to an ER resident protein resulted in export (Votsmeier and Gallwitz, 2001). Furthermore, peptides containing the wild-type Sys1p export signal DxE200 could compete for binding the Sec23/24p complex of the COPII coat, whereas the peptide with the mutant signal could not (Miller et al., 2003). We engineered reporter proteins comprised of CPY*, the membrane-spanning domain from the single membrane-spanning protein Fus1p and either the wild-type (S = DxE200) or ER export deficient (s′ = AxA200) Sys1p cytosolic domain. The resulting proteins, CFS and CFs′, were used to investigate the hypothesis that a competent ER export signal can result in trafficking of a misfolded protein from the ER, whereas the absence of a competent signal would contribute to a protein remaining in the ER. CFS and CFs′ both received N-linked glycosylation in the ER as expected and achieved the correct membrane orientation (unpublished data). We anticipated CFS, with an intact cytosolic ER export signal, would traffic efficiently to the Golgi, whereas CFs′ with an impaired ER exit signal, would traffic to a lesser extent. CFS was found to receive ∼10-fold more α-1,6-mannosylation than CFs′ (Figure 2). This finding supports the hypothesis that a competent ER export signal can play a significant role in the ER exit and Golgi delivery of misfolded substrates.
Figure 2.
Delivery of misfolded proteins to the Golgi is dependent on ER exit signals. KHY517 cells expressing CFS (pAC812) or CFs′ (pAC815) were radiolabeled for 15 min and chased for 10 min, and CFS or CFs′ were immunoprecipitated with anti-CPY antibodies. Samples were solubilized, diluted, and reimmunoprecipitated with either anti-CPY or anti-α-1,6-mannose antibodies followed by treatment with Endo H. Graphic representation of the α-1,6-mannose modification of CFS and CFs′ is shown.
CFS Efficiently Exits the ER and Is Transported to the Vacuole, Whereas CFs′ Remains in the ER as an ERAD Substrate Degraded by the Proteasome
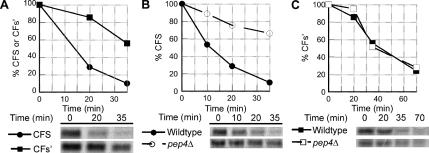
The turnover kinetics of CFS and CFs′ was examined and found to differ significantly, with CFS degraded three- to four-fold more rapidly than CFs′ (Figure 3A). ER exit of misfolded proteins could result in vacuolar delivery, and delivery of CFS to the proteolytically charged vacuole could account for faster turnover. Degradation of CFS and CFs′ was examined in wild-type cells and cells lacking vacuolar proteases (pep4Δ). CFS was strongly stabilized in pep4Δ cells (Figure 3B), whereas CFs′ was unaffected (Figure 3C), demonstrating that CFS is delivered to the vacuole, whereas CFs′ is not. ER exit of CFS was not due to an ER overload response, as observed in virally infected cells synthesizing very large amounts of cargo in the ER, as CFS and CFs′ were both under transcriptional control of the PRC1/CPY promoter and expressed to the same relatively low level.
Figure 3.
ER exit of misfolded proteins and subsequent vacuolar delivery and degradation depends on ER exit signal efficiency. (A) Wild-type (KHY298) cells expressing CFS (pAC812) or CFs′ (pAC815) were subjected to pulse-chase analysis as described. Graphic representations of the degradation of CFS and CFs′ are shown. (B and C) Wild-type (KHY298) and pep4Δ (KHY401) cells expressing CFS (pAC812) or CFs′ (pAC815) were subjected to pulse-chase analysis. Graphic representations of the degradation of CFS (B) and CFs′ (C) in wild-type and pep4Δ cells are shown.
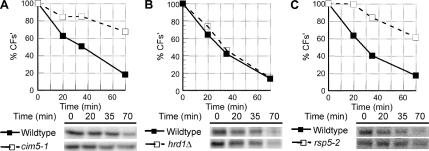
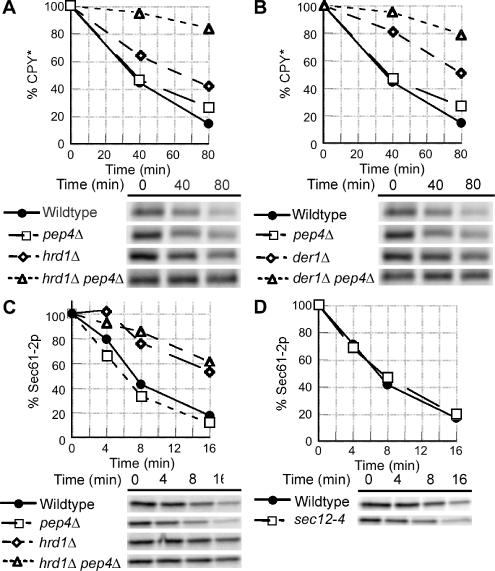
Like many misfolded proteins that remain in the ER, CFs′ is likely retrotranslocated and degraded by the 26S proteasome (Ellgaard and Helenius, 2003; McCracken and Brodsky, 2003). Degradation of CFs′ was significantly stabilized in cim5-1 cells that have reduced proteasomal activity (Ghislain et al., 1993), confirming CFs′ is an ERAD substrate (Figure 4A). The turnover of CFs′ is independent of the ERAD E3 ubiquitin ligases Hrd1p and Doa10p as CFs′ degradation was unaffected in hrd1Δ, doa10Δ, and hrd1Δ doa10Δ cells (Figure 4B and unpublished data). Previous work demonstrated misfolded proteins can be ubiquitinated by Rsp5p (Haynes et al., 2002), a ubiquitin ligase known to act on ER membrane substrates (Hoppe et al., 2000). Turnover of CFs′ was Rsp5p-dependent, as CFs′ degradation was significantly impaired in rsp5-2 cells at the restrictive temperature compared with wild-type cells (Figure 4C; Gitan and Eide, 2000; Haynes et al., 2002). In wild-type cells, CFS is degraded three- to four-fold faster than CFs′ (Figure 3A), likely due to rapid delivery of CFS to the vacuole where it is swiftly degraded. However most of CFs′ remains in the ER and is subject to the complex, multistep processes of ERAD.
Figure 4.
CFs′ is an ERAD substrate and degraded by the proteasome. (A) Wild-type (KHY516) and cim5-1 (KHY517) cells expressing CFs′ (pAC815) were subjected to pulse-chase analysis as described. The degradation rate of CFs′ in wild-type and cim5-1 cells is shown. (B) Degradation of CFs′ was determined in wild-type (KHY298) and hrd1Δ (KHY299) cells. (C) Wild-type (KHY741) and rsp5-2 (KHY747) cells expressing CFs′ (pAC815) were grown at 24°C and shifted to the restrictive temperature (38°C) for 20 min before pulse-chase analysis. Graphic representations of the degradation of CFs′ in wild-type and rsp5-2 cells are shown.
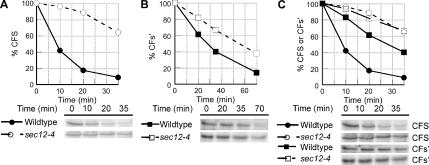
Given the vacuolar-dependent degradation of CFS, blocking ER exit using a sec12-4 conditional allele would be expected to prevent vacuolar delivery and therefore impair degradation of CFS. We examined the effect of blocking ER exit on CFS degradation and found CFS was significantly stabilized in sec12-4 cells at the restrictive temperature (Figure 5A), similar to the effect seen in pep4Δ cells (Figure 3B). The turnover of CFs′ was relatively unaffected when ER exit was blocked in sec12-4 cells (Figure 5B), compared with CFS.
Figure 5.
ER exit of misfolded proteins depends on ER exit signal efficiency. (A and B) Wild-type (KHY298) and sec12-4 (KHY388) cells expressing CFS (pAC812) or CFs′ (pAC815) were grown at 24°C and shifted to the restrictive temperature (34.5°C) for 15 min before pulse-chase analysis. Samples were treated with Endo H before SDS-PAGE analysis. Graphic representations of the degradation of CFS (A) and CFs′ (B) in wild-type and sec12-4 cells are shown. (C) Overlay of graphic representations of the degradation of CFS and CFs′ in wild-type and sec12-4 cells from data shown in A and B.
Inactivation of ERAD Results in the Increased Exit of Some Misfolded Proteins from the ER
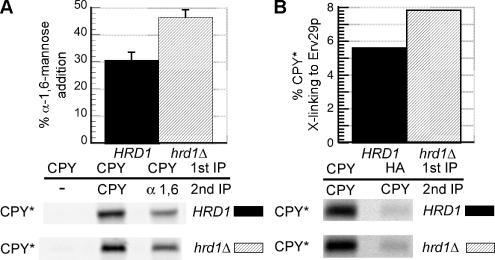
The observation that some CPY* binds Erv29p and exits the ER, whereas the remainder is degraded by ERAD, implies that within the ER, a common pool of CPY* is subject to two competing pathways, ERAD and ER exit. We therefore predicted that inactivating ERAD would result in more CPY* available for ER exit. Inactivation of the HRD/DER pathway, a well-characterized component of ERAD (Hiller et al., 1996; Knop et al., 1996; Bordallo et al., 1998; Friedlander et al., 2000; Gardner et al., 2000; Wilhovsky et al., 2000; Bays et al., 2001), resulted in increased Golgi delivery of CPY*, as evidenced by a ∼50% increase in α-1,6-mannosylation of CPY* (Figure 6A). Moreover, the interaction between Erv29p and CPY* is enhanced in hrd1Δ cells, as demonstrated by a ∼38% increase in cross-linking of CPY* to Erv29p (Figure 6B). Increased transport of CPY* to post-ER compartments could result in vacuolar delivery and degradation. In cells with functional ERAD, little to no CPY* was delivered to the vacuole, as no stabilization of the degradation of CPY* was observed in pep4Δ cells (Figure 7, A and B). However, stabilization of CPY* in hrd1Δ pep4Δ and der1Δ pep4Δ cells was significantly greater than that in either hrd1Δ or der1Δ cells alone (Figure 7, A and B). These data support the hypothesis that disabling ERAD results in increased ER exit and subsequent vacuolar delivery of CPY*.
Figure 6.
Inactivation of ERAD results in increased Golgi delivery of CPY* by allowing greater interaction with Erv29p. (A) HRD1 (KHY252) and hrd1Δ cells (KHY265) expressing CPY* were radiolabeled for 20 min and chased for 15 min, and CPY* was immunoprecipitated. The samples were then reimmunoprecipitated with either anti-CPY or anti-α-1,6-mannose antibodies followed by treatment with Endo H. Graphic representation of the α-1,6-mannose modification of CPY* in HRD1 and hrd1Δ cells is shown. The data were plotted as mean values with SDs of three independent experiments, and representative gels are shown. (B) erv29Δ (KHY270) and erv29Δ hrd1Δ (KHY279) cells expressing Erv29p-HA (pAC530) were radiolabeled for 10 min before cross-linking and immunoprecipitation with anti-CPY or anti-HA antibodies followed by reimmunoprecipitation with anti-CPY antibodies. Graphic representation of the extent of CPY* binding to Erv29p in erv29Δ (KHY270) and erv29Δ hrd1Δ (KHY279) cells is shown.
Figure 7.
Inactivation of ERAD results in increased trafficking of CPY* from the ER, but not Sec61-2p. (A) Wild-type (KHY163), pep4Δ (KHY252), hrd1Δ (KHY171), and hrd1Δ pep4Δ (KHY265) cells were grown at 30°C and radiolabeled for 10 min and chased. Samples were removed at the times indicated, cell extracts were prepared, and CPY* was immunoprecipitated. The immunoprecipitated protein was then digested with Endo H before SDS-PAGE analysis. Graphic representations of the degradation of CPY* are shown. (B) Wild-type (KHY163), pep4Δ (KHY252), der1Δ (KHY169), and der1Δ pep4Δ (KHY264) cells were grown at 30°C, radiolabeled, and chased. Aliquots were harvested at times indicated and treated as described above. Graphic representations of the degradation of CPY* are shown. (C) Wild-type (KHY163), pep4Δ (KHY583), hrd1Δ (KHY171), and hrd1Δ pep4Δ (KHY662) cells expressing Sec61-2p-HA (pAC460) were grown at 30°C, radiolabeled, and chased. Aliquots were harvested at times indicated and treated as described. Graphic representations of the degradation of Sec61-2p-HA are shown. (D) Wild-type (KHY163) and sec12-4 (KHY306) cells expressing Sec61-2p-HA (pAC460) were grown at 24°C and shifted to the restrictive temperature (34.5°C) for 15 min before radiolabeling and chase. Samples were removed at the times indicated, cell extracts were prepared, and Sec61-2p-HA was immunoprecipitated. Graphic representations of the degradation of Sec61-2p-HA in wild-type and sec12-4 cells are shown.
We also examined the effect of inactivating ERAD on another misfolded protein, Sec61-2p. Contrary to CPY*, Sec61-2p does not exit the ER upon ERAD inactivation, as evidenced by vacuolar-independent degradation in ERAD-deficient cells (Figure 7C). ER-resident proteins such as correctly folded Sec61p are expected to lack ER exit signals, and hence misfolded Sec61-2p would also lack an exit signal. Consistent with this, we found the degradation of Sec61-2p was independent of ER-Golgi trafficking as blocking ER exit in sec12-4 cells had no effect on the turnover of Sec61-2p (Figure 7D), whereas such a transport block did impair CPY* degradation (Caldwell et al., 2001; Vashist et al., 2001). These data suggest that misfolded proteins with an existing propensity to traffic from the ER (CPY*) can exit to a greater extent when ERAD is disabled; yet other misfolded proteins that fail to traffic from the ER (Sec61-2p) do not appear to exit the ER upon ERAD inactivation. This distinction suggests ER exit is not simply a consequence for all aberrant proteins in ERAD-deficient cells but is instead is substrate dependent.
DISCUSSION
The model that ER export signals play a role in trafficking of misfolded proteins from the ER is substantiated by our findings utilizing export signals on either side of the membrane (the luminal Erv29p-dependent signals found in wtCPY and CPY* and the cytosolic signal of CFS). Erv29p binds wtCPY and is required for efficient trafficking of wtCPY from the ER to the Golgi (Belden and Barlowe, 2001; Caldwell et al., 2001). Erv29p also binds misfolded CPY* and is required for transport of CPY* to the Golgi as the extent of Golgi specific α-1,6-mannosylation of CPY* was reduced in the absence of Erv29p. The analyses of CFS and CFs′, which differ only by the presence or absence of a cytosolic ER export signal, further support the role of ER export signals in ER-Golgi trafficking of misfolded proteins. CFS efficiently reaches the Golgi, as evidenced by α-1,6-mannosylation, and is subsequently degraded in the vacuole. Blocking ER exit prevents vacuolar delivery and significantly stabilizes CFS degradation. Conversely, without an efficient export signal, little CFs′ reaches the Golgi or vacuole. The turnover of CFs′ is relatively independent of ER-Golgi trafficking and is instead subject to the complex, multistep processes of ERAD. Negating this difference by maintaining both CFS and CFs′ in the ER in sec12-4 cells rendered them “equivalent” substrates and resulted in slowing of CFS degradation to equal that of CFs′ (Figure 5C). These data support the idea that misfolded proteins exit the ER by the same mechanisms responsible for trafficking of wild-type proteins, which likely involves presentation of an intact ER export signal despite the misfolding of the aberrant protein.
Misfolded proteins within the ER need not be classified into two mutually exclusive groups: 1) those incapable of ER exit and consequently degraded solely by ERAD and 2) misfolded proteins capable of ER exit that are degraded independently of ERAD. More likely, there will be a range of degradative outcomes for many misfolded proteins involving ERAD and vacuolar/lysosomal degradation or secretion. At one end of the spectrum are substrates such as Sec61-2p, which are solely degraded by ERAD. Contrary to Sec61-2p, CFS contains a strong cytosolic exit signal, exits the ER rapidly and is degraded in the vacuole in an ERAD-independent manner. CPY* lies between these two ends of the spectrum and is degraded primarily by ERAD but maintains the ability to exit the ER via Erv29p, resulting in a portion of the CPY* population trafficking from the ER.
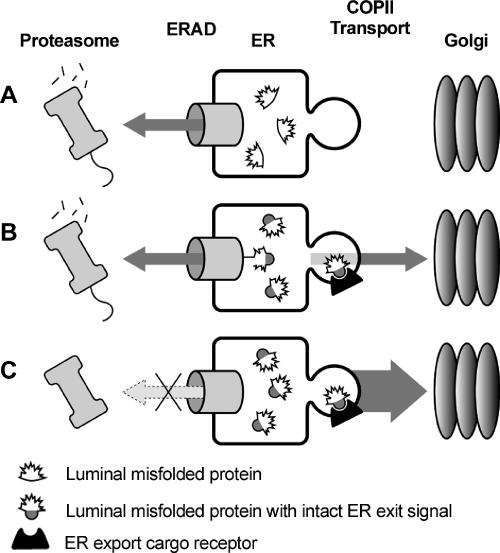
The relative export signal strength of any aberrant protein versus the affinity for ERAD machinery would likely determine what fraction, if any, of a misfolded substrate exits the ER (Figure 8). Thus misfolded proteins with ER export signals are likely subject to a dynamic interplay in which both ERAD and ER exit “compete” for binding of misfolded proteins (Figure 8B). Inactivating one mechanism (ERAD) can result in the greater dependency on the other (ER exit; Figure 8C). Inactivating ERAD enhanced ER export of CPY* that contains an ER export signal. As predicted, however, ERAD inactivation did not cause export of Sec61-2p that likely lacks a functional ER export signal. This is an important consideration for those seeking possible therapeutic approaches involving inactivating ERAD in anticipation of exporting a partially active substrate.
Figure 8.
ERAD and ER exit can compete for misfolded substrates. (A) Misfolded proteins lacking ER exit signals remain in the ER and are degraded by ERAD. (B) Misfolded proteins with intact ER exit signals are engaged by ERAD machinery for dislocation and proteasomal degradation. However, these misfolded proteins can also exit the ER via interaction with ER exit receptors (for soluble substrates) or via direct interaction of cytosolic signals with COPII components (for membrane-spanning substrates). (C) Inactivating ERAD results in greater ER exit of misfolded proteins containing ER export signals.
Could export of misfolded proteins be simply due to a loss of active retention? Does the inactivation of ERAD lead to loss of retention, resulting in escape of misfolded proteins to distal compartments? Evidence argues against this proposal. 1) If ERAD inactivation causes a loss of active retention and therefore ER export, then Sec61-2p would be expected to “passively” exit the ER when ERAD is inactivated. Yet Sec61-2p is not further stabilized in a hrd1Δ pep4Δ strain relative to a hrd1Δ strain (Figure 7C). 2) ER export of CPY* in ERAD-deficient cells remains dependent on Erv29p (Figure 6, A and B). If ERAD inactivation (hrd1Δ) causes a loss of active retention and misfolded proteins then passively exit, inactivation of active transport machinery (erv29Δ) in hrd1Δ cells would have no additional effect on CPY* degradation. However, the degradation of CPY* is greatly stabilized in hrd1Δ erv29Δ and hrd1Δ sec12-4 cells, suggesting that vacuolar delivery occurs via an active and export receptor-dependent manner (Haynes et al., 2002). If CPY* delivery to the vacuole in hrd1Δ cells is due to loss of ER retention, then the removal of Erv29p should not affect this process.
The observation that a number of misfolded proteins exit the ER, but do not reach the vacuole implies a complex fate of the misfolded proteins reaching the Golgi. It appears some misfolded proteins that traffic to the Golgi may return to the ER (Vashist et al., 2001; Haynes et al., 2002), though the mechanism for retrograde trafficking of misfolded proteins has not been determined. Increased Golgi delivery of misfolded proteins may potentially saturate the return mechanism, resulting in enhanced delivery of misfolded proteins to distal compartments, such as the vacuole. Trafficking to the Golgi is not essential for substrate degradation but may confer an advantage, which is lost when exit is blocked, thereby slowing the rate of turnover in trafficking mutants. The nature of this advantage remains to be determined but may involve a Golgi attained modification that increases the affinity for ERAD components or alternatively results in delivery to a specialized ER subcompartment.
Alternative models have been proposed to account for why efficient degradation of some misfolded proteins requires ER exit (Vashist et al., 2001; Kostova and Wolf, 2003; Vashist and Ng, 2004). However, these models do not mechanistically explain why only some misfolded proteins are affected when trafficking is blocked or why the extent of the effect varies between different misfolded proteins. One model proposes the slowed turnover of some substrates upon cessation of trafficking is an indirect effect of ER perturbation(s) (Kostova and Wolf, 2003). How such perturbation results in a significant impact on CFS degradation, some effect on CPY*, a minor impact on CFs′, and no effect on Sec61-2p or unassembled Vph1p remains unclear (Hill and Stevens, 1994; Caldwell et al., 2001; Vashist et al., 2001; Taxis et al., 2002). ERAD inactivation was also proposed to cause ER-Golgi morphological changes that result in CPY* “escape” (Kostova and Wolf, 2003). Why such morphological changes would increase CPY* binding by Erv29p is uncertain (Figure 6B). Another model proposes that the basis for either ER retention or ER-Golgi trafficking of misfolded proteins is dependent upon the subcellular location of the mutated residue(s), relying on distinct and temporally ordered cytosolic and luminal checkpoints termed ERAD-C and ERAD-L (Vashist and Ng, 2004). The ERAD-L/C model is unclear as to what machinery is responsible for exporting only misfolded substrates with luminal mutations but not substrates with cytosolic mutations. Furthermore, why would this export machinery be distinct from that which exports correctly folded proteins (Vashist et al., 2001)? Finally, if the cell purposely harnesses the advantage of exporting luminally misfolded substrates, then why not export all misfolded substrates? The anterograde trafficking aspects of the ERAD-C/L model involves unidentified ERAD components for specific targeting of misfolded proteins with luminal mutations to the Golgi. Contrary to this model we find rather than deliberate targeting of aberrant proteins to the Golgi by ERAD components, ER exit signals play a role in the ER exit of misfolded proteins. The chimeric ERAD-L substrates used to formulate the ERAD-C/L model contain correctly encoded cytosolic domains of a plasma membrane protein likely to contain an ER exit signal (Vashist and Ng, 2004). In contrast, the nontrafficking Ste6*/Ste6-166 based ERAD-C substrates contain a large deletion of the cytosolic domain, which likely removes or affects presentation of the ER exit signal, and could account for them remaining in the ER (Vashist and Ng, 2004).
Loss or deletion of the ER export signal, or disruption of signal presentation, likely contributes to misfolded proteins remaining in the ER, providing a passive mechanism to prevent ERAD substrates from exiting. Furthermore, chaperones binding to a misfolded domain on either side of the membrane could sterically interfere with incorporation into COPII vesicles or exit signal presentation and account for a failure to exit. The absence of functional export signals on misfolded proteins provides an attractive model for why such proteins remain in the ER but does not dismiss that an active retention system may also exist and in fact the two processes may coexist.
ACKNOWLEDGMENTS
We thank Cole Haynes, Kathryn Hill, Chris Kincaid, Stephen King, and Tom Menees for assistance and critical reading of the manuscript; Drs. Dieter Gallwitz and Howard Riezman for reagents; and Dr. Randy Schekman for suggestions regarding fusion construction. M.M.K. was supported by the UMKC Chancellor's Fellowship. This work was supported by National Institutes of Health Grant GM55848.
Abbreviations used:
- CFS
CPY* luminal domain/Fus1p transmembrane domain/Sys1p cytosolic domain
- CFs′
CPY* luminal domain/Fus1p transmembrane domain/sys1p (AxA200) cytosolic domain
- CPY
carboxypeptidase Y
- wtCPY
correctly folded carboxypeptidase Y
- CPY*
mutant carboxypeptidase Y
- ER
endoplasmic reticulum
- ERAD
ER-associated degradation
- ERQC
ER quality control
- gpαf
glycosylated pro-α-factor
- wtPrA
correctly folded proteinase A
- PrA*
mutant PrA.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E06-08-0696) on November 17, 2006.
REFERENCES
- Aridor M., Weissman J., Bannykh S., Nuoffer C., Balch W. E. Cargo selection by the COPII budding machinery during export from the ER. J. Cell Biol. 1998;141:61–70. doi: 10.1083/jcb.141.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlowe C. Signals for COPII-dependent export from the ER: what's the ticket out? Trends Cell Biol. 2003;13:295–300. doi: 10.1016/s0962-8924(03)00082-5. [DOI] [PubMed] [Google Scholar]
- Bays N. W., Gardner R. G., Seelig L. P., Joazeiro C. A., Hampton R. Y. Hrd1p/Der3p is a membrane-anchored ubiquitin ligase required for ER-associated degradation. Nat. Cell Biol. 2001;3:24–29. doi: 10.1038/35050524. [DOI] [PubMed] [Google Scholar]
- Belden W. J., Barlowe C. Role of Erv29p in collecting soluble secretory proteins into ER-derived transport vesicles. Science. 2001;294:1528–1531. doi: 10.1126/science.1065224. [DOI] [PubMed] [Google Scholar]
- Bordallo J., Plemper R. K., Finger A., Wolf D. H. Der3p/Hrd1p is required for endoplasmic reticulum-associated degradation of misfolded lumenal and integral membrane proteins. Mol. Biol. Cell. 1998;9:209–222. doi: 10.1091/mbc.9.1.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodsky J. L., McCracken A. A. ER protein quality control and proteasome-mediated protein degradation. Semin. Cell Dev. Biol. 1999;10:507–513. doi: 10.1006/scdb.1999.0321. [DOI] [PubMed] [Google Scholar]
- Caldwell S. R., Hill K. J., Cooper A. A. Degradation of endoplasmic reticulum (ER) quality control substrates requires transport between the ER and Golgi. J. Biol. Chem. 2001;276:23296–23303. doi: 10.1074/jbc.M102962200. [DOI] [PubMed] [Google Scholar]
- Coughlan C. M., Walker J. L., Cochran J. C., Wittrup K. D., Brodsky J. L. Degradation of mutated bovine pancreatic trypsin inhibitor in the yeast vacuole suggests post-endoplasmic reticulum protein quality control. J. Biol. Chem. 2004;279:15289–15297. doi: 10.1074/jbc.M309673200. [DOI] [PubMed] [Google Scholar]
- Deshaies R. J., Sanders S. L., Feldheim D. A., Schekman R. Assembly of yeast Sec proteins involved in translocation into the endoplasmic reticulum into a membrane-bound multisubunit complex. Nature. 1991;349:806–808. doi: 10.1038/349806a0. [DOI] [PubMed] [Google Scholar]
- Dunn R., Hicke L. Domains of the Rsp5 ubiquitin-protein ligase required for receptor-mediated and fluid-phase endocytosis. Mol. Biol. Cell. 2001;12:421–435. doi: 10.1091/mbc.12.2.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellgaard L., Helenius A. Quality control in the endoplasmic reticulum. Nat. Rev. Mol. Cell Biol. 2003;4:181–191. doi: 10.1038/nrm1052. [DOI] [PubMed] [Google Scholar]
- Franzusoff A., Schekman R. Functional compartments of the yeast Golgi apparatus are defined by the sec7 mutation. EMBO J. 1989;8:2695–2702. doi: 10.1002/j.1460-2075.1989.tb08410.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedlander R., Jarosch E., Urban J., Volkwein C., Sommer T. A regulatory link between ER-associated protein degradation and the unfolded-protein response. Nat. Cell Biol. 2000;2:379–384. doi: 10.1038/35017001. [DOI] [PubMed] [Google Scholar]
- Gardner R. G., Swarbrick G. M., Bays N. W., Cronin S. R., Wilhovsky S., Seelig L., Kim C., Hampton R. Y. Endoplasmic reticulum degradation requires lumen to cytosol signaling. Transmembrane control of Hrd1p by Hrd3p. J. Cell Biol. 2000;151:69–82. doi: 10.1083/jcb.151.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghislain M., Udvardy A., Mann C. S. cerevisiae 26S protease mutants arrest cell division in G2/metaphase. Nature. 1993;366:358–362. doi: 10.1038/366358a0. [DOI] [PubMed] [Google Scholar]
- Gitan R. S., Eide D. J. Zinc-regulated ubiquitin conjugation signals endocytosis of the yeast ZRT1 zinc transporter. Biochem. J. 2000;346(Pt 2):329–336. [PMC free article] [PubMed] [Google Scholar]
- Graham L. A., Hill K. J., Stevens T. H. Assembly of the yeast vacuolar H+-ATPase occurs in the endoplasmic reticulum and requires a Vma12p/Vma22p assembly complex. J. Cell Biol. 1998;142:39–49. doi: 10.1083/jcb.142.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahm M. S., Chung B. H. Refolding and purification of yeast carboxypeptidase Y expressed as inclusion bodies in Escherichia coli. Protein Expr. Purif. 2001;22:101–107. doi: 10.1006/prep.2001.1418. [DOI] [PubMed] [Google Scholar]
- Hampton R. Y., Gardner R. G., Rine J. Role of 26S proteasome and HRD genes in the degradation of 3-hydroxy-3-methylglutaryl-CoA reductase, an integral endoplasmic reticulum membrane protein. Mol. Biol. Cell. 1996;7:2029–2044. doi: 10.1091/mbc.7.12.2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes C. M., Caldwell S., Cooper A. A. An HRD/DER-independent ER quality control mechanism involves Rsp5p-dependent ubiquitination and ER-Golgi transport. J. Cell Biol. 2002;158:91–101. doi: 10.1083/jcb.200201053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes C. M., Titus E. A., Cooper A. A. Degradation of misfolded proteins prevents ER-derived oxidative stress and cell death. Mol. Cell. 2004;15:767–776. doi: 10.1016/j.molcel.2004.08.025. [DOI] [PubMed] [Google Scholar]
- Hermosilla R., Oueslati M., Donalies U., Schonenberger E., Krause E., Oksche A., Rosenthal W., Schulein R. Disease-causing V(2) vasopressin receptors are retained in different compartments of the early secretory pathway. Traffic. 2004;5:993–1005. doi: 10.1111/j.1600-0854.2004.00239.x. [DOI] [PubMed] [Google Scholar]
- Hill J. E., Myers A. M., Koerner T. J., Tzagoloff A. Yeast/E. coli shuttle vectors with multiple unique restriction sites. Yeast. 1986;2:163–167. doi: 10.1002/yea.320020304. [DOI] [PubMed] [Google Scholar]
- Hill K., Cooper A. A. Degradation of unassembled Vph1p reveals novel aspects of the yeast ER quality control system. EMBO J. 2000;19:550–561. doi: 10.1093/emboj/19.4.550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill K. J., Stevens T. H. Vma21p is a yeast membrane protein retained in the endoplasmic reticulum by a di-lysine motif and is required for the assembly of the vacuolar H(+)-ATPase complex. Mol. Biol. Cell. 1994;5:1039–1050. doi: 10.1091/mbc.5.9.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiller M. M., Finger A., Schweiger M., Wolf D. H. ER degradation of a misfolded luminal protein by the cytosolic ubiquitin-proteasome pathway. Science. 1996;273:1725–1728. doi: 10.1126/science.273.5282.1725. [DOI] [PubMed] [Google Scholar]
- Hong E., Davidson A. R., Kaiser C. A. A pathway for targeting soluble misfolded proteins to the yeast vacuole. J. Cell Biol. 1996;135:623–633. doi: 10.1083/jcb.135.3.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoppe T., Matuschewski K., Rape M., Schlenker S., Ulrich H. D., Jentsch S. Activation of a membrane-bound transcription factor by regulated ubiquitin/proteasome-dependent processing. Cell. 2000;102:577–586. doi: 10.1016/s0092-8674(00)00080-5. [DOI] [PubMed] [Google Scholar]
- Jarosch E., Taxis C., Volkwein C., Bordallo J., Finley D., Wolf D. H., Sommer T. Protein dislocation from the ER requires polyubiquitination and the AAA-ATPase Cdc48. Nat. Cell Biol. 2002;4:134–139. doi: 10.1038/ncb746. [DOI] [PubMed] [Google Scholar]
- Johnson A. E., van Waes M. A. The translocon: a dynamic gateway at the ER membrane. Annu. Rev. Cell Dev. Biol. 1999;15:799–842. doi: 10.1146/annurev.cellbio.15.1.799. [DOI] [PubMed] [Google Scholar]
- Knop M., Finger A., Braun T., Hellmuth K., Wolf D. H. Der1, a novel protein specifically required for endoplasmic reticulum degradation in yeast. EMBO J. 1996;15:753–763. [PMC free article] [PubMed] [Google Scholar]
- Kostova Z., Wolf D. H. For whom the bell tolls: protein quality control of the endoplasmic reticulum and the ubiquitin-proteasome connection. EMBO J. 2003;22:2309–2317. doi: 10.1093/emboj/cdg227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuehn M. J., Herrmann J. M., Schekman R. COPII-cargo interactions direct protein sorting into ER-derived transport vesicles. Nature. 1998;391:187–190. doi: 10.1038/34438. [DOI] [PubMed] [Google Scholar]
- Le A., Graham K. S., Sifers R. N. Intracellular degradation of the transport-impaired human PiZ alpha 1-antitrypsin variant. Biochemical mapping of the degradative event among compartments of the secretory pathway. J. Biol. Chem. 1990;265:14001–14007. [PubMed] [Google Scholar]
- Lee M. C., Miller E. A., Goldberg J., Orci L., Schekman R. Bi-directional protein transport between the ER and Golgi. Annu. Rev. Cell Dev. Biol. 2004;20:87–123. doi: 10.1146/annurev.cellbio.20.010403.105307. [DOI] [PubMed] [Google Scholar]
- Lilley B. N., Ploegh H. L. A membrane protein required for dislocation of misfolded proteins from the ER. Nature. 2004;429:834–840. doi: 10.1038/nature02592. [DOI] [PubMed] [Google Scholar]
- Longtine M. S., McKenzie A., 3rd, Demarini D. J., Shah N. G., Wach A., Brachat A., Philippsen P., Pringle J. R. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Malkus P., Jiang F., Schekman R. Concentrative sorting of secretory cargo proteins into COPII-coated vesicles. J. Cell Biol. 2002;159:915–921. doi: 10.1083/jcb.200208074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manolson M. F., Proteau D., Preston R. A., Stenbit A., Roberts B. T., Hoyt M. A., Preuss D., Mulholland J., Botstein D., Jones E. W. The VPH1 gene encodes a 95-kDa integral membrane polypeptide required for in vivo assembly and activity of the yeast vacuolar H(+)-ATPase. J. Biol. Chem. 1992;267:14294–14303. [PubMed] [Google Scholar]
- McCracken A. A., Brodsky J. L. Evolving questions and paradigm shifts in endoplasmic-reticulum-associated degradation (ERAD) Bioessays. 2003;25:868–877. doi: 10.1002/bies.10320. [DOI] [PubMed] [Google Scholar]
- Miller E. A., Beilharz T. H., Malkus P. N., Lee M. C., Hamamoto S., Orci L., Schekman R. Multiple cargo binding sites on the COPII subunit Sec24p ensure capture of diverse membrane proteins into transport vesicles. Cell. 2003;114:497–509. doi: 10.1016/s0092-8674(03)00609-3. [DOI] [PubMed] [Google Scholar]
- Nishimura N., Balch W. E. A di-acidic signal required for selective export from the endoplasmic reticulum. Science. 1997;277:556–558. doi: 10.1126/science.277.5325.556. [DOI] [PubMed] [Google Scholar]
- Otte S., Barlowe C. The Erv41p-Erv46p complex: multiple export signals are required in trans for COPII-dependent transport from the ER. EMBO J. 2002;21:6095–6104. doi: 10.1093/emboj/cdf598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otte S., Barlowe C. Sorting signals can direct receptor-mediated export of soluble proteins into COPII vesicles. Nat. Cell Biol. 2004;6:1189–1194. doi: 10.1038/ncb1195. [DOI] [PubMed] [Google Scholar]
- Rabinovich E., Kerem A., Frohlich K. U., Diamant N., Bar-Nun S. AAA-ATPase p97/Cdc48p, a cytosolic chaperone required for endoplasmic reticulum-associated protein degradation. Mol. Cell. Biol. 2002;22:626–634. doi: 10.1128/MCB.22.2.626-634.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson N. G., Hamaker S. A., Patriub V., Aster J. C., Morton C. C. Subcellular localisation, secretion, and post-translational processing of normal cochlin, and of mutants causing the sensorineural deafness and vestibular disorder, DFNA9. J. Med. Genet. 2003;40:479–486. doi: 10.1136/jmg.40.7.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose M. D., Broach J. R. Cloning genes by complementation in yeast. Methods Enzymol. 1991;194:195–230. doi: 10.1016/0076-6879(91)94017-7. [DOI] [PubMed] [Google Scholar]
- Schubert U., Anton L. C., Gibbs J., Norbury C. C., Yewdell J. W., Bennink J. R. Rapid degradation of a large fraction of newly synthesized proteins by proteasomes. Nature. 2000;404:770–774. doi: 10.1038/35008096. [DOI] [PubMed] [Google Scholar]
- Sikorski R. S., Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer T., Jentsch S. A protein translocation defect linked to ubiquitin conjugation at the endoplasmic reticulum. Nature. 1993;365:176–179. doi: 10.1038/365176a0. [DOI] [PubMed] [Google Scholar]
- Swanson R., Locher M., Hochstrasser M. A conserved ubiquitin ligase of the nuclear envelope/endoplasmic reticulum that functions in both ER-associated and Matalpha2 repressor degradation. Genes Dev. 2001;15:2660–2674. doi: 10.1101/gad.933301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taxis C., Vogel F., Wolf D. H. ER-golgi traffic is a prerequisite for efficient ER degradation. Mol. Biol. Cell. 2002;13:1806–1818. doi: 10.1091/mbc.01-08-0399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas B. J., Rothstein R. Elevated recombination rates in transcriptionally active DNA. Cell. 1989;56:619–630. doi: 10.1016/0092-8674(89)90584-9. [DOI] [PubMed] [Google Scholar]
- Vashist S., Kim W., Belden W. J., Spear E. D., Barlowe C., Ng D. T. Distinct retrieval and retention mechanisms are required for the quality control of endoplasmic reticulum protein folding. J. Cell Biol. 2001;155:355–368. doi: 10.1083/jcb.200106123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vashist S., Ng D. T. Misfolded proteins are sorted by a sequential checkpoint mechanism of ER quality control. J. Cell Biol. 2004;165:41–52. doi: 10.1083/jcb.200309132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Votsmeier C., Gallwitz D. An acidic sequence of a putative yeast Golgi membrane protein binds COPII and facilitates ER export. EMBO J. 2001;20:6742–6750. doi: 10.1093/emboj/20.23.6742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Matteson J., An Y., Moyer B., Yoo J. S., Bannykh S., Wilson I. A., Riordan J. R., Balch W. E. COPII-dependent export of cystic fibrosis transmembrane conductance regulator from the ER uses a di-acidic exit code. J. Cell Biol. 2004;167:65–74. doi: 10.1083/jcb.200401035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe R., Riezman H. Differential ER exit in yeast and mammalian cells. Curr. Opin. Cell Biol. 2004;16:350–355. doi: 10.1016/j.ceb.2004.06.010. [DOI] [PubMed] [Google Scholar]
- Wilhovsky S., Gardner R., Hampton R. HRD gene dependence of endoplasmic reticulum-associated degradation. Mol. Biol. Cell. 2000;11:1697–1708. doi: 10.1091/mbc.11.5.1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y., Meyer H. H., Rapoport T. A. The AAA ATPase Cdc48/p97 and its partners transport proteins from the ER into the cytosol. Nature. 2001;414:652–656. doi: 10.1038/414652a. [DOI] [PubMed] [Google Scholar]
- Ye Y., Shibata Y., Yun C., Ron D., Rapoport T. A. A membrane protein complex mediates retro-translocation from the ER lumen into the cytosol. Nature. 2004;429:841–847. doi: 10.1038/nature02656. [DOI] [PubMed] [Google Scholar]