Abstract
Autophagy is a catabolic pathway for the degradation of cytosolic proteins or organelles and is conserved among all eukaryotic cells. The hallmark of autophagy is the formation of double-membrane cytosolic vesicles, termed autophagosomes, which sequester cytoplasm; however, the mechanism of vesicle formation and the membrane source remain unclear. In the yeast Saccharomyces cerevisiae, selective autophagy mediates the delivery of specific cargos to the vacuole, the analog of the mammalian lysosome. The transmembrane protein Atg9 cycles between the mitochondria and the pre-autophagosomal structure, which is the site of autophagosome biogenesis. Atg9 is thought to mediate the delivery of membrane to the forming autophagosome. Here, we characterize a second transmembrane protein Atg27 that is required for specific autophagy in yeast. Atg27 is required for Atg9 cycling and shuttles between the pre-autophagosomal structure, mitochondria, and the Golgi complex. These data support a hypothesis that multiple membrane sources supply the lipids needed for autophagosome formation.
INTRODUCTION
Cells must be able to respond to changes in the environment. One process that cells use to respond to internal or external stress is autophagy. Autophagy is a degradative process responsible for the rapid degradation of damaged or unnecessary organelles and a large portion of the cytosol in the lysosome/vacuole lumen (Reggiori and Klionsky, 2002; Klionsky, 2004). In addition, autophagy is involved in cellular remodeling, development, and aging and also plays a role in preventing a range of diseases including some types of cancer and neurodegeneration (reviewed in Shintani and Klionsky, 2004; Levine and Klionsky, 2004). Autophagy is conserved among all eukaryotic cells. The hallmark of the autophagic process is the sequestration of cytoplasm into a double-membrane vesicle called the autophagosome, which then docks and fuses with the lysosome/vacuole, releasing the inner vesicle into the lysosome/vacuole lumen for degradation (Reggiori and Klionsky 2002; Klionsky, 2004).
Autophagy can be a selective or a nonselective process. Studies in the yeast Saccharomyces cerevisiae have provided two examples of selective autophagy that morphologically and mechanistically overlap with nonselective autophagy. First, when cells are shifted from growth in oleic acid, a condition in which peroxisomes are essential, to a preferred carbon source, the now superfluous peroxisomes are selectively degraded by a mechanism termed pexophagy (Dunn et al., 2005). Another example is seen with import of the resident vacuolar hydrolase, aminopeptidase I (Ape1), which is targeted to the vacuole through the cytoplasm-to-vacuole targeting (Cvt) pathway. Both the Cvt pathway and pexophagy are examples of specific autophagy where the cargo, precursor Ape1 (prApe1), or peroxisomes are enwrapped in double-membrane vesicles and then transported from the cytosol directly to the vacuole lumen. In contrast to autophagy, which is induced by starvation, the Cvt pathway is biosynthetic and is constitutive in vegetative conditions.
More than 25 novel AuTophaGy-related (ATG) genes in the yeast S. cerevisiae have been identified through genetic screens of yeast mutants blocked in one of these pathways (Klionsky et al., 2003). Studies of Atg components have provided some insight into the molecular basis for the autophagy process; however, there are many questions that remain to be addressed. One of the most intriguing questions is the origin of the membrane for the double-membrane Cvt vesicles or autophagosomes. Recent data suggest that Atg9 may mark the membrane that is donated to the forming sequestering vesicles (Noda et al., 2000; Reggiori et al., 2004a). Atg9 localizes to mitochondria and cycles between this compartment and the pre-autophagosomal structure (PAS), the site of organization for Cvt vesicle and autophagosome formation (Tucker et al., 2003). Recently, we showed that Atg23 and the actin cytoskeleton are needed for anterograde delivery of Atg9 to the PAS, whereas Atg1, Atg13, Atg2, Atg18, and the phosphatidylinositol (PtdIns) 3-kinase, Vps34, are required for retrograde movement (Reggiori et al., 2004a, 2005a). In this report we show that Atg27 is another transmembrane protein required for Atg9 cycling. The localization and transit pattern of the Atg27 protein is similar to that of Atg9 and is dependent on the latter protein. Atg27, originally named Etf1, was identified as a PtdIns(3) phosphate-binding protein that is a downstream effector of Vps34 (Wurmser and Emr, 2002). Because of the correction of a sequencing error in the Saccharomyces Genome Database, the true full-length Atg27 contains 75 additional amino acids at the N terminus relative to Etf1. We discovered that Atg27 is a type I transmembrane protein with an N-terminal signal sequence, resulting in a topology opposite to that reported for Etf1. Atg27 function is required for specific types of autophagy and for efficient bulk autophagy.
MATERIALS AND METHODS
Strains and Media
The S. cerevisiae strain (BY4742) knockout library was purchased from ResGen (Invitrogen, Carlsbad, CA). The yeast strains used in this study are listed in Table 1. For gene disruptions, the entire coding regions were replaced with the S. cerevisiae TRP1, LEU2, HIS3; Kluyveromyces lactis URA3, LEU2; Schizosaccharomyces pombe HIS5; or the Escherichia coli kanr gene using PCR primers containing ∼45 bases identical to the flanking regions of the open reading frames. For the PCR-based integration of the 3xHA, green fluorescent protein (GFP), and RFP tags at the 3′ end of ATG27, ATG20, ATG9, VRG4, and PEX14 genes, pFA6a-3HA-TRP1, pFA6a-GFP-HIS3, pFA6a-GFP-KanMX, and pFA6a- mRFP-TRP1 were used as templates to generate strains expressing fusion proteins under the control of their own promoters (Longtine et al., 1998; Campbell and Choy, 2002). PCR verification and prApe1 processing were used to verify the functionality of all the fusion proteins.
Table 1.
Yeast strains used in this study
| Strain | Genotype | Reference |
|---|---|---|
| atg1Δ | BY4742; atg1Δ::KAN | ResGena |
| BY4742 | MATα his3Δ leu2Δ lys2Δ ura3Δ | ResGen |
| FRY143 | SEY6210; pep4Δ::LEU2 vps4Δ::TRP1 | Cheong et al. (2005) |
| HAY572 | TN124 atg1Δ::URA3 | Abeliovich et al.(2003) |
| IRA001 | BY4742; PEX14-GFP::HIS3 | Reggiori et al. (2005a) |
| IRA002 | BY4742; PEX14-GFP::HIS3 atg1Δ::URA3K.l. | Reggiori et al. (2005a) |
| JGY3 | SEY6210; atg18Δ::HIS5 S.p. | Stromhaug et al.(2004) |
| JHY28 | SEY6210; pep4Δ::LEU2 vps4Δ::TRP1 atg1Δ::HIS5 S.p. | This study |
| JLY43 | SEY6210; ATG9-GFP::KAN atg27Δ::HIS5 S.p. | This study |
| JLY44 | SEY6210; ATG9-GFP::HIS5 S.p. | This study |
| JLY45 | SEY6210; ATG9-GFP::HIS5 S.p. atg1Δ::URA3 | This study |
| JLY47 | SEY6210; ATG9-GFP::KANatg1Δ::URA3atg27Δ::HIS5 S.p. | This study |
| SEY6210 | MATα his3Δ leu2Δ lys2Δ ura3Δ | Robinson et al. (1988) |
| TN121 | MATa leu2-3,112 trp1 ura3-52 pho8::pho8Δ60 pho13Δ::URA3 | Noda et al.(1995) |
| TN124 | MATaleu2-3,112 ura3-52 trp1 pho8::pho8Δ60 pho13Δ::URA3 | Noda et al. (1995) |
| TVY1 | SEY6210; pep4Δ::LEU2 | Gerhardt et al. (1998) |
| Vps34tsf | SEY6210; vps34Δ::TRP1 pvps34tsf::URA3 | Stack et al. (1995) |
| WHY001 | SEY6210; atg1Δ::HIS5 S.p. | Shintani et al. (2002) |
| WLY1 | SEY6210; ATG27–3xHA::TRP1 | This study |
| WLY2 | SEY6210; atg27Δ::TRP1 | This study |
| WLY3 | TN121; atg27Δ::TRP1 | This study |
| WLY5 | SEY6210; ATG27-GFP::HIS3 | This study |
| WLY6 | SEY6210; ATG27-GFP::HIS VRG4-RFP::TRP1 | This study |
| WLY8 | SEY6210; pep4Δ::LEU2 vps4Δ::TRP1 atg27Δ::HIS3 | This study |
| WLY11 | SEY6210; atg1Δ::URAK.l.ATG27-GFP::HIS3 | This study |
| WLY18 | SEY6210; atg13Δ::LEU2ATG27-GFP::HIS3 | This study |
| WLY27 | BY4742; PEX14-GFP::HIS3 Atg27Δ::URA3 K.l. | This study |
| WLY33 | SEY6210; pep4Δ::LEU2 vam3Δ::URA3 atg27Δ::HIS3 pvam3ts414 | This study |
| WLY36 | SEY6210; pep4Δ::LEU2 vam3Δ::URA3 pvam3ts414 | This study |
| WLY40 | TN124; atg27Δ::KAN | This study |
| WLY41 | SEY6210; atg1Δ::LEU2 K.l. atg9Δ::KAN ATG27-GFP::HIS3 RFP-Ape1::LEU2 | This study |
| WLY49 | JLY45; atg27ΔC::TRP1 | This study |
| WLY50 | SEY6210; vps34Δ::TRP1 pvps34tsf::URA3 ATG27-GFP::HIS3 RFP-APE1::LEU2 | This study |
| WLY51 | SEY6210; vps34Δ::TRP1 pvps34tsf::URA3 | This study |
| WLY52 | SEY6210; atg14Δ::TRP1 ATG27-GFP::HIS3 | This study |
| WLY70 | JGY3; ATG27-GFP::TRP1 | This study |
| WLY74 | SEY6210; pep4Δ::LEU2 vam3Δ::URA3 atg1Δ::HIS5 pvam3ts414 | This study |
| WLY78 | SEY6210; atg2Δ::HIS5 ATG27-GFP::TRP1 | This study |
a Invitrogen, Carlsbad, CA.
Cells were grown in YPD (1% yeast extract, 2% peptone, 2% glucose) or synthetic minimal medium (SMD; 0.67% yeast nitrogen base, 2% glucose, supplemented with the appropriate amino acids and vitamins). For starvation experiments, cells were shifted to synthetic medium lacking nitrogen (SD-N; 0.17% yeast nitrogen base without amino acids and ammonium sulfate, but containing glucose).
Plasmids and Constructions
The carboxyl-terminal hemagglutinin (HA) fusion of Atg27 [pATG27-3xHA(416)] was made by PCR amplification of the ATG27 ORF and upstream 500 base pairs of genomic DNA, followed by ligation into the pRS416–3xHA plasmid. The plasmid pATG27K188-193A-3xHA(416), pATG27G105N-3HA(416), pATG27V17P-3HA(416), pATG27Q155N-3HA(416), and pATG27D176TD181T-3HA(416) were made using the QuikChange Site-directed Mutagenesis Kit (Stratagene, La Jolla, CA). The vectors for the gene fusion to SUC2 have been described previously (Klionsky et al., 1988). To make the ATG27-SUC2 fusion construct, the ATG27 gene was amplified from S. cerevisiae genomic DNA by PCR and cloned as a BamHI fragment. This procedure was used to generate an Atg27 N-terminal 28 amino acids-invertase fusion. The pP4I-23 and pP4I-137 (Klionsky et al., 1988), pCuGFP-AUT7(416) (Kim et al., 2001a), pATG1K54A (Abeliovich et al., 2003), and pRFP-APE1(414) (pPS130; Stromhaug et al., 2004) plasmids have been described previously. Two plasmids, pAtg27-HA(424) and pATG27K188-193A-HA(424) used for the Pho8Δ60 assay, were cloned from the pATG27–3xHA(416) and pATG27K188-193A-3xHA(416) plasmids, respectively, into the pRS424 empty vector using SacI and SalI sites.
Protein Extraction and Immunoblot Analysis
S. cerevisiae strains were generally grown at 30°C to the early midlog phase in YPD or SMD media. Cells were harvested and treated with 10% trichloroacetic acid (TCA) on ice for 20 min. After centrifugation at 16,000 × g for 5 min, cell pellets were washed with 100% acetone and air-dried. The dry cell pellets were resuspended in MURB buffer (50 mM Na2HPO4, 25 mM MES, pH 7.0, 1% SDS, 3 M urea, 0.5% β-mercaptoethanol, 1 mM NaN3, and 0.05% bromophenol blue), disrupted by vortex with an equal volume of glass beads for 5 min, and then heated at 70°C for 10 min. Aliquots (OD600 = 0.2) were resolved by SDS-PAGE and probed with appropriate antiserum.
Invertase Assay
Cells expressing the Atg27 signal sequence-invertase hybrid protein were grown to early midlog phase in SMD-URA medium and then washed in 10 mM NaN3. Cultures were divided into two aliquots, one for measuring the total activity and the other for secreted activity. Cell lysate preparation was performed as described previously (Klionsky et al., 1988). The invertase enzyme assay was done as described previously (Goldstein and Lampen, 1975), with minor modifications: We used μM glucose/min/OD600 for the unit of invertase activity, instead of using μM glucose/min/mg.
Endoglycosidase H Treatment
WLY2 cells harboring 3xHA-tagged Atg27G105N were grown to late midlog phase (OD600 = 1.0), TCA-precipitated, subjected to centrifugation at 16,000 × g, and washed once with acetone. The pellet was air-dried, and then glass beads and 100 μl elution buffer (1% SDS, 0.1 M Tris-HCl, pH 7.5, 1% 2-mercaptoethanol) were added, and the pellet was resuspended by sonication, followed by heating at 95°C for 5 min. After cooling, 900 μl of Endo H buffer (0.15 M citric acid, pH 5.5) was added, and the resuspension was subjected to centrifugation at 15,000 × g for 30 s. The lysate was then split into two equal portions and 10 μl of 100 mM PMSF and protease inhibitor cocktail were added to each tube. Endo H (10 mU) was then added to the mixture, which was incubated at 37°C overnight, boiled for 3 min, and subjected to immunoblotting.
Cell Labeling and Immunoprecipitation
Cells were grown to early midlog phase in SMD media. Ten milliliters of cells were harvested, and the cells were labeled in 200 μl SMD with 20 μCi [35S]-Trans label for 10 min at 30°C. For a nonradioactive chase, 1 ml SMD containing 0.2% yeast extract and 2 mM cysteine and methionine was added. At each indicated time point, samples were collected, precipitated with 10% TCA for 20 min on ice, and then spun down at 16,000 × g for 5 min at 4°C. The pellets were washed twice with 1 ml acetone, and the cell extracts were prepared and immunoprecipitated as described previously (Harding et al., 1995).
Microscopy
For fluorescence microscopy, yeast cells were grown in YPD or SMD selective media or starved in SD-N before imaging. Cells were visualized with a DeltaVision Spectris microscope (Applied Precision, Issaquah, WA) fitted with differential interference contrast optics and Photometrics CoolSNAP HQ camera (Roper Scientific, Tucson, AZ). The images were deconvolved with softWoRx software (Applied Precision). Electron microscopy was performed as described previously (Kaiser and Schekman, 1990).
Additional Assays
For the GFP-Atg8 processing assay, yeast strains harboring the GFP-Atg8 plasmid [pGFP-Aut7(414)] were grown to early midlog phase in SMD medium lacking auxotrophic amino acids and then shifted to SD-N medium for 4 h. At each indicated time point, 1 ml of cell culture was removed and TCA-precipitated. The protein extracts were resolved by SDS-PAGE and probed with anti-GFP mAb (Covance Research Products, Berkeley, CA).
The alkaline phosphatase assay to measure Phο8Δ60 activity, Pex14-GFP processing to monitor pexophagy, the protease protection assay, and the subcellular fractionation have been described previously (Harding et al., 1995; Noda et al., 1995; Kim et al., 2001b; Scott et al., 2001; Tucker et al., 2003; Reggiori et al., 2005a). The sucrose density gradient used to fractionate Atg27 was generated as described previously (Reggiori et al., 2005b) with minor modifications (1 ml each of 18, 22, 26, 30, 34, 38, 42, 46, 50, and 54% sucrose).
RESULTS
Atg27 Contains an N-terminal Signal Sequence
We identified atg27Δ in a screen for mutants defective in processing of the Cvt pathway cargo protein precursor aminopeptidase I (prApe1; Nice et al., 2002 and our unpublished results). The ETF1 gene was simultaneously identified in a screen based on a missorting phenotype that was synthetic with a vps34ts mutant (Wurmser and Emr, 2002); however, the subsequent analysis of ETF1 and its gene product were influenced by a sequencing error originally present in the Saccharomyces Genome Database. ETF1 is allelic with ATG27, and the corrected full-length Atg27 protein is predicted to contain a signal sequence at the N terminus, based on the SignalP program (http://www.cbs.dtu.dk/services/SignalP/). If the additional amino acids at the N terminus function as an authentic signal sequence, it would likely result in a membrane topology different from that published previously. Accordingly, we undertook a careful analysis of Atg27 biosynthesis.
There are three possible models of Atg27 topology (Figure 1A). In model I, Atg27 has no signal peptide; with only a single transmembrane domain, Atg27 is in a type II orientation, in agreement with the prediction reported previously (Wurmser and Emr, 2002). The corrected full-length Atg27 with an N-terminal signal sequence is shown in models II and III. In model II, the signal sequence of Atg27 is not cleaved, resulting in a protein with two transmembrane domains; both the N and C termini of Atg27 face the cytosol. In contrast, if model III is correct the signal sequence of Atg27 is cleaved giving a type I membrane protein topology.
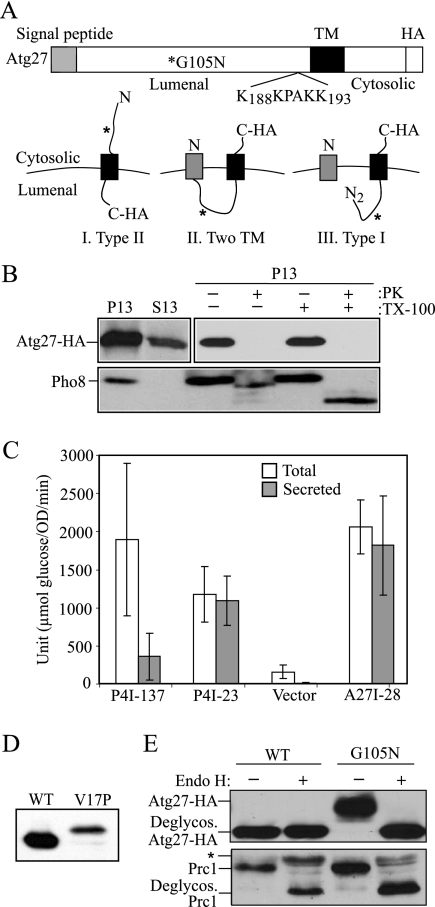
Figure 1.
Atg27 is a type I transmembrane protein. (A) Schematic drawing of full-length Atg27. The full-length Atg27 protein contains 271 amino acids. Analysis of the primary amino acid sequence using the SignalP program indicates that Atg27 has a signal sequence (residues 1-19) and a transmembrane (TM) region (residues 199-221) according to TMHMM-prediction of helices in proteins (http://www.cbs.dtu.dk/services/TMHMM-2.0/), in a type I membrane topology. The putative PtdIns(3)phosphate-binding site (residues 188–193, KKPAKK) from the previous report (Wurmser and Emr, 2002) is indicated. Three possible membrane topologies for Atg27 are shown: I, Type II transmembrane orientation; II, two transmembrane domains; and III, Type I transmembrane protein. The asterisk marks the mutation introduced to replace glycine at position 105 with asparagine, creating a glycosylation site. (B) The C terminus of Atg27 is exposed to the cytosol. Atg27-HA (WLY1) and pep4Δ (TVY1) cells were converted into spheroplasts and then osmotically lysed. The cell lysates were centrifuged at 13,000 × g for 10 min to generate the pellet (P13) and supernatant (S13) fractions. The P13 fractions then were resuspended and subjected to treatment with 1% Triton X-100, proteinase K, both or neither on ice for 30 min. The lysates were then TCA-precipitated and analyzed by SDS-PAGE and Western blot. (C) The N-terminal 28 amino acids of Atg27 are able to direct invertase secretion. Cells expressing invertase fusion proteins (P4I-137, P4I-23, empty vector [pSEYC306], or A27I-28) were grown to early log phase. The cells were collected and subjected to an invertase activity assay as described in Materials and Methods. (D) The signal sequence of Atg27 is cleaved. Cells expressing Atg27-HA (WT) or Atg27V17P-HA (V17P) from the pAtg27–3xHA(416) or pAtg27V17P-3xHA(416) plasmids were grown to early log phase. The protein extracts were analyzed by Western blot and probed with monoclonal anti-HA antibody. (E) The lumenal region of Atg27 translocates into the ER. Cells expressing Atg27-HA (WT) and Atg27G105N-HA (G105N) were grown to early log phase. The cell lysates were subjected to endoglycosidase H treatment as described in Materials and Methods. After resolution by SDS-PAGE, the samples were analyzed by Western blot and probed with antibodies against Prc1 and HA, separately. The positions of glycosylated and deglycosylated forms of both proteins are indicated. The asterisk indicates cross-reacting bands.
To distinguish among these models, we performed a protease protection assay (Figure 1B). Yeast spheroplasts were osmotically lysed under conditions that retain the integrity of subcellular compartments and separated into supernatant and pellet fractions, and the pellet fractions were treated with exogenous protease. To monitor Atg27, we tagged the C terminus with the HA epitope; the resulting protein was functional (data not shown). As a control, we followed the cleavage of the vacuole membrane protein Pho8. Pho8 was found only in the pellet fraction, indicating efficient separation of the cytosol from the membrane. The cytosolic tail of Pho8 was protease-accessible in the absence of Triton X-100, whereas the lumenally oriented propeptide was cleaved only when the vacuolar membrane was solubilized by detergent, verifying that the vacuole, and presumably other osmotically sensitive compartments, were intact after spheroplast lysis. Atg27 was found in the P13 fraction as expected for a membrane-associated protein (Figure 1B); however, the C-terminal HA tag of Atg27 was cleaved by protease both in the absence and the presence of detergent. This result suggested that the C terminus of Atg27 faced the cytosol, ruling out the topology predicted by model I (Figure 1A).
To test the functionality of the Atg27 signal sequence, we performed an assay for invertase secretion. Invertase lacking its native signal sequence remains in the cytosol, whereas replacement with an endogenous signal sequence restores secretion to the periplasm (Klionsky et al., 1988). We constructed an ATG27-SUC2 fusion (A27I-28) containing the amino-terminal 28 amino acids of Atg27 fused to invertase lacking its endogenous signal sequence. As controls, we examined two previously characterized Pep4-invertase chimeras. P4I-23 and P4I-137 contain the Pep4 signal sequence (23 amino acids) and N-terminal propeptide including the vacuolar targeting sequence (137 amino acids), respectively (Klionsky et al., 1988). The P4I-23 chimeric protein was efficiently secreted into the periplasmic space, whereas P4I-137 was efficiently retained within the cell, in agreement with previous data (Figure 1C). To have a comparable expression level with controls, an overexpressed A27I-28 construct was used in this experiment; expression from the endogenous ATG27 promoter was below a practical level of detection by this assay. The yeast strain expressing A27I-28 exhibited almost complete secretion of invertase activity (Figure 1C). After normalizing to the vector control, 95% of the invertase activity was secreted with the A27I-28 chimera, suggesting that the N-terminal 28 amino acids can function as a signal sequence.
To determine whether the N-terminal signal sequence of Atg27 is cleaved, we used a molecular genetic approach. According to the SignalP program, there is a signal sequence cleavage site between alanine at position 19 and leucine at position 20 in Atg27. A proline residue at the −3 amino acid position would disrupt recognition of the signal sequence cleavage site (von Heijne, 1984). We replaced the valine with a proline residue at the −3 position, creating Atg27V17P, and examined the effect on signal sequence cleavage. In the case of model II (Figure 1A) in which the signal sequence of Atg27 is not cleaved, the wild-type Atg27 and Atg27V17P would show a similar mobility after SDS-PAGE; however, if the signal sequence is normally cleaved as shown in model III, Atg27V17P would be ∼2 kDa larger than the wild-type Atg27. Protein extracts were prepared from cells expressing Atg27-HA and Atg27V17P-HA, and the position of Atg27 was monitored by Western blot. The molecular mass of Atg27V17P was ∼2 kDa larger than that of wild-type Atg27 (Figure 1D), suggesting that the predicted cleavage site was disrupted by the V17P mutation. These data suggest that Atg27 contains a signal sequence that is cleaved in normal cellular conditions.
As a final assessment of the functionality of the Atg27 putative signal sequence we examined the Atg27 topology by introducing a glycosylation site in the predicted lumenal region; Atg27 lacks endogenous glycosylation sites. Accordingly, glycosylation would confirm that the introduced sites had gained access to the lumen of the endoplasmic reticulum (ER). We introduced a canonical Asn-X-Thr N-linked glycosylation site by mutating glycine at position 105 to asparagine in the HA-tagged Atg27 plasmid. atg27Δ cells expressing Atg27G105N were grown to early log phase and harvested, and cellular proteins were evaluated by Western blot using an antibody against HA. Atg27G105N-HA showed a slower mobility after SDS-PAGE compared with wild-type Atg27-HA (Figure 1E). The shift in molecular mass corresponded to ∼2–3 kDa, which would fit with the increase expected from the addition of a single glycosyl side chain. To confirm that the change in migration was due to glycosylation, we treated the lysates with endoglycosidase H. After endoglycosidase H treatment, the mobility of Atg27G105N-HA was restored to that of wild-type Atg27-HA. As a control, Prc1 (carboxypeptidase Y, a vacuolar hydrolase known to be glycosylated) was examined from the same cell lysates and showed similar results indicating that the molecular mass shift of Atg27G105N was due to the addition of sugar molecules. These results suggested that the major soluble domain of Atg27 translocated into the ER lumen. Taken together, these data indicate that Atg27 is a type I transmembrane protein with an N-terminal signal sequence. This topology is the opposite of that predicted from the previous studies (Wurmser and Emr, 2002).
Atg27 Is Required for the Cvt Pathway and Pexophagy and for Efficient Bulk Autophagy
Previously, Atg27 was reported to be specifically involved in the Cvt pathway but not bulk autophagy (Wurmser and Emr, 2002). Because of the previously mentioned sequencing error, we decided it was important to reinvestigate the phenotype of the atg27Δ mutation. We deleted the full-length ATG27 gene and examined the role of Atg27 in the Cvt pathway by pulse-chase analysis of prApe1 processing (Figure 2A). After delivery to the vacuole, the propeptide of prApe1 is removed resulting in a convenient molecular mass shift that can be followed to monitor delivery to the vacuole (Klionsky et al., 1992). Yeast strains were grown in SMD selective media, pulse-labeled with [35S]methionine/cysteine for 10 min, and then subjected to a nonradioactive chase for 2 h at 30°C. Ape1 was immunoprecipitated from the cell lysates and then analyzed by SDS-PAGE and autoradiography. In a control atg1Δ strain defective in the Cvt pathway, prApe1 processing was blocked, whereas in wild-type cells, prApe1 was processed as expected (Figure 2A). In the atg27Δ strain, even after a 2-h chase, prApe1 remained unprocessed. These data confirm that Atg27 is required for the Cvt pathway.
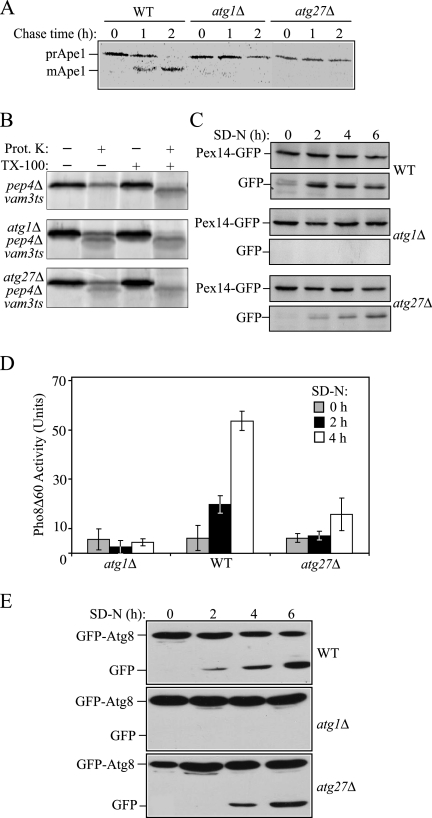
Figure 2.
The atg27Δ mutant is defective for autophagy-related pathways. (A) atg27Δ cells are defective in the Cvt pathway. Wild-type (WT; SEY6210), atg1Δ (WHY001), and atg27Δ (WLY2) cells were pulse-labeled for 10 min and subjected to a nonradioactive chase for 2 h. At the indicated time points cells were collected and TCA-precipitated. The cell lysates were immunoprecipitated with anti-Ape1 serum, resolved by SDS-PAGE, and then subjected to autoradiography. The positions of prApel and mApel are indicated. (B) Atg27 functions in the vesicle formation and/or completion step. Spheroplasts from the wild-type (pep4Δ vam3ts; WLY36) strain or this same strain harboring the atg1Δ (WLY74) or atg27Δ (WLY33) deletions were incubated at 37°C for 20 min, pulse-labeled with [35S]methionine/cysteine for 10 min, and then subjected to a nonradioactive chase for 27 min. The spheroplasts were osmotically lysed and separated into low-speed pellet and supernatant fractions after 5000 × g centrifugation. The pellet fractions that contained prApe1 were treated with proteinase K in the presence or absence of 0.2% Triton X-100. The resulting samples were immunoprecipitated with Ape1 antiserum and resolved by SDS-PAGE. (C) Atg27 is required for efficient pexophagy. Pex14-GFP (WT; IRA001), Pex14-GFP atg1Δ (IRA002), and Pex14-GFP atg27Δ (WLY27) strains were grown in oleic acid–containing medium to induce peroxisome proliferation and shifted to starvation medium. Protein extracts were prepared from cells at each indicated time point, resolved by SDS-PAGE, and probed with monoclonal anti-GFP antibody. The positions of Pex14-GFP and free GFP are indicated. (D) atg27Δ has an intermediate autophagy defect. atg1Δ (HAY572), wild-type (TN124), and atg27Δ (WLY3) cells expressing Pho8Δ60 were shifted from SMD to SD-N medium for 4 h. Samples at the indicated time points were collected, and protein extracts were assayed for alkaline phosphatase activity. The result represents the mean of three separate experiments, and the error bars represent the SD. (E) Wild-type (WT; SEY6210), atg1Δ (WHY001), and atg27Δ (WLY2) strains harboring a plasmid expressing GFP-Atg8 [pGFP-Aut7(414)] were grown in SMD lacking auxotrophic amino acids and shifted to SD-N. Aliquots were removed at the indicated time points. Protein extracts were prepared and resolved by SDS-PAGE. After Western blot, the membranes were probed with anti-GFP antibody.
The Cvt and autophagy pathways can be broken down into several steps: induction, vesicle formation and completion, docking and fusion of the vesicle with the vacuole, breakdown of the cargo, and recycling. Most of the Atg proteins are involved in the vesicle formation step. To determine whether Atg27 acts during vesicle formation, we performed an Ape1 protease-sensitivity assay. If prApe1 is enclosed in a completed vesicle, the potentially protease-sensitive propeptide domain would be protected from exogenously added protease. Alternatively, if Atg27 is required for vesicle formation and/or completion, prApe1 would be sensitive to the protease, and the resulting cleavage would result in a molecular mass shift. To block the delivery of prApe1 to the vacuole and subsequent prApe1 processing within this organelle, we used a pep4Δ vam3ts strain that is defective for fusion of vesicles with the vacuole at the nonpermissive temperature and that cannot process the propeptide within the vacuole lumen. Spheroplasts from the wild-type (pep4Δ vam3ts) strain and from this strain deleted for the ATG1 or ATG27 gene were incubated at 37°C for 20 min to inactivate the Vam3 protein. The cells were then pulse-labeled with [35S]methionine/cysteine for 10 min, followed by a nonradioactive chase reaction. After osmotic lysis the low-speed pellet fractions, which contained prApe1, were subjected to proteinase K treatment in the presence or absence of detergent. In the wild-type cells, prApe1 was protected from exogenously added protease and was digested only after the membrane was disrupted with detergent (Figure 2B). In atg1Δ cells, a portion of prApe1 was sensitive to the protease in the absence of detergent, reflecting a defect in vesicle formation and/or completion. Similarly, in the atg27Δ strain, an equivalent fraction of prApe1 was protease-accessible independent of detergent addition, indicating that the prApe1 was not completely enwrapped by the membrane. This result suggests that Atg27 functions in the vesicle formation/completion step.
Next, we extended our study to examine the role of Atg27 in the specific degradation of peroxisomes, termed pexophagy, and bulk or nonspecific autophagy. To test the role of Atg27 in pexophagy, we monitored the degradation of the peroxisomal integral membrane protein Pex14. The C terminus of Pex14 was chromosomally tagged with GFP. The induction of pexophagy results in delivery of peroxisomes into the vacuoles and Pex14 degradation, whereas the GFP moiety remains relatively stable within the vacuole lumen. Thus, the appearance of free GFP correlates with pexophagy (Reggiori et al., 2005a). Pexophagy was induced as described in Materials and Methods. In wild-type cells expressing Pex14-GFP, peroxisomes were delivered into the vacuoles upon pexophagy induction, represented by the appearance of free GFP (Figure 2C). No free GFP was detected in the atg1Δ strain, indicating that the assay reflects an autophagic process. Pex14-GFP underwent processing in atg27Δ cells; however, there was a kinetic delay relative to the wild-type strain, indicating that Atg27 is required for efficient pexophagy.
In the previous study (Wurmser and Emr, 2002), atg27Δ cells were reported to process prApe1 after induction of autophagy by rapamycin. Analysis of prApe1 processing is not sufficient for monitoring autophagy, however, because prApe1 is a specific marker; import in starvation conditions still utilizes specificity components including the receptor Atg19 (Scott et al., 2001). Therefore, we performed additional experiments to test the role of Atg27 in nonspecific autophagy. To make a quantitative measurement of autophagy, we utilized Pho8Δ60, a truncated version of the vacuolar alkaline phosphatase, Pho8. Pho8Δ60, lacking the N-terminal transmembrane domain, is unable to enter the ER and accumulates in the cytosol; it is only delivered into the vacuole through autophagy (Noda et al., 1995). Once inside the vacuole this protein is cleaved and becomes enzymatically active. Thus, Pho8Δ60 activity serves as a marker for bulk cytosolic autophagy. The Pho8Δ60 activity was measured in wild-type, atg1Δ, and atg27Δ cells (Figure 2D). As expected, atg1Δ cells that are defective in autophagy showed no increase of Pho8Δ60 activity after autophagy was induced. Wild-type cells showed Pho8Δ60 activity that increased after 2–4 h of starvation, whereas atg27Δ cells induced Pho8Δ60 activity to <50% of the wild-type level. The partial induction of Pho8Δ60 activity suggested that autophagy occurred in atg27Δ cells but not as efficiently as in wild-type cells.
The partial block in nonspecific autophagy led us to further analyze the role of Atg27 through another biochemical approach, GFP-Atg8 processing. Atg8 is an ubiquitin-like protein that is conjugated to phosphatidylethanolamine (Kirisako et al., 1999; Huang et al., 2002) and is one of two Atg proteins that remain associated with the completed autophagosome membrane. Similar to Pex14-GFP, Atg8 is degraded after delivery into the vacuole, whereas the GFP moiety is again relatively stable (Shintani et al., 2002). Wild-type, atg1Δ, and atg27Δ cells were transformed with a plasmid-based GFP-Atg8, grown in SMD medium and then subjected to nitrogen starvation. At various time points, aliquots were removed and TCA-precipitated and then subjected to Western blot using anti-GFP antibody (Figure 2E). In wild-type cells, the amount of free GFP increased over time during starvation, representing a functional autophagy pathway, whereas in autophagy-defective atg1Δ cells, no free GFP was detected. In the atg27Δ mutant, there was a kinetic delay in free GFP accumulation compared with the wild-type strain. This result agreed with the Pho8Δ60 analysis and further suggested that the atg27Δ mutant had an intermediate autophagy defect.
The atg27Δ mutant had an intermediate autophagy defect as examined by Pho8Δ60 activity and the GFP-Atg8 processing assays under starvation conditions (Figure 2, D and E). Two possibilities may explain the phenotype of atg27Δ: a reduction of either autophagosome size or autophagosome number. To determine the role for Atg27 in autophagosome biogenesis, we used electron microscopy to examine the ultrastructure of the autophagic bodies accumulated in the absence of Atg27 (Figure 3A). Autophagosomes are double-membrane vesicles. After the outer membrane of an autophagosome fuses with the vacuole limiting membrane, the single-membrane inner vesicles, now termed autophagic bodies, are released into the vacuolar lumen where they are degraded in a Pep4-dependent manner. In pep4Δ cells, which lack vacuolar proteinase A activity, the breakdown of autophagic bodies is blocked, allowing them to be visualized by electron microscopy. To eliminate the background vesicles that are delivered into the vacuole through the multivesicular body pathway, we deleted the VPS4 gene (Reggiori et al., 2004b). Wild-type, atg1Δ, and atg27Δ strains additionally harboring pep4Δ vps4Δ double mutations were grown in YPD to early log phase and then shifted to SD-N for 5 h and prepared for electron microscopy as described in Materials and Methods.
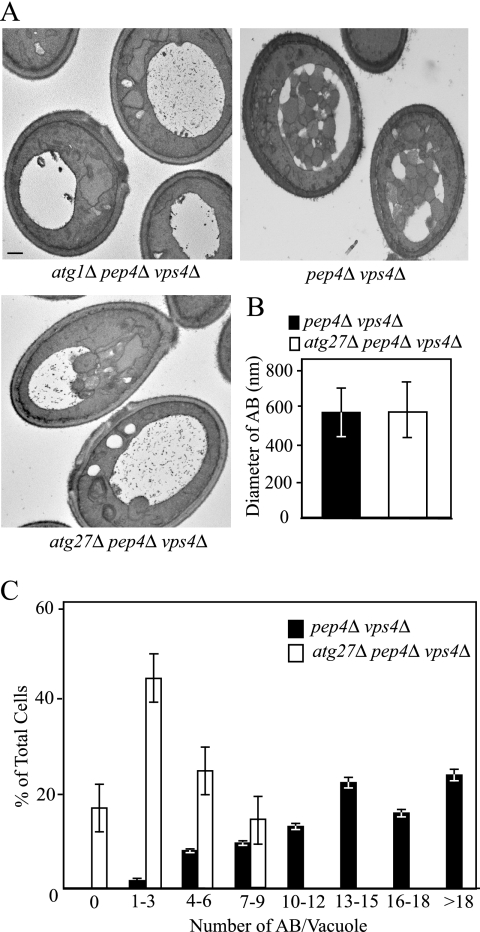
Figure 3.
The atg27Δ mutant generated fewer autophagosomes. (A) The wild-type (pep4Δ vps4Δ; FRY143), atg1Δ (JHY28), and atg27Δ (WLY8) strains were grown to early log phase in YPD, shifted to SD-N for 5 h to induce autophagy, and examined by electron microscopy as described in Materials and Methods. Bar, 0.5 μm. (B) Quantification of the diameter of autophagic bodies (AB). To quantify the size of the accumulated ABs inside the vacuole, the diameter of ABs with a clear membrane boundary was measured. The number of ABs counted was 50 for the atg27Δ strain and 67 for wild type. (C) Quantification of autophagic body accumulation. To quantify the number of ABs accumulated, the number of autophagic bodies was counted in cells containing similar-sized vacuoles.
Wild-type (pep4Δ vps4Δ) cells showed numerous autophagic bodies, with 13–15 autophagic bodies in 22% of the vacuoles and an average of 14.62 ± 5.31 per vacuole after 5-h starvation (Figure 3, A and C; n = 63 vacuoles). The control atg1Δ pep4Δ vps4Δ cells, defective in autophagy, did not accumulate autophagic bodies as expected. The atg27Δ pep4Δ vps4Δ cells accumulated a reduced number of these structures, with 1–3 autophagic bodies in 44% of the vacuoles and an average of 3.13 ± 2.62 per vacuole (n = 77 vacuoles). To determine whether atg27Δ pep4Δ vps4Δ cells accumulated normal-sized autophagic bodies, we quantified their size in wild-type and atg27Δ pep4Δ vps4Δ cells by measuring the diameter (Figure 3B). We found that the autophagic bodies accumulated in wild-type and atg27Δ pep4Δ vps4Δ cells were similar in diameter, suggesting that atg27Δ pep4Δ vps4Δ cells produced normal sized autophagosomes. Taken together, the reduction of the autophagic body number in addition to the biochemical data indicate that the decreased efficiency of nonspecific autophagy in the atg27Δ mutant was not due to a structural defect in autophagosome formation but rather a kinetic delay in the process.
Atg27 Cycles among the PAS, Mitochondria, and Golgi Complex
It has been shown that Atg27 localizes to multiple unidentified perivacuolar punctate structures (Wurmser and Emr, 2002). To gain insight into the function of Atg27, we decided to examine the structures at which Atg27 resides in vivo by using fluorescence microscopy. A common feature of Atg proteins is that they appear to transiently localize at the PAS. The function of the PAS is not clear, but it has been implicated in the formation of Cvt vesicles and autophagosomes (Suzuki et al., 2001; Kim et al., 2002). We hypothesized that Atg27 may have the same subcellular distribution as most other Atg proteins. To monitor the PAS localization of Atg27, we generated a functional C-terminal GFP fusion at the ATG27 chromosomal locus. An Atg27-GFP strain harboring an RFP-Ape1 plasmid was grown to the early log phase and then visualized by fluorescence microscopy (top panel of Figure 4A). Atg27-GFP was distributed in several subcellular punctate structures, only one of which colocalized with the PAS marker RFP-Ape1. Next, we extended our study by examining the colocalization of organelle markers with the non-PAS population of Atg27. Among the organelle markers that we examined, Atg27-GFP partially colocalized with the mitochondrial (MitoFluor Red) and Golgi complex markers (Vrg4-RFP; middle and bottom panels of Figure 4A). Atg27-GFP was found to not colocalize with the ER or peroxisomes (data not shown).
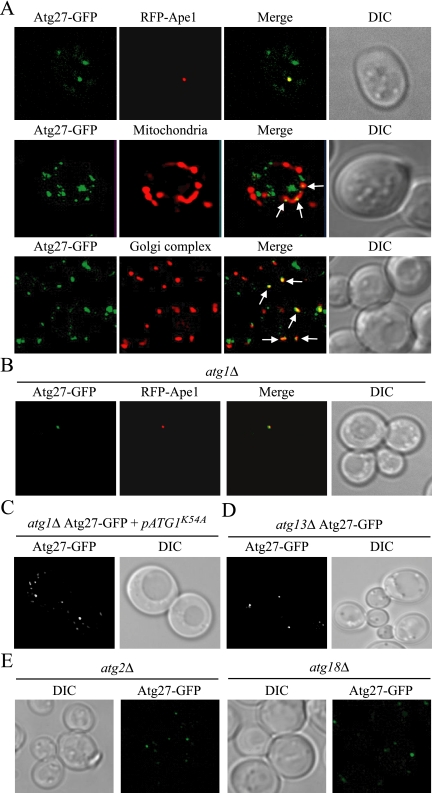
Figure 4.
Atg27 cycles among the PAS, mitochondria, and Golgi complex. (A) Atg27 localizes to the PAS and partially to the mitochondria and Golgi. The strain expressing chromosomally tagged Atg27-GFP (WLY5) carrying either a PAS marker [pRFP-APE1(414)] or expressing chromosomally tagged Vrg4-RFP (Golgi complex marker; WLY6) were grown to early log phase or nitrogen starved for 3 h before imaging. For mitochondrial staining, the Atg27-GFP culture was incubated for 30 min in the presence of 1 μM MitoFluor Red 589 (Molecular Probes, Eugene, OR). The arrows indicate the colocalization of Atg27-GFP with mitochondria or the Golgi complex. (B) Atg27 is restricted to the PAS in atg1Δ cells. The chromosomally tagged Atg27-GFP atg1Δ strain (WLY11) carrying a PAS marker [pRFP-APE1(414)] was grown in selective medium to early log phase and then visualized by fluorescence microscopy. (C and D) The absence of Atg13, but not reduced Atg1 kinase activity, affected Atg27 localization. (C) The chromosomally tagged Atg27-GFP atg1Δ strain carrying an Atg1 kinase mutant (ATG1K54A) plasmid and (D) the Atg27-GFP atg13Δ strain (WLY18) were grown in selective SMD medium to OD600 = 0.8 and analyzed by fluorescence microscopy. Essentially identical results were obtained when cells were incubated in starvation medium. (E) Atg2 and Atg18 are required for efficient Atg27 retrograde cycling from the PAS. The chromosomally tagged Atg27-GFP atg2Δ strain (WLY78) and Atg27-GFP atg18Δ strain (WLY70) were grown in SMD medium and fixed with 1.5% formaldehyde in 50 mM potassium phosphate buffer (pH 8) for 30 min before imaging. DIC, differential interference contrast.
Most Atg proteins can only be detected at the PAS. Two exceptions, Atg9 and Atg23, show unique localization in multiple punctate structures other than the PAS (Tucker et al., 2003; Reggiori et al., 2004a), similar to the Atg27 distribution. Both Atg9 and Atg23 cycle between the PAS and mitochondria (Reggiori et al., 2004a). This cycling is dependent on the Atg1-Atg13 complex (Reggiori et al., 2004a). Atg1 is a serine/threonine kinase, which may play an important role in regulating the switch between the Cvt pathway and autophagy. In atg1Δ cells, both Atg9 and Atg23 are restricted to the PAS (Reggiori et al., 2004a). We visualized Atg27-GFP distribution in atg1Δ cells to see if Atg27 showed a similar trafficking pattern (Figure 4B). The majority of Atg27 was restricted to the PAS in the absence of Atg1, as indicated by colocalization with RFP-Ape1 in both growing and starvation conditions, but occasionally with some additional very faint punctate structures in the cytosol. This PAS restriction could be reversed and the wild-type localization restored by expressing a plasmid encoding wild-type Atg1 (data not shown). Moreover, a recent study showed that Atg1 kinase activity is required for Atg23 cycling in growing conditions, but not for Atg9 cycling (Reggiori et al., 2004a). A plasmid containing an ATG1 mutant with reduced kinase activity (Atg1K54A; Kamada et al., 2000) was introduced into the Atg27-GFP atg1Δ strain. Similar to Atg9, Atg27 distribution was not affected when the Atg1 kinase activity was reduced in both growing and starvation conditions (Figure 4C).
The regulatory function of Atg1 is regulated through protein–protein interactions with several other Atg proteins (Kamada et al., 2000). Therefore, we examined the effect on Atg27 localization of mutants deleted for Atg1-interacting proteins. With the exception of Atg13, deletion of other Atg proteins we screened had no effect on Atg27 distribution (Figure 4D and data not shown). Similar to atg1Δ, most of the Atg27-GFP was restricted to the PAS in atg13Δ cells under both growing and starvation conditions. Taken together, these data show that Atg27 cycles between the PAS and the non-PAS pool in an Atg1-Atg13 complex–dependent manner, but normal Atg1 kinase activity is not required for Atg27 trafficking.
The results with the atg1Δ and atg13Δ strains indicated that Atg27 cycling was dependent on the Atg1-Atg13 complex, similar to Atg9. To further examine the cycling pattern of Atg27, we tested the effect on Atg27 movement of other factors that are required for Atg9 trafficking. In the absence of either Atg2 or Atg18, Atg9 is restricted to the PAS (Reggiori et al., 2004a). Atg27 distribution in the atg2Δ and atg18Δ strains was also confined to one strong dot, although additional faint dots could also be detected (Figure 4E). Thus, Atg27 appears to have a less stringent requirement for components involved in retrograde cycling. Along these lines, Atg27 cycling was independent of Atg14 (see Figure 7B) in contrast to Atg9. The actin cytoskeleton is required for Atg9 anterograde movement; Atg9 does not accumulate at the PAS in the atg1ts strain treated with latrunculin A at the nonpermissive temperature (Reggiori et al., 2005a). We were unable to determine the requirement of actin filaments in Atg27 cycling, however, because of a delayed PAS accumulation phenotype seen with Atg27-GFP in the atg1ts strain; atg27Δ cells lost viability after treatment with latrunculin A before we could assay the movement of Atg27-GFP.
Figure 7.
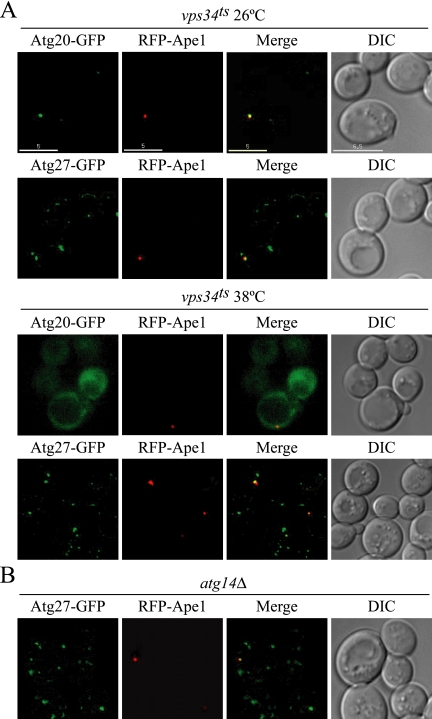
Binding to PtdIns(3)P is not required for Atg27 function. (A) Atg27 localization is not affected by Vps34. Chromosomally tagged Atg20-GFP and Atg27-GFP strains expressing plasmid-based vps34ts and RFP-Ape1 (WLY50 and WLY51, respectively) were grown in selective SMD medium to OD600 = 0.8 at 26°C or shifted to 38°C for 12 min before imaging. (B) Atg14 does not affect Atg27 PAS localization. The Atg27-GFP atg14Δ strain (WLY52) expressing RFP-Ape1 was grown in selective SMD medium and visualized by fluorescence microscopy. Bar, 5 μm.
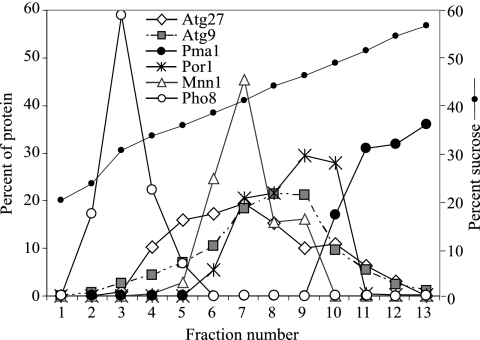
Finally, to extend our analysis of Atg27 localization, a P13 pellet fraction from the Atg27-HA strain was separated on a sucrose density gradient as described in Materials and Methods (Figure 5). After centrifugation at 176,000 × g for 18 h at 4°C, 13 fractions were collected from the top to the bottom and subjected to immunoblot analysis. The vacuole membrane marker Pho8 was concentrated in the top fractions, whereas the plasma membrane marker, Pma1, was in the bottom fractions. In agreement with a previous report (Reggiori et al., 2005b), the P13 population of Atg9 cofractionated with mitochondria (Por1). Atg27-HA was distributed through several fractions with the peak concentration in fraction 7. A population of Atg27 cofractionated with mitochondria (Por1) and the Golgi complex (Mnn1), supporting the fluorescence data suggesting that Atg27 resides in these membrane compartments.
Figure 5.
Atg27 localizes to mitochondria and the Golgi complex. The Atg27-HA (WLY1) strain was grown in YPD to OD600 = 1.0 and converted into spheroplasts. The spheroplasts were osmotically lysed and separated into pellet (P13) and supernatant (S13) fractions after a 13,000 × g centrifugation. The P13 pellet fraction was separated on a sucrose density gradient (18–54%) and centrifuged for 18 h at 176,000 × g as described in Materials and Methods. A total of 13 fractions were collected from the top to the bottom of the gradient and were subjected to immunoblot analysis with antiserum against Atg27-HA, Atg9, Pma1 (plasma membrane), Por1 (mitochondria), Mnn1 (Golgi complex), and Pho8 (vacuole).
Atg27 Is Required for Atg9 Cycling from the Mitochondria to the PAS
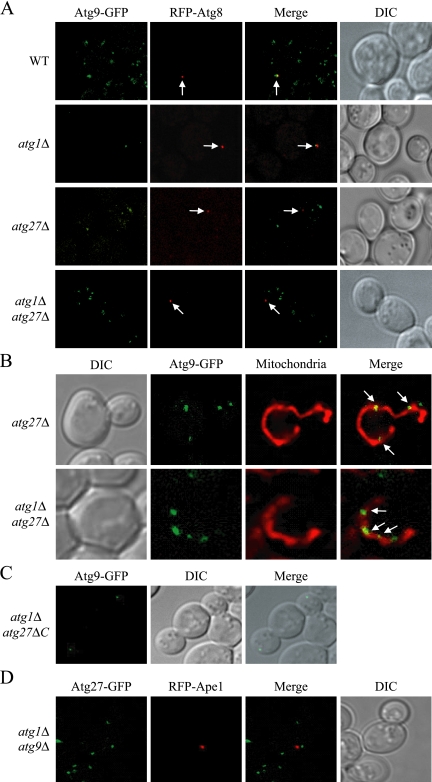
Atg9, Atg23, and Atg27 have a common phenotype involving the PAS and multiple additional punctate dots. Atg23 is needed for delivery of Atg9 to the PAS (J. E. Legakis, W.-L. Yen, and D. J. Klionsky, unpublished results). To examine whether Atg27 is required for Atg9 localization before Atg1 function, we performed an epistasis analysis termed the transport of Atg9 after knocking out ATG1 (TAKA) assay (Cheong et al., 2005). In wild-type cells Atg9 displayed multiple punctate dots, one of which localized to the PAS, indicated by colocalization with the PAS marker RFP-Atg8 (Figure 6A). In an atg1Δ strain, Atg9 was restricted to the PAS, as previously shown (Reggiori et al., 2004a; Figure 6A). In both atg27Δ and atg1Δ atg27Δ double mutant cells, Atg9 localized to multiple punctate dots, with none of the dots corresponding to the PAS marker, indicating that Atg9 was unable to reach the PAS. The colocalization of Atg9 with MitoFluor Red confirmed that a population of Atg9 was restricted to the mitochondria in the atg27Δ and atg1Δ atg27Δ double mutants (Figure 6B). This result suggested that Atg27 functions before Atg1 in Atg9 cycling and is required for Atg9 movement from the mitochondria to the PAS.
Figure 6.
Atg27 functions before Atg1 and is required for Atg9 cycling. (A) Atg27 is required for Atg9 anterograde trafficking. The chromosomally tagged Atg9-GFP (WT; JLY44), Atg9-GFP atg1Δ (JLY45), Atg9GFP atg27Δ (JLY43), and Atg9-GFP atg1Δ atg27Δ (JLY47) strains expressing plasmid-based RFP-Atg8 were grown to OD600 = 0.8 in selective SMD medium and visualized by fluorescence microscopy. Arrows locate the PAS marker RFP-Atg8. (B) Atg9 localizes to mitochondria in the atg27Δ and atg1Δ atg27Δ mutants. The chromosomally tagged Atg9-GFP atg27Δ (JLY43) and Atg9-GFP atg1Δ atg27Δ (JLY47) strains were grown in SMD complete medium, and mitochondria were stained by incubating for 30 min in the presence of 1 μM MitoFluor Red 589. (C) The C-terminal cytosolic portion of Atg27 is not required for Atg9 cycling. Chromosomally tagged Atg9-GFP atg1Δ atg27ΔC (WLY49) cells were grown to early-log phase and collected for fluorescence microscopy. (D) Atg9 is required for Atg27 cycling from the non-PAS structures to the PAS. The chromosomally tagged Atg27-GFP atg1Δ atg9Δ strain (WLY41) expressing chromosomally tagged RFP-Ape1 was grown to early log phase in selective SMD medium before imaging. DIC, differential interference contrast.
The type I topology suggests that the majority of Atg27 is localized on the lumenal side of the membrane, whereas Atg23 is a cytosolic protein. To determine whether the cytosolic tail of Atg27 is required for the movement of Atg9, we tested the effect of an Atg27 truncation on Atg9 cycling. We generated a version of Atg27 lacking the C-terminal cytosolic tail, Atg27ΔC. In atg1Δ atg27ΔC cells, Atg9 was restricted in one dot similar to the localization in the atg1Δ strain (Figure 6C). This result indicates that the cytosolic C terminus of Atg27 is not required for anterograde movement of Atg9 to the PAS.
In light of the data that Atg27 is required for Atg9 cycling, we decided to test the requirement of Atg9 for Atg27 movement. In atg9Δ and atg1Δ atg9Δ cells, Atg27 localized to multiple punctate structures similar to the wild-type Atg27 distribution except that none of the dots corresponded with the PAS marker (Figure 6D and data not shown). This result indicated that Atg9 functions in Atg27 cycling before Atg1 and is required for Atg27 movement from the non-PAS structures (mitochondria and Golgi complex) to the PAS.
Atg27 Localization Does Not Require Vps34 Function
In the yeast S. cerevisiae, Vps34 is the only PtdIns 3-kinase. Vps34 is found in two distinct tetrameric complexes named complex I and complex II (Kihara et al., 2001). Vps34 kinase complex I generates PtdIns(3)P at the PAS and is essential for both the Cvt pathway and autophagy (Kihara et al., 2001; Kim et al., 2002; Nice et al., 2002). Etf1 was originally identified as a PtdIns(3)P binding protein, which functions as a Vps34 downstream effector specifically involved in the Cvt pathway (Wurmser and Emr, 2002). Given the type I membrane protein topology that we demonstrated (Figure 1), the proposed PtdIns(3)P-binding site in Etf1/Atg27 would not be exposed to the cytosol, yet PtdIns(3)P is generally limited to the cytosolic face of membranes. To examine the requirement for PtdIns(3)P in Atg27 localization, we analyzed a temperature-sensitive Vps34 (vps34ts) mutant (Figure 7A). Atg20, a phox homology domain-containing protein required for Cvt pathway function, binds to PtdIns(3)P and requires Vps34 for its localization (Nice et al., 2002) and served as a control. In vps34ts cells grown at permissive temperature (26°C), Atg20-GFP showed one strong dot localized at the PAS indicated by the colocalization with RFP-Ape1. After shifting to 38°C, a nonpermissive temperature, for 12 min, Atg20-GFP dissociated from the membrane and was dispersed throughout the cytosol. In contrast, Atg27-GFP distribution was similar in both wild-type and vps34ts mutant cells at permissive as well as nonpermissive temperatures. Moreover, Atg27 still localized to the PAS even after Vps34 was inactivated. To further confirm that the PAS localization of Atg27 does not require binding to PtdIns(3)P, we examined the localization of Atg27 in an atg14Δ background. Atg14, one of the components in Vps34 complex I, is required to localize the Vps34 complex to the PAS (Obara et al., 2006). In the atg14Δ mutant, Atg27-GFP distribution was not affected, and the chimera still localized to the PAS (Figure 7B). Because Atg27 localization was not affected by inactivation of Vps34 or deletion of ATG14, we concluded that Atg27 does not bind PtdIns(3)P or at least does not require binding for its localization.
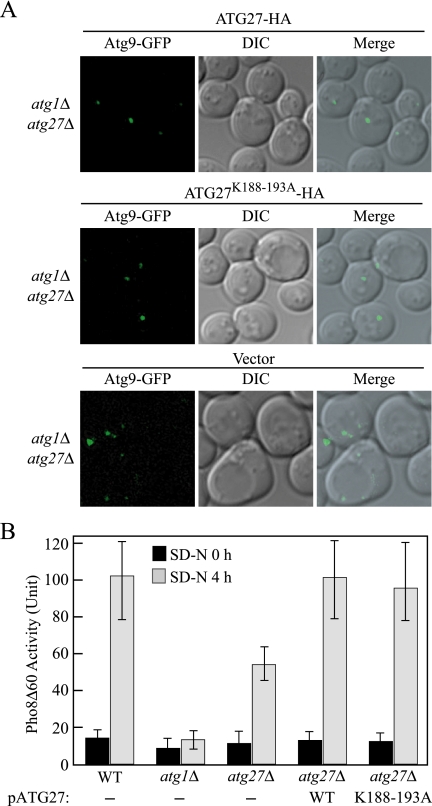
To further test our conclusion, we mutated the previously proposed putative PtdIns(3)P binding site on Etf1/Atg27 (residues 188-193; Wurmser and Emr, 2002) and tested the function of the mutant Atg27K188-193A. First, we examined the complementation of the plasmid-based Atg27K188-193A by pulse-chase analysis of prApe1 import in the atg27Δ background. Both wild-type Atg27 and Atg27K188-193A constructs driven by the endogenous promoter showed ∼85% complementation of the prApe1 import defect of atg27Δ cells after a 2-h chase (data not shown), indicating that this construct was functional for the Cvt pathway. We also tested the complementation of the Atg27K188-193A mutant by the TAKA assay (Figure 8A). Plasmids encoding either wild-type ATG27 or ATG27K188-193A but not the empty vector allowed Atg9 cycling from the non-PAS pool to the PAS in the atg1Δ atg27Δ background under both growing (Figure 8A) and starvation conditions (data not shown). To test the function of the Atg27K188-193A mutant in autophagy, we examined nonspecific uptake of the cytosolic marker Pho8Δ60. We transformed the atg27Δ Pho8Δ60 strain with a plasmid encoding either wild-type ATG27 or ATG27K188-193A. The Pho8Δ60 activity of the strains expressing Atg27 and Atg27K188-193A were essentially identical to that of the wild-type strain (Figure 8B). This result supports the conclusion that PtdlIns(3)P binding by Atg27 is not required for Atg27 function.
Figure 8.
Mutation of the Atg27 putative PtdIns(3)P binding site has no effect on function. (A) The Atg27K188-193A mutant does not affect Atg9 cycling to the PAS. The chromosomally tagged Atg9-GFP atg1Δ atg27Δ strain (JLY47) expressing either plasmid-based Atg27–3xHA, Atg27K188-193A-3xHA or vector were grown in selective SMD medium and visualized by fluorescence microscopy. DIC, differential interference contrast. (B) Wild-type (TN124), atg1Δ (HAY572), and atg27Δ (WLY40) strains and the atg27Δ strain expressing plasmid-based wild-type Atg27-HA or Atg27K188-193A-HA were shifted from SMD to SD-N medium for 4 h. Samples were colleted at the indicated time points, and protein extracts were assayed for Pho8Δ60-dependent alkaline phosphatase activity. The results represent the mean of three separate experiments and the error bars represent the SD.
DISCUSSION
Atg27 was characterized as a type II transmembrane protein (Wurmser and Emr, 2002); however, this assessment was based on an incorrect sequence that had been present in the Saccharomyces Genome Database. The corrected full-length Atg27 includes an N-terminal extension that contains a signal sequence, which would typically result in an orientation opposite to the one reported. In the present study we showed that Atg27 is in fact a type I transmembrane protein (Figure 1). Also in the original description, Atg27 was reported to be required for the Cvt pathway but not autophagy. That conclusion was based solely on an analysis of prApe1 processing in conditions that induce autophagy (Wurmser and Emr, 2002); however, more recent studies have shown that monitoring prApe1 is not sufficient to assess autophagic capacity (Cheong et al., 2005). Our analysis of the atg27Δ mutant phonotype demonstrated that Atg27 is required for efficient autophagy and pexophagy (Figures 2 and 3).
One of the major questions about the process of autophagy is the source of the lipid that is used for formation of autophagosomes and the mechanism used for lipid movement to the vesicle assembly site. Unlike most endomembrane trafficking processes in which the vesicles bud from a pre-existing membrane surface, the double-membrane of the autophagosome is thought to form de novo and likely involves an expansion process that necessitates multiple membrane delivery events. Until now, Atg9 was the only potential candidate to help us understand the mechanism and the source of the membrane for autophagosome formation. Recent studies show that Atg9 cycles between the PAS and the mitochondria (Reggiori et al., 2005b). This characteristic, coupled with it being an integral membrane protein, make Atg9 a potential carrier bringing membrane to the autophagosome formation site. These studies also suggest that the mitochondria are part of the membrane source for the double-membrane vesicles. In this report, we show that Atg27 is the second transmembrane protein that is involved in vesicle formation in the Cvt pathway and autophagy. We found that Atg27 localizes to the mitochondria and the Golgi complex. Similar to Atg9, Atg27 cycles between mitochondria and the PAS, as well as the Golgi complex, and possibly other unknown structures. These characteristics of Atg27 lead us to suggest that Atg27 may also mark the membrane source that is donated to the forming vesicles. The localization of Atg27 to the Golgi complex fits with previous studies implicating this organelle as another source of membrane for the forming vesicles (Reggiori et al., 2004b).
To gain insight on how lipid is recruited to the sequestering vesicles, we investigated the mechanisms that regulate Atg9 cycling. This is a complex process and several components have been shown to be required for Atg9 cycling between the PAS and the mitochondria, including the Atg1-Atg13 complex, Atg2, Atg18, and the PtdIns 3-kinase complex I (Reggiori et al., 2004a). In contrast, the Atg components that are required for Atg9 anterograde transport from the mitochondria to the PAS have not yet been identified; however, we have shown that the actin cytoskeleton is involved in this step (Reggiori et al., 2005a). We have recently discovered that Atg27, Atg23, and Atg11 are all required for Atg9 anterograde trafficking (Figure 6; J. E. Legakis, W.-L. Yen, and D. J. Klionsky, unpublished results; He et al., 2006). The requirement for Atg27 in Atg9 cycling suggests that these two proteins may interact with each other. The finding that Atg9 interacts with Atg27 by yeast two-hybrid analysis, and affinity isolation supports this idea (J. E. Legakis, W.-L. Yen, and D. J. Klionsky, unpublished results).
Etf1/Atg27 was originally identified as a PtdIns(3)P-binding protein, and a putative PtdIns(3)P-binding site was proposed in the previous study; upon mutation of the proposed site, the mutant lost the ability to bind this phosphoinositide (Wurmser and Emr, 2002). From the detailed topological characterization, we found that this putative binding site resides in the lumenal portion of the protein. Because PtdIns(3)P is present on the cytosolic face of membranes, it is highly unlikely that a binding site is in the lumen. Our studies suggest that Atg27 does not bind to PtdIns(3)P via this lumenal domain and that the presence of PtdIns(3)P does not influence the localization of Atg27 (Figures 7 and 8). Moreover, mutation of this site did not affect the function of Atg27 in either the Cvt pathway or autophagy (Figure 8B and data not shown). Therefore, we concluded that Atg27 is probably not a PtdIns(3)P-binding protein and that at any rate, binding to this phosphoinositide does not play a role in its function.
Atg27 is the second transmembrane Atg protein that is required for the formation of the sequestering vesicle in autophagy-related pathways. The topology of Atg27 indicates that the majority of the protein would be present within the intermembrane space between the autophagosome inner and outer vesicle membrane or in the lumenal space during autophagosome formation. The specific function of this domain is not known. Further analysis of Atg27 may provide more information regarding the membrane sources in autophagy and the cycling of proteins that are proposed to deliver membrane to the site of autophagosome formation.
ACKNOWLEDGMENTS
We thank Dr. Kay Hofmann (Bioinformatics Group, Miltenyi Biotec GmbH, Cologne, Germany) and Dr. Scott Emr (University of California, San Diego) for providing information about the frameshift error in the Atg27 sequence. D.J.K. is supported by National Institutes of Health Public Health Service Grant GM53396.
Abbreviations used:
- Ape1
saminopeptidase I
- Cvt
cytoplasm to vacuole targeting
- ER
endoplasmic reticulum
- GFP
green fluorescent protein
- PAS
pre-autophagosomal structure
- prApe1
precursor Ape1
- PtdIns
phosphatidylinositol.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E06-07-0612) on December 4, 2006.
REFERENCES
- Abeliovich H., Zhang C., Dunn W. A., Jr, Shokat K. M., Klionsky D. J. Chemical genetic analysis of Apg1 reveals a non-kinase role in the induction of autophagy. Mol. Biol. Cell. 2003;14:477–490. doi: 10.1091/mbc.E02-07-0413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell T. N., Choy F. Y. Expression of two green fluorescent protein variants in citrate-buffered media in Pichia pastoris. Anal. Biochem. 2002;311:193–195. doi: 10.1016/s0003-2697(02)00409-8. [DOI] [PubMed] [Google Scholar]
- Cheong H., Yorimitsu T., Reggiori F., Legakis J. E., Wang C.-W., Klionsky D. J. Atg17 regulates the magnitude of the autophagic response. Mol. Biol. Cell. 2005;16:3438–3453. doi: 10.1091/mbc.E04-10-0894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn W. A., Jr, Cregg J. M., Kiel J.A.K.W., van der Klei I. J., Oku M., Sakai Y., Sibirny A. A., Stasyk O. V., Veenhuis M. Peoxphagy: the selective autophagy of peroxisomes. Autophagy. 2005;1:75–83. doi: 10.4161/auto.1.2.1737. [DOI] [PubMed] [Google Scholar]
- Gerhardt B., Kordas T. J., Thompson C. M., Patel P., Vida T. The vesicle transport protein Vps33p is an ATP-binding protein that localizes to the cytosol in an energy-dependent manner. J. Biol. Chem. 1998;273:15818–15829. doi: 10.1074/jbc.273.25.15818. [DOI] [PubMed] [Google Scholar]
- Goldstein A., Lampen J. O. β-D-fructofuranoside fructohydrolase from yeast. Methods Enzymol. 1975;42:504–511. doi: 10.1016/0076-6879(75)42159-0. [DOI] [PubMed] [Google Scholar]
- Harding T. M., Morano K. A., Scott S. V., Klionsky D. J. Isolation and characterization of yeast mutants in the cytoplasm to vacuole protein targeting pathway. J. Cell Biol. 1995;131:591–602. doi: 10.1083/jcb.131.3.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He C., Song H., Yorimitsu T., Monastyrska I., Yen W.-L., Legakis J. E., Klionsky D. J. Recruitment of Atg9 to the pre-autophagosomal structure by Atg11 is essential for selective autophagy in budding yeast. J. Cell Biol. 2006 doi: 10.1083/jcb.200606084. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W.-P., Scott S. V., Kim J., Klionsky D. J. The itinerary of a vesicle component, Aut7p/Cvt5p, terminates in the yeast vacuole via the autophagy/Cvt pathways. J. Biol. Chem. 2000;275:5845–5851. doi: 10.1074/jbc.275.8.5845. [DOI] [PubMed] [Google Scholar]
- Kaiser C. A., Schekman R. Distinct sets of SEC genes govern transport vesicle formation and fusion early in the secretory pathway. Cell. 1990;61:723–733. doi: 10.1016/0092-8674(90)90483-u. [DOI] [PubMed] [Google Scholar]
- Kamada Y., Funakoshi T., Shintani T., Nagano K., Ohsumi M., Ohsumi Y. Tor-mediated induction of autophagy via an Apg1 protein kinase complex. J. Cell Biol. 2000;150:1507–1513. doi: 10.1083/jcb.150.6.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kihara A., Noda T., Ishihara N., Ohsumi Y. Two distinct Vps34 phosphatidylinositol 3-kinase complexes function in autophagy and carboxypeptidase Y sorting in Saccharomyces cerevisiae. J. Cell Biol. 2001;152:519–530. doi: 10.1083/jcb.152.3.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Huang W.-P., Klionsky D. J. Membrane recruitment of Aut7p in the autophagy and cytoplasm to vacuole targeting pathways requires Aut1p, Aut2p, and the autophagy conjugation complex. J. Cell Biol. 2001a;152:51–64. doi: 10.1083/jcb.152.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Huang W.-P., Stromhaug P. E., Klionsky D. J. Convergence of multiple autophagy and cytoplasm to vacuole targeting components to a perivacuolar membrane compartment prior to de novo vesicle formation. J. Biol. Chem. 2002;277:763–773. doi: 10.1074/jbc.M109134200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Kamada Y., Stromhaug P. E., Guan J., Hefner-Gravink A., Baba M., Scott S. V., Ohsumi Y., Dunn W. A., Jr, Klionsky D. J. Cvt9/Gsa9 functions in sequestering selective cytosolic cargo destined for the vacuole. J. Cell Biol. 2001b;153:381–396. doi: 10.1083/jcb.153.2.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirisako T., Baba M., Ishihara N., Miyazawa K., Ohsumi M., Yoshimori T., Noda T., Ohsumi Y. Formation process of autophagosome is traced with Apg8/Aut7p in yeast. J. Cell Biol. 1999;147:435–446. doi: 10.1083/jcb.147.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky D. J. Autophagy. Georgetown, TX: 2004. Landes Bioscience. [Google Scholar]
- Klionsky D. J., Banta L. M., Emr S. D. Intracellular sorting and processing of a yeast vacuolar hydrolase: proteinase A propeptide contains vacuolar targeting information. Mol. Cell Biol. 1988;8:2105–2116. doi: 10.1128/mcb.8.5.2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky D. J., Cueva R., Yaver D. S. Aminopeptidase I of Saccharomyces cerevisiae is localized to the vacuole independent of the secretory pathway. J. Cell Biol. 1992;119:287–299. doi: 10.1083/jcb.119.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky D. J. A unified nomenclature for yeast autophagy-related genes. Dev. Cell. 2003;5:539–545. doi: 10.1016/s1534-5807(03)00296-x. [DOI] [PubMed] [Google Scholar]
- Levine B., Klionsky D. J. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev. Cell. 2004;6:463–477. doi: 10.1016/s1534-5807(04)00099-1. [DOI] [PubMed] [Google Scholar]
- Longtine M. S., McKenzie A., III, Demarini D. J., Shah N. G., Wach A., Brachat A., Philippsen P., Pringle J. R. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Nice D. C., Sato T. K., Stromhaug P. E., Emr S. D., Klionsky D. J. Cooperative binding of the cytoplasm to vacuole targeting pathway proteins, Cvt13 and Cvt20, to phosphatidylinositol 3-phosphate at the pre-autophagosomal structure is required for selective autophagy. J. Biol. Chem. 2002;277:30198–30207. doi: 10.1074/jbc.M204736200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda T., Kim J., Huang W.-P., Baba M., Tokunaga C., Ohsumi Y., Klionsky D. J. Apg9p/Cvt7p is an integral membrane protein required for transport vesicle formation in the Cvt and autophagy pathways. J. Cell Biol. 2000;148:465–480. doi: 10.1083/jcb.148.3.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda T., Matsuura A., Wada Y., Ohsumi Y. Novel system for monitoring autophagy in the yeast Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 1995;210:126–132. doi: 10.1006/bbrc.1995.1636. [DOI] [PubMed] [Google Scholar]
- Obara K., Sekito T., Ohsumi Y. Assortment of phosphatidylinositol 3-kinase complexes—Atg14p directs association of complex I to the pre-autophagosomal structure in Saccharomyces cerevisiae. Mol. Biol. Cell. 2006;17:1527–1539. doi: 10.1091/mbc.E05-09-0841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggiori F., Klionsky D. J. Autophagy in the eukaryotic cell. Eukaryot. Cell. 2002;1:11–21. doi: 10.1128/EC.01.1.11-21.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggiori F., Monastyrska I., Shintani T., Klionsky D. J. The actin cytoskeleton is required for selective types of autophagy, but not nonspecific autophagy, in the yeast Saccharomyces cerevisiae. Mol. Biol. Cell. 2005a;16:5843–5856. doi: 10.1091/mbc.E05-07-0629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggiori F., Tucker K. A., Stromhaug P. E., Klionsky D. J. The Atg1-Atg13 complex regulates Atg9 and Atg23 retrieval transport from the pre-autophagosomal structure. Dev. Cell. 2004a;6:79–90. doi: 10.1016/s1534-5807(03)00402-7. [DOI] [PubMed] [Google Scholar]
- Reggiori F., Wang C.-W., Nair U., Shintani T., Abeliovich H., Klionsky D. J. Early stages of the secretory pathway, but not endosomes, are required for Cvt vesicle and autophagosome assembly in Saccharomyces cerevisiae. Mol. Biol. Cell. 2004b;15:2189–2204. doi: 10.1091/mbc.E03-07-0479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggiori F., Shitani T., Nair U., Klionsky D. J. Atg9 cycles between mitochondria and the pre-autophagosomal structure in yeasts. Autophagy. 2005b;1:101–109. doi: 10.4161/auto.1.2.1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson J. S., Klionsky D. J., Banta L. M., Emr S. D. Protein sorting in Saccharomyces cerevisiae: isolation of mutants defective in the delivery and processing of multiple vacuolar hydrolases. Mol. Cell Biol. 1988;8:4936–4948. doi: 10.1128/mcb.8.11.4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott S. V., Guan J., Hutchins M. U., Kim J., Klionsky D. J. Cvt19 is a receptor for the cytoplasm-to-vacuole targeting pathway. Mol. Cell. 2001;7:1131–1141. doi: 10.1016/s1097-2765(01)00263-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shintani T., Klionsky D. J. Autophagy in health and disease: a double-edged sword. Science. 2004;306:990–995. doi: 10.1126/science.1099993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shintani T., Huang W.-P., Stromhaug P. E., Klionsky D. J. Mechanism of cargo selection in the cytoplasm to vacuole targeting pathway. Dev. Cell. 2002;3:825–837. doi: 10.1016/s1534-5807(02)00373-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stack J. H., DeWald D. B., Takegawa K., Emr S. D. Vesicle-medicated protein transport: regulatory interactions between the Vps14 protein kinase and the Vps34 PtdIns 3-kinase essential for protein sorting to the vacuole in yeast. J. Cell Biol. 1995;129:321–334. doi: 10.1083/jcb.129.2.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stromhaug P. E., Reggiori F., Guan J., Wang C.-W., Klionsky D. J. Atg21 is a phosphoinositide binding protein required for efficient lipidation and localization of Atg8 during uptake of aminopeptidase I by selective autophagy. Mol. Biol. Cell. 2004;15:3553–3566. doi: 10.1091/mbc.E04-02-0147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K., Kirisako T., Kamada Y., Mizushima N., Noda T., Ohsumi Y. The pre-autophagosomal structure organized by concerted functions of APG genes is essential for autophagosome formation. EMBO J. 2001;20:5971–5981. doi: 10.1093/emboj/20.21.5971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker K. A., Reggiori F., Dunn W. A., Jr, Klionsky D. J. Atg23 is essential for the cytoplasm to vacuole targeting pathway and efficient autophagy but not pexophagy. J. Biol. Chem. 2003;278:48445–48452. doi: 10.1074/jbc.M309238200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Heijne G. How signal sequences maintain cleavage specificity. J. Mol. Biol. 1984;173:243–251. doi: 10.1016/0022-2836(84)90192-x. [DOI] [PubMed] [Google Scholar]
- Wurmser A. E., Emr S. D. Novel PtdIns(3)P-binding protein Etf1 functions as an effector of the Vps34 PtdIns 3-kinase in autophagy. J. Cell Biol. 2002;158:761–772. doi: 10.1083/jcb.200112050. [DOI] [PMC free article] [PubMed] [Google Scholar]