Abstract
As for most cell–cell fusion events, the molecular details of membrane fusion during yeast mating are poorly understood. The multipass membrane protein Prm1 is the only known component that acts at the step of bilayer fusion. In its absence, mutant mating pairs lyse or arrest in the mating reaction with tightly apposed plasma membranes. We show that deletion of FIG 1, which controls pheromone-induced Ca2+ influx, yields similar cell fusion defects. Although extracellular Ca2+ is not required for efficient cell fusion of wild-type cells, cell fusion in prm1 mutant mating pairs is dramatically reduced when Ca2+ is removed. This enhanced fusion defect is due to lysis. Time-lapse microscopy reveals that fusion and lysis events initiate with identical kinetics, suggesting that both outcomes result from engagement of the fusion machinery. The yeast synaptotagmin orthologue and Ca2+ binding protein Tcb3 has a role in reducing lysis of prm1 mutants, which opens the possibility that the observed role of Ca2+ is to engage a wound repair mechanism. Thus, our results suggest that Prm1 and Fig1 have a role in enhancing membrane fusion and maintaining its fidelity. Their absence results in frequent mating pair lysis, which is counteracted by Ca2+-dependent membrane repair.
INTRODUCTION
Membrane fusion is an essential process, affording the dynamic communication between membrane-bounded organelles in all eukaryotic cells. Membrane vesicles constantly pinch off one membrane and fuse with another, providing transport shuttles between distinct compartments. For each fusion event, lipid bilayers have to be brought into tight contact so that lipids can flow between two apposing bilayers, leading to their union. Current models suggest that first a lipid stalk forms between the apposing monlayers of the two bilayers, leading to a state called “hemifusion” in which the apposed monolayers are continuous, yet the monolayers on the opposite side of the membrane remain distinct. The hemifusion state is then resolved by establishing a fusion pore through the center of the stalk (Jahn et al., 2003). These events are catalyzed by specific fusases, the best characterized fusases being the family of soluble N-ethylmaleimide-sensitive factor attachment protein receptor (SNARE) proteins that mediate intracellular transport vesicle delivery, and viral fusion proteins that mediate entry of enveloped viruses into cells by fusion with the plasma membrane or endocytic membranes (Weber et al., 1998; Jahn et al., 2003; Kielian and Rey, 2006).
Biological membranes do not fuse spontaneously because of a large energy barrier that must be overcome by dehydration and destabilization of the apposed membranes. Both viral fusases and SNAREs are thought to overcome this barrier by forming tight coiled-coil interactions that bring membrane anchors from each membrane in proximity, thereby squeezing out water and distorting the packing of membrane lipids to allow fusion (Sollner, 2004). For other membrane fusion events, such as cell–cell fusion, the players and the mechanism have remained largely elusive.
Cell–cell fusion events occur during sperm–egg fusion in fertilization, syncytia formation during development such as myoblast fusion to form myotubes, tumorigenesis (Chen and Olson, 2005), and the mating of haploid yeast cells to form diploid cells (White and Rose, 2001). A common mechanism for cell–cell fusion has not been elucidated, but all characterized fusion mechanisms are thought to involve integral plasma membrane proteins, which bring bilayers into tight apposition and distort them sufficiently to promote lipid flow between them (Jahn et al., 2003).
A few integral membrane proteins have been described to promote cell–cell fusion, yet it is not clear what relative contributions they provide to the membrane fusion step. EFF-1, for example, is necessary for epidermal cell fusion, which serves in Caenorhabditis elegans to form a continuous syncytium (Mohler et al., 2002). EFF-1 is a type-I plasma membrane protein and localizes to fusion zones. Importantly, expression of EFF-1 in cells that would not normally fuse is sufficient to cause cell–cell fusion, strongly implicating EEF-1 as a core part of the fusion machinery (Podbilewicz et al., 2006). In myoblast fusion, numerous integral membrane proteins are important for cell migration and adhesion; yet, cytoplasmic proteins (such as Ants and Rols) also play important roles and interact with fusion-relevant membrane proteins (Taylor, 2002). The tetraspanin CD9 is required in mouse eggs for fertilization, suggesting that specialized membrane domains may be assembled for fusion by tetraspanins (Kaji et al., 2000; Hemler, 2001).
The fusion of haploid yeast cells of opposite mating types provides a genetically tractable model system to study cell–cell fusion. Diploid formation is a multistep process requiring pheromone secretion and sensing, cell cycle arrest, cell polarization toward the mating partner, cell–cell agglutination, cell wall remodeling so that the two plasma membranes can touch, plasma membrane fusion to form a fused mating pair, and finally karyogamy (White and Rose, 2001).
Cell polarization is induced upon pheromone binding to its cognate receptor that activates a trimeric G protein and allows cells to polarize cell growth and secretion toward their mating partner by forming a shmoo. Interestingly, the plasma membrane is also polarized as it becomes highly enriched in ergosterol and lipids containing long-chain bases in shmoo tip (Bagnat and Simons, 2002). Ultrastructural analyses of shmooing cells and fusing mating pairs revealed clusters of densely staining vesicles under the shmoo tip and at the zone of cell fusion. Fus1 has been implicated in the focusing of the vesicle clusters to the shmoo tip; in its absence mating pair fusion is defective and arrests before cell wall removal (Gammie et al., 1998). Cells lacking FIG 1 show subtle polarization and fusion defects, which can be enhanced by removing calcium and suppressed using higher calcium concentrations (Erdman et al., 1998; Muller et al., 2003a). FIG 1 encodes a four-spanning membrane protein that is required for a peak of calcium influx induced by pheromone and for rapid death in a fraction of cells exposed to high concentrations of pheromone (Erdman et al., 1998; Muller et al., 2003a; Zhang et al., 2006). Deletion of FIG 1 also prevents filamentation of yeast growing in the presence of butanol, further demonstrating a link between Fig1 and polarized growth (Lorenz et al., 2000). Two additional genes, FUS2 and RVS161, are required for cell wall removal. Vesicle clustering still occurs in their absence (Gammie et al., 1998).
After cell wall removal, membrane–membrane contact is initiated, and fusion rapidly ensues. The first gene, PRM1, participating in the bilayer fusion step per se was initially identified using a bioinformatics approach (Heiman and Walter, 2000). Prm1 is a multipass membrane protein only expressed in the mating context and is required for efficient membrane fusion. Prm1 localizes to the cell surface at the fusion zone in mating pairs. In its absence, 40% remain arrested as unfused mating pairs (prezygotes), with plasma membranes closely apposed but unfused (Heiman and Walter, 2000; Jin et al., 2004). Unfused mating pairs exhibit cytoplasmic bubbles, in which the two apposed plasma membrane are pushed at the zone of contact into one or the other cell of the unfused mating pair (Heiman and Walter, 2000). A second outcome of mating in the absence of PRM1 is cell lysis. An additional 20% of prm1Δ × prm1Δ mating pairs lyse. Lysis depends on membrane contact, as further removing FUS1, an upstream gene that promotes cell wall removal, suppresses mating pair lysis (Jin et al., 2004). This contact-dependent lysis event is molecularly distinct from Pkc1-regulated lysis that mating pairs can undergo due to rapid cell wall remodeling during shmoo formation or premature cell wall removal between cells of a mating pair (Philips and Herskowitz, 1997; Jin et al., 2004). In contrast to Pkc1-regulated lysis, lysis of mating prm1Δ mutant cells cannot be suppressed by growing cells on osmotic support. These observations suggest that lysis occurs as a consequence of the engagement of a defective membrane fusion machine. Fusion pores in Δprm1 × Δprm1 mating pairs have a small decrease in the initial permeance, further supporting a role for Prm1 of the fusion machinery (Nolan et al., 2006).
Here, we show that Fig1 is required for efficient membrane fusion during yeast mating and that Ca2+ depletion increases lysis of fig1Δ × fig1Δ and prm1Δ × prm1Δ mating pairs. Lysis occurs with identical kinetics as cell–cell fusion initiation, strengthening the hypothesis that mating pair lysis is an off-pathway outcome caused by engagement of defective cell–cell fusion machinery. We identify the yeast synaptotagmin homologue Tcb3 as a mediator of Ca2+-dependent lysis prevention.
MATERIALS AND METHODS
Media and Yeast Strains
Synthetic complete (SC), and complex (YPD) media were prepared and supplemented with 2% glucose by using reagents from Difco (Detroit, MI) and Sigma-Aldrich (St. Louis, MO). Synthetic growth media lacking calcium were prepared similarly using yeast nitrogen base (YNB) without calcium chloride (Bio 101, Vista, CA) and further treatment of the complete media with resin-bound 1,2-bis(2-aminophenoxy)ethane-N,N,N′,N′-tetraacetic acid (BAPTA) calcium sponge (Invitrogen, Carlsbad, CA). All strains used in this study (Table 1) are derivatives of wild-type strain W303. Gene replacements were generated with the polymerase chain reaction (PCR) transformation technique (Longtine et al., 1998) and confirmed by PCR.
Table 1.
Yeast strains
| Strain | Genotype | Source |
|---|---|---|
| MHY426 | MATaprm1Δ::HIS3 | Walter laboratory |
| MHY191 | MATα prm1Δ::HIS3 with pcGFP (CEN, URA3, GFP) | Walter laboratory |
| PAY239 | MATafig1Δ::KanMX4 | This work |
| PAY248 | MATα fig1Δ::KanMX4 pcGFP MATa | This work |
| PAY251 | MATaprm1Δ::HIS3 fig1Δ::KanMX4 | This work |
| PAY254 | MATα prm1Δ::HIS3fig1Δ::KanMX4 pcGFP | This work |
| PAY261 | MATa FIG1-GFP::HIS3 | This work |
| PAY262 | MATα FIG1-GFP::HIS3 | This work |
| AEY9 | MATafus1Δ::KanMX4 | This work |
| AEY1 | MATα fus1Δ::KanMX4 with pcGFP | This work |
| AEY10 | MATafus2Δ::KanMX4 | This work |
| AEY2 | MATα fus2Δ::KanMX4 with pcGFP | This work |
| PAY579 | MATatcb1Δ::HIS3 tcb2Δ::cgTRP1atcb3Δ::HIS3 | This work |
| PAY580 | MATα tcb1Δ::HIS3tcb2Δ::cgTRP1atcb3Δ::HIS3 pcGFP | This work |
| PAY581 | MATaprm1Δ::cgLEU2atcb1Δ::HIS3tcb2Δ::cgTRP1atcb3Δ::HIS3 | This work |
| PAY582 | MATα prm1Δ::cgLEU2atcb1Δ::HIS3tcb2Δ::cgTRP1atcb3Δ::HIS3 pcGFP | This work |
| PAY606 | MATaprm1Δ::cgLEU2atcb1Δ::HIS3 | This work |
| PAY607 | MATα prm1Δ::cgLEU2atcb1Δ::HIS3 pcGFP | This work |
| PAY610 | MATaprm1Δ::cgLEU2atcb3Δ::HIS3 | This work |
| PAY614 | MATα prm1Δ::cgLEU2atcb3Δ::HIS3 pcGFP | This work |
| PAY623 | MATaprm1Δ::HIS3tcb2Δ::cgLEU2a | This work |
| PAY624 | MATα prm1Δ::cgLEU2atcb2Δ::cgTRP1a pcGFP | This work |
a cgTRP1 and cgLEU2 indicate Candida glabrata TRP1 and LEU2 genes, respectively.
Quantitative Cell Fusion Assays
In a standard assay, cells of opposite mating types, with the MATα strain expressing soluble cytosolic green fluorescent protein (GFP), were grown to mid-log phase. An equal number of cells of each mating type were mixed and vacuumed to a nitrocellulose filter. The filter was placed cell-side up on either YPD or supplemented YPD plates and then incubated for 3 h at 30°C. Cells were scraped off the filter, fixed in 4% paraformaldehyde, incubated at 4°C overnight, and inspected by fluorescence microscopy. To quantify cell lysis, mating mixtures were scraped and stained either with 0.02% methylene blue or 0.008% trypan blue for 15 min at 30°C. Methylene blue-stained cells were directly imaged by light microscopy, and trypan blue cells were washed and fixed in 4% paraformaldehyde before fluorescence microscopy analysis. Both methods yielded indistinguishable results. For liquid media cell fusion assays, cells of opposite mating types were grow as described above and then mixed (0.3 OD/mating type) before filtering onto 12-mm transwells (Corning Life Sciences, Acton, MA). Transwells were placed on chambers with 1 ml of synthetic media and covered with 300 μl of same media. After 3.5 h at 30°C, mating mixtures were treated for quantification of lysis and fixed as described above.
Fluorescence Microscopy
Fluorescence and differential interference contrast (DIC) microscopy was performed using an Axiovert 200M microscope (Carl Zeiss, Jena Germany), equipped with an X-cite 120 mercury arc lamp (EXFO) and an Orca ER camera (Hamamatsu, Bridgewater, NJ). Image-Pro (Media Cybernetics, Silver Spring, MD) or MetaMorph (Molecular Devices, Sunnyvale, CA) were used for data collection. Time-lapse microscopy was performed as described previously (Jin et al., 2004), with a few modifications. In brief, cells derived from preincubated mating mixtures were mounted on agarose pads, which contained 1.8% agarose in SC media. A coverslip was placed on top of this pad and sealed using VALAP or nail polish. Mating was followed at room temperature. Analysis of whole cell fluorescence during lysis was done using ImageJ (http://rsb.info.nih.gov/ij/).
Electron Microscopy
Cells of opposite mating type were treated as described above for quantitative cell fusion assays. Mating mixtures were scraped off, fixed, and processed as described previously (Heiman and Walter, 2000). Briefly, cells were fixed in 1% glutaraldehyde, 0.2% paraformaldehyde, and 0.04 M potassium phosphate, pH 7, washed, and then incubated in 2% KMnO4. After dehydration in ethanol, cells were prepared for embedding, replacing ethanol with propylene oxide. Embedding was performed using graded concentrations of resin (32% Epon, 18% Araldite, 34% dodecenyl succinic anhydride, and 16% nadic methyl anhydride; Ted Pella, Redding, CA) mixed with propylene oxide, followed by overnight infiltration with pure resin. Then, cells were transferred to resin containing 2% benzyl dimethyl amine (Ted Pella), and incubated at 70°C for 1 d. Sections of 90 nm thickness were cut, stained with lead citrate (Ted Pella), and imaged with an electron microscope (EM400; Philips, FEI Co., Hillsboro, OR).
RESULTS
FIG 1 Has a Role in the Membrane Fusion Step of Yeast Mating
To identify additional mutants that are defective at the step of membrane fusion, we asked whether mating of mutants bearing deletion of nonessential genes results in unfused mating pairs (prezygotes) that display cytoplasmic bubbles as observed in prm1Δ × prm1Δ matings. Bubble formation is indicative of successful cell wall degradation without plasma membrane fusion. To this end, we screened genes that were identified by a bioinformatics approach as pheromone-induced membrane proteins (Heiman and Walter, 2000). Indeed, mating cells bearing deletions of the most highly pheromone-induced candidate gene FIG 1 yielded an accumulation of unfused mating pairs and approximately 1/10 of those showed the bubble phenotype (Figure 1A). This observation suggests that Fig1, like Prm1, has a role in promoting membrane fusion during yeast mating. FIG 1 was originally identified as a pheromone induced gene that encodes a membrane protein with four potential transmembrane domains (Erdman et al., 1998).
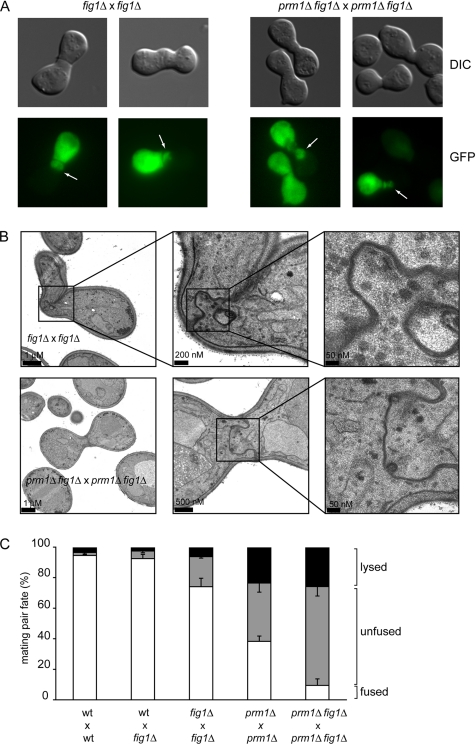
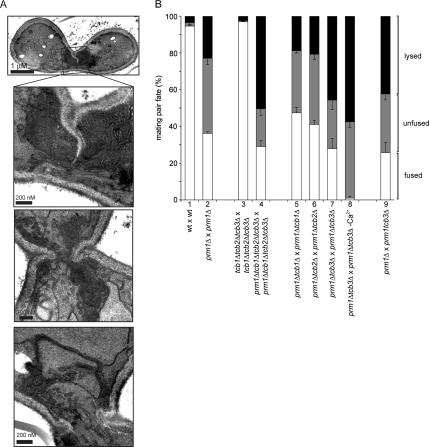
Figure 1.
PRM1 and FIG 1 promote the membrane fusion step during mating. (A) Unfused fig1Δ × fig1Δ mating pairs form bubbles. MATα cells carrying cytoplasmic GFP were mixed with MATa cells on nitrocellulose filters and incubated on YPD plates for 3 h at 30°C. Fixed mating mixtures were then imaged by DIC and fluorescence microscopy. Arrows point to mating pair bubbles. (B) Bubbles in unfused fig1Δ × fig1Δ mating pairs contain closely apposed membranes. The ultrastructural detail of fig1Δ × fig1Δ mating pairs was determined as described in A with mating mixtures processed as described in Materials and Methods. The panels show three different magnifications for each mating pair. The lower magnification picture in the bottom panel corresponds to a different section of the same mating pair. (C) PRM1 and FIG 1 act through different pathways to promote cell fusion. Quantitative cell fusion and lysis assays were performed as described in Materials and Methods. All deletion genotypes were tested for both mating types; the results were indistinguishable. Error bars indicate SEs for four independent experiments with 300 mating pairs scored for each case.
The cytoplasmic bubbles in unfused mating pairs resulting from fig1Δ × fig1Δ crosses were indistinguishable in appearance by fluorescent microscopy from the bubbles seen in prm1Δ × prm1Δ mating pairs (Figure 1A). Examination of thin sections of fig1Δ × fig1Δ unfused mating pair bubbles by transmission electron microscopy confirmed that the cell wall between the mating partners was removed over extended areas with membranes protruding into one mating partner (Figure 1B). In these regions, the plasma membranes of both cells were in close, evenly spaced apposition (∼10 nm), as reported previously for prm1Δ × prm1Δ unfused mating pairs (Heiman and Walter, 2000).
We quantified cell fusion efficiency by using a microscopy assay, imaging mating pairs with one partner expressing soluble cytosolic GFP. Fused mating pairs are easily distinguished from unfused pairs because GFP diffuses throughout the entire mating pair. In addition, we stained cells in each mating reaction with a vital dye, which allowed us to score mating-induced cell lysis. Unilateral matings in which FIG 1 was deleted in one of either mating type led to only minor, insignificant mating defects (Figure 1C; wt × fig1Δ). By contrast, cell fusion was reduced by ∼25% in a bilateral cross where both mating partners lacked FIG 1 (Figure 1C; fig1Δ × fig1Δ; ∼20% unfused and ∼6% lysed). The fusion defect of fig1Δ × fig1Δ mating reactions was weaker than the 60% effect typically observed in prm1Δ × prm1Δ mating reactions (Figure 1C; prm1Δ × prm1Δ; 35% unfused and 25% lysed). Note that a significant fraction of the increased mating failure in prm1Δ × prm1Δ mating reactions is due to cell lysis.
The mild cell fusion phenotype of fig1Δ × fig1Δ mating reactions suggests that PRM1 is still functional in these strains. This notion is corroborated by the synthetic phenotype observed for prm1Δ fig1Δ double mutants. Compared with the prm1Δ × prm1Δ mating reaction (∼40% fused mating pairs), the prm1Δ fig1Δ × prm1Δ fig1Δ double mutant mating reaction suffers a marked reduction (∼10% fused mating pairs; Figure 1C). This fourfold decrease in fusion efficiency is the result of the accumulation of more unfused mating pairs and not of increased lysis.
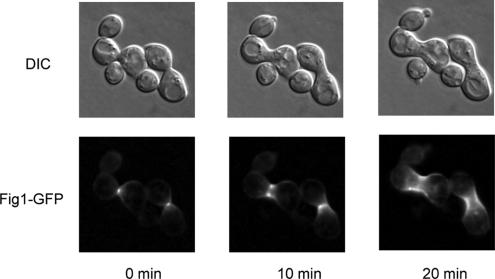
Unfused mating pairs in prm1Δ fig1Δ × prm1Δ fig1Δ mating reactions exhibited bubbles, indistinguishable from those seen in the single mutants both in abundance and in morphology as assessed by fluorescence and electron microscopy (Figure 1, A and B). Like Prm1, Fig1 is enriched in the shmoo tip and in the fusion zone of mating pairs (Figure 2). After fusion, Fig1-GFP remains localized as a collar around the neck of the zygotes. Together, the localization and functional data are consistent with a role of Fig1 in the membrane fusion event.
Figure 2.
Fig1 localizes at the site of cell fusion. Cells of opposite mating types bearing the FIG 1–GFP fusion gene were mixed, preincubated on filters for 1 h on YPD at 30°C, and mounted on agarose complete media pads for imaging.
The Residual Fusion Activity in prm1Δ × prm1Δ Mating Reactions Requires Extracellular Ca2+
Fig1 was recently described as regulator of pheromone-induced Ca2+ influx (Muller et al., 2003a). This observation prompted us to explore the possibility that the membrane fusion reaction in cell–cell fusion might require Ca2+. As shown in Figure 3A, fusion of wild-type cells was completely insensitive to the Ca2+ chelator EGTA. By contrast, we discovered to our surprise that the residual fusion observed in prm1Δ × prm1Δ, and prm1Δ fig1Δ × prm1Δ fig1Δ mating reactions was significantly inhibited when Ca2+ was removed from the media by addition of EGTA. In the presence of EGTA, the production of fused mating pairs was reduced 10-fold in the prm1Δ × prm1Δ and prm1Δ fig1Δ × prm1Δ fig1Δ fusion reactions, and it was reduced to a lesser degree (1.4-fold) in the fig1Δ × fig1Δ fusion reaction (Figure 2A). Fusion efficiency of a fus1Δ × fus1Δ mating reaction where cell fusion is blocked at the cell wall remodeling step (McCaffrey et al., 1987; Trueheart et al., 1987; Gammie et al., 1998) was not sensitive to Ca2+ chelation, suggesting that extracellular Ca2+ removal affects one or more steps after cell wall remodeling.
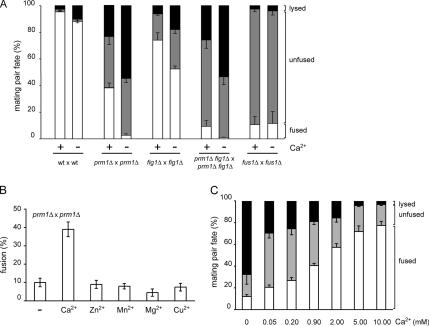
Figure 3.
Extracellular Ca2+ suppresses cell lysis in prm1Δ × prm1Δ mating reactions. (A) Fusion of prm1Δ × prm1Δ mating pairs is highly sensitive to EGTA. Mating mixtures were incubated on YPD plates with (Ca2+, −) or without (Ca2+, +) 20 mM EGTA. (B) Sensitivity of prm1Δ × prm1Δ mating pairs to EGTA is due to a lack of Ca2+. Liquid media cell fusion assays were performed in synthetic media treated with resin-bound BAPTA and supplemented either with water (−) or divalent cation salts at concentrations found in synthetic complete media: 900 μM calcium chloride, 1 μM zinc sulfate, 10 μM manganese sulfate, 1 mM magnesium sulfate, and 1 μM copper sulfate. (C) Extracellular calcium suppresses the prm1Δ × prm1Δ mating defect. Liquid media cell fusion assays were performed as in B by using media supplemented with calcium chloride as indicated. Error bars indicate SEs for at least three independent experiments with 300 mating pairs scored per experiment.
Interestingly, the reduction in cell fusion efficiency of the prm1Δ × prm1Δ, and prm1Δ fig1Δ × prm1Δ fig1Δ fusion reactions in the absence of Ca2+ was due almost exclusively to an increase in cell lysis (Figure 3A). For the fig1Δ × fig1Δ fusion reaction, the reduction in fusion efficiency in the absence of Ca2+ was due to increases in both cell lysis and accumulation unfused mating pairs.
To confirm that the sensitivity of the mating reaction to EGTA was indeed due to Ca2+ removal rather than chelation of some other divalent cation, we developed a quantitative mating assay using liquid growth media (see Materials and Methods). The cell fusion efficiency of prm1Δ × prm1Δ mating pairs was the same in synthetic media using the liquid assay as in our standard mating assay on YPD plates. We then removed Ca2+ from the synthetic media by incubation with a BAPTA-resin. Cell fusion dropped to levels comparable to those observed in 20 mM EGTA-YPD (Figure 3B), and an equivalent increase in mating pair lysis was measured. On readdition of different divalent cations such as Ca2+, Mn2+, Zn2+, Mg2+, and Cu2+, only Ca2+ suppressed the prm1Δ × prm1Δ fusion defect (Figure 3B). Surprisingly, higher levels of extracellular Ca2+ (2–10 mM) alleviate the prm1Δ × prm1Δ fusion defect even further (Figure 3C). These assays performed with a wide range of Ca2+ concentrations show a direct relationship between the extracellular concentration of Ca2+, cell fusion, and reduction of lysis. We therefore conclude that Ca2+ helps prevent cell lysis and promotes fusion, and that it is required in prm1Δ × prm1Δ mating pairs after cell wall removal.
Fusion and EGTA-induced Lysis Occur with Similar Kinetics
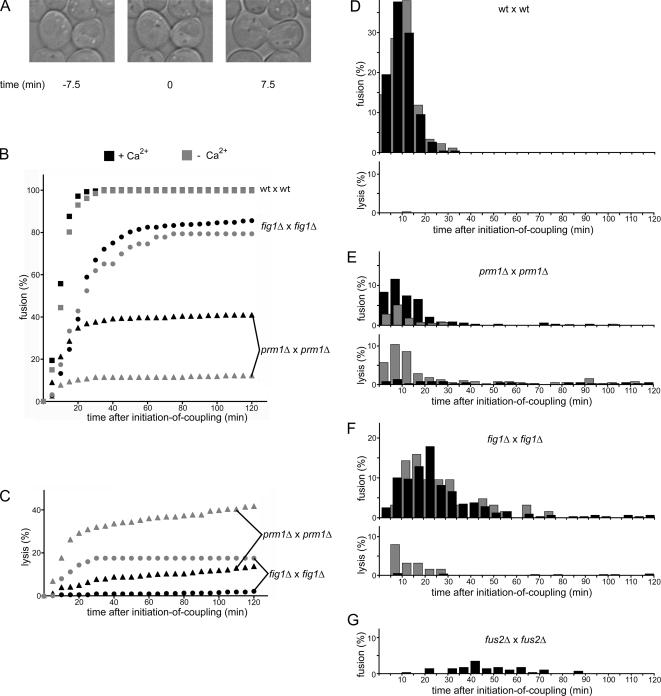
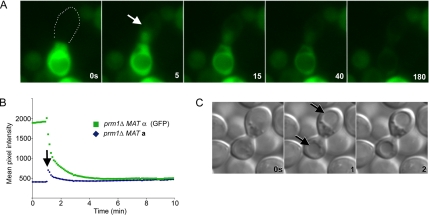
The analyses described so far suggest that mating-induced cell lysis and cell fusion are linked events. According to this notion, lysis would result from the initiation of a fusion event that fails to go to completion. One prediction of this scenario is that fusion and lysis events should occur with similar time courses. To test this prediction, we collected kinetic data using time-lapse microscopy to determine when fusion and lysis events occur in the lifetime of a mating pair. We followed cell fusion by imaging mating mixtures on agar slips at 2.5-min intervals for 3 h. Because mating pairs form asynchronously in a mating mixture, we established a reference time point to make comparison between different mating pairs possible. To this end, we defined for each mating pair a time-zero point marking the “onset-of-coupling” as the moment at which a mating pair is formed (Figure 4A, middle). At this point, the cells have begun to agglutinate their cell wall with that of a mating partner (a mature mating pair with complete cell wall agglutination is seen in the third panel). We scored fusion as the mixing of cytoplasmic GFP, and lysis as both loss of turgor pressure in the mating pair and loss of cytoplasmic GFP (Jin et al., 2004).
Figure 4.
Kinetics of fusion and lysis initiation in mating populations. The fate of individual mating pairs in a mating mixture was followed by time-lapse microscopy. (A) Example of the determination of initiation of coupling. The picture in the middle, showing the two cells initiating contact in a polarized manner, serves as a time-zero reference point for the mating pair. (B) Plot showing the progression of the fusion outcome as a function of time in the presence (black symbols) or absence (gray symbols) of Ca2+. (C) Plot showing the progression of the lysis outcome as a function of time. (D–G) Fusion and lysis events were binned into 5-min time windows and normalized to indicate the percentage of the mating pair population that would undergo the indicated event within these time windows. Black bars represent events in the presence of Ca2+; gray bars represent events in the absence of Ca2+. All data shown are the result of three independent experiments. No lysis was observed for fus2Δ × fus2Δ. The lower rate of lysis (compared with the endpoint assays shown in Figures 1 and 2) is likely due to performing the mating reaction at room temperature instead of at 30°C.
After wild-type cells form a mating pair, fusion rapidly ensued. Every observed mating pair fused (n = 231) and 97% of these fusion events occurred within the first 20 min after onset-of-coupling (t1/2 = 9.5 min; Figure 3B). When fusion events were binned and plotted over time (Figure 4D, top), the distribution fitted to a Gaussian curve (r = 0.99), and the distribution of the same reaction carried out in EGTA was superimposable, indicating that Ca2+ removal affected neither kinetics nor extent of wild-type cell fusion. Moreover, we observed virtually no mating-induced lysis under either condition for wild type cells; only ∼0.3% of wild-type mating pairs lysed in the presence of EGTA (Figure 4D, bottom).
Although only 40% of prm1Δ × prm1Δ mating pairs fused, those that did so followed nearly identical kinetics as wild-type cells: 85% of fusion-destined mating pairs fused within the first 20 min after onset-of-coupling (t1/2 = 10 min; Figure 4B). Unlike wild-type cells, a few prm1Δ × prm1Δ mating pairs fused significantly later than 20 min after onset-of-coupling (Figure 4E, top). More than half of such late-fusing mating pairs extended a bubble early in the life of the mating pair, suggesting that these events resulted from an impaired membrane fusion step rather than delayed cell wall removal. In agreement with the results shown in Figure 3, prm1Δ × prm1Δ fusion was antagonized by EGTA. However, the rate at which fusion-destined mating pairs fused was not delayed by Ca2+ removal, as 84% of prm1Δ × prm1Δ fusion events on EGTA occurred within the first 20 min after onset-of-coupling (t1/2 = 9.5 min; Figure 3B). Because we estimate that the error of determining the time of onset of coupling is ±2 min, these t1/2 values are the same within error.
Under normal conditions, i.e., in the presence of Ca2+, prm1Δ × prm1Δ mating pairs lysed at a steady, slow rate over the 2-h time frame of the experiment (Figure 4C, black triangles, and E, bottom, black bars). By contrast, we observed a dramatically changed kinetic profile when lysis was enhanced by Ca2+ removal with EGTA. Under these conditions, lysis followed biphasic kinetics. Interestingly, all of the additional lysis due to Ca2+ removal could be accounted for in an initial rapid burst phase. The t1/2 for the burst phase (8.5 min) closely matched that for fusion of wild-type (wt) × wt and prm1Δ × prm1Δ cells observed in the presence or absence of Ca2+. Lysis continued at later time points with slow kinetics indistinguishable from those seen in the presence of Ca2+ (Figure 4C, gray triangles). Thus, prm1Δ × prm1Δ mating pairs have a tendency to lyse with low frequency in the presence of Ca2+, but lyse frequently during the time window in which fusion takes place in the absence of Ca2+.
Although the fusion defect measured at a late time point after onset of coupling in fig1Δ × fig1Δ fusion reactions was not as strong as that observed for prm1Δ × prm1Δ fusion reactions, fig1Δ × fig1Δ mating pairs fused with significantly delayed kinetics (t1/2 = 21 min) compared with the t1/2 of both wt × wt and prm1Δ × prm1Δ mating pairs (Figure 4B, circles, and F, top). Only 45% of fusion events occurred within the first 20 min after onset of coupling, and the presence or absence of Ca2+ did not affect the kinetics of the reaction. Similar to prm1Δ × prm1Δ mating reactions, fig1Δ × fig1Δ mating pair lysis occurred at a slow rate both in the presence and absence of Ca2+, and a rapid burst phase of increased lysis paralleling the window of fig1Δ × fig1Δ fusion was superimposed on the slow phase in the absence of Ca2+ (Figure 4, C and F). The prm1Δ fig1Δ × prm1Δ fig1Δ double mutant fusion reaction exhibited identical delays in fusion and lysis as fig1Δ × fig1Δ mating kinetics (Supplemental Figure S1).
As a basis for comparison, we also characterized the behavior of mutants that have cell wall remodeling defects. When bilateral crosses of fus1Δ × fus1Δ and fus2Δ × fus2Δ were performed, we observed an even more apparent delay in the initiation of fusion (t1/2 = 45 and 35 min, respectively; Figure 4G; data not shown), indicating that cell wall-remodeling mutants cause a large delay in fusion, unlike the prm1Δ and fig1Δ mutants analyzed above.
Cell Lysis and Cytoplasmic Mixing Occur Synchronously
In a few of the lysed mating pairs in the time-course experiments shown in Figure 4, we observed transient spreading of GFP from the MATα cell in which it was expressed into the MATa mating partner, indicating that fusion and cytoplasmic mixing preceded or was simultaneous with lysis (Jin et al., 2004). We therefore recorded high time resolution movies to resolve content mixing and fusion. The results in five of five movies recorded were identical. Selected time frames of a representative movie imaging prm1Δ × prm1Δ mating pairs in the presence of EGTA at 5-s intervals are shown in Figure 5. Content mixing was first evident in the 5-s frame as monitored by GFP spreading (Figure 5, A and B, arrows; Supplemental Movie 1). In all cases, we observed that lysis initiated synchronously in the same frame as monitored by the diminution of overall GFP fluorescence in the mating pair and the rounding-up of the vacuole. The cytoplasmic GFP slowly diffused from the mating pair over the next 3 min.
Figure 5.
Lysis occurs simultaneously in each cell of a mating pair and is concomitant with cytoplasmic mixing. Lysis of prm1Δ × prm1Δ mating pairs in media lacking Ca2+ was imaged with fast time resolution. (A) Cytoplasmic GFP in a MATα cell spread into the MATa cell after initiation of lysis at 5 s (white arrow). Note that GFP has spread throughout the entire MATa cell in all frames subsequent to the 0-s frame, demonstrating cytoplasmic continuity. White dots represent the boundaries of the MATa cell. (B) Quantification of average pixel intensity in each mating partner over time; initiation of lysis is indicated by the black arrow. (C) DIC images of a lysing mating pair. Evidence of lysis is seen in the 1 s frame as rounded vacuoles (black arrows).
We also scored the synchrony of lysis between each cell of a mating pair by monitoring vacuolar morphology. When mating pairs lyse, the vacuoles become rounded and static (Figure 5A and Supplemental Movie 1) and loss of turgor results in high-contrast vacuole profiles (Jin et al., 2004). Figure 5C shows synchronous lysis of two cells of a prm1Δ × prm1Δ mating pair in the absence of Ca2+. The change in vacuole morphology in both the MATa and MATα cell is apparent in the 1-s frame, which is likely indicative of the synchronous loss of turgor pressure. In each of 11 lysis events analyzed this way, lysis of both cells occurred within a 2-s time window.
A Yeast Synaptotagmin Homolog, TCB3, Dampens prm1Δ × prm1Δ Mating Pair Lysis
Lysis of mating pairs is a result of plasma membrane disruption, and, as we have shown here, low extracellular Ca2+ concentrations enhance the penetrance of lysis, whereas higher concentrations suppress it. As shown in Figure 6A, prm1Δ × prm1Δ mating pairs in EGTA showed a remarkable abundance of membranes accumulating in the zone of cell–cell fusion/lysis, suggesting that membrane vesicles are recruited there yet in the absence of Ca2+ do not get consumed. These observations are reminiscent of repair mechanisms that have been described for damaged membranes in numerous systems and have been shown to require extracellular Ca2+ (Yawo and Kuno, 1985; Steinhardt et al., 1994). In mammalian cells, for example, membrane wound repair can be mediated by Ca2+ triggered exocytosis of lysosomes, and synaptotagmin VII has been suggested as a potential Ca2+ sensor for this regulated exocytosis event (Reddy et al., 2001). A family of yeast proteins, Tcb1, Tcb2, and Tcb3, shares similar domain architectures with synaptotagmins (Schulz and Creutz, 2004). These proteins contain predicted transmembrane helices, followed by multiple C2 (Ca2+ binding) domains.
Figure 6.
TCB3 prevents mating pairs lysis in the absence of PRM1. (A) In the absence of Ca2+, prm1Δ × prm1Δ mating pairs show extensive membrane accumulation in the zone of cell fusion. Mating mixtures were processed as described in Materials and Methods. The panels show the zone of cell–cell contact for three different mating pairs. (B) Deletions of TCB1, TCB2, and TCB3 were tested for their effect on cell fusion and lysis during mating. Crosses are labeled as MATα × MATa. Error bars indicate SEs for at least three independent experiments with 300 mating pairs scored per experiment.
To test whether Tcb1, Tcb2, and/or Tcb3 have a role in suppressing lysis during yeast mating, we deleted the genes encoding these proteins in prm1Δ and isogenic wild-type cells and assayed unilateral and bilateral crosses for fusion defects as described above. Deletion of all three TCB genes in a prm1Δ background resulted in a greater than twofold increase in mating pair lysis (Figure 6B, bar 4), whereas it caused no significant defect in wild-type cells (Creutz et al., 2004). The degree of the enhanced lysis defect in prm1Δ × prm1Δ TCB-deleted mating pairs (∼50%) was equivalent to that observed in prm1Δ × prm1Δ mating pairs in the absence of Ca2+ (Figure 3A, bar 4). Moreover, it was exclusively due to the deletion of TCB3 (Figure 6B, bar 7), whereas deletion of TCB1 and TCB2 had no effect (Figure 6B, bars 5 and 6). The enhanced lysis defect was undiminished if only one of the prm1Δ cells was missing TCB3, regardless of which mating type lacked the gene (Figure 6B, bar 9; data not shown). Both cells of a mating pair lyse even if TCB3 is deleted only in one of the mating partners. The lysis defect of prm1Δ tcb3Δ × prm1Δ tcb3Δ mating pairs was only slightly enhanced upon removal of Ca2+ (Figure 6B, bar 8), consistent with the notion that to suppress lysis Tcb3 is an important target of Ca2+. It is likely, however, that there are other targets of Ca2+ because in regard to fusion efficiency prm1Δ tcb3Δ × prm1Δ tcb3Δ mating pairs do not completely phenocopy the mating defect of prm1Δ × prm1Δ mating pairs in the absence of Ca2+.
DISCUSSION
Before this work, Prm1 was the only protein known to act at the plasma membrane fusion step during yeast mating. Here, we expand the cast of players in membrane fusion with the characterization of Fig1. FIG 1 was initially identified as a pheromone-induced gene with roles in cell polarization and fusion (Erdman et al., 1998). Here, we show that Fig1 localizes to the zone of cell–cell contact and that deletion of FIG 1 results in a membrane fusion defect after cell wall removal, as indicated by ultrastructural analyses and the formation of cytoplasmic bubbles that are bounded by tightly apposed plasma membranes from both mating partners. Moreover, bilateral deletion of FIG 1 significantly enhances the fusion defects observed in prm1Δ × prm1Δ mating pairs. Unlike wild-type and prm1Δ × prm1Δ mating pairs, fig1Δ × fig1Δ mating pairs are delayed in initiating of fusion but the delay is shorter than that observed in fus1Δ × fus1Δ mating pairs and, unlike fus1Δ × fus1Δ mating pairs, fig1Δ × fig1Δ mating pairs still cluster vesicles at the zone of cell fusion (Aguilar, unpublished data). Although fig1Δ × fig1Δ mating reactions are sensitive to the removal of Ca2+, Fig1 most likely has fusion promoting roles independent of Ca2+ influx because removal of extracellular calcium from wild-type mating pairs does not result in mating defects. As a member of the Claudin superfamily, Fig1 may share functional properties with tight junction proteins and possibly help to arrange the fusion machinery by holding membranes in proximity (Van Itallie and Anderson, 2004; Zhang et al., 2006).
Albeit severely compromised, some residual fusion activity remains in the absence of Prm1 and Fig1. This observation suggests that either 1) Prm1 and Fig1 are important yet nonessential components of the fusion machinery or that 2) an alternate Prm1- and Fig1-independent fusion pathway(s) can compensate for their absence. Currently available data do not allow us to distinguish between these possibilities.
Nonproductive mating pairs that fail to fuse in the absence of Prm1 and/or Fig1 can either lyse or remain unfused with their plasma membranes in close apposition. It was previously suggested that the observed cell lysis may be a direct result of engagement of the cell fusion machinery and possibly be intrinsically linked to the mechanism of lipid bilayer fusion (Jin et al., 2004); the results presented here support this view. In Table 2, we describe the relationship between the two phenotypes of mating pairs lacking Prm1 and/or Fig1 by quantifying to values, the “activity” and “fidelity” of the membrane fusion machinery. We define activity as the probability of engagement of an active membrane fusase, which can lead to either fusion or lysis. Only 62% of prm1Δ × prm1Δ mating pairs engage a fusase, compared with 99% for wild-type mating pairs (Table 2). We define fidelity as the probability that cells in a mating pair will survive after engagement of a fusase. Fidelity declines from 96% in wild-type mating pairs to 63 and 43% in mating pairs missing Prm1, and Prm1 and Fig1 in both mating partners, respectively. Thus, only 37% of cells missing Prm1 and Fig1 engage the fusion machinery and of those that do only 43% survive.
Table 2.
Influence of Ca2+ on activity and fidelity values
| Activity |
Fidelity |
|||
|---|---|---|---|---|
| +Ca2+ | −Ca2+ | +Ca2+ | −Ca2+ | |
| wt × wt | 99.0 | 98.3 | 96.3 | 94.9 |
| prm1Δ × prm1Δ | 62.0 | 56.5 | 62.7 | 7.1 |
| fig1Δ × fig1Δ | 80.3 | 70.3 | 92.2 | 74.5 |
| prm1Δfig1Δ × prm1Δfig1Δ | 37.4 | 43.6 | 42.7 | 1.2 |
In contrast to fusase activity, fusase fidelity is sensitive to the state of extracellular Ca2+. Whereas we found no requirement for Ca2+ during mating of wild-type cells, fidelity values for mating reactions carried out in the absence of Ca2+ drop to 7 and 1.2% for prm1Δ × prm1Δ and prm1Δ fig1Δ × prm1Δ fig1Δ mating pairs, respectively. Thus, Ca2+ masks the true extent of the fidelity defects of prm1 and fig1 mutant fusion machines, but it is important only in the context of the defective fusion machine in the mutant cells.
Our results support in multiple ways a functional coupling of lysis to the engagement of the fusion machine: First, by removing Ca2+ to favor lysis, we observe that the timing of lysis events is the same as the timing of fusion. Second, we demonstrate that the two cells of a mating pair lyse synchronously, as expected for events at the interface between both cells in a mating pair. Third, mixing of cytoplasmic contents occurs concomitant with the initiation of lysis. This implies that lysis is initiated as fusion is catalyzed, most simply explained by hypothesizing a common machinery for the two outcomes. It is possible, for example, that a defective fusion machinery may not contain the fusion zone properly or correctly resolve unstable membrane intermediates, leading to mating pair lysis. Indeed, recent models of bilayer fusion (Muller et al., 2003b) pose that membrane fusionlysis is not a failsafe process: formation of the lipid stalk favors the formation of holes adjacent to the stalk in each of the two engaged membranes. Thus, it is conceivable that the very act of bilayer membrane fusion can cause membrane rupture and cell lysis—unless the fusion zone is contained by accessory proteins of the fusion machinery. Prm1 and Fig1 could play such a role, for example, by providing a molecular fence that corrals the fusion zone and prevents the catastrophic spread of local membrane damage. Corralling also could serve an instructive role helping organizing the activity of the fusion machine, thus explaining the reduced fusion activity in mating reactions of cells lacking Prm1 and Fig1.
In this light, an attractive explanation for the Ca2+ effect in the mutant cells is that the mutations enhance lysis, which is counteracted by Ca2+-dependent membrane repair mechanisms, thus influencing the fusion–lysis balance by rescuing potential lysis events. This would explain why Ca2+ is not required during wild-type mating reactions where Prm1 and Fig1 prevent lysis events from occurring. We provide evidence that Tcb3, a yeast synaptotagmin orthologue, may function as a Ca2+ sensor in a membrane repair pathway operating during this process. Deletion of TCB3 mimics the lysis increase observed in prm1Δ × prm1Δ mating pairs upon Ca2+ depletion. This model also would explain the accumulation of membranes in prm1Δ × prm1Δ mating pairs upon Ca2+ depletion.
Although attractive, this model leaves many interesting questions to be solved: For example it does not explain all of the observed effects that Ca2+ exerts on membrane fusion. In particular, the observation that high Ca2+ concentrations partially suppress the defects of prm1Δ × prm1Δ mating pairs suggests that Ca2+ at high concentrations also may promote the fusion of apposed membranes, perhaps by directly interacting with membrane lipids as seen in membrane fusions assays of pure lipid vesicles (Duzgunes et al., 1981; Ellens et al., 1985). Alternatively, other yet to be identified Ca2+ sensors in addition to Tcb3 may participate in the fusion process.
Supplementary Material
ACKNOWLEDGMENTS
We thank Tobias Walther, Tomas Aragon, Max Heiman, and the members of the Walter laboratory for many invaluable suggestions and discussion, and Saskia Neher, Sebastian (Seabass) Bernales, and Alexei Korennykh for helpful comments on the manuscript. This work was supported in part by postdoctoral fellowships from the Fundación Antorchas and Damon Runyon Cancer Research Fundation to P.A. and by grants from the National Institutes of Health to P.W. P.W. is an investigator of the Howard Hughes Medical Institute.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E06-09-0776) on December 6, 2006.

 The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
REFERENCES
- Bagnat M., Simons K. Cell surface polarization during yeast mating. Proc. Natl. Acad. Sci. USA. 2002;99:14183–14188. doi: 10.1073/pnas.172517799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen E. H., Olson E. N. Unveiling the mechanisms of cell-cell fusion. Science. 2005;308:369–373. doi: 10.1126/science.1104799. [DOI] [PubMed] [Google Scholar]
- Creutz C. E., Snyder S. L., Schulz T. A. Characterization of the yeast tricalbins: membrane-bound multi-C2-domain proteins that form complexes involved in membrane trafficking. Cell Mol. Life Sci. 2004;61:1208–1220. doi: 10.1007/s00018-004-4029-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duzgunes N., Wilschut J., Fraley R., Papahadjopoulos D. Studies on the mechanism of membrane fusion. Role of head-group composition in calcium- and magnesium-induced fusion of mixed phospholipid vesicles. Biochim. Biophys. Acta. 1981;642:182–195. doi: 10.1016/0005-2736(81)90148-6. [DOI] [PubMed] [Google Scholar]
- Ellens H., Bentz J., Szoka F. C. H+- and Ca2+-induced fusion and destabilization of liposomes. Biochemistry. 1985;24:3099–3106. doi: 10.1021/bi00334a005. [DOI] [PubMed] [Google Scholar]
- Erdman S., Lin L., Malczynski M., Snyder M. Pheromone-regulated genes required for yeast mating differentiation. J. Cell Biol. 1998;140:461–483. doi: 10.1083/jcb.140.3.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gammie A. E., Brizzio V., Rose M. D. Distinct morphological phenotypes of cell fusion mutants. Mol. Biol. Cell. 1998;9:1395–1410. doi: 10.1091/mbc.9.6.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heiman M. G., Walter P. Prm1p, a pheromone-regulated multispanning membrane protein, facilitates plasma membrane fusion during yeast mating. J. Cell Biol. 2000;151:719–730. doi: 10.1083/jcb.151.3.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemler M. E. Specific tetraspanin functions. J. Cell Biol. 2001;155:1103–1107. doi: 10.1083/jcb.200108061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahn R., Lang T., Sudhof T. C. Membrane fusion. Cell. 2003;112:519–533. doi: 10.1016/s0092-8674(03)00112-0. [DOI] [PubMed] [Google Scholar]
- Jin H., Carlile C., Nolan S., Grote E. Prm1 prevents contact-dependent lysis of yeast mating pairs. Eukaryot. Cell. 2004;3:1664–1673. doi: 10.1128/EC.3.6.1664-1673.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaji K., Oda S., Shikano T., Ohnuki T., Uematsu Y., Sakagami J., Tada N., Miyazaki S., Kudo A. The gamete fusion process is defective in eggs of Cd9-deficient mice. Nat. Genet. 2000;24:279–282. doi: 10.1038/73502. [DOI] [PubMed] [Google Scholar]
- Kielian M., Rey F. A. Virus membrane-fusion proteins: more than one way to make a hairpin. Nat. Rev. Microbiol. 2006;4:67–76. doi: 10.1038/nrmicro1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine M. S., McKenzie A., 3rd, Demarini D. J., Shah N. G., Wach A., Brachat A., Philippsen P., Pringle J. R. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Lorenz M. C., Cutler N. S., Heitman J. Characterization of alcohol-induced filamentous growth in Saccharomyces cerevisiae. Mol. Biol. Cell. 2000;11:183–199. doi: 10.1091/mbc.11.1.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCaffrey G., Clay F. J., Kelsay K., Sprague G. F., Jr Identification and regulation of a gene required for cell fusion during mating of the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 1987;7:2680–2690. doi: 10.1128/mcb.7.8.2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohler W. A., Shemer G., del Campo J. J., Valansi C., Opoku-Serebuoh E., Scranton V., Assaf N., White J. G., Podbilewicz B. The type I membrane protein EFF-1 is essential for developmental cell fusion. Dev. Cell. 2002;2:355–362. doi: 10.1016/s1534-5807(02)00129-6. [DOI] [PubMed] [Google Scholar]
- Muller E. M., Mackin N. A., Erdman S. E., Cunningham K. W. Fig1p facilitates Ca2+ influx and cell fusion during mating of Saccharomyces cerevisiae. J. Biol. Chem. 2003a;278:38461–38469. doi: 10.1074/jbc.M304089200. [DOI] [PubMed] [Google Scholar]
- Muller M., Katsov K., Schick M. A new mechanism of model membrane fusion determined from Monte Carlo simulation. Biophys. J. 2003b;85:1611–1623. doi: 10.1016/S0006-3495(03)74592-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolan S., Cowan A. E., Koppel D. E., Jin H., Grote E. FUS1 regulates the opening and expansion of fusion pores between mating yeast. Mol. Biol. Cell. 2006;17:2439–2450. doi: 10.1091/mbc.E05-11-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philips J., Herskowitz I. Osmotic balance regulates cell fusion during mating in Saccharomyces cerevisiae. J. Cell Biol. 1997;138:961–974. doi: 10.1083/jcb.138.5.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podbilewicz B., Leikina E., Sapir A., Valansi C., Suissa M., Shemer G., Chernomordik L. V. The C. elegans developmental fusogen EFF-1 mediates homotypic fusion in heterologous cells and in vivo. Dev. Cell. 2006;11:471–481. doi: 10.1016/j.devcel.2006.09.004. [DOI] [PubMed] [Google Scholar]
- Reddy A., Caler E. V., Andrews N. W. Plasma membrane repair is mediated by Ca(2+)-regulated exocytosis of lysosomes. Cell. 2001;106:157–169. doi: 10.1016/s0092-8674(01)00421-4. [DOI] [PubMed] [Google Scholar]
- Schulz T. A., Creutz C. E. The tricalbin C2 domains: lipid-binding properties of a novel, synaptotagmin-like yeast protein family. Biochemistry. 2004;43:3987–3995. doi: 10.1021/bi036082w. [DOI] [PubMed] [Google Scholar]
- Sollner T. H. Intracellular and viral membrane fusion: a uniting mechanism. Curr. Opin. Cell Biol. 2004;16:429–435. doi: 10.1016/j.ceb.2004.06.015. [DOI] [PubMed] [Google Scholar]
- Steinhardt R. A., Bi G., Alderton J. M. Cell membrane resealing by a vesicular mechanism similar to neurotransmitter release. Science. 1994;263:390–393. doi: 10.1126/science.7904084. [DOI] [PubMed] [Google Scholar]
- Taylor M. V. Muscle differentiation: how two cells become one. Curr. Biol. 2002;12:R224–R228. doi: 10.1016/s0960-9822(02)00757-1. [DOI] [PubMed] [Google Scholar]
- Trueheart J., Boeke J. D., Fink G. R. Two genes required for cell fusion during yeast conjugation: evidence for a pheromone-induced surface protein. Mol. Cell. Biol. 1987;7:2316–2328. doi: 10.1128/mcb.7.7.2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Itallie C. M., Anderson J. M. The molecular physiology of tight junction pores. Physiology. 2004;19:331–338. doi: 10.1152/physiol.00027.2004. [DOI] [PubMed] [Google Scholar]
- Weber T., Zemelman B. V., McNew J. A., Westermann B., Gmachl M., Parlati F., Sollner T. H., Rothman J. E. SNAREpins: minimal machinery for membrane fusion. Cell. 1998;92:759–772. doi: 10.1016/s0092-8674(00)81404-x. [DOI] [PubMed] [Google Scholar]
- White J. M., Rose M. D. Yeast mating: getting close to membrane merger. Curr. Biol. 2001;11:R16–R20. doi: 10.1016/s0960-9822(00)00036-1. [DOI] [PubMed] [Google Scholar]
- Yawo H., Kuno M. Calcium dependence of membrane sealing at the cut end of the cockroach giant axon. J. Neurosci. 1985;5:1626–1632. doi: 10.1523/JNEUROSCI.05-06-01626.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N. N., Dudgeon D. D., Paliwal S., Levchenko A., Grote E., Cunningham K. W. Multiple signaling pathways regulate yeast cell death during the response to mating pheromones. Mol. Biol. Cell. 2006;17:3409–3422. doi: 10.1091/mbc.E06-03-0177. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.