Abstract
Nucleolin is one of the most abundant protein in the nucleolus and is a multifunctional protein involved in different steps of ribosome biogenesis. In contrast to animals and yeast, the genome of the model plant Arabidopsis thaliana encodes two nucleolin-like proteins, AtNUC-L1 and AtNUC-L2. However, only the AtNUC-L1 gene is ubiquitously expressed in normal growth conditions. Disruption of this AtNUC-L1 gene leads to severe plant growth and development defects. AtNUC-L1 is localized in the nucleolus, mainly in the dense fibrillar component. Absence of this protein in Atnuc-L1 plants induces nucleolar disorganization, nucleolus organizer region decondensation, and affects the accumulation levels of pre-rRNA precursors. Remarkably, in Atnuc-L1 plants the AtNUC-L2 gene is activated, suggesting that AtNUC-L2 might rescue, at least partially, the loss of AtNUC-L1. This work is the first description of a higher eukaryotic organism with a disrupted nucleolin-like gene and defines a new role for nucleolin in nucleolus structure and rDNA chromatin organization.
INTRODUCTION
Biogenesis of ribosomes takes place in the most prominent subnuclear structure: the nucleolus. Here, the tandem rRNA genes encoding the 18S, 5.8S, and 25S structural rRNAs are transcribed by RNA polymerase I as a precursor or pre-rRNA, which is then processed into mature forms that assemble with ribosomal proteins to form ribosome particles (for review, see Lam et al., 2005). The driving force for the assembly of the nucleolus is transcription of the tandem rRNA gene repeats, as has been shown conclusively by the assembly of nucleoli at active rRNA genes integrated at ectopic locations (Oakes et al., 1998, and references therein). The synthesis of rRNA is completely inhibited during mitosis, when nucleoli disassemble and the rDNA is arranged in a partially condensed form at the nucleolus organizer regions (NOR). When rRNA synthesis resumes at the end of mitosis, nucleoli reassemble (Hernandez-Verdun, 2005).
In interphase cells, the nucleolus is formed by three basic components: the fibrillar center (FC), the dense fibrillar component (DFC), and the granular component (GC). Occasionally, depending on cell type and stage, other structures such as vacuoles can also be observed in plant nucleolus (Gonzalez-Melendi et al., 2001). It has been accepted for some time that ribosome biosynthesis occurs in a vectorial pattern within the nucleolus. The transcription of pre-rRNA probably occurs at the border of the FC and DFC; the early processing steps of pre-rRNA occur in the DFC; and the later processing and RNA modification steps, together with the formation of preribosomal particles is observed in the GC (Thiry and Lafontaine, 2005; Raska et al., 2006).
Several nonribosomal nucleolar proteins as well as many small nucleolar ribonucleolar protein complexes (snoRNPs) have been implicated in the different steps of rRNA production and ribosome assembly (Fromont-Racine et al., 2003). Nucleolin is one of the most abundant nonribosomal proteins in the nucleolus, where it plays a key role in the different steps involved in ribosome biogenesis, including RNA polymerase (Pol) I transcription and processing of pre-rRNA (Ginisty et al., 1998; Roger et al., 2003), assembly and nucleocytoplasmic transport of ribosome particles (Bouvet et al., 1998). Nucleolin and nucleolin-like proteins have been described in a large variety of organisms, including Chinese hamster, mouse, rat, human, chicken, frog, yeast, and plants (for review, see Ginisty et al., 1999). In these protein sequences, three structural and multifunctional domains have been identified: the acidic N-terminal region, the central region that contains two or four RNA RRM domains, and the C-terminal region or GAR domain (Ginisty et al., 1999).
Mutations of genes encoding nucleolin homologue in budding (NSR1) and fission (GAR2) yeast produce severe effects on cell growth and disrupt balanced production of the small and large ribosomal subunits (Lee et al., 1992; Gulli et al., 1995). Moreover, it has been shown that the nucleolar structure in gar2 cells is dramatically reorganized and unusual structures are detected (Leger-Silvestre et al., 1997). However, mutations that abolish nucleolin gene expression have not yet been reported in a higher organism. Thus, most of our knowledge on the function of this protein in higher eukaryotic cells comes from in vitro studies using acellular systems, microinjection experiments, or overexpression of a nucleolin transgene in Xenopus oocytes (Yanagida et al., 2001; Roger et al., 2002, 2003). Only two reports have described the effect of reduction of nucleolin in higher organisms. These reports revealed that reduction of the nucleolin by RNA interference (RNAi) in human cells increases expression of p53 protein (Takagi et al., 2005) and inhibits RNA polymerase I transcription (Angelov et al., 2006).
To obtain information about the role of nucleolin in vivo in higher eukaryotes, we have characterized an Arabidopsis thaliana mutant plant with a disrupted nucleolin-like gene. In contrast to animals and yeast, the A. thaliana genome encodes two nucleolin-like proteins, AtNUC-L1 and AtNUC- L2, but only the former is ubiquitously expressed and seems to play the role of its animal counterpart. We show that disruption of the AtNUC-L1 gene affects plant growth and development and demonstrate that AtNUC-L1 gene expression is required to preserve the ultrastructure of the nucleolus and NOR condensation. Remarkably, in Atnuc-L1 plants, the AtNUC-L2 gene is expressed and rescues, at least partially, AtNUC-L1 disruption. The role and biological significance of these two genes are discussed.
MATERIALS AND METHODS
Plant Growth Conditions and Mutant Isolation
All lines were derived from A. thaliana Columbia (Col 0) ecotype. Seeds were first sowed on Murashige and Skoog medium (MS) or on soil and left 2 d at 4°C to synchronize. Plants were then grown either under continuous light (MS plants) or under a 16:8-h light/dark cycle (soil plants). Seeds corresponding to Atnuc-L1 (SALK_053590 and SALK_502764) plants lines were obtained from the Nottingham Arabidopsis Stock Center (http://nasc.life.nott.ac.uk). Seeds of T-DNA insertion line SALK_053590 that have been backcrossed to Col 0 were used for further analysis.
AtNUC-L1 and AtNUC-L2 Cloning
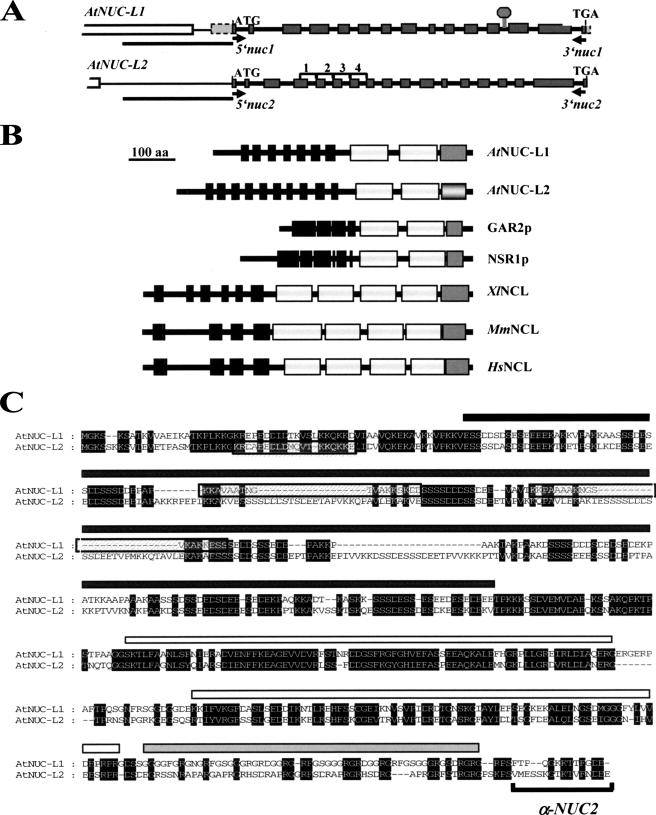
Cloning of cDNA sequences encoding AtNUC-L1 (At1g48920) and AtNUC-L2 (At3g18610) was performed by reverse transcription-polymerase chain reaction (RT-PCR) with total RNA isolated from 15-d-old A. thaliana wild-type (WT) and Atnuc-L1 plants, respectively. Primers 5′nuc1 and 3′nuc1 to amplify AtNUC-L1 and 5′nuc2 and 3′nuc2 to amplify AtNUC-L2 are located at the beginning of the ATG start and TGA stop codons (Figure 1A).
Figure 1.
The A. thaliana genome encodes two nucleolin-like proteins. (A) Diagram of AtNUC-L1 and L2 genes from the ATG start to the TGA stop codons. Gray boxes correspond to exons separated by fourteen introns. The T-DNA insertion in the Atnuc-L1 plants is indicated by a gray diamond. Position of primers 5′nuc1 and 3′nuc1 used to detect AtNUC-L1 transcripts are indicated by black arrows. White boxes shows At1g48930 and At3g18600 genes, localized upstream of AtNUC-L1 and AtNUC-L2, respectively. Black bars under each gene show the 1.1- and 1.0-kbp DNA sequence fused to GUS reporter genes construct. Brackets 1–4 show tandemly repeated sequences in the AtNUC-L2 gene. (B) Schematic representation of nucleolin and nucleolin-like proteins from A. thaliana (AtNUC-L1 and L2), Schizosaccharomyces pombe (GAR2p), Saccharomyces cerevisiae (NSR1p), X. laevis (XlNCL), Mus musculus (MmNCL), and Homo sapiens (HsNCL). The black boxes correspond to the acidic regions in the N-terminal domain, the white boxes represent the RRM domains, and the dark gray boxes represent the GAR domain. The light gray box in the AtNUC-L2 sequence indicates the less conserved GAR domain. (C) Amino acid sequence alignment of AtNUC-L1 and AtNUC-L2 proteins. Conserved amino acids are black shaded. The black rectangle shows the acidic N-terminal domain, the white rectangles show the two RRM domains, and the gray rectangle the putative GAR domain. The putative nuclear localization signals of AtNUC-L1 and AtNUC-L2 are boxed. Specific antibodies α-NUC2, were prepared against the peptide sequence located in the C-terminal domain.
Methods Related to RNA
Total RNA was extracted from 15-d-old A. thaliana plants by using TRIzol reagent (GE Healthcare, Little Chalfont, Buckinghamshire, United Kingdom). After treatment with RQ1 RNase-free DNase (Promega, Madison, WI) to eliminate contaminant DNA, first-strand cDNA synthesis was performed on 5 μg of total RNA by using the ProSTAR first-strand RT-PCR kit following the manufacturer's instructions (Stratagene, La Jolla, CA). To ensure that the amount of the amplified products remains in linear proportion, a semiquantitative RT-PCR reaction of 25 cycles was performed using a PTC-200 (MJ Research, Watertown, MA).
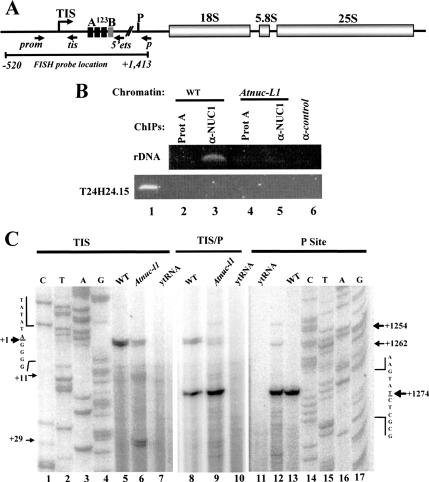
Primer extension analysis was performed using total RNA and specific 5′-end labeled primers as previously described (Saez-Vasquez et al., 2004). Primers used were tis for detection of primary pre-rRNA precursor (+104 nucleotides from transcription initiation site [TIS]) and p (+1362 nucleotides from TIS) for detection of pre-rRNA cleaved at the P site (Figure 9A). Dideoxy sequencing reactions were performed using the fmol DNA Cycle Sequencing System (Promega) with a pGem-3Z plasmid vector containing the A. thaliana rDNA sequences from −520 to +1940 (Gruendler et al., 1989).
Figure 9.
AtNUC-L1 binds rDNA and affects rate of processing of pre-rRNA. (A) Diagram of an A. thaliana rDNA unit containing the 18S, 5.8S, and 25S rRNA genes. TIS at +1 and primary cleavage site (P) at +1275 are indicated. The A123B box (from +131 to +183), indicates the rDNA binding site of the U3snoRNP complex containing nucleolin-like protein in plants cruciferous. Blacks arrows show position of primers prom and 5′ets used for ChIPs experiments; and primers tis and p used to measure RNA Pol I transcription and pre-rRNA processing respectively. (B) Chromatin isolated from WT and Atnuc-L1 plants was incubated either with protein A only (lanes 2 and 4) or with α-NUC1-conjugated to protein A (lanes 3 and 5) or with unrelated antibodies (lane 6). Immunoprecipitated DNA was analyzed by PCR to detect rDNA (top) or T24H24.15 control (bottom) genes. Lane 1 corresponds to PCR amplification using chromatin isolated from WT plants to verify amplification of T24H24.15 gene. (C) Primer extension experiments were performed using total RNA extracted from WT and Atnuc-L1 plants and primer tis (lanes 5 and 6) or p (lanes 12 and 13) or tis and p together (lanes 8 and 9). Lanes 7 and 10 and 11 correspond to control reactions using yeast tRNA. Lanes 1–4 and 14–17 show DNA sequencing reactions used to accurately map transcription initiation site and pre-RNA processing at the P site, respectively.
-β Glucuronidase (GUS), -Green Fluorescent Protein (GFP), and Histology Studies
The 1.1-kilobase pair (kbp) sequence of the AtNUC-L1 gene promoter was amplified from genomic DNA by using primers p5′nuc1 and p3′nuc1 and fused to a GUS reported gene in the vector pCAMBIA1381 (CAMBIA, Canberra, Australia). A. thaliana plants were transformed with Agrobacterium tumefaciens GV3101 by using the “floral-dip” method (Clough and Bent, 1998). GUS activity staining was performed according to Sessions et al. 1999.
Gateway cassettes with AtNUC-L1 and -L2 coding sequences were generated by polymerase chain reaction (PCR) following instructions by Invitrogen (Carlsbad, CA). Resulting PCR product was introduced first into pDNR221 (Invitrogen) and then cloned by recombination into pK7FWG2 to produce AtNUC-L1(L2)::GFP plasmid. For transient expression, 5 μg of plasmid was coated to 1.6-μm gold particles (Bio-Rad, Hercules, CA) according to the Bio-Rad transformation. The onion epidermal layers were transfected using the PDS-1000/He biolistic transformation system (Bio-Rad), and fluorescence was observed 24 h later.
For cytological analysis, plant tissues were fixed overnight at 4°C with 2.5% formaldehyde (37% stock solution), 2.5% glutaraldehyde (25% stock solution), 0.1 M sodium phosphate, pH 7.2, rinsed twice in the same buffer, and then dehydrated through a graded ethanol series before being embedded in Technovit 7100 resin (Kulzer, Wehrheim, Germany). Sections of 4 μm were obtained with a RM 2255 microtome (Leica, Heerbruss, Switzerland) and stained for 15 s with 1% toluidine blue solution containing 1% of sodium tetraborate.
All microscopic images—GUS, GFP, and toluidine blue staining—were observed using a Zeiss Axioskop 2 microscope and recorded using a Leica DC 300 FX digital camera (Leica).
Electron Microscopy, Immunofluorescence, and Immunogold Techniques
Immunofluorescence was performed using 4- and 8-d-old (WT and Atnuc-L1, respectively) A. thaliana seedlings following the procedures for immunocytological and structural analysis described previously (Gonzalez-Camacho and Medina, 2005). Treated seedlings were incubated with α-NUC1 diluted 1:1000 or with α-NUC2 diluted 1:250 for 16 h at 37°C and then with fluorochrome A-488–conjugated goat anti-rabbit IgG (Sigma-Aldrich, St. Louis, MO) diluted 1:100, for 3 h at 37°C. Finally, the samples were mounted in anti-fading mounting medium containing 4,6-diamidino-2-phenylindole (DAPI) solution. Preparations were visualized with a Leica TCS SP2 confocal laser scanning microscope and recorded with a Photometrics digital camera (CE 200A; Leica).
For electron microscopy experiments, 4- and 8-d-old A. thaliana roots were fixed with either 3% glutaraldehyde (ultrastructural studies) or with 4% paraformaldehyde and 0.5% glutaraldehyde (immunogold experiments). To obtain a higher resolution of the ultrastructure of nucleolus, fixed A. thaliana roots were additionally acetylated and methylated with a 5:1 methanol/acetic acid solution as described previously (Testillano et al., 1995). Then, the tissues were dehydrated and embedded in LR White resin (London Resin, Bershire, United Kingdom). For immunogold, ultrathin sections were mounted on Formvar-coated nickel grids and incubated with α-NUC1 diluted 1:100 for 1 h 30 min, at room temperature (RT). After several washes to remove excess of primary antibodies, grids were incubated with goat-anti rabbit IgG coupled with 10-nm colloidal gold particles (Sigma-Aldrich) diluted 1:50 for 1 h, at RT. In all electron microscopical experiments grids were counterstained with uranyl acetate and lead citrate and examined with a Jeol 1200 transmission electron microscope (Jeol, Tokyo, Japan).
In both immunofluorescence and immunogold experiments, negative controls, consisting of the omission of the first antibody were performed.
Western Blotting
Plant material (0.2 g) was homogenized and extracted in 2 ml of 50 mM Tris-Cl, pH 8, 150 mM NaCl, 10 mM EDTA, 50 mM NaF, 1% NP-40, 0.5% sodium desoxycholate, 0.1% SDS, 1 mM phenylmethylsulfonyl fluoride, 10 mM β-mercaptoethanol, and 20 μl of anti-protease cocktail (Sigma-Aldrich). The extracts were cleared by centrifugation at 13,000 × g for 15 min and conserved at −80°C. SDS-PAGE, and Western blot was performed as described previously (Saez-Vasquez et al., 2004). The membranes were hybridized with a 1:10,000 dilution of α-NUC1 or with a 1:5000 dilution of α-NUC2. Rabbit polyclonal antibodies against C-terminal peptide sequence of AtNUC-L2 (Figure 1C) were customer made by NeoMPS (Strasbourg, France).
Chromatin Immunoprecipitation (ChIP) and Fluorescence In Situ Hybridization (FISH)
ChIP was performed as described in Gendrel et al. 2005. To amplify rDNA promoter sequences, we used primers 5′prom (−110 nucleotides from TIS) and 3′ets (+219 nucleotides from TIS) (Figure 9). The T24H24.15 gene, used as control, encodes a phosphofrutokinase β-subunit, and the primers used for gene amplification are described in Gendrel et al. 2005.
FISH experiments were performed according to Schubert et al. 2001. The probe containing intergenic spacer and 5′-external transcribed sapcer (ETS) rDNA sequences cloned in pGem-T was amplified by PCR by using universal primers T7/M13R and by supplying biotin-16-dUTP (Roche, Meylan, France) to the reaction. The biotin-labeled probe was detected using avidin-conjugated with Texas Red (1:500; Vector Laboratories, Burlingame, CA) followed by goat anti-avidin conjugated with biotin (1:100; Vector Laboratories) and avidin-Texas Red (1:500). Slides were mounted using Vectashield (Vector Laboratories) mounting medium with 1.5 μg/ml DAPI and then observed by fluorescence microscopy, by using an AxioPlan2 microscope (Carl Zeiss, Le Pecq, France). Images were acquired using the Axiocam MRm camera (Carl Zeiss) and the software Axiovision 4.2 (Carl Zeiss). Images were further processed and enhanced using Adobe Photoshop software (Adobe Systems, Mountain View, CA).
Sequencing Analysis and Primers
All clones used in this study were sequenced with a model 3100DNA sequencer and an ABI PRISM Big Dye Terminator Cycle Sequencing Ready Reaction kit (Applied Biosystems, Foster City, CA). Database searches were performed at the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov) and the Arabidopsis Information Resources (http://www.arabidopsis.org). Primers used in this work are described in Supplemental Table S1.
Primers
All primers used in this study were customer made by Eurogentec (Seraing, Belgium). Primers were as follows: 5′nuc1, 5′-ATGGGAAAGTCTAAATCCCGCACC-3′; 3′nuc1, 5′-CTACTCGTCACCGAAGGTAGTC-3′; 5′nuc2, 5′-ATGGGCAAGTCTAGTAAGAAATCC-3′; 3′nul2, 5′-CTACTCTTCATCATTAAAGACCGTC-3′; 5′act2, 5′-GTTAGCAACTGGGATGATA-3′; 3′act2, 5′-CAGCACCAATCGTGATGACTTGCCC-3′; tis, 5′-CCTCGTGCCGATATCCGATACCATCCC-3′; p, 5′-CATCAATCGTTCCAACTAATCTAC-3′; prom, 5′-CCAAGCTTGTATCCTTATGATGCATGC-3′; 5′ets, 5′-GAGAACTGCTGAGAAAACTCGG-3′; p5′nuc1, 5′-CATAACCAAGTCAATCCTTG-3′; and p3′nuc1, 5′-CCATGGGGAGAACTGAGAAAGAGACGAC-3.
RESULTS
The A. thaliana Genome Contains Two Nucleolin-like Protein Genes
We initiated the molecular and functional characterization of two genes encoding nucleolin-like proteins in A. thaliana, namely, AtNUC-L1 and AtNUC-L2. These two genes are located in duplicated regions of chromosome 1 and chromosome 3, respectively (http://wolfe.gen.tcd.ie/athal/dup). However, AtNUC-L1 and AtNUC-L2 genes show divergent genomic organization (Figure 1A). From the starting ATG to the TGA stop codon, the AtNUC-L1 gene is 3480 nucleotides and contains 15 exons, whereas the AtNUC-L2 gene is 3553 nucleotides long and contains 18 exons. Interestingly, AtNUC-L2 contains four tandemly repeated sequences, consisting of the second part of exon 4 and the following intron (Supplemental Figures 1A and S1A). These sequences are also found in the nucleolin-like sequence described in pea (Tong et al., 1997).
The predicted AtNUC-L1 and AtNUC-L2 proteins are 557 and 636 amino acids, respectively, and share 49% identity (Figure 1C). Both have a tripartite structure as described for nucleolin in animals and Gar2p/Nsr1p proteins in yeast (Figure 1B). However, in contrast to nucleolin in vertebrates, which have four RRM domains, the A. thaliana sequences contain two RRM domains as in NSR1p/GAR2p proteins.
AtNUC-L1 contains seven stretches of aspartic acid (D), glutamic acid (E), and serine (S) residues, compared with 10 in AtNUC-L2 (Figure 1, B and C). Coding sequences for at least two of these additional acidic stretches in AtNUC-L2 are located within exons in the first and fourth tandemly repeated sequences described above. Moreover, it is noteworthy that the potential AtNUC-L2-GAR domain is much less conserved than AtNUC-L1-GAR domain compared with the nucleolin -GAR domain from other species (Figure 1C). The putative GAR domain of AtNUC-L1 is 75% rich in glycine and arginine residues compared with a 47% in the AtNUC-L2 sequence. In animals, GAR domains are ∼88% rich in glycine and arginine, suggesting a functional conservation of the GAR domain only in AtNUC-L1. Furthermore, the N-terminal region of AtNUC-L1 contains two potential bipartite nuclear localization signals (NLSs), whereas there is only one NLS in AtNUC-L2 (Figure 1C).
Additional plant nucleolin-like proteins were identified by Blast searches by using as query the entire AtNUC-L1 and AtNUC-L2 protein sequences. Phylogenetic analysis showed that nucleolin-like sequences from A. thaliana, rice, onion, maize, tobacco, alfalfa, and pea form a plant-specific group (Supplemental Figure S2). This shows that plant genes diverged from a common ancestor very early during plant evolution and also reveals that the two genes in rice and A. thaliana result from separate and recent duplication events. This is also supported by results indicating that nucleolin homologues of alfalfa are encoded by a small multigene family (Bogre et al., 1996). Unfortunately only one sequence is available.
In conclusion, A. thaliana AtNUC-L1 and AtNUC-L2 genes encode similar but distinct nucleolin-like proteins, homologous to NSR1p and GAR2p proteins in fungi.
The AtNUC-L1 Gene Is Constitutively Expressed but Not AtNUC-L2
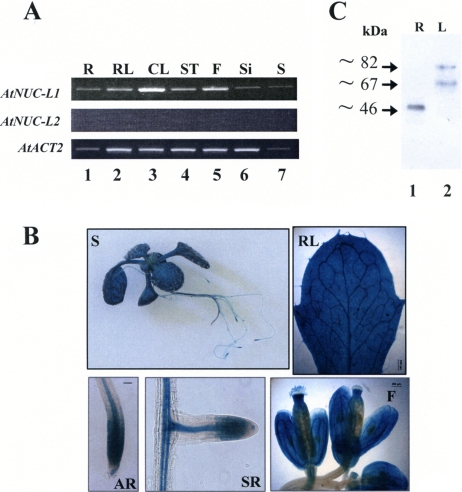
To test expression of AtNUC-L1 and AtNUC-L2, we performed a semiquantitative RT-PCR analysis. As shown in Figure 2A, primers 5′nuc1 and 3′nuc1 amplify a band of ∼1750 kbp in roots (R), rosette leaf (RL), cauline leaf (CL), stem (ST), flowers (F), siliques (Si), and seeds (S) (top, lanes 1–7). We confirmed by sequencing that this band corresponds to specific AtNUC-L1 transcripts and not to genomic sequences, which produce an ∼3.5 kbp band containing intron sequences (data not shown). In contrast, PCR amplification using primers 5′nuc2 and 3′nuc2 did not detect AtNUC-L2 cDNA transcripts in any of these samples (Figure 2A, middle, lanes 1–7). PCR reactions using specific primers to amplify ACTIN2 transcripts were used to verify amounts of cDNA in each reaction (Figure 2A, bottom, lanes 1–7).
Figure 2.
AtNUC-L1 gene and protein expression in A. thaliana plants. (A) RT-PCR analysis of AtNUC-L1 (top) and AtNUC-L2 (middle) gene expression in roots (R), rosettes leaves (RL), cauline leaves (CL), stem (ST), flowers (F), siliques (Si), and seeds (S). AtACT2 (bottom) gene expression was analyzed as a PCR control to evaluate the amount of cDNA used in each reaction. (B) Analysis of the AtNUC-L1 promoter activity in A. thaliana plants transformed with a AtNUC-L1:GUS construct. The GUS staining is visualized in 3-wk-old S, RL, apical root (AR), secondary root (SR), and F. (C) Western blot analyses of AtNUC-L1 protein expression in R (lane 1) and leaves using α-NUC-L1 antibodies.
To test AtNUC-L1 and AtNUC-L2 promoter activity, we transformed A. thaliana plants with a plasmid construct containing the GUS reporter gene fused to either the AtNUC-L1 or AtNUC-L2 promoter sequence (Figure 2B). We observed that in plants transformed with the AtNUC-L1::GUS construct, staining was detected in all tissues and organs tested, including seedlings grown in vitro (S), RL, apical roots (AR), secondary roots (SR) and F. In contrast, in plants transformed with the AtNUC-L2::GUS construct, we did not detect GUS staining in any part of the plant (data not shown).
Next, to determine the presence of the AtNUC-L1 protein, we performed a Western blot analysis using protein extracts prepared from different plant tissues. As shown in Figure 2C, antibodies against AtNUC-L1, α-NUC1 (Saez-Vasquez et al., 2004), cross-reacted with three distinct polypeptides, ∼82, ∼67, and ∼46 kDa, in A. thaliana protein extracts. Both the ∼82- and the ∼67-kDa polypeptides were detected in leaves (lane 2), whereas the ∼46-kDa polypeptide was detected only in roots (lane 1). These three polypeptides also were detected in seedling (Figure 4), flower, stem, and green siliques (data no shown). Nevertheless, none of the observed polypeptides correspond to the 59-kDa expected size of AtNUC-L1. We could show that the smaller 67- and 46-kDa bands derive from proteolysis of the 87-kDa protein (Supplemental Figure S4). This is similar to nucleolin, which in animals has aberrant migration on SDS-PAGE and is highly susceptible to proteolysis (Bouche et al., 1984).
Figure 4.
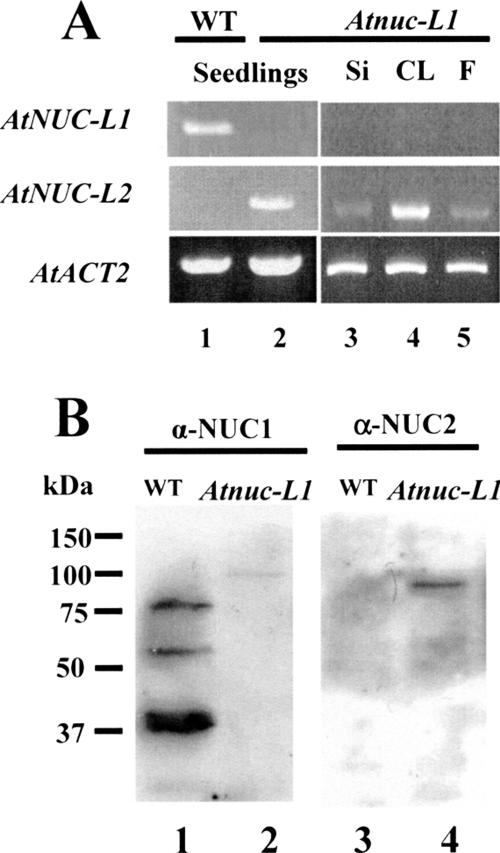
Disruption of AtNUC-L1 induces expression of AtNUC-L2. (A) RT-PCR reaction using cDNA prepared from RNA isolated from 15-d-old seedlings (lanes 1 and 2), siliques (Si; lane 3), cauline leaves (CL; lane 4), and flowers (F; lane 5) (B). Whole cell protein extract from WT and Atnuc-L1 plants were fractionated on SDS-PAGE and hybridized either with α-NUC1 (lanes 1 and 2) or with α-NUC2 antibodies (lanes 3 and 4).
In conclusion, these results demonstrate that under normal growth and development conditions, only the AtNUC- L1 gene is expressed and the AtNUC-L2 gene promoter is not active.
AtNUC-L1 Localizes Preferentially in the DFC of the Nucleolus
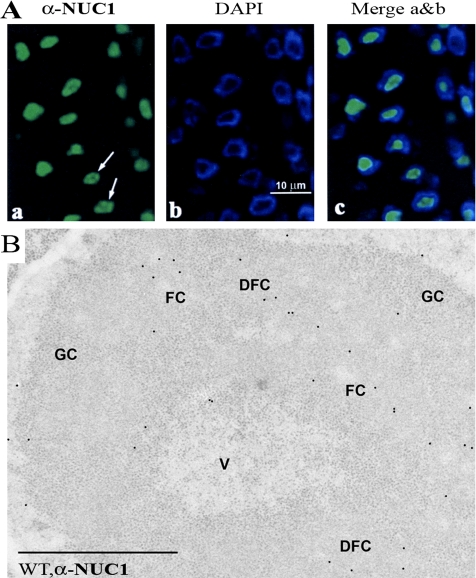
To test whether AtNUC-L1 localizes in the nucleolus of A. thaliana plants, we used an immunofluorescence and immunogold labeling approach. After incubation of A. thaliana root meristematic cells with α-NUC1, the immunofluorescence labeling seemed specifically concentrated in the nucleolus (Figure 3, left, and Supplemental Figure S5). DAPI staining was visualized around the nucleolus because the concentration of intranucleolar chromatin, which is mostly in a decondensed state, makes it undetectable with the light microscope (Figure 3, middle). The merged confocal image clearly shows the nucleolar specificity of the AtNUC-L1 localization (Figure 3, right). However, AtNUC-L1 did not seem to be evenly distributed in the nucleolus. This feature was more evident in those nucleoli showing a lower labeling intensity (Figure 3A, arrows). We did not see nucleoplasmic or cytoplasmic labeling.
Figure 3.
Nucleolar localization of AtNUC-L1. (A) Immunofluorescence localization of AtNUC-L1 in A. thaliana root meristematic cells fixed with paraformaldehyde/dimethyl sulfoxide and incubated with α-NUC1 antibodies, observed with the confocal microscope. The images correspond to a single optical section, obtained with a Z-step of 0.3 μm. Labeling is shown concentrated in the nucleolus (a), but it does not seem evenly distributed through the whole nucleolar area (arrows). Nucleoplasm labeled by DAPI is observed as a ring around the dark unstained nucleolus (b), and the merged image shows that the immunofluorecent labeling localizes precisely in the nucleolus, unstained by DAPI (panel c). (B) Immunogold electron microscopic localization of AtNUC-L1 in A. thaliana root meristematic cells. AtNUC-L1 is observed to localize in the DFC, relatively near FCs. The interior of FCs seems devoid of gold particles, whereas the GC and the nucleolar vacuole (V) show a very scarce labeling, the same as nucleoplasm. Bar, 1 μm.
To establish precisely the intranucleolar distribution of AtNUC-L1 protein, we used immunogold electron microscopical detection (Figure 3B). We observed that in the nucleolus, the AtNUC-L1 labeling was preferentially concentrated in the DFC surrounding fibrillar centers (FCs). No gold particles were observed within FCs, and very little labeling was detected in the granular component (GC) and in the nucleolar vacuole (V). Some AtNUC-L1 labeling was also detected in the nonnucleolar region of the nucleus. However, gold particles were detected neither in the cytoplasm nor in the organelles, demonstrating that AtNUC-L1 localizes exclusively in the nucleus and nucleolus of A. thaliana cells.
Disruption of AtNUC-L1 by a T-DNA Insertion Induces AtNUC-L2 Expression
To investigate the functional role of AtNUC-L1 in vivo, an Atnuc-L1 homozygous line was isolated. In these mutant line (SALK_053590), expression of the AtNUC-L1 gene was disrupted by a T-DNA insertion shown in Figure 1A.
RT-PCR analysis using primers 5′nuc1 and 3′nucl does not detect AtNUC-L1 transcripts in Atnuc-L1 seedlings or in the different plant tissues tested (Figure 4A, top, lanes 2–5). Remarkably, RT-PCR reactions using primers 5′nuc2 and 3′nuc2 amplified a band of ∼1.9 kbp in Atnuc-L1 but not in the WT plants (Figure 4A, middle, lanes 1 and 2). Cloning and sequencing confirmed that this band is the product of amplification of specific AtNUC-L2 transcripts and not from genomic sequence, which produces an ∼3.6-kbp band containing intron sequences (data not shown). The AtNUC-L2 transcripts are detectable in all tissues and plant organs tested, including siliques, leaves, and flowers (lanes 3–5), revealing constitutive expression of AtNUC-L2 gene in this plant. RT-PCR reactions using specific primers to ACTIN2 transcripts were used to verify amounts of cDNA in each reaction (Figure 4A, bottom, lanes 1–5). Analysis of another mutant line (SALK_502764) displayed also AtNUC-L2 transcription induction (data not shown).
To determine the presence of the AtNUC-L2 protein, we performed Western blot analysis using antibodies against AtNUC-L1 (Figure 4B). This antibody was raised against the central domain of AtNUC-L1, which is 61% identical to AtNUC-L2 domain, and we expected a cross-reaction. Indeed, α-NUC1 detected an ∼100-kDa polypeptide in Atnuc- L1 protein extracts (lane 2) that it is not present in the WT protein extract (lane 1). This result was confirmed using a specific antibody raised against C-terminal peptide of AtNUC-L2. This antibody is specific to AtNUC-L2 and does not recognize AtNUC-L1 (Figure 4B, lane 3). These antibodies also detected the ∼100-kDa polypeptide only in the Atnuc-L1 plants (Figure 4B, lane 4) but not in the WT plants (lane 3), confirming that this polypeptide corresponds to AtNUC-L2 protein. The expected molecular mass of AtNUC-L2 is 69 kDa, which does not match the size of the detected ∼100-kDa polypeptide. As we mentioned, this can be explained by the atypical migration of nucleolin and nucleolin-like proteins in SDS-PAGE. It is also interesting to observe that α-NUC2 did not detect smaller polypeptides as did α-NUC1 in WT plants (Figure 4B, lane 1).
AtNUC-L2 Is a Nucleolar Protein and Localizes in the Nucleolus of Atnuc-L1 Plants
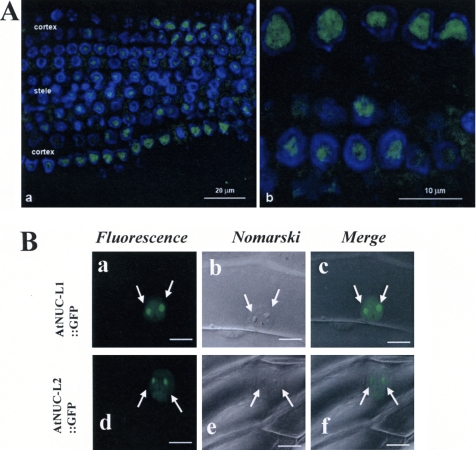
To obtain some insight on the role of AtNUC-L2 in the Atnuc-L1 plants, we established its cellular localization in Atnuc-L1 plants by immunofluorescence. Using α-NUC2 antibodies, we observed a nucleolar signal in root meristematic cells of AtNUC-L1 plants (Figure 5A). However, a larger view of the root meristem revealed different signal intensities in the cellular layer with a more intense signal in the peripheral cortex and a weaker signal in the central stele (Figure 5A, a). These two lateral regions of the meristem are characterized by a differential proliferative activity, which is higher in the cortex (Scheres, 2001).
Figure 5.
AtNUC-L2 is a nucleolar protein. (A) Immunofluorescent localization of AtNUC-L2 in Atnuc-L1 plants. The images represent single optical sections, obtained using a Z-step of 0.3 μm. In a, a panoramic overview of the root is displayed, showing the differential distribution of the labeling in the different parts of the root, the cortex showing a more intense labeling, and the stele a fainter immunostaining. In b, a higher magnification is shown, with nucleoli being the major target of the antibody. (B) Nucleolar localization of AtNUC-L1::GFP and AtNUC-L2::GFP fusion protein in transfected onion epidermis cells. Arrows point the two nucleolus visualized by the GFP fluorescence (a and d). Nucleolus can be easily observed by Nomarski (b and e), and they colocalize with the GFP in the merge images (c and f).
To confirm the nucleolar localization of AtNUC-L2 in WT cells, we transformed epidermal onion cells by bombardment using an AtNUC-L2::GFP plasmid. As shown in Figure 5B, in transiently transformed onion cells the AtNUC- L2::GFP fusion protein was found in the nucleus, in a diffuse pattern through the nucleoplasm, but it was concentrated in the nucleolus, visualized by two intense GFP fluorescent signals (Figure 5B, d). The nucleus and nucleolus of onion cells are easily visualized by Nomarski (Figure 5B, e), and the GFP fluorescence colocalizes with these structures (Figure 5B, f). Similar results were observed in onion cells transformed with an AtNUC-L1::GFP plasmid used as a control (Figure 5B, a–c).
AtNUC-L1 Disruption Affects Plant Growth and Development
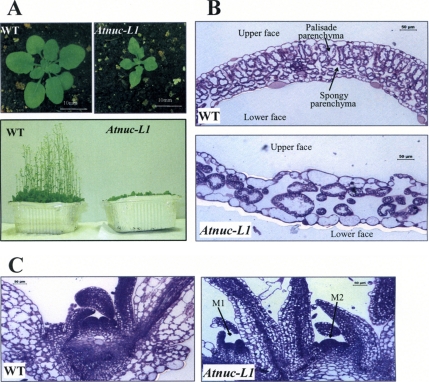
Atnuc-L1 plants grew slower than WT plants and the newly emerged leaves were smaller, pointed, irregular shaped, and scrunched (Figure 6). However, when these plants were grown further, the pointed shape of later leaves was less severe, but they remained smaller than WT leaves (Figure 6A). We observed also major phenotypic changes in flower, sepals, anthers and siliques (data not shown). It is also interesting to mention that Atnuc-L1 plants produced a reduced number of seeds. Analysis of the second mutant line (SALK_502764) displayed similar growing and developmental phenotypes (data not shown).
Figure 6.
AtNUC-L1 gene disruption affects growth and plant development. (A) A. thaliana WT and Atnuc-L1 plants grown on soil 4 (top) or 6 (bottom) wk under a 16:8 (L:D)-h cycle. (B and C) Transverse section of first leaves (B) and apical meristem (C) from WT and Atnuc-L1 plants stained with toluidine blue. Arrows M1 and M2 show two meristems in Atnuc-L1 plants. Bars, 50 μm.
To further analyze the Atnuc-L1 phenotype, we performed cytological analysis of primary leaves and young seedlings grown on MS media (Figure 6, B and C). Transversal sections of the first leaf revealed that whereas control leaves have the typical leaf structure of WT dicotyledonous plants, the Atnuc-L1 leaves exhibited significant reduction in the number of cells. We further observed changes in size and form of cells that create considerable disorganization in every cell layer (Figure 6B). Moreover, it was amazing to observe that at the same developmental stage as WT plants (4 d old), most of the Atnuc-L1 plants (7 d old) displayed two apical meristems (Figure 6C), but only one meristem was fully developed during plant growth.
AtNUC-L1 Gene Disruption Causes Nucleolus Disorganization of Atnuc-L1 Plants
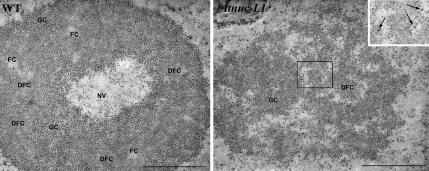
Given the nucleolar localization of AtNUC-L1, we examined the nucleolar structure of the Atnuc-L1 cells. As shown in Figure 7, a distinct morphological change was observed in the mutant Atnuc-L1 compared with the wild-type cells. In the Atnuc-L1 plants, the nucleolus was completely disorganized. The nucleolus was often seen as an open “reticulate” nucleolar structure with many interstices that seem to communicate with the nucleoplasm. Subnucleolar structures show profound changes, such as the disappearance of FCs, strong reduction of the DFC as a distinct component, and the alteration of the GC which, in the mutant nucleolus, seems formed by loosely packed granules embedded in a matrix (compare WT and Atnuc-L1 images in Figure 7). Furthermore, a second type of granules, other than those of the GC, can be identified in the mutant nucleolus. They show a larger size and occur either as single entities or loosely packed in clusters of a few of them. They are often found at the nucleolar interstices, peripherally located with respect to the masses of granular (or fibrillo-granular) component. We have identified these granules as the so-called “nucleolar perichromatin-like granules” (NPGs), which were previously described in the nucleolus of cells treated with some inhibitors of RNA metabolism (Puvion-Dutilleul et al., 1983; Risueno and Medina, 1986). Occasionally, nucleoli of Atnuc- L1 cells lost their spherical shape and become more elongated (data not shown). Moreover, the characteristic nucleolar vacuole observed in G2 and sometimes in G1 of WT cells disappeared in the Atnuc-L1 cells.
Figure 7.
AtNUC-L1 gene disruption affects ultrastructure of the nucleolus. The WT nucleolus is formed by some masses of DFC, which contain FCs in their interior and that are surrounded by GC. These components seem organized forming a cortex surrounding a central nucleolar “vacuole” (NV) in which granules similar to those of the GC can be identified. In Atnuc-L1 plants, this nucleolar organization is lost. Small masses of DFC can hardly be identified, and the bulk of the nucleolus is composed by a component reminiscent of the GC, but with the granules more loosely packed and embedded in a matrix. Numerous interstices occur, in some cases connected with the nucleoplasm. In the border between the GC-like component and the interstices, some granules can be observed. They are larger than the usual nucleolar GC granules and seem either isolated or forming small loose clusters (see in the inset a magnification of the area contained in the square). They have been identified as NPGs (arrows in the inset). Bar, 1 μm.
In conclusion, these observations clearly show that AtNUC-L1 is required for nucleolus structure in A. thaliana.
AtNUC-L1 Gene Disruption Induces rDNA Heterochromatin Decondensation
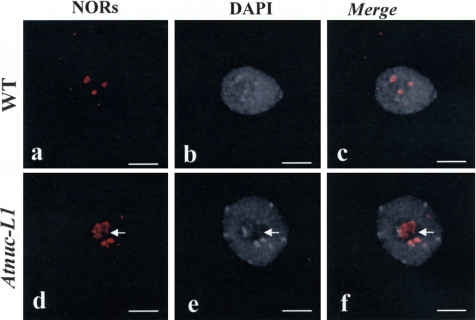
To determine the effect of the absence of AtNUC-L1 on the state of rDNA chromatin structure, WT and Atnuc-L1–derived NORs were examined in interphase nuclei by using a FISH probe containing intergenic spacer and 5′-ETS rDNA sequences (Figure 9). In A. thaliana cells, four rDNA loci located at the tips of chromosomes 2 and 4 are found in diploid nucleus. As shown in Figure 8, we detected two to four NOR fluorescence signals in WT interphase nuclei, in agreement with Pontes et al. 2003, due to association of homologous NORs. FISH analysis of the WT nuclei revealed a high condensation of NOR sequences and shows a tight colocalization with heterochromatin structure counterstained with DAPI (Figure 8, a–c). In the Atnuc-L1 background (Figure 8, d–f), we observed an increase in the number of NOR signals (52% of the nuclei analyzed displayed more than four NOR signals; see Supplemental Table S1), rDNA sequences seem to be decondensed in all nuclei when compared with WT. Moreover, in the Atnuc-L1 context, we observed that some FISH signals no longer colocalize with heterochromatin structure (arrows, panel d, e and f). This indicates that condensed NORs in WT plants acquire some euchromatic features in the Atnuc-L1 context. Finally, we could not see a significant difference in the number and size of centromeres between the DAPI labeled nuclei in WT and Atnuc-L1 plants, indicating that decondensation observed in Atnuc-L1 plants has occurred in the NORs.
Figure 8.
NOR condensation is affected in Atnuc-L1 plants. FISH in nuclei of WT (a–c) and Atnuc-L1 (d–f) seedlings. Left, FISH using rRNA gene probe shown in Figure 9A (a and d); middle, chromatin counterstained with DAPI (b and e); and right, superposition of a and b (c) and d and e (f) images. Arrows shows FISH signals that do not colocalize with heterochromatin. Bar, 5 μm.
In conclusion, these results show that AtNUC-L1 helps to maintain rDNA chromatin structure.
AtNUC-L1 Gene Disruption Affects rRNA Synthesis
We have previously reported that in cruciferous plants, a nucleolin-like protein is a component of a large U3snoRNP complex that binds 5′-ETS rDNA sequence (Caparros-Ruiz et al., 1997) and accurately cleaves pre-rRNA at the primary processing site (P site) in vitro (Saez-Vasquez et al., 2004).
To demonstrate that AtNUC-L1 binds rDNA in vivo, chromatin from WT and Atnuc-L1 plants was extracted and immunoprecipitated with antibodies against AtNUC-L1. A PCR reaction using primers prom and 5′ets (shown in Figure 9A) detected rDNA only in ChIP fractions obtained from chromatin extracted from WT plants (Figure 9B, lane 3). We did not detect rDNA amplification in ChIP fractions obtained from Atnuc-L1 plants chromatin (Figure 9B, lane 5), indicating that α-NUC1 specifically immunoprecipitated AtNUC-L1–bound rDNA chromatin. No or faint bands were detected in the PCR reactions by using either chromatin incubated with protein A alone (Figure 9B, lanes 2 and 4) or with unrelated antibodies (Figure 9B, lane 6). We did not detect any band when the PCR reaction was performed using the same ChIP fraction and primers specific to the T24H24.15 gene used to demonstrate specific interaction of AtNUC-L1 with the rDNA (Figure 9B, lanes 2–6). Thus, this result shows that AtNUC-L1 specifically binds rDNA chromatin in WT plants and suggests that this interaction may be required for rDNA chromatin organization.
To investigate whether synthesis of pre-rRNA was affected in the Atnuc-L1 plants, we measure the accumulation of primary pre-rRNA precursor produced by RNA Pol I and processed pre-rRNA at the primary cleavage site (P site). Primer tis maps the transcription initiation site accurately, TIS (Figure 9C, lanes 5–7; Saez-Vasquez and Pikaard, 1997) and allows quantification of pre-rRNA precursors, whereas primer p accurately maps the primary cleavage site, P (Figure 9C, lanes 11–13; Saez-Vasquez et al., 2004) and allows quantification of processed pre-rRNA precursors. To compare the ratio between primary pre-rRNA and cleaved pre-rRNA (TIS/P) in WT and Atnuc-L1 plants, tis and p primers were added simultaneously to the same primer extension reactions (Figure 9C, lanes 8–10). In these conditions, we observed that the TIS/P ratio was 1:4 in WT plants (Figure 9C, lane 8) and 1 :24 in Atnuc-L1 plants (Figure 9C, lane 9), signifying an increase of approximately sixfold of pre-rRNA cleaved at the P site in the Atnuc-L1 plants. Moreover, in the Atnuc-L1 plants, primer extension experiments using primer tis also gave a detectable signal at +11 and +29 nucleotides downstream from the normal start site (+1) (Figure 9C, lane 6). Note that even if we detect a stronger TIS signal in the WT than in Atnuc-L1 plants (Figure 9C, compare lanes 5 and 6), the +11 and +29 signals are not detectable at all in the control plants. Similarly, the p primer also produced at least two additional distinct signals in Atnuc-L1 plants, upstream from the P site: at +1254 and at +1262 (Figure 9C, lane 12). Interestingly, the amount of 18S, 5.8S, and 25S rRNA final products does not seem to be affected in the Atnuc-L1 plants (data not shown).
Together, these observations demonstrate that AtNUC-L1 disruption affects processing (and maybe also transcription) of pre-rRNA not only at the P site but also at other putative and/or abnormal processing sites.
DISCUSSION
The results described here provide new data to understand the in vivo role of nucleolin-like proteins not only in plants but also in higher eukaryotes. We report that in contrast to mammals and yeast, the A. thaliana genome encodes two nucleolin like proteins, AtNUC-L1 and AtNUC-L2, which are probably derived from a recent genome duplication event (Supplemental Figures 1 and S2). The data presented here also reveal that under normal growth conditions, only the AtNUC-L1 gene is expressed, suggesting that AtNUC-L1 protein is the functional homologue of nucleolin and NSR1/GAR2 proteins (Figures 2 and 3).
Additionally, we show that disruption of the AtNUC-L1 gene provokes changes in growth and plant development (Figure 6). Interestingly, the retarded growth, pointed leaf, and cellular disorganization phenotype observed in the Atnuc-L1 plants is reminiscent of phenotypes reported in two mutated ribosomal protein gene loci, RPS13 (Ito et al., 2000) and RPS18 (Van Lijsebettens et al., 1994), suggesting a functional relationship between AtNUC-L1 and some early steps of ribosome assembly. Indeed, we have identified RPS13 and RPS18 associated with a large U3snoRNP complex containing nucleolin-like protein (our unpublished data), which is involved in early processing of pre-rRNA in B. oleracea (Saez-Vasquez et al., 2004). Moreover, it has been also shown that these two RP proteins bind 18S rRNA and are 40S early ribosome assembly proteins in yeast (Xiang and Lee, 1989). Nevertheless, despite all these observations, it is likely that the phenotype observed in Atnuc-L1 can also be due to an effect on one or several of the multifunctional roles described for nucleolin in eukaryotes (Ginisty et al., 1999; Srivastava and Pollard, 1999).
AtNUC-L1 is localized in the nucleolus in the DFC surrounding FC at the site of rRNA transcription (which occurs in the border zone between FC and DFC; Gonzalez-Melendi et al., 2001, and references therein); we also demonstrate that AtNUC-L1 disruption has a major effect on nucleolus structure. We have observed that the nucleolus is completely disorganized in the Atnuc-L1 plant, reminiscent of the ultrastructural changes observed in the yeast nucleolus of gar2 (Leger-Silvestre et al., 1997). Among other changes, the typical “large vacuole”, well known in the nucleolus of plants (Shaw and Jordan, 1995), disappears in the Atnuc-L1 plants. Interestingly, AtNUC-L1 does not localize in the nucleolar vacuole (Figure 3), and the absence of this structure could be due to early ribosome assembly steps requiring AtNUC-L1 as mentioned above.
Decondensation of NOR in Atnuc-L1 plants is remarkable. Chromatin is generally considered as a prerequisite for transcriptional activation, in such a way that the amount of decondensed chromatin is taken as a marker of the gene expression activity, either real or potential. The decondensation of rDNA has no discernible effect on rRNA mature transcripts level, as observed in the HDA6 mutant (Earley et al., 2006). One explanation would be that increased rRNA gene transcription is undetectable. Alternatively, the number of active rRNA genes may change in Atnuc-L1 mutant without changing the overall level of transcription. In S. cerevisiae, the number of active rRNA genes can change more than twofold without changing steady-state rRNA transcript levels owing to compensatory changes in the average number of RNA polymerases engaged in transcription of each gene (French et al., 2003).
The increased NOR chromatin decondensation in Atnuc- L1 plant nucleoli could be also considered contradictory with the reduced amount of pre-rRNA precursors observed in Figure 9. A possible interpretation may be the uncoupling of mechanisms controlling transcription and early processing of pre-rRNA (Gallagher et al., 2004; Saez-Vasquez et al., 2004). Indeed, the data presented in Figure 9 provide evidence that AtNUC-L1 gene disruption induces changes in the ratio of primary pre-rRNA and processed pre-rRNA at the P site. We cannot establish if the lesser amount of pre-rRNA is due to inhibition of RNA pol I transcription or a higher rate of pre-rRNA processing at the P site. However, it is probably that both transcription and processing of pre-rRNA are affected in Atnuc-L1. Indeed, in vitro assays and microinjection experiments in X. laevis oocytes showed a role of nucleolin in both transcription of pre-rRNA (Roger et al., 2002) and in the first step of pre-rRNA processing (Ginisty et al., 1998). Moreover, our results also show that AtNUC-L1 binds the rDNA promoter in vivo; consequently, we expect that the absence of AtNUC-L1 in mutant plants might also affect rDNA condensation and gene transcription. These observations are supported by a recent study demonstrating that Xenopus laevis nucleolin modifies chromatin structure and facilitates RNA Pol II transcription (Angelov et al., 2006).
Our results show that AtNUC-L1 is not only involved in the primary cleavage site of pre-rRNA, but probably also in other processing events in vivo (Figure 9). It is possible that the absence of AtNUC-L1 in Atnuc-L1 plants has an effect on correct folding of nascent pre-rRNA required for the different pre-rRNA processing events (Allain et al., 2000). In this respect, the observation of the so-called NPGs in the modified nucleolar structure of mutant plant cells could be a sign of an altered pre-rRNA processing (Puvion-Dutilleul et al., 1983; Risueno and Medina, 1986).
A remarkable result of this work is the transcriptional activation of AtNUC-L2 gene in Atnuc-L1 plants. This activation suggests that AtNUC-L2 protein might rescue the AtNUC-L1 gene disruption (Figures 4 and 5). We discarded the possibility of partial complementation by a truncated AtNUC-L1 protein, because we detected neither shorter AtNUC-L1 transcripts encoding the acidic domain and the first RRM of AtNUC-L1 (based on the T-DNA insertion position) nor smaller polypeptides. Our data demonstrate that AtNUC-L2 localizes in the nucleolus and associates with large protein complexes, as does AtNUC-L1 (Supplemental Figures 5A and S3). Moreover, we demonstrate that the nucleolar localization of AtNUC-L2 is not the result of the nucleolar disorganization in Atnuc-L1 plants. Indeed, AtNUC-L2 also localized in the nucleoli of onion cells, suggesting a nucleolar role for AtNUC-L2 in WT plants (Figure 5B). Finally, the possibility that AtNUC-L2 might replace AtNUC-L1 is supported by unsuccessful attempts to obtain Atnuc-L1 and Atnuc-L2 double mutation, which is probably lethal. However, despite all these observations, the activation of AtNUC-L2 is nonnatural in Atnuc-L1 plants and raises the major question about the biological role of this second nucleolin-like gene in A. thaliana. Transcriptome analysis revealed that 333 genes (of ∼25,000 gst; http://www.catma.org/) are deregulated in the Atnuc-L1 plants, i.e., their expression is enhanced or repressed by more than 1.66 times (Hilson et al., 2004). The AtNUC-L2 gene is ranked 124th among 148 up-regulated genes. We know that the Atnuc-L1 mutant lines are stressed plants; indeed, transcriptome analysis revealed that several stress related genes are also activated in the Atnuc-L1 plants (data not shown). Therefore, it is tempting to propose that in Atnuc-L1 the AtNUC-L2 gene becomes activated to accomplish its role in a stressed plant, but in the absence of the AtNUC-L1, the induced AtNUC-L2 protein is recruited to the set of AtNUC- L1 associated factors to accomplish at least some of the AtNUC-L1 roles. These functions are probably achieved partially as a consequence of the structural differences between AtNUC-L1 and AtNUC-L2, in particular the less conserved GAR domain and the extended acidic N-terminal domain. So far, the analysis of an A. thaliana plant mutant line with a disrupted AtNUC-L2 gene has not revealed any particular phenotype when it is grown in the same conditions as control plants.
How AtNUC-L1 controls AtNUC-L2 expression? Transcriptional and or posttranscriptional mechanisms may be involved. This is one of the questions we are trying to answer. Thus far, although the role of AtNUC-L2 remains speculative and requires additional experimentation to establish its role in plant cells; our data indicate that conserved AtNUC-L1 plays an important role in chromatin regulation of rDNA genes, nucleolus structure, and growth in plants and possibly in other higher eukaryotes. Studies to establish more precisely the mechanisms involved in these major structural changes are the next challenge.
Supplementary Material
ACKNOWLEDGMENTS
We thank M. Delseny for supporting this work and our fellow laboratory member for stimulating discussion. We also thank R. Cooke for comments and corrections of English and J. R. Pages for computer assistance in artwork. We thank also J. P. Renou for help with the microarrays experiments. This work was supported by the Centre National de la Recherche Scientifique; by a grant to J.S.-V. and F.J.M. from PAI “Programme d'Actions Integrées franco-espagnol,” Picasso 11398VF (French team) and HF 2005-0209 (Spanish team); by a grant from the French Ministry of Research (ACI BCMS 04-5486); and by the Spanish “Plan Nacional de I+D+I” (ESP2003-09475-C02-02). The CATMA program 2005 allowed transcriptome analysis of Atnuc-L1. F.P. is supported by a fellowship from Ministèrede l'Enseignement et de la Recherche.
Footnotes
This article was published online ahead of print in MBC in Press(http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E06-04-0304) on November 15, 2006.
 The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
REFERENCES
- Allain F. H., Bouvet P., Dieckmann T., Feigon J. Molecular basis of sequence-specific recognition of pre-ribosomal RNA by nucleolin. EMBO J. 2000;19:6870–6881. doi: 10.1093/emboj/19.24.6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelov D., et al. Nucleolin is a histone chaperone with FACT-like activity and assists remodeling of nucleosomes. EMBO J. 2006;25:1669–1679. doi: 10.1038/sj.emboj.7601046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogre L., Jonak C., Mink M., Meskiene I., Traas J., Ha D. T., Swoboda I., Plank C., Wagner E., Heberle-Bors E., Hirt H. Developmental and cell cycle regulation of alfalfa nucMs1, a plant homolog of the yeast Nsr1 and mammalian nucleolin. Plant Cell. 1996;8:417–428. [PMC free article] [PubMed] [Google Scholar]
- Bouche G., Caizergues-Ferrer M., Bugler B., Amalric F. Interrelations between the maturation of a 100 kDa nucleolar protein and pre rRNA synthesis in CHO cells. Nucleic Acids Res. 1984;12:3025–3035. doi: 10.1093/nar/12.7.3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouvet P., Diaz J. J., Kindbeiter K., Madjar J. J., Amalric F. Nucleolin interacts with several ribosomal proteins through its RGG domain. J. Biol. Chem. 1998;273:19025–19029. doi: 10.1074/jbc.273.30.19025. [DOI] [PubMed] [Google Scholar]
- Caparros-Ruiz D., Lahmy S., Piersanti S., Echeverria M. Two ribosomal DNA-binding factors interact with a cluster of motifs on the 5′ external transcribed spacer, upstream from the primary pre-rRNA processing site in a higher plant. Eur. J. Biochem. 1997;247:981–989. doi: 10.1111/j.1432-1033.1997.00981.x. [DOI] [PubMed] [Google Scholar]
- Clough S. J., Bent A. F. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Earley K., Lawrence R. J., Pontes O., Reuther R., Enciso A. J., Silva M., Neves N., Gross M., Viegas W., Pikaard C. S. Erasure of histone acetylation by Arabidopsis HDA6 mediates large-scale gene silencing in nucleolar dominance. Genes Dev. 2006;20:1283–1293. doi: 10.1101/gad.1417706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- French S. L., Osheim Y. N., Cioci F., Nomura M., Beyer A. L. In exponentially growing Saccharomyces cerevisiae cells, rRNA synthesis is determined by the summed RNA polymerase I loading rate rather than by the number of active genes. Mol. Cell Biol. 2003;23:1558–1568. doi: 10.1128/MCB.23.5.1558-1568.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromont-Racine M., Senger B., Saveanu C., Fasiolo F. Ribosome assembly in eukaryotes. Gene. 2003;313:17–42. doi: 10.1016/s0378-1119(03)00629-2. [DOI] [PubMed] [Google Scholar]
- Gallagher J. E., Dunbar D. A., Granneman S., Mitchell B. M., Osheim Y., Beyer A. L., Baserga S. J. RNA polymerase I transcription and pre-rRNA processing are linked by specific SSU processome components. Genes Dev. 2004;18:2506–2517. doi: 10.1101/gad.1226604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendrel A. V., Lippman Z., Martienssen R., Colot V. Profiling histone modification patterns in plants using genomic tiling microarrays. Nat. Methods. 2005;2:213–218. doi: 10.1038/nmeth0305-213. [DOI] [PubMed] [Google Scholar]
- Ginisty H., Amalric F., Bouvet P. Nucleolin functions in the first step of ribosomal RNA processing. EMBO J. 1998;17:1476–1486. doi: 10.1093/emboj/17.5.1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginisty H., Sicard H., Roger B., Bouvet P. Structure and functions of nucleolin. J. Cell Sci. 1999;112:761–772. doi: 10.1242/jcs.112.6.761. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Camacho F., Medina F. J. The nucleolar structure and the activity of NopA100, a nucleolin-like protein, during the cell cycle in proliferating plant cells. Histochem. Cell Biol. 2005;125:139–153. doi: 10.1007/s00418-005-0081-1. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Melendi P., Wells B., Beven A. F., Shaw P. J. Single ribosomal transcription units are linear, compacted Christmas trees in plant nucleoli. Plant J. 2001;27:223–233. doi: 10.1046/j.1365-313x.2001.01091.x. [DOI] [PubMed] [Google Scholar]
- Gruendler P., Unfried I., Pointner R., Schweizer D. Nucleotide sequence of the 25S-18S ribosomal gene spacer from Arabidopsis thaliana. Nucleic Acids Res. 1989;17:6395–6396. doi: 10.1093/nar/17.15.6395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulli M. P., Girard J. P., Zabetakis D., Lapeyre B., Melese T., Caizergues-Ferrer M. gar2 is a nucleolar protein from Schizosaccharomyces pombe required for 18S rRNA and 40S ribosomal subunit accumulation. Nucleic Acids Res. 1995;23:1912–1918. doi: 10.1093/nar/23.11.1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez-Verdun D. Nucleolus in the spotlight. Cell Cycle. 2005;4:106–108. doi: 10.4161/cc.4.1.1355. [DOI] [PubMed] [Google Scholar]
- Hilson P., et al. Versatile gene-specific sequence tags for Arabidopsis functional genomics: transcript profiling and reverse genetics applications. Genome Res. 2004;14:2176–2189. doi: 10.1101/gr.2544504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T., Kim G. T., Shinozaki K. Disruption of an Arabidopsis cytoplasmic ribosomal protein S13-homologous gene by transposon-mediated mutagenesis causes aberrant growth and development. Plant J. 2000;22:257–264. doi: 10.1046/j.1365-313x.2000.00728.x. [DOI] [PubMed] [Google Scholar]
- Lam Y. W., Trinkle-Mulcahy L., Lamond A. I. The nucleolus. J. Cell Sci. 2005;118:1335–1337. doi: 10.1242/jcs.01736. [DOI] [PubMed] [Google Scholar]
- Lee W. C., Zabetakis D., Melese T. NSR1 is required for pre-rRNA processing and for the proper maintenance of steady-state levels of ribosomal subunits. Mol. Cell Biol. 1992;12:3865–3871. doi: 10.1128/mcb.12.9.3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leger-Silvestre I., Gulli M. P., Noaillac-Depeyre J., Faubladier M., Sicard H., Caizergues-Ferrer M., Gas N. Ultrastructural changes in the Schizosaccharomyces pombe nucleolus following the disruption of the gar2+ gene, which encodes a nucleolar protein structurally related to nucleolin. Chromosoma. 1997;105:542–552. doi: 10.1007/BF02510491. [DOI] [PubMed] [Google Scholar]
- Oakes M., Aris J. P., Brockenbrough J. S., Wai H., Vu L., Nomura M. Mutational analysis of the structure and localization of the nucleolus in the yeast Saccharomyces cerevisiae. J. Cell Biol. 1998;143:23–34. doi: 10.1083/jcb.143.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontes O., Lawrence R. J., Neves N., Silva M., Lee J. H., Chen Z. J., Viegas W., Pikaard C. S. Natural variation in nucleolar dominance reveals the relationship between nucleolus organizer chromatin topology and rRNA gene transcription in Arabidopsis. Proc. Natl. Acad. Sci. USA. 2003;100:11418–11423. doi: 10.1073/pnas.1932522100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puvion-Dutilleul F., Nicoloso M., Bachellerie J. P. Altered structure of ribosomal RNA transcription units in hamster cells after DRB treatment. Exp. Cell Res. 1983;146:43–52. doi: 10.1016/0014-4827(83)90322-1. [DOI] [PubMed] [Google Scholar]
- Raska I., Shaw P. J., Cmarko D. Structure and function of the nucleolus in the spotlight. Curr. Opin. Cell Biol. 2006;18:325–334. doi: 10.1016/j.ceb.2006.04.008. [DOI] [PubMed] [Google Scholar]
- Risueno M. C., Medina F. J. The nucleolar structure in plant cells. Revis. Biol. Celular. 1986;7:1–154. [PubMed] [Google Scholar]
- Roger B., Moisand A., Amalric F., Bouvet P. Repression of RNA polymerase I transcription by nucleolin is independent of the RNA sequence that is transcribed. J. Biol. Chem. 2002;277:10209–10219. doi: 10.1074/jbc.M106412200. [DOI] [PubMed] [Google Scholar]
- Roger B., Moisand A., Amalric F., Bouvet P. Nucleolin provides a link between RNA polymerase I transcription and pre-ribosome assembly. Chromosoma. 2003;111:399–407. doi: 10.1007/s00412-002-0221-5. [DOI] [PubMed] [Google Scholar]
- Saez-Vasquez J., Caparros-Ruiz D., Barneche F., Echeverria M. A plant snoRNP complex containing snoRNAs, fibrillarin, and nucleolin-like proteins is competent for both rRNA gene binding and pre-rRNA processing in vitro. Mol. Cell Biol. 2004;24:7284–7297. doi: 10.1128/MCB.24.16.7284-7297.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saez-Vasquez J., Pikaard C. S. Extensive purification of a putative RNA polymerase I holoenzyme from plants that accurately initiates rRNA gene transcription in vitro. Proc. Natl. Acad. Sci. USA. 1997;94:11869–11874. doi: 10.1073/pnas.94.22.11869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres B. Plant cell identity. The role of position and lineage. Plant Physiol. 2001;125:112–114. doi: 10.1104/pp.125.1.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert I., Fransz P. F., Fuchs J., de Jong J. H. Chromosome painting in plants. Methods Cell Sci. 2001;23:57–69. [PubMed] [Google Scholar]
- Sessions A., Weigel D., Yanofsky M. F. The Arabidopsis thaliana MERISTEM LAYER 1 promoter specifies epidermal expression in meristems and young primordia. Plant J. 1999;20:259–263. doi: 10.1046/j.1365-313x.1999.00594.x. [DOI] [PubMed] [Google Scholar]
- Shaw P. J., Jordan E. G. The nucleolus. Annu. Rev. Cell Dev. Biol. 1995;11:93–121. doi: 10.1146/annurev.cb.11.110195.000521. [DOI] [PubMed] [Google Scholar]
- Srivastava M., Pollard H. B. Molecular dissection of nucleolin's role in growth and cell proliferation: new insights. FASEB J. 1999;13:1911–1922. [PubMed] [Google Scholar]
- Takagi M., Absalon M. J., McLure K. G., Kastan M. B. Regulation of p53 translation and induction after DNA damage by ribosomal protein L26 and nucleolin. Cell. 2005;123:49–63. doi: 10.1016/j.cell.2005.07.034. [DOI] [PubMed] [Google Scholar]
- Testillano P. S., Gonzalez-Melendi P., Ahmadian P., Risueno M. C. The methylation-acetylation method: an ultrastructural cytochemistry for nucleic acids compatible with immunogold studies. J. Struct. Biol. 1995;114:123–139. doi: 10.1006/jsbi.1995.1012. [DOI] [PubMed] [Google Scholar]
- Thiry M., Lafontaine D. L. Birth of a nucleolus: the evolution of nucleolar compartments. Trends Cell Biol. 2005;15:194–199. doi: 10.1016/j.tcb.2005.02.007. [DOI] [PubMed] [Google Scholar]
- Tong C. G., Reichler S., Blumenthal S., Balk J., Hsieh H. L., Roux S. J. Light regulation of the abundance of mRNA encoding a nucleolin-like protein localized in the nucleoli of pea nuclei. Plant Physiol. 1997;114:643–652. doi: 10.1104/pp.114.2.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Lijsebettens M., Vanderhaeghen R., De Block M., Bauw G., Villarroel R., Van Montagu M. An S18 ribosomal protein gene copy at the Arabidopsis PFL locus affects plant development by its specific expression in meristems. EMBO J. 1994;13:3378–3388. doi: 10.1002/j.1460-2075.1994.tb06640.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang R. H., Lee J. C. Identification of proteins crosslinked to RNA in 40S ribosomal subunits of Saccharomyces cerevisiae. Biochimie. 1989;71:1201–1204. doi: 10.1016/0300-9084(89)90024-2. [DOI] [PubMed] [Google Scholar]
- Yanagida M., Shimamoto A., Nishikawa K., Furuichi Y., Isobe T., Takahashi N. Isolation and proteomic characterization of the major proteins of the nucleolin-binding ribonucleoprotein complexes. Proteomics. 2001;1:1390–1404. doi: 10.1002/1615-9861(200111)1:11<1390::AID-PROT1390>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.