Abstract
The Saccharomyces cerevisiae basic leucine zipper transcription factor Hac1p is synthesized in response to the accumulation of unfolded polypeptides in the lumen of the endoplasmic reticulum (ER), and it is responsible for up-regulation of ∼5% of all yeast genes, including ER-resident chaperones and protein-folding catalysts. Hac1p is one of the most short-lived yeast proteins, having a half-life of ∼1.5 min. Here, we have shown that Hac1p harbors a functional PEST degron and that degradation of Hac1p by the proteasome involves the E2 ubiquitin-conjugating enzyme Ubc3/Cdc34p and the SCFCdc4 E3 complex. Consistent with the known nuclear localization of Cdc4p, rapid degradation of Hac1p requires the presence of a functional nuclear localization sequence, which we demonstrated to involve basic residues in the sequence 29RKRAKTK35. Two-hybrid analysis demonstrated that the PEST-dependent interaction of Hac1p with Cdc4p requires Ser146 and Ser149. Turnover of Hac1p may be dependent on transcription because it is inhibited in cell mutants lacking Srb10 kinase, a component of the SRB/mediator module of the RNA polymerase II holoenzyme. Stabilization of Hac1p by point mutation or deletion, or as the consequence of defects in components of the degradation pathway, results in increased unfolded protein response element-dependent transcription and improved cell viability under ER stress conditions.
INTRODUCTION
In eukaryotic cells, the endoplasmic reticulum (ER) is the portal for newly synthesized proteins destined for all compartments of the exocytic and endocytic pathways as well as for the cell surface and the extracellular space. Transport along the exocytic pathway requires that passenger proteins are correctly folded and assembled (Gething et al., 1986; for review, see Ellgaard and Helenius, 2003), a process that is assisted by molecular chaperones and folding catalysts resident in the ER lumen (for review, see van Anken and Braakman, 2005). The concentrations of these components in the ER are adjusted according to need by the unfolded protein response (UPR; Kozutsumi et al., 1988; Mori et al., 1992; for review, see Kaufman, 1999; Ma and Hendershot, 2001; Patil and Walter, 2001; Schröder and Kaufman, 2005). The UPR regulates the transcription of ∼5% of the open reading frames in the Saccharomyces cerevisiae genome, many of which encode proteins with functions involved in diverse processes, including protein translocation, glycosylation, folding and degradation, lipid/inositol metabolism, vesicular trafficking, vacuolar protein sorting, and cell wall biogenesis (Travers et al., 2000).
The yeast unfolded protein response (UPR) involves two unique participants, the Ire1p transmembrane receptor kinase/endonuclease (Cox et al., 1993; Mori et al., 1993), and the basic leucine zipper (bZip) transcription factor Hac1p (Cox et al., 1996; Mori et al., 1996), which transactivates genes bearing UPRE elements (Mori et al., 1992, 1998; Patil and Walter, 2004). HAC1 mRNA is synthesized constitutively as a precursor bearing a 252-nucleotide intron that blocks translation as the result of base pairing with a sequence in the 5′-untranslated region of the mRNA (Chapman and Walter, 1997; Kawahara et al., 1997; Ruegsegger et al., 2001). This intron is removed by the endoribonuclease activity of the C terminus of Ire1p (Cox et al., 1996; Kawahara et al., 1997; Sidrauski and Walter, 1997), which is activated after oligomerization of the receptor in response to the accumulation of unfolded proteins in the ER and trans-autophosphorylation of the kinase domain (Shamu and Walter, 1996; Welihinda and Kaufman, 1996). The resulting HAC1 mRNA exons are joined by the Rlg1p ligase to produce the mature, efficiently translated mRNA (Sidrauski et al., 1996; Kawahara et al., 1997). This process brings together sequences in the first exon that encode a potential nuclear localization signal and the DNA binding domain with sequences in the second exon that encode the transcriptional activator domain (TAD) (Mori et al., 2000).
Previous studies on the regulation of the UPR have largely focused on events that initiate the synthesis of Hac1p. However, mechanisms that regulate the rate of turnover of Hac1p will also be crucial in determining the cellular concentration of the active transcription factor and thus the magnitude of the stress response. The concentration of Hac1p should also affect the scope of the response, because different classes of UPR-regulated genes are transactivated at different threshold levels of Hac1p (Leber et al., 2004). The rate of degradation of Hac1p will also determine how quickly the response is terminated once the ER stress is removed. In this study, we have investigated the sequence elements and cellular machinery that contribute to the very rapid rate of turnover of Hac1p (t1/2 of 1–2 min; Kawahara et al., 1997; Chapman and Walter, 1997).
MATERIALS AND METHODS
Strains and Media
The yeast strains used in this study are listed in Table 1. Cells were grown in YP medium containing glucose (YPD) or synthetic complete media lacking appropriate amino acids (Kaiser et al., 1994). Strains containing a temperature-sensitive mutation were grown at the permissive temperature (23°C) before incubation under nonpermissive conditions (37°C). The NCY1810 Δhac1 strain (DL1783, hac1Δ::kanMX6) was generated by direct replacement of the HAC1 open reading frame with the kanMX6 cassette (Guldener et al., 1996). The NCY1811 Δhac1 Δsrb10 strain (KMY1045, srb10Δ::His3MX6) was generated by direct replacement of the SRB10 open reading frame with His3MX6 cassette as described by Longtine et al. (1998). LHY100 (KMY2105 mpk1Δ::HIS3) was constructed by single-step gene replacement (Rothstein, 1991) by using the HIS3 gene amplified from plasmid pRS313 (Sikorski and Hieter, 1989) with primers incorporating the first and the last 50 nucleotides of the MPK1 open reading frame. LHY102 (KMY2005 pkc1Δ::LEU2) was also constructed by single step gene replacement by using plasmid pL924 (Levin et al., 1990). In all cases, clones containing the desired deletion were obtained after growth on the appropriate selective medium. The genotypes were confirmed by polymerase chain reaction (PCR).
Table 1.
Yeast strains
| Strain | Genotype | Reference or source |
|---|---|---|
| W303-1a | MATa his3-11 leu2-3 ura3-1 ade2-1 trp1-1 can1-100 | B. Futcher |
| can1-1 | W303 but can1-1 | Ghislain et al. (1993) |
| DL376 | MATa his4 leu2-3,112 ura3-52 trp1-1 can1rpkc1Δ::LEU2 | Levin and Bartlett-Heubusch (1992) |
| DL455 | MATa his4 leu2-3,112 ura3-52 trp1-1 can1rmpk1Δ:TRP1 | Lee et al. (1993) |
| DL1783 | MATa his4 leu2-3,112 ura3-52 trp1-1 can1r | Lee and Levin (1992) |
| JN1 | MATα ura3, his3, leu2, lys2, trp1, gal4, GAL1-LacZ reporter | Muratani et al. (2005) |
| JN5 | JN1 but srb10Δ::kanMX6 | Muratani et al. (2005) |
| KMY1005 | MATα leu2-3,112 ura3-52 his3-Δ200 trp-901 lys2-801 | Mori et al. (1996) |
| KMY1045 | KMY1005 but hac1Δ::TRP1 | Mori et al.(1996) |
| KMY2005 | MATα leu2-3,112 ura3-52 his3Δ200 trp1Δ901 lys2-801 sec53-6 gal1 suc2 | Mori et al.(1996) |
| KMY2105 | MATα leu2-3,112 ura3-52 his3Δ200 trp1Δ901 lys2-801sec53-6 gal1 suc2 | Kawahara et al.(1997) |
| UPRE-lacZ::URA3 | ||
| KMY2145 | KMY2105 but hac1Δ::TRP1 | Kawahara et al.(1997) |
| L40 | MATα leu2-3,112 ura3-1 his3-Δ200 15 trp1-901 ade2-1lys2-801am | Hollenberg et al. (1995) |
| URA3::(lexAOP)8-lacZ LYS2::(lexAop)4-HIS3 | ||
| LHY100 | KMY2105 but mpk1Δ::HIS3 | This study |
| LHY102 | KMY2005 but pkc1Δ::LEU2 | This study |
| MT668* | W303 but cdc4-1 | Patton et al.(1998) |
| MT670 | W303 but cdc34-1 | Willems et al. (1996) |
| MT871 | W303 but cdc53-1 | Willems et al. (1996) |
| NCY1810 | DL1783 but hac1Δ::kanMX6 | This study |
| NCY1811 | KMY1045 but srb10Δ::His3MX61r | This study |
| PY1 | 15Daub(wt): a bar1Δ ura3Δns ade1 his2 leu2-3,112 trp1-1 | Kaiser et al. (2000) |
| PY283* | PY1 but met30-6::KANR | Kaiser et al. (2000) |
| Y80 | MATaade2-1his3-11,15leu2-3,112trp1-1ura3-1 | Bai et al. (1996) |
| Y552 | Y80 but skp1-11 | Bai et al. (1996) |
| Y554 | Y80 but skp1-12 | Bai et al. (1996) |
| BY4742 | MATα his3Δ1leu2Δ0lys2Δ0 ura3Δ0 can1-100 | Research Genetics |
| RG16902* | BY4742 but grr1Δ::kanMX6 | Research Genetics |
| RG12708* | BY4742 but hrt3Δ::kanMX6 | Research Genetics |
| RG11133* | BY4742 but skp2Δ::kanMX6 | Research Genetics |
| RG11856* | BY4742 but dia1Δ::kanMX6 | Research Genetics |
| RG11597* | BY4742 but rax1Δ::kanMX6 | Research Genetics |
| RG13343* | BY4742 but cos111Δ::kanMX6 | Research Genetics |
| RG14065* | BY4742 but locus YDR131C deleted with kanMX6 | Research Genetics |
| RG14173* | BY4742 but locus YLR224W deleted with kanMX6 | Research Genetics |
| RG11276* | BY4742 but locus YJL149W deleted with kanMX6 | Research Genetics |
| RG13578* | BY4742 but locus YDR219C deleted with kanMX6 | Research Genetics |
| RG11221* | BY4742 but rcy1Δ::kanMX6 | Research Genetics |
| RG14611* | BY4742 but rtf1Δ::kanMX6 | Research Genetics |
| RG13072* | BY4742 but locus YBL046W deleted with kanMX6 | Research Genetics |
| RG13665* | BY4742 but locus YDR306C deleted with kanMX6 | Research Genetics |
| RG12298* | BY4742 but rev7Δ::kanMX6 | Research Genetics |
| RG17172* | BY4742 but locus YBR280C deleted with kanMX6 | Research Genetics |
| RG15277* | BY4742 but flm1Δ::kanMX6 | Research Genetics |
| RG15261* | BY4742 but locus YLR352W deleted with kanMX6 | Research Genetics |
| RG11982* | BY4742 but ela1Δ::kanMX6 | Research Genetics |
| RG15308* | BY4742 but bdf1Δ::kanMX6 | Research Genetics |
| BY4739 | MATα leu2Δ0lys2Δ0ura3Δ0 | Research Genetics |
| RG10482* | BY4739 but ufo1Δ::kanMX6 | Research Genetics |
* F-box mutants.
Plasmid Constructions and Oligonucleotide-directed Mutagenesis
All plasmid manipulations were carried out using standard protocols (Sambrook et al., 1989). References to the sources or details of the construction of the various plasmids used in this study can be found in the Supplemental Material. Descriptions of the procedures used to make HAC1 deletion and point mutants can also be found in the Supplemental Material, and the oligonucleotides used in this work are listed in Supplemental Table S1. DNA sequence analysis was used to confirm the introduction of mutations and to rule out the possibility that additional alterations had been introduced by the mutagenesis or cloning procedures.
PESTfind Analysis of Hac1p Sequence
The PESTfind server (emb1.bcc.univie.ac.at/embnet/tools/bio/PESTfind) was used to identify possible PEST sequences within Hac1p.
Protein Synthesis Shutoff Assay
S. cerevisiae cells were grown overnight to OD600 0.6 at 30 or 23°C (for temperature-sensitive yeast strains) in 50 ml of YPD media or SC-Leu, or SC-Trp-Ura. To examine the turnover of Hac1p, tunicamycin (Calbiochem. San Diego, CA) was added to a final concentration of 5 μg/ml to induce the UPR, and the culture was incubated for another 90 min. In temperature-sensitive yeast strains, cultures were shifted from 23°C to the nonpermissive temperature of 37°C for 30 min. An 8-ml aliquot was removed as the zero-time sample and transferred to a tube containing 100 μl of 20% sodium azide (Sigma-Aldrich, St. Louis, MO) and 1 ml of dimethyl sulfoxide (Sigma-Aldrich), mixed by inversion, and snap-frozen in liquid nitrogen. Cycloheximide (Sigma-Aldrich) was added to a final concentration of 1 mg/ml to the bulk culture to halt protein synthesis, and incubation was continued. Samples were collected at various times between 1 and 30 min after the addition of cycloheximide and snap-frozen as described above. Samples were thawed in a 4°C water-bath (∼45 min) before pelleting at 4°C (10 min; 3000 rpm) and storage at −80°C. To examine the turnover of Ura3/HAp fusion proteins, the zero-time point sample was collected before adding cycloheximide and aliquots were collected at 30-, 60-, 90-, and 150-min time points.
Cell Extracts and Immunoblotting
Unless otherwise indicated protein extracts from yeast cells were made in EZ buffer (60 mM Tris, pH 6.8, 10% [vol/vol] glycerol, 2% [wt/vol] SDS, and 5% [vol/vol] β-mercaptoethanol) by boiling for 10 min, followed by mixing and centrifugation (13,000 rpm for 10 min) (Muratani et al., 2005). Protein concentrations were determined using the Bradford protein assay kit (Bio-Rad, Hercules, CA). Volumes of extract containing equal amounts of total protein (∼80 μg) were boiled in sample buffer for 10 min and then loaded on either an 8 or 12% SDS gel and submitted to polyacrylamide gel electrophoresis (PAGE). For immunoblot analysis, the proteins were transferred onto nitrocellulose (Protran; Whatman Schleicher and Schuell, Keene, NH) by wet transfer in the Mini Trans-Blot Cell (Bio-Rad) and blocked with 5% skim milk in Tris-buffered saline containing 0.1% Tween 20. Detection of wild-type Hac1p and the various mutant Hac1p proteins was performed using a polyclonal anti-Hac1p antibody (Kawahara et al., 1997) or an anti-Hac1pi tail antibody (Cox and Walter, 1996). Analysis of Ura3/HA and Hac1/HA fusion proteins used anti-hemagglutinin (HA) tag antibodies, (Muratani et al., 2005), whereas detection of green fluorescent protein (GFP) fusion proteins used polyclonal anti GFP antibodies (a gift from Pamela Silver, Dana-Farber Institute, Boston, MA). Subsequently, the membranes were probed with monoclonal anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (a gift from Trevor Lithgow, University of Melbourne, Melbourne, Australia) as a control for protein loading. The proteins were detected using enhanced chemiluminescence reagents (Pierce Chemical, Rockford, IL or Roche Diagnostics, Indianapolis, IN) according to the manufacturers' instructions. Densitometry was carried out using ImageQuant 4.5 software (Media Cybernetics, Silver Spring, MD).
Growth Assays
Yeast cells were grown at 30°C in appropriate media until OD600 reached 0.6. Cultures were then diluted with water to OD600 of 0.04, and a series of 1:10 serial dilutions in water were prepared. For each dilution, a 5-μl aliquot was spotted onto solid media (YPD and YPD supplemented with either 0.5 or 1.0 μg/ml tunicamycin for ER stress survival assays, and SC lacking tryptophan or SC lacking both tryptophan and uracil for uracil prototrophy assays) and incubated at 30°C until colonies formed (2–3 d).
Fluorescence Microscopy
Yeast cells expressing GFP fusion proteins were prepared for visualization as follows: log-phase cultures (OD600 of 1.0–1.5) were grown at 30°C in synthetic medium in the absence of methionine to induce expression from the MET25 promoter of the pGFP-Nfus vector. Cells were fixed by adding 1/10 volume formaldehyde (standard stock solution is 37%) directly to the medium and by incubating for at least 30 min. Cells were harvested and washed twice with 0.1 M potassium phosphate, pH 7.5, and then twice with 1× phosphate-buffered saline (PBS), and then resuspended in PBS (200 μl per 10 ml of culture). Coverslips were coated with poly-l-lysine (Sigma-Aldrich) and allowed to dry after which 20 μl of the cell suspension was smeared evenly on the coverslip and allowed to dry. The coverslip was then inverted onto a slide with 10 μl of Mowiol (Calbiochem) containing 4,6-diamidino-2-phenylindole (DAPI) dye (Sigma-Aldrich). Cells were viewed at room temperature with an Axioplan2 microscope (Carl Zeiss, Thornwood, NY) equipped with an AxioCam Mrm digital camera. Picture analysis was performed using Axiovision2 software.
Two-Hybrid Analysis
Yeast two-hybrid assays were carried out using pACT encoding the Gal4 transcriptional activation domain (AD) alone as a negative control or Gal4AD fused with CDC4 sequences encoding residues (339-779) (Drury et al., 1997) as bait to test the interaction of wild-type and mutant Hac1p sequences with Cdc4p substrates. pBTM116 and pBTM116-CDC6(1–47), which encode the LexA DNA binding domain (BD) alone or fused to residues 1–47 of Cdc6p (Drury et al., 1997), were used as negative and positive prey plasmids, respectively. pBTM116–HAC1 fusion constructs were generated as described in the Supplemental Material. pACT and pBTM116 constructs were transformed into S. cerevisiae strain L40 (Hollenberg et al., 1995). Protein–protein interaction assays were performed by growing transformants on nitrocellulose filters before lysing the cells and measuring β-galactosidase activity as described by Xie et al. (1993).
Osmotic and UPR Stress Treatments
For experiments using osmotic modulation, cells were grown to mid-logarithmic phase (OD600 of 0.8–1.0) at 23°C (for KMY2015 sec53 cells) or 30°C (for SEC53+ cells) in either YPD or selective media supplemented with 1 M sorbitol. To apply osmotic stress, cell cultures were diluted with 4 parts of osmotic diluents: 0.4% glucose (hypotonic) or 0.4% glucose + 1M sorbitol (isotonic) made up in water or selective medium and prewarmed to the temperature of the subsequent incubation. Where appropriate, tunicamycin was added to the diluents to a final concentration of 5 μg/ml to initiate UPR stress. For sec53 cells, ER stress was imposed either by incubation at the semipermissive temperature of 30°C or by combining incubation at 30°C with tunicamycin treatment. Control cells (no ER stress) were incubated at 23°C (sec53 cells) or at 30°C (SEC53+ cells) in the absence of tunicamycin. At the end of each incubation period cells were placed on ice, pelleted, and frozen in liquid N2. All further manipulations were done at 0–4°C.
β-Galactosidase Assay
Cells were lysed using glass beads and the extracts assayed for β-galactosidase activity as described previously (Kaiser et al., 1994). Protein concentrations were determined using the Bio-Rad DC protein assay kit, and β-galactosidase activity was defined as units per milligram of protein where 1 U results in OD420 of 0.001/min.
In Vivo Labeling and Immunoprecipitation
35S Labeling and Immunoprecipitation of Hac1p.
Yeast cells were grown to OD600 of 0.5–0.8 at 23°C. Cells were resuspended in low sulfate medium at a density of 3 OD600 units/ml and incubated for 30 min at 23°C before 4-ml aliquots were taken for incubation at 23°C (control) or 30°C (ER stress) for a further 60 min, at which point they were pulse labeled with 150 μCi of [35S]Promix labeling mix (GE Healthcare, Little Chalfont, Buckinghamshire, United Kingdom) for 5 min and harvested as described previously (Kawahara et al., 1997). Cell extracts were prepared as described previously (Cox et al., 1997), and equal amounts of protein (70–100 μg) were taken for immunoprecipitation (IP) in a total volume of 500 μl. IP reactions were precleared using preimmune serum and 50 μl of 10% (vol/vol) Pansorbin (Calbiochem), and Hac1p was precipitated with 5 μl of anti-Hac1p antiserum and 50 μl of protein A-Sepharose (50%, vol/vol). Washings followed established protocols (Franzusoff et al., 1991). Hac1p was eluted from beads in Laemmli loading buffer and separated by SDS-PAGE. After treatment with Amplify (GE Healthcare), gels were dried and subjected to fluorography.
32P Labeling and Immunoprecipitation of Hac1p.
Yeast cells were grown in high phosphate medium to OD600 of 0.8 at 23°C, washed into low phosphate medium at a density of OD600 of 0.8, and incubated at 23°C for a further 4 h (Shamu and Walter, 1996). The OD600 was adjusted to 0.8, and 4 ml of cells was labeled with 100 μCi of [32P]orthophosphoric acid/ml at 23°C for 1 h, and then the cells were divided into two aliquots and placed at either 23 or 30°C for a further 2 h. Cells were harvested in the presence of phosphatase inhibitors (including 50 nM calyculin A; Shamu and Walter, 1996) and extracts were prepared (Cox et al., 1993). Volumes of cell lysates were adjusted to 160 μl, of which 100 μl was used in each IP reaction in a total volume of 500 μl with the addition of phosphatase inhibitors (Shamu and Walter, 1996). The concentration of protein in the lysates was determined using the Bio-Rad DC protein assay kit. After immunoprecipitation (see above), eluted proteins were separated on SDS-PAGE gels, dried, and subjected to autoradiography. Parallel cultures of cells grown under identical conditions but without the addition of radiolabel were harvested in the presence of phosphatase inhibitors, and proteins were extracted for analysis by immunoblotting with anti-Hac1p.
RESULTS
Hac1p Is Degraded by the Proteasome
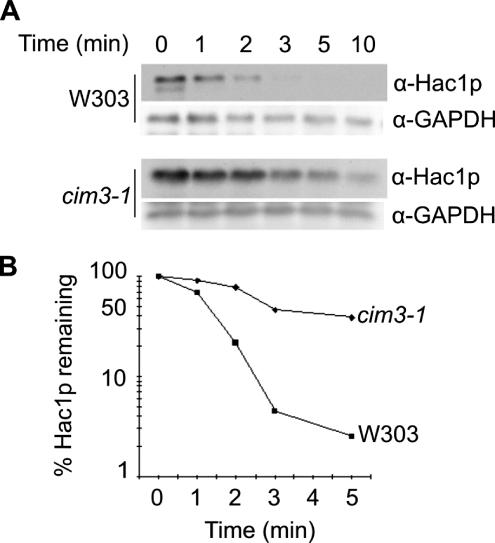
In eukaryotic cells, most rapidly turned over proteins are degraded via the ubiquitin–proteasome pathway (Varshavsky, 1997; Hershko and Ciechanover, 1998; Ciechanover et al., 2000). We tested the stability of Hac1p in an S. cerevisiae strain carrying a temperature-sensitive mutation in the CIM3 (SUG1/RPT6) gene, which encodes an “AAA” ATPase of the 19S regulatory subunit of the proteasome (Ghislain et al., 1993). A protein synthesis shutoff assay in which the decay of Hac1p is measured after treatment of cells with cycloheximide was used to compare the half-life of the protein in cim3–1ts cells with that in parental W303 cells. Because Hac1p is synthesized only in cells undergoing ER stress, cells grown to OD600 of ∼0.6 at the permissive temperature of 23°C were first treated with tunicamycin to induce the UPR as the result of accumulation of abnormally glycosylated and malfolded secretory precursors in the ER (Elbein, 1987; Kozutsumi et al., 1988). After 90 min, the cultures were shifted to the nonpermissive temperature of 37°C, and 30 min later the cycloheximide shutoff assay was performed as described in Materials and Methods. As shown in Figure 1, A and B, the half-life of Hac1p in the parental W303 cells was 1–1.5 min, consistent with the values of 1–2 min obtained previously with other yeast strains by using either protein shutoff or pulse-chase techniques (Chapman and Walter, 1997; Kawahara et al., 1997). By contrast, in cim3–1ts cells the rate of degradation of Hac1p was significantly decreased, indicating the involvement of the proteasome in the rapid turnover of Hac1p.
Figure 1.
Hac1p stability is regulated via the 26S proteasome. (A) cim3–1 cells and parental W303 cells were grown to OD600 of 0.6 at 23°C in YPD media. The UPR stressor tunicamycin was then added to a final concentration of 5 μg/ml to induce synthesis of Hac1p, and incubation was continued for 90 min before the cultures were shifted to the nonpermissive temperature of 37°C. Cycloheximide (final concentration 1 mg/ml) was added to the culture to halt protein synthesis, and samples were collected at the indicated chase times, snap-frozen and treated as described in Materials and Methods. Cell extracts were then prepared and protein concentrations were measured. Samples containing 80 μg of cellular protein were analyzed by SDS-PAGE and immunoblotted first with α-Hac1p then with α-GAPDH antibodies. (B) The plots were generated from the data shown in A by using ImageQuant 4.5 software.
Substrate proteins are marked for recognition and degradation by the 26S proteasome by attachment of monoubiquitin to a lysine residue followed by assembly of a tetraubiquitin chain via linkage through lysine 48 of ubiquitin (Thrower et al., 2000). To investigate the role of the 11 lysines in Hac1p in ubiquitin-mediated degradation, we analyzed the stability and capacity to confer ER stress resistance of Hac1p mutants in which one or more of the lysine residues were substituted by arginine. These experiments are described in full in the Supplemental Material. The data presented in Supplemental Figure S1 show that none of the mutations significantly affected the stability of the protein, suggesting that as previously observed for other target proteins such as Gcn4p (Kornitzer et al., 1994) and Sic1p (Petroski and Deshaies, 2003), no single lysine residue is the sole required attachment point for ubiquitylation-mediating degradation.
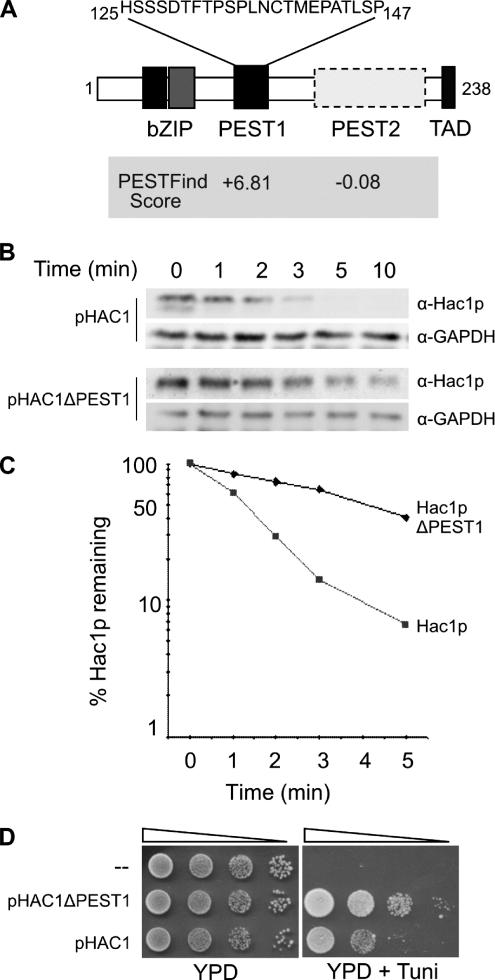
Hac1p Harbors a PEST Degron
Many proteins that undergo rapid turnover contain sequence identifiers that target the protein for degradation (Laney and Hochstrasser, 1999). Inspection of the Hac1p sequence failed to identify any common destruction motifs such as the D-box and KEN-box found in cyclins and other cell cycle-regulated proteins (Yamano et al., 1998; Vodermaier, 2004). The PESTfind algorithm (Rogers et al., 1986) was then used to analyze the Hac1p sequence for potential PEST motifs, which are hydrophilic, enriched in proline, glutamate, serine, and threonine, and at least 12 amino acids in length (Rogers et al., 1986; Rechsteiner and Rogers, 1996). The PESTfind program identified one high-confidence PEST sequence (score +6.81) located between residues 125 and 147 of Hac1p (Figure 2A), whose existence had previously been noted by Cox and Walter (1996), and one low-confidence sequence (residues 167–217, score −0.08; location boxed in Figure 2A). Oligonucleotide-directed mutagenesis was used to delete each potential PEST sequence from the HAC1 coding sequence. Restriction fragments encompassing each deletion were inserted into the CEN-based pHAC1 expression vector, replacing the corresponding wild-type sequences. Vectors encoding the wild-type or PEST1- or PEST2-deleted Hac1 proteins were transfected into KMY1045 (Δhac1) cells, and the resulting transformants were grown at 30°C to OD600 of 0.6 and then treated with tunicamycin to induce the synthesis of Hac1p before the cycloheximide shutoff assay was performed to compare the rates of turnover of the wild-type and mutant Hac1 proteins. As shown in Figure 2, B and C, the half-life of exogenously expressed wild-type Hac1p in KMY1045 cells (∼1.2 min) is essentially identical to that observed for endogenous Hac1p in W303 cells (Figure 1). However, deletion of the PEST1 motif caused a significant increase in the half-life of the protein (to ∼4 min), indicating that a degradation signal (degron) located within residues 125-147 of Hac1p plays an important role in the rapid turnover of the protein. By contrast, deletion of the low-scoring PEST2 sequence had no significant effect on the half-life of Hac1p (data not shown). Consistent with its greater stability, Δhac1 cells expressing the PEST1-deleted Hac1p protein from the pHAC1ΔPEST1 vector were more resistant to ER stress than were those expressing wild-type Hac1p from pHAC1 (Figure 2D).
Figure 2.
Hac1p stability is dependent on the presence of a functional PEST degron. (A) Schematic diagram indicating regions in the active (238-amino acid) form of Hac1p and showing the scores of potential PEST regions identified using the PESTfind program. (B) Synthesis shutoff assays were performed essentially as described in the legend to Figure 1, except that KMY1045 (Δhac1) cells expressing wild-type or ΔPEST Hac1 proteins were grown at and the assay was performed at 30°C. (C) Plots showing the amount of Hac1p remaining compared with that present at the zero time point before the addition of cycloheximide were generated from the data shown in B by using ImageQuant 4.5 software. (D) Serial 10-fold dilutions of KMY1045 (Δhac1) cells transformed with pHAC1 or pHAC1ΔPEST were spotted on YPD media alone or YPD media containing 1.0 μg/ml tunicamycin (to test for sensitivity to ER stress).
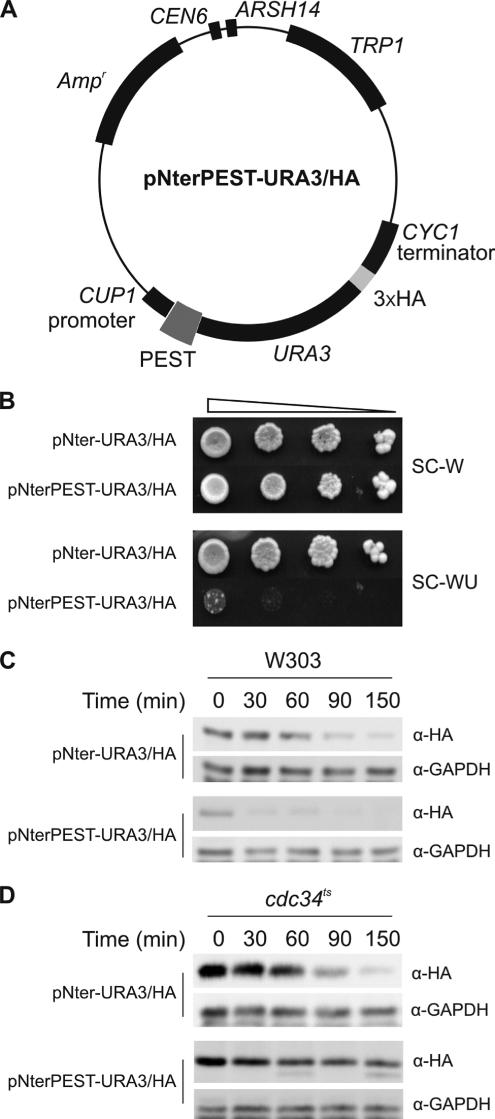
To determine whether the Hac1p PEST1 (hereinafter called PEST) degron is able to target a reporter protein for rapid degradation, we constructed a vector capable of expressing an HA-tagged Ura3 protein, and then inserted HAC1 nucleotides encoding a sequence spanning the PEST degron (residues 122-158) in frame between the ATG initiation codon and the sequences encoding Ura3/HAp (Figure 3A). W303 (trp1 ura3) cells expressing the Ura3/HA protein were equally viable in synthetic media containing or lacking uracil (Figure 3B), demonstrating that the presence of the HA tag at the C terminus of Ura3p had no significant effect on the ability of the enzyme to support uracil biosynthesis. By contrast, cells expressing the PEST-Ura3/HA protein were unable to grow in medium lacking uracil (Figure 3B). Synthesis shutoff assays (Figure 3C) showed that the Ura3/HA fusion protein has a half-life of ∼60 min (Figure 3C, top). Consistent with its failure to support cell viability in the absence of uracil, the PEST-Ura3/HA fusion protein was much less stable, being almost completely degraded by the 30-min time point (Figure 3C, bottom). Interestingly, the PEST–Ura3/HA fusion protein, unlike the Ura3/HA protein, was very significantly stabilized at the nonpermissive temperature in MT670 cells, which have a temperature-sensitive defect in the Ubc3p/Cdc34p E2 ubiquitin-conjugating enzyme (Figure 3D). This is the same E2 enzyme that we show below to mediate degradation of Hac1p. Because other degradation signals target a similar Ura3/HA fusion protein to different E2 enzymes such as Ubc6p and/or Ubc7p (Gilon et al., 1998), our data suggest that residues 122–158 of Hac1p contain a specific recognition signal for Ubc3p/Cdc34p.
Figure 3.
Hac1p PEST degron confers proteolytic instability on Ura3p. (A) The pNterPEST-URA3/HA plasmid was generated as described in the Supplemental Material to check the degron activity of the Hac1p PEST sequence. (B) Serial 10-fold dilutions of W303 cells transformed with pNter-URA3/HA (empty vector) or pNterPEST-URA3/HA were plated on SC media lacking tryptophan (to select for retention of the plasmid) or SC media lacking both tryptophan and uracil (to test for cell viability in the absence of uracil). (C) Synthesis shutoff assays were performed as described in the legend to Figure 2B but over the time course 0–150 min on W303 cells transformed with pNter-URA3/HA or pNterPEST-URA3/HA. (D) Synthesis shutoff assays were performed as described in C but on MT670 (cdc34ts) cells transformed with pNter-URA3/HA or pNterPEST-URA3/HA.
Degradation of Hac1p Involves the E2 Ubiquitin-conjugating Enzyme Ubc3/Cdc34p
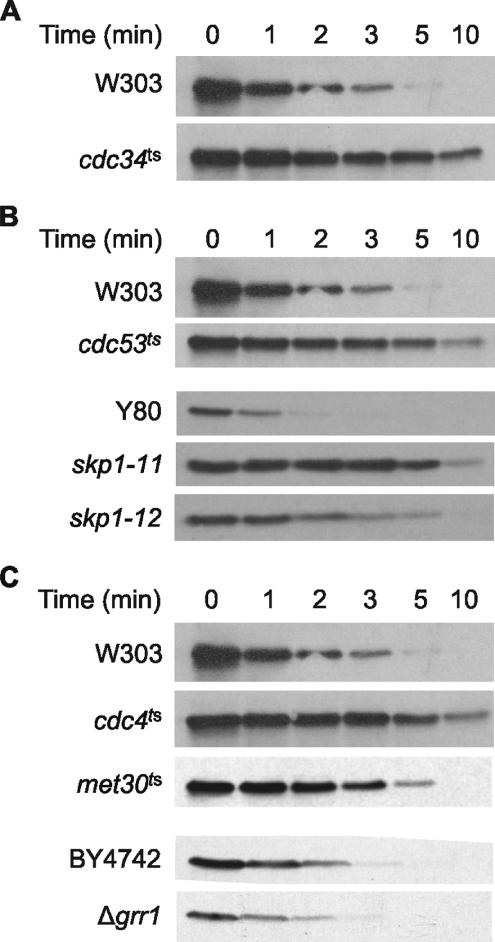
Ubiquitylation of proteins for targeting to the proteasome requires a multienzyme system involving E1, E2, and E3 enzymes, respectively, for the activation, conjugation and ligation of ubiquitin (for review, see Ciechanover et al., 2000; Pickart and Eddins, 2004). In S. cerevisiae, a single gene encodes the E1 ubiquitin-activating enzyme. However, 11 different genes encode E2 ubiquitin-conjugating enzymes (Pickart and Eddins, 2004). To identify which E2 enzyme(s) is involved in the turnover of Hac1p, we first tested the stability of the protein in a set of deletion strains that variously lack one (UBC1, UBC2, UBC4, UBC5, UBC7, UBC8) or two (UBC1 and UBC4, UBC2 and UBC4, UBC6 and UBC7) of the nonessential E2 genes that have been demonstrated to play a role in ubiquitin-mediated degradation (Chen et al., 1993). No significant differences were observed in the rate of turnover of Hac1p in these strains compared with the parental W303 strain (data not shown). We then analyzed the stability of Hac1p in MT670 cells, which as noted above have a temperature-sensitive defect in the essential UBC3/CDC34 gene. As shown in Figure 4A, and as anticipated from our data with the PEST–Ura3/HA fusion protein (Figure 3D), the cdc34ts defect caused significant stabilization of Hac1p.
Figure 4.
Proteolytic turnover mediated by the Hac1p degron requires components of the SCFCdc4 pathway. (A) Hac1p protein is stabilized in MT670 cells temperature sensitive for the E2 ubiquitin-conjugating enzyme Ubc3 (Cdc34p). Synthesis shutoff assays on MT670 and parental W303 cells were performed as described in the legend to Figure 1A. (B) The Cdc53 cullin and Skp1 core components of the SCFCdc4 E3 ubiquitin ligase complex are required for rapid turnover of Hac1p. Synthesis shutoff assays on MT871 (cdc53ts), Y552 (skp1-11ts), and Y554 (skp1-12ts) cells and their respective parental W303 and Y80 strains were performed as described in the legend to Figure 1A. (C) The Cdc4 F-box protein is required for rapid turnover of Hac1p. Synthesis shutoff assays on MT668 (cdc4ts) and PY283 (met30ts) and their respective parental W303 and PY1 strains were performed as described in the legend to Figure 1A. Assays on deletion strain grr1Δ and its BY4742 parent strain were performed as described in the legend to Figure 2B.
Hac1p Is Stabilized in Yeast Mutants Defective in Components of the SCFCdc4 E3 Complex
E3 enzymes or enzyme-complexes bind E2 enzymes and provide the specificity of ubiquitin-dependent proteolysis by recognizing particular substrates through dedicated interaction domains (for review, see Ciechanover et al., 2000; Weissman, 2001; Pickart and Eddins, 2004; Cardozo and Pagano, 2004). In S. cerevisiae, the Ubc3/Cdc34 E2 enzyme functions in conjunction with SCF E3 complexes, which contain the RING domain protein Rbx1p in addition to the Cdc53/Cul1 scaffold protein, the Skp1 adaptor protein, and an F-box protein that recruits substrates via a specific protein–protein interaction domain, such as a WD40 or leucine-rich domain (for review, see Deshaies, 1999; Willems et al., 2004). We confirmed the involvement of an SCF complex in the degradation of Hac1p by measuring the stability of endogenous Hac1p in S. cerevisiae cells bearing temperature-sensitive defects in the Cdc53 or Skp1 proteins, which are core components of yeast SCF complexes (Deshaies, 1999; Xu et al., 2003; Willems et al., 2004; Vodermaier, 2004). As shown in Figure 4B, the half-life of Hac1p was increased in cdc53ts and skp1-11ts cells compared with that in the parental cells to an extent very similar to that observed for the cdc34ts mutation. skp1-12ts cells also displayed slower turnover of Hac1p, but the effect was less than that observed with the skp1-11ts allele.
S. cerevisiae cells contain at least 21 F-box–containing proteins (Willems et al., 2004). Of these, four (Cdc4p, Grr1p, Met30p, and Dsg1/Mdm30p) are components of SCF complexes required for degradation of various cell cycle regulatory proteins and transcription factors (for review, see Deshaies, 1999; Willems et al., 2004; Muratani et al., 2005). Proteomic approaches showed that a further three (Ydr131c, Yjl149w, and Ylr097c) form complexes with Cdc53p (Willems et al., 1999). To identify the F-box protein(s) required for rapid degradation of Hac1p, we analyzed the stability of Hac1p in cells having temperature-sensitive mutations in the CDC4 or MET30 genes, or lacking the GRR1, DSG1, Ydr131c, Yjl149w, or HRT3 genes, or lacking both the MET30 and MET4 genes. We also analyzed Hac1p stability in an additional 11 deletion strains (Table 1) lacking genes encoding F-box motif-containing proteins that have not yet been shown to form SCF complexes. Of the 19 mutants examined, only strain MT668 (temperature sensitive for Cdc4p) displayed any difference in the stability of Hac1p compared with its parent cell line (Figure 4C; data not shown). At the nonpermissive temperature, the half-life of Hac1p in cdc4ts cells was increased to ∼5 min, a value similar to that observed with the cdc34ts, cdc53ts, and skp1-11ts mutants. We therefore concluded that rapid degradation of Hac1p involves the SCFCdc4 E3 complex.
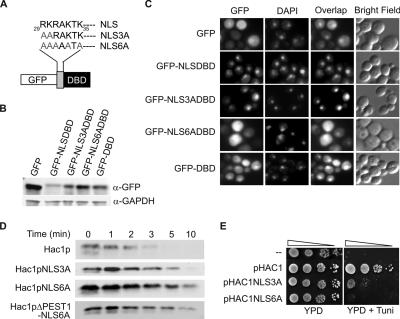
Degradation of Hac1p Correlates with its Nuclear Localization
Consistent with the finding that the cellular localization of Cdc4p is exclusively nuclear (Blondel et al., 2000), the degradation of the SCFCdc4-targeted substrates Far1p and Gcn4p requires their presence in the nucleus (Blondel et al., 2000; Pries et al., 2002). To determine whether rapid turnover of Hac1p also requires its nuclear localization, we first examined the function of a potential nuclear localization sequence (NLS) located at or near the N-terminal end of the bZIP domain of the protein (Figure 5A). This sequence, 29RKRAKTK35, contains a cNLS consensus motif (KR/KxR/K; Fontes et al., 2000). Site-directed mutagenesis using the QuikChange PCR method (Wang and Malcolm, 1999) was used to substitute alanine residues for either the first two or all five basic residues of the NLS to generate the NLS3A and NLS6A mutations (Figure 5A). A ΔNLS mutation that removed all seven residues of the putative cNLS also was constructed. The mutated sequences were incorporated either into the pGFP-NLSDBD vector that encodes a GFP fusion protein containing the NLS and DNA binding domain (DBD) sequences (amino acids 29–65) of Hac1p, or into the pHAC1 or pHAC1ΔPEST vectors that encode the full-length or ΔPEST Hac1 proteins (see Materials and Methods). The DBD sequences were included in the GFP fusion proteins to test the possibility that additional basic sequences within the DNA binding domain contribute to nuclear import.
Figure 5.
Hac1p degradation correlates with nuclear localization. (A) Wild-type and mutant versions of an NLS present near the N terminus of Hac1p. (B) Extracts of W303 cells expressing unfused GFP (empty vector) or fusion proteins containing wild-type or mutant versions of the Hac1 NLSDBD sequence were analyzed by immunoblotting using α-GFP and α-GAPDH antibodies. (C) W303 cells expressing unfused GFP (empty vector) or fusion proteins containing wild-type or mutant versions of the Hac1 NLSDBD sequence were analyzed by fluorescence microscopy. (D) Synthesis shutoff assays were performed as described in Figure 2A on KMY1045 (hac1Δ) cells transformed with pHAC1, pHAC1NLS3A, pHAC1NLS6A, pHAC1ΔPEST, and pHAC1NLS6AΔPEST. (E) Serial 10-fold dilutions of KMY1045 (Δhac1) cells transformed with pHAC1, pHAC1NLS3A, and pHAC1NLS6A were spotted on YPD media alone or YPD media containing 0.5 μg/ml tunicamycin (to test for sensitivity to ER stress).
The parental pGFP-Nfus plasmid and the pGFP-NLSDBD vectors were introduced into W303 yeast cells, and the expression levels of GFP and the various fusion proteins were analyzed by immunoblotting with an anti-GFP antibody (Figure 5B), whereas the intracellular localization of GFP was analyzed by confocal microscopy (Figure 5C). As shown in the left-hand panel of the top row of Figure 5C, cells expressing unfused GFP displayed both cytoplasmic and nuclear fluorescence, with the exception of a dark patch that does not colocalize with the nucleus (shown by DAPI staining in the middle panels), but probably corresponds to the vacuole. When the wild-type NLSDBD sequence was attached to GFP, the fusion protein was efficiently targeted to the nucleus (Figure 5C, second row), confirming that this sequence does indeed contain a functional NLS. In comparison, cells expressing the GFP–NLS3ADBD fusion protein displayed a relatively small increase in cytoplasmic fluorescence (Figure 5C, third row), whereas those expressing the GFP–NLS6ADBD fusion protein displayed a greatly reduced proportion of fluorescence in the nucleus. The residual nuclear fluorescence occurred at a level only slightly higher than that in the surrounding cytoplasm and very similar to that exhibited in cells expressing either unfused GFP (Figure 5C, top row) or the GFP–DBD fusion protein (Figure 5C, bottom row). We therefore concluded that substitution by alanine of the first two basic residues of the RKRAKTK sequence only slightly reduced NLS activity, whereas substitution of all the basic residues in this sequence, or deletion of the whole sequence, completely disrupted its ability to promote nuclear localization of GFP. The basic residues in the remainder of the DBD sequence do not seem to contribute any NLS activity.
We then tested the stability of full-length Hac1 proteins containing the same alanine substitution mutations using synthesis shutoff assays (Figure 5D), which demonstrated that the NLS3A and NLS6A mutants have significantly longer half-lives than the wild-type Hac1 protein, with that of the NLS6A mutant approaching that of the ΔPEST mutant. The degree of stabilization of the NLS mutants correlated with the severity of the nuclear localization defect, supporting the hypothesis that localization of Hac1p in the nucleus is necessary for its SCFCdc4-dependent turnover. The combination of the ΔPEST and NLS6A mutations did not further increase the stability of Hac1p above that seen for the NLS6A mutation alone, consistent with the majority of the protein being localized in the cytoplasm where it would not be available for SCFCdc4-dependent degradation. Finally, we tested the capacity of Hac1 proteins containing the alanine substitution mutations to support the viability of cells under ER stress conditions. Expression of wild-type Hac1p from the pHAC1 vector fully complemented the sensitivity of KMY1045 (Δhac1) cells to the presence of 0.5 μg/ml tunicamycin (compare top two rows of Figure 5E). However, cells expressing the NLS3A and NLS6A mutant proteins displayed considerably reduced viability under ER stress conditions, with the degree of sensitivity to tunicamycin correlating inversely with both the extent of nuclear localization and the stability of the mutant proteins.
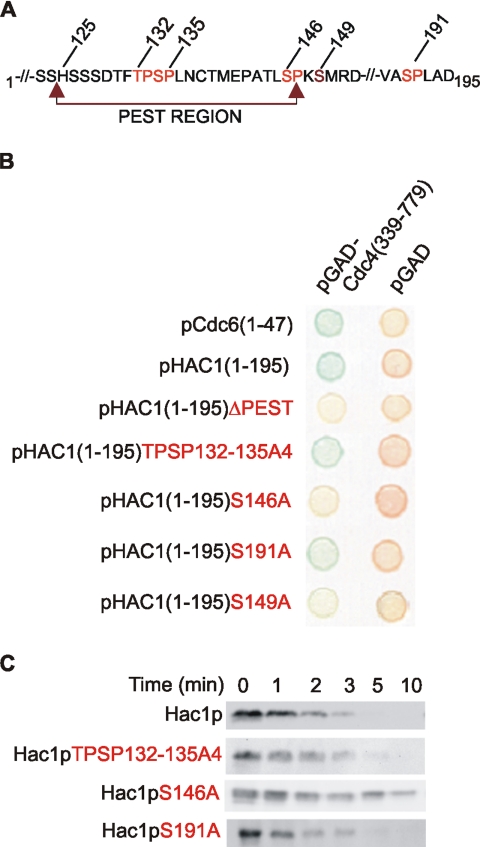
The PEST-dependent Interaction of Hac1p with Cdc4p Requires Ser146 and Ser149
To confirm that Hac1p forms a complex with Cdc4p, we fused the N-terminal 195 amino acids of Hac1p (Figure 6A), which include the PEST degron but not the transactivation domain (residues 221–238; Mori et al., 2000), to the LexA DNA binding domain and compared its ability to interact in a two-hybrid experiment with a fragment of Cdc4p (residues 339-779) that contains WD40 repeats (Fong et al., 1986; Drury et al., 1997). A known binding partner, the N-terminal 47 amino acids of Cdc6p (Drury et al., 1997; Perkins et al., 2001), was used as a positive control. The data shown in Figure 6B (rows 1 and 2) demonstrate that the N-terminal 195 residues of Hac1p, like the N-terminal 47 amino acids of Cdc6p, interacted strongly with the WD40 repeat-containing fragment of Cdc4p, indicating that this region of Hac1p can act as a Cdc4-interaction domain in vivo. Deletion of the Hac1 PEST degron abrogated the interaction (Figure 6B, row 3), supporting our earlier conclusion that the PEST sequence is necessary for Cdc4p-mediated degradation of Hac1p.
Figure 6.
Interaction between Hac1p and Cdc4p is dependent on the presence of the Hac1 PEST sequence, and, in particular, Ser146 and Ser149. (A) Partial polypeptide sequence of Hac1p showing the PEST degron and putative Cdc4 binding sites. (B) W303 cells cotransformed with plasmids pGAD or pGAD-Cdc4(339-779) and plasmids containing LexA (DBD) fusions were assayed for β-galactosidase activity as indicated by development of blue coloration. The column on the left shows cells expressing the GAD–Cdc4(339-779) fusion protein. The column on the right shows cells expressing GAD alone. Cells also expressed LexA DBD fused to various mutant forms of Hac1p (residues 1-195) or Cdc6p (residues 1-47) (as a positive control). (C) Synthesis shutoff assays were performed as described in Figure 2A on KMY1045 (Δhac1) cells transformed with pHAC1 or pHAC1-based vectors encoding the indicated Hac1p mutations.
All known substrates of SCFCdc4 must be phosphorylated before they can be ubiquinated (for review, see Deshaies, 1997; Willems et al., 1999, 2004), and the WD40 repeat domain of Cdc4 binds with high-affinity to a consensus phosphopeptide motif I/L-I/L/P-pT-P-{K/R}4 (where {} indicates disfavored residues) called the Cdc4 Phospho-Degron or CPD (Nash et al., 2001). It is noteworthy that the CPD motif contains the minimal consensus sequence S/T-P for phosphorylation by both cyclin-dependent kinases (CDKs) and mitogen-activated protein (MAP) kinases (Nigg, 1995), and indeed members of these kinase families are required for SCFCdc4-dependent degradation of several different substrates (for review, see Willems et al., 2004). Examination of the sequence of Hac1p revealed four S/T-P motifs, of which three (T132P, S134P, and S146P) lie within the PEST degron and are each embedded in the sequence context of a putative CPD (Figure 3A). Substitution by alanines of residues 132–135, which removes the first two motifs, had no effect on the capacity of LexA-Hac1(1-195) to interact with GAD-Cdc4(339-779) in the two-hybrid assay (Figure 6B, row 4). However, substitution of Ser146 by alanine completely inhibited the interaction (Figure 6B, row 5), indicating that the sequence TLS146PKSMR is a functional CPD, despite the presence of two “disfavored” basic residues in the +2 and +5 positions of the motif. It is noteworthy that this CPD does not lie entirely within the PEST1 sequence defined by the PESTfind algorithm (Figure 2A), but the deletion of residues 125–147 that proved this sequence to contain a functional degron (Figure 2B) removed five of the eight residues of the CPD, including the S146-P phosphorylation site. Importantly, the complete CPD was included in the slightly longer sequence (residues 122–158) that promoted the degradation of the PEST–Ura3/HA fusion protein (Figure 3C). Consistent with Ser191 not lying within a CPD motif, its substitution by alanine had no effect on the Hac1–Cdc4 interaction (Figure 6B, row 6). When the half-lives of full-length Hac1 proteins containing the same alanine substitution mutations were measured using the synthesis shutoff assay and compared with those of the wild-type protein and the PEST deletion mutant, we found that their stabilities showed an inverse correlation with their capacities to interact with Cdc4p (Figure 6C). Thus, the instability of the TPSP132-135A and S191A mutants resembled that of wild-type Hac1p, whereas the stability of the S146A mutant was increased to an extent very similar to that of the PEST deletion mutant, confirming the role of the CPD in rapid turnover of Hac1p and the importance of Ser146 in this process.
Included within the Hac1 CPD is a consensus protein kinase C (PKC) phosphorylation site, S149MR, which had come to our attention previously because osmotic shock conditions that activate Pkc1p, the sole member of the PKC family in S. cerevisiae (Levin et al., 1990), cause an increase in the steady-state level of Hac1p (see below). Substitution of Ser149 by alanine increased the half-life of Hac1p to ∼5 min (Dyke, 2003). Introduction of the S149A mutation into the LexA–Hac1(1-195) fusion protein caused a very significant but not complete inhibition of its interaction with GAD-Cdc4 (Figure 6B, row 7), indicating that phosphorylation of S149 may play an important but not absolutely essential role in the recognition of the CPD by Cdc4p.
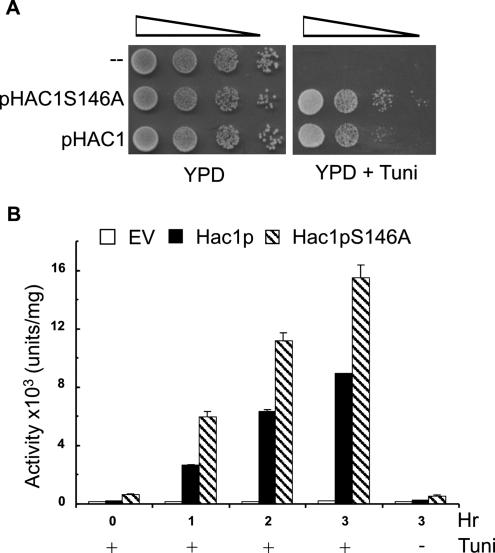
The S146A Mutation in the SCFCdc4 Binding Site Results in Increased Cell Viability and UPRE-dependent Transcription under ER Stress Conditions
Two assays demonstrated the effect of the S146A mutation in improving the function of Hac1p in vivo. First, KMY1045 (Δhac1) cells transformed with a single-copy vector encoding the Hac1pS146A mutant were ∼10-fold less sensitive to ER stress caused by growth in the presence of 1 μg/ml tunicamycin than cells transformed with the same vector encoding wild-type Hac1p (Figure 7A). The magnitude of this increased resistance to ER stress is very similar to that observed with the ΔPEST mutation (Figure 2D). Second, the S146A mutant displayed 1.8- to 2-fold higher capacity than the wild-type protein in inducing the expression of β-galactosidase from the UPRE-CYC1-lacZ reporter construct (Figure 7B). This relatively modest increase is consistent with the finding that the KAR2 gene, the origin of the 22 base pairs UPRE used in the UPRE-CYC1-lacZ reporter construct, is a member of a class of UPR-target genes whose transcription does not increase linearly in proportion to the concentration of Hac1p (Leber et al., 2004). This is probably because Hac1p binds to the UPRE sequences of these genes with high affinity, so that binding is at or near saturation level at the concentration of Hac1p that accumulates under normal ER stress conditions. Other classes of UPR target genes require higher concentrations of Hac1p to achieve maximal levels of transcription of their target proteins (Leber et al., 2004). The 10-fold increase in ER stress resistance observed in the experiments shown in Figures 2D and 7A probably reflects the effect of stabilization of Hac1p by the ΔPEST and S146A mutations on the transcription of target genes belonging to these latter classes.
Figure 7.
S146A mutation in the SCFCdc4 binding site results in increased cell viability and UPRE-dependent transcription under ER stress conditions. (A) Serial 10-fold dilutions of Δhac1 cells transformed with pHAC1 or pHAC1S146A were spotted on YPD media alone or YPD media containing 1.0 μg/ml tunicamycin (to test for sensitivity to ER stress). (B) NCY1810 (Δhac1) cells cotransformed with the pMCZY multicopy UPRE-CYC1-LacZ reporter construct (Mori et al., 1993) and the pRS315 empty vector or pHAC1-based plasmids encoding wild-type Hac1p or the S146A Hac1p mutant were grown overnight at 30°C in SC-Leu/Ura–selective media until the OD600 reached 0.8. Incubation of the cultures was then continued in the presence (+) or absence (−) of 5 μg/ml tunicamycin for the time intervals (0–3 h) indicated. Protein extracts were prepared, and assays of β-galactosidase activity were carried out as described in Materials and Methods. Comparisons of the transcriptional activities (units per milligram of total cell protein) of the three strains were performed in triplicate with at least two independent transformants. The results are presented as mean ± SD.
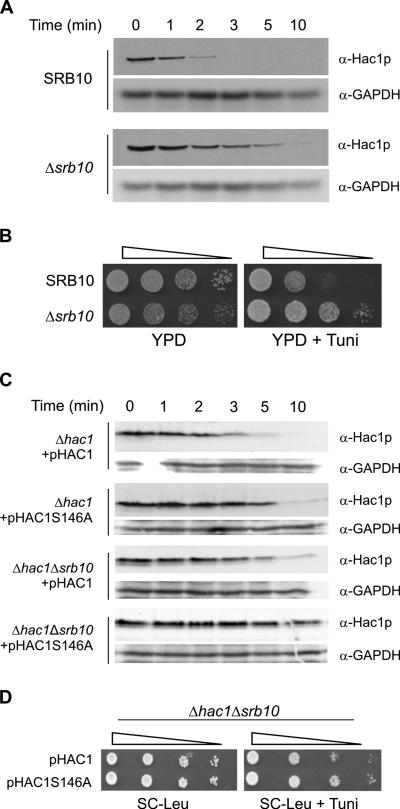
Srb10 Kinase Activity Is Essential for Efficient Degradation of Hac1p
In yeast, Cdc4p-dependent degradation of CDK inhibitors like Sic1p and Far1p and the DNA replication regulator Cdc6p require the activity of the cell cycle CDK Cdc28p (Henchoz et al., 1997: Verma et al., 1997; Elsasser et al., 1999). In contrast, degradation of the transcriptional activator Gcn4 involves its phosphorylation by Srb10 or Pho85 kinases (Meimoun et al., 2000; Chi et al., 2001). We used a Δsrb10 strain to test whether the Srb10 kinase, which together with its activator the Srb11 cyclin is a component of the SRB mediator complex (Myer and Young, 1998), is required for degradation of Hac1p. As shown in Figure 8A, synthesis shutoff assays demonstrated that the half-life of Hac1p was significantly increased in JN5 (Δsrb10) cells compared with that in parental JN1 cells. Δsrb10 cells were also significantly (∼50-fold) less sensitive to ER stress than the parental cells (Figure 8B). To determine whether effects of the S146A mutation and the absence of the Srb10 kinase are additive, we deleted the SRB10 gene from KMY1045 (Δhac1) cells to generate the NCY1811 (Δhac1Δsrb10) strain and then compared the stability of exogenous wild-type and S146A Hac1 proteins in the Δhac1 and Δhac1Δsrb10 cells. Consistent with the earlier experiments (Figures 6C and 8A), each of the single S146A or Δsrb10 mutations increased the stability of the Hac1 protein (Figure 8C, compare the first three sets of panels). The combination of the two mutations resulted in even greater stability (Figure 8C, bottom), indicating that Srb10 kinase-mediated phosphorylation of S146 is not the sole pathway of activation of turnover of Hac1p. This additive effect of the S146A and Δsrb10 mutations on the stability of Hac1p was reflected in Δhac1Δsrb10 cells expressing Hac1pS146A being significantly less sensitive to ER stress than those expressing wild-type Hac1p (Figure 8D).
Figure 8.
Srb10 kinase activity is essential for efficient degradation of Hac1p. (A) Synthesis shutoff assays were performed as described in the legend to Figure 1A on JN5 (Δsrb10) and parental JN1 cells. (B) Serial 10-fold dilutions of Δsrb10 and parental cells were spotted on YPD media alone or YPD media containing 1.0 μg/ml tunicamycin to test for sensitivity to ER stress. (C) Synthesis shutoff assays were performed as described in the legend to Figure 2B on NCY1811 (Δhac1 Δsrb10) and parental KMY1045 (Δhac1) cells transformed with pHAC1 or pHAC1S146A. (D) Serial 10-fold dilutions of Δhac1 Δsrb10 cells transformed with pHAC1 or pHAC1S146A were spotted on SC-Leu media alone or SC-Leu media containing 1.0 μg/ml tunicamycin.
Activation of the Cell Integrity MAP Kinase Pathway Stabilizes Hac1p
In a separate study we recently showed that overexpression of components of the cell integrity (CI) MAP kinase signal transduction pathway, which regulates cell integrity by up-regulating cell wall synthesis in response to weakening or stretching of the cell wall (Gustin et al., 1998; Heinisch et al., 1999), suppresses the transcriptional defect of a reporter gene controlled by a point mutated UPRE element (Helfenbaum, unpublished data). This raised the possibility of a previously unrecognized interaction between the UPR and CI responses. To investigate how activation of the CI pathway by hypotonic shock affects the UPR, KMY2105 sec53ts cells containing an integrated UPRE-lacZ reporter gene were grown to mid-logarithmic phase in medium containing 1 M sorbitol at the permissive temperature of 23°C. sec53ts cells are defective in phosphomannomutase (Kepes and Schekman, 1988) and activate the UPR at the semipermissive temperature of 30°C due to the accumulation of abnormally glycosylated and malfolded secretory precursors in the ER (Normington et al., 1989). The cells were then treated with osmotic diluents as described in the legend to Figure 9A and simultaneously subjected to UPR stress by raising the incubation temperature to 30°C. Control cultures were treated with osmotic diluents but were incubated at 23°C. At various times (0–3 h) after dilution, aliquots of the culture were collected for assay of β-galactosidase activity. Figure 9A shows that the UPR, measured by expression of β-galactosidase from the reporter gene, was accelerated when cells were transferred into hypotonic conditions at the same time as UPR stress was applied. Hypotonic shock does not itself activate the UPR in the absence of ER stress, because no induction of β-galactosidase activity was observed if the cells were maintained at 23°C throughout the experiment. Essentially identical effects of hypotonic shock were obtained using SEC+ cells grown at 30°C with the UPR being induced using tunicamycin in the absence of any temperature shift (data not shown).
Figure 9.
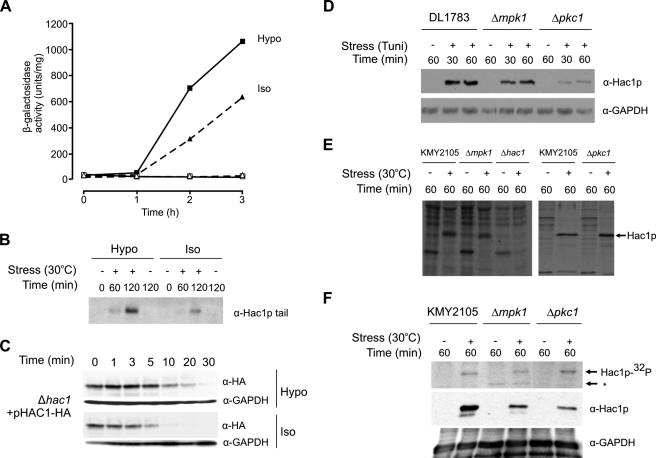
Modulation of the UPR by osmotic shock involves stabilization of Hac1p. (A) KMY2105 cells (sec53ts cells containing a chromosomally integrated UPRE-lacZ reporter; Kawahara et al., 1997) were grown to mid-logarithmic phase at 23°C in 20% YPD medium supplemented with 1 M sorbitol. Aliquots of the culture were then subjected to UPR stress under isotonic (triangles) or hypotonic (squares) shock conditions by dilution with 4 parts of prewarmed (30°C, closed symbols) osmotic diluents (0.4% glucose for hypotonic, 0.4% glucose + 1 M sorbitol for isotonic). Zero-time samples were removed to ice immediately and incubation was continued for 3 h at 30°C. Control cultures (open symbols) were treated with osmotic diluents but were incubated at 23°C. At various times (0–3 h) after dilution, aliquots of the cultures were collected for assay of β-galactosidase activity as described in Materials and Methods. The results (units per milligram of total cell protein) are presented as mean ± SD based on duplicate determinations in two separate experiments. (B) KMY2105 cells transformed with pHAC1B were grown in SC-Leu supplemented with 1 M sorbitol. The cells were treated as described for A and then incubated for 0–120 min before aliquots were collected and the cells snap-frozen in liquid nitrogen before extracts were prepared essentially as described by Cox et al. (1993). Hac1p protein was detected by immunoblotting using anti-Hac1p-tail antibody. (C) KMY1045 cells transformed with pHAC1-HA were grown to OD600 of 0.6 in SC-Leu supplemented with 1 M sorbitol. The culture was then subjected to UPR stress for 60 min by using dithiothreitol (DTT; 6 mM final concentration) to prevent correct disulfide bond formation. Aliquots of the culture were then subjected to isotonic or hypotonic shock conditions by diluting with 4 volumes of prewarmed (30°C) osmotic diluents containing cycloheximide (SC-Leu media for hypotonic and SC-Leu supplemented with 1 M sorbitol for isotonic; final concentration of cycloheximide 1 mg/ml). Zero-time samples were removed and snap-frozen in liquid nitrogen as described in Materials and Methods. Incubation was continued at 30°C with further samples taken at the indicated time points and snap-frozen as described above. Cell extracts were then prepared and analyzed by immunoblotting with anti-HA tag and then anti-GAPDH as described in Materials and Methods. (D) DL1783 (parental), DL455 (Δmpk1), and DL376 (Δpkc1) cells were grown to OD600 of 0.8 at 30°C in selective medium supplemented with 1 M sorbitol. The cultures were then divided in two and the cells incubated for a further 1 h in the presence (+) or absence (−) of tunicamycin. Aliquots were collected at the indicated times and cell extracts were prepared as described in B before Hac1p proteins were detected by immunoblotting using anti-Hac1p antibody. The filters were then stripped and reprobed using anti-GAPDH antibodies. Then, 100 μg of total cell protein was loaded in each lane. (E) KMY2105, LHY100 (Δmpk1), KMY2145 (Δhac1), and LHY102 (Δpkc1) cells were grown at 23°C in selective medium (left) or in selective medium supplemented with 1 M sorbitol (right). Cells were labeled for 5 min with 35S[methionine and cysteine] after induction of UPR stress (30°C for 60 min) or after incubation at the permissive temperature of 23°C. Hac1p immunoprecipitated from cell lysates was subjected to SDS-PAGE and detected by fluorography. (F) KMY2105, LHY100 (Δmpk1), and LHY102 (Δpkc1) cells grown in selective medium supplemented with 1 M sorbitol were labeled with [32P]orthophosphoric acid as described in Materials and Methods before UPR stress was imposed for 60 min by raising the temperature to 30°C. Control cultures were maintained at 23°C. Top, Hac1p was immunoprecipitated from protein extracts prepared as described by Cox and Walter (1996) and separated by SDS-PAGE. The gels were dried and autoradiographed. Hac1p-32P indicates Hac1p identified by autoradiography, whereas the asterisk indicates a nonspecific band not reproducibly observed in other experiments. Middle and bottom, parallel cultures were treated as described above but not radiolabeled. Whole cell lysates (100 μg/lane) were analyzed by immunoblotting, first using anti-Hac1p (middle) and then anti-GAPDH antibodies (bottom).
To determine whether the time course of UPR-induced accumulation of Hac1p was similarly affected by osmotic conditions, cell extracts collected in an experiment similar to that shown in Figure 9A were immunoblotted with an antibody specific for the C terminus of Hac1p, which is encoded by the HAC1 3′ exon and translated only from spliced HAC1 mRNA (Cox and Walter, 1996). Figure 9B shows that Hac1p accumulated faster and to higher levels in hypotonic shock conditions compared with isotonic conditions. To investigate to what extent stabilization of Hac1p might contribute to this increased accumulation of Hac1p, we then used synthesis shutoff assays to analyze the effect of hypotonic shock on the half-life of the protein. As shown in Figure 9C, the half-life of Hac1p was significantly increased under hypotonic shock conditions.
Interestingly, the sequences surrounding Ser146 and Ser149 in Hac1p make these residues potential sites of phosphorylation by the Pkc1p and/or Mpk1p kinases, which are important components of the CI signaling pathway (Levin et al., 1990; Lee and Levin, 1992). However, Pkc1p- or Mpk1p-mediated phosphorylation of these residues should activate (in Ser146) or stimulate (in Ser149) degradation of Hac1p, which is inconsistent both with activation of the CI pathway causing stabilization of Hac1p and with deletion of PKC1 or MPK1 causing significantly decreased accumulation of Hac1p after UPR stress, both under normal osmotic conditions (Figure 9D) and after hypotonic shock (Helfenbaum, unpublished data). This decreased accumulation was not due to decreased synthesis of Hac1p in the deletion strains (Figure 9E). Because deletion of either kinase resulted in increased overall phosphorylation of Hac1p (Figure 9F), we think that Pkc1p and or Mpk1p may inhibit or activate, respectively, other kinases or phosphatases that act upon Hac1p.
DISCUSSION
Many transcription factors are among the most rapidly degraded cellular proteins and their high rates of turnover provide tight coupling of their transcriptional activity to their continued synthesis. Previous studies on the regulation of the UPR have concentrated on the initiation of signaling, which results in the splicing of constitutively expressed HAC1 precursor mRNA by a nonconventional mechanism that releases the blockage of translation of Hac1p (for review, see Kaufman, 1999; Patil and Walter, 2001). Hac1p has been known for some time to be an extremely short-lived transcriptional activator (Chapman and Walter, 1997; Kawahara et al., 1997), but to date the sequence elements in Hac1p and the cellular machinery that facilitate its turnover had not been characterized. In this study, we demonstrated that Hac1p harbors a functional PEST sequence (residues 125–147) that is required for its rapid degradation. Furthermore, we showed that proteolysis of Hac1p by the proteasome involves the E2 ubiquitin-conjugating enzyme Ubc3/Cdc34p and the SCFCdc4 E3 complex. Consistent with the known nuclear localization of the F box protein Cdc4p, rapid degradation of Hac1p requires the presence of a nuclear localization sequence, whose function depends upon basic residues in the sequence 29RKRAKTK35. Two-hybrid analysis demonstrated that the interaction of Hac1p with a WD40 repeat-containing fragment of Cdc4p required the PEST sequence, and in particular Ser146, which is present within a sequence, TLS146PKSMR, that fits the CPD motif (Nash et al., 2001). Ser146 is likely to be a site of phosphorylation by the kinase Srb10, whose activity was shown to be necessary for efficient degradation of Hac1p. Interaction of Hac1p with the Cdc4p fragment was partially inhibited by alanine substitution of S149, which lies just within the CPD. S149 is not present in the context of a CPD motif and is not a substrate for proline-directed kinases such as Srb10. Thus, the reduced interaction with Cdc4 observed with the S149A mutant is not due to the loss of an additional or alternative Cdc4 binding site. It is possible that phosphorylation of S149 by an as yet unknown kinase stimulates formation of the Cdc4 phospho degron by potentiating phosphorylation of S146 by Srb10 (or another kinase).
The regulation of turnover of Hac1p shares many features with that of the transcription factor Gcn4p, which in addition to its involvement as a master regulator of diverse responses to starvation and stress signals (Hinnebusch and Natarajan, 2002), has recently been shown to cooperate with Hac1p in activating UPRE-induced genes (Patil et al., 2004). Like Hac1p, Gcn4p is a bZIP protein whose cellular levels are regulated both by translational control of synthesis (Hinnebusch, 1984) and by degradation (Kornitzer et al., 1994). Like Hac1p, it is a highly unstable protein whose degradation by the proteasome requires Cdc34p and SCFCdc4, to which it is targeted by phosphorylation of one or more S/TP motifs by Srb10 or Pho85 kinases (Meimoun et al., 2000; Chi et al., 2001). Srb10 is implicated in the phosphorylation of Gcn4p during their encounter at promoter regions, whereas Pho85 is a regulator of Gcn4p in response to the availability of amino acids (for review, see Hinnebusch and Natarajan, 2002).
The presence in Hac1p of two potential phosphorylation sites in the CPD provides the opportunity for physiological regulation of its turnover. SCF complexes are known to couple protein kinase signaling pathways to the control of protein abundance. Petroski and Deshaies (2005) have reviewed three different modes of regulation for SCF-dependent proteolysis. The “AND” mode used for degradation of cyclin E by human SCFCdc4 requires phosphorylation by both GSK3 and CDK kinases (Welcker et al., 2003). The “nanoswitch” or “threshold” mode of regulation is exemplified by SCFCdc4-mediated degradation of the yeast Sic1p CDK inhibitor, which requires phosphorylation by Cdc28p of any six of nine potential sites that are present within suboptimal CDP sequences (Nash et al., 2001; Orlicky et al., 2003). Finally, SCFCdc4-mediated degradation of Gcn4 illustrates the “OR” mode of control because it requires CPD phosphorylation by either Srb10 or Pho85 (Meimoun et al., 2000; Chi et al., 2001). Recently, another OR mode, involving a protein having two or more degrons that are regulated by different SCF complexes, has been described for Gal4p (Muratani et al., 2005). In this case, SCFGrr1 facilitates turnover of Gal4p at a relatively slow rate during yeast cell growth in the presence of a noninducing carbon source, whereas SCFDsg1/Mdm30 mediates rapid degradation of a differentially phosphorylated form of Gal4p during growth in the presence of galactose (Muratani et al., 2005). Regulation of Hac1p degradation does not involve the AND or nanoswitch modes, because it contains only a single CPD that does not require simultaneous activation by more than one kinase, although as discussed above, phosphorylation of a second nearby site may stimulate the interaction of the CPD with Cdc4p. We have not ruled out the possibility that the classical “OR” mode of regulation for SCF-dependent proteolysis may apply to Hac1p, with one or more additional kinases capable of targeting Ser146 to activate SCFCdc4- mediated degradation of Hac1p.
The PEST/CPD degron that we have identified in Hac1p does not fully account for the extremely rapid rate of degradation of Hac1p. Point mutation or deletion of this degron, defects in components of the SCFCdc4 complex and deletion of the Srb10 kinase all cause significant stabilization of Hac1p. However, even after the increase in half-life from ∼1 to 4–5 min, Hac1p is still a rapidly turned over protein. In a separate study, we have identified a second degron in Hac1p that is colocalized with the transactivation domain at the C terminus of the protein (Pal, Dyke, and Gething, unpublished data).
There are many examples of unstable transcription factors in which there is overlap between TAD and degron sequences (Salghetti et al., 2000; Muratani and Tansey, 2003). Such overlap is thought to contribute to the coupling of the activity of transcription factors with their proteasome-mediated destruction and various models have been proposed to describe how activator degradation might limit or stimulate transcription (Salghetti et al., 2000; Lipford and Deshaies, 2003; Muratani and Tansey, 2003; Lipford et al., 2005). Thus, degradation might function to limit how long any single activator remains bound at a promoter, facilitating reprogramming of transcriptional patterns, or it might be required to remove spent activators that would otherwise block continued initiation of transcription. Alternatively, degradation may be linked to prior ubiquitin-mediated “licencing” of transcription factor activity. In Hac1p, not only is the C-terminal degron colocalized with the TAD, but the function of the PEST degron/CPD, which is distant from the TAD in the primary sequence of the protein, may also be dependent on transcription because it is inhibited in cell mutants lacking Srb10 kinase, a component of the SRB/mediator module of the RNA polymerase II holoenzyme.
Modulation of the rate of degradation of Hac1p would be of little consequence if it did not affect the pattern of transcription of UPRE-regulated genes. Transactivation of a reporter construct that is under the control of the UPRE derived from the KAR2 (BiP) gene (Mori et al., 1993) was increased approximately twofold by blockage of SCFCdc4-mediated degradation of Hac1p. Transactivation of other UPR-controlled genes is likely to be up-regulated to much greater extents, because KAR2 belongs to a subclass of UPRE-controlled genes whose transcription seems to be maximally activated at low concentrations of Hac1p (Leber et al., 2004). As noted by Leber et al. (2004), target genes of this class likely have promoters that are saturated by the lower amount of Hac1p and thus reach full activation more readily. For a variety of other UPR targets, induction continues to increase as Hac1p levels increase; lower concentrations of Hac1p are inadequate for full stimulation of these genes, which may have lower affinity for Hac1p. Consistent with this, cell viability under ER stress conditions was increased up to 50-fold by point mutation or deletion of the Hac1p phospho degron, or as the consequence of defects in components of the SCFCdc4-dependent proteasomal degradation pathway. Leber et al. (2004) also commented that because genes respond differentially to Hac1p levels, regulation of HAC1 mRNA abundance can be used as a gene-specific gain control for target activation. We propose that regulation of Hac1p stability could serve a similar purpose, especially if other transcription factors, such as Gcn4p (Patil et al., 2004) or UMF (Leber et al., 2004), that collaborate with Hac1p to up-regulate the UPR are simultaneously stabilized. That the same SCFCdc4 machinery is used for turnover of Hac1p and Gcn4p provides the opportunity for their coregulation via physiological inputs that target this machinery.
We recently identified a physiological condition that could regulate the UPR via stabilization of Hac1p by showing that overexpression of components of the cell integrity (CI) Pkc1p–MAP kinase signaling pathway suppress the transcriptional defect of a reporter gene controlled by a point mutated UPRE element (Helfenbaum, unpublished data). Activation of the CI pathway, which responds to weakening or stretching of the cell wall and functions to activate the synthesis and delivery of cell wall components (Gustin et al., 1998; Heinisch et al., 1999), increases both the rate of ER stress-mediated induction of HAC1 mRNA splicing (Helfenbaum, unpublished data) and the stability of Hac1p, suggesting that the multicopy suppression occurred because the increased level of accumulation of Hac1p compensated for its reduced affinity for the point mutated UPRE element. Analysis of the roles played by the CI pathway kinases indicated that Pkc1p was necessary for acceleration of HAC1 mRNA splicing under hypotonic shock conditions that activate CI signaling (Helfenbaum, unpublished data), whereas both Pkc1p and Mpk1p were required for stabilization of Hac1p even under normal (isotonic) conditions. Although it is intriguing that the sequence surrounding Ser146 in Hac1p provides phosphorylation motifs for both Pkc1p and Mpk1p, our data are not compatible with one or both of these kinases playing a direct role in regulating degradation of Hac1p by targeting this residue. Phosphorylation of Ser146 should activate SCF-mediated degradation of Hac1p; however, CI pathway activation of Pkc1p and Mpk1p stabilizes Hac1p and accumulation of Hac1p is significantly decreased in Δpkc1 or Δmpk1 cells. Furthermore, the overall level of phosphorylation of Hac1p is increased, rather than decreased, in Δpkc1 and Δmpk1 cells, suggesting that a complex interplay between kinases and phosphatases is likely to be involved in the regulation of phosphorylation of Hac1p. In the specific context of regulation of SCFCdc4-dependent degradation of Hac1p, it is possible that the CI kinases act indirectly by inhibiting other kinase(s), such as Srb10, that phosphorylate Ser146 to target Hac1p for destruction, or by activating a phosphatase that reverses phosphorylation at Ser146.
Supplementary Material
ACKNOWLEDGMENTS
We thank Mark Hochstrasser and Antony Cooper for yeast E2 mutant strains; Bruce Futcher, Wade Harper, and Peter Kaiser for yeast E3 mutant strains; John Diffley for the yeast strain and plasmids used for yeast two-hybrid analysis; David Levin for CI kinase deletion strains; and Trevor Lithgow, Kazu Mori, Peter Walter, and Pam Silver for gifts of antibodies. We also thank Masafumi Muratani and Simone Salghetti for technical advice and helpful discussions. This work was supported by grants to M.-J.G. from the Australian Research Council and to W.P.T. from the National Institutes of Health (GM-067728).
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E06-04-0304) on November 15, 2006.
 The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
REFERENCES
- Bai C., Sen P., Hofmann K., Ma L., Goebl M., Harper J. W., Elledge S. J. SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-Box. Cell. 1999;86:263–274. doi: 10.1016/s0092-8674(00)80098-7. [DOI] [PubMed] [Google Scholar]
- Blondel M., Galan J. M., Chi Y., Lafourcade C., Longaretti C., Deshaies R. J., Peter M. Nuclear-specific degradation of Far1 is controlled by the localization of the F-box protein cdc4. EMBO J. 2000;19:6085–6097. doi: 10.1093/emboj/19.22.6085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardozo T., Pagano M. The SCF ubiquitin ligase: insights into a molecular machine. Nat. Rev. Mol. Cell Biol. 2004;6:739–751. doi: 10.1038/nrm1471. [DOI] [PubMed] [Google Scholar]
- Carrano A. C., Eytan E., Hershko A., Pagano M. SKP2 is required for ubiquitin-mediated degradation of the CDK inhibitor p27. Nat. Cell Biol. 1999;1:193–199. doi: 10.1038/12013. [DOI] [PubMed] [Google Scholar]
- Chapman R. E., Walter P. Translational attenuation mediated by an mRNA intron. Curr. Biol. 1997;7:850–859. doi: 10.1016/s0960-9822(06)00373-3. [DOI] [PubMed] [Google Scholar]
- Chen P., Johnson P., Sommer T., Jentsch S., Hochstrasser M. Multiple ubiquitin-conjugating enzymes participate in the in vivo degradation of the yeast MAT-alpha2 repressor. Cell. 1993;74:357–369. doi: 10.1016/0092-8674(93)90426-q. [DOI] [PubMed] [Google Scholar]
- Chi Y., Huddleston M. J., Zhang X., Young R. A., Annan R. S., Carr S. A., Deshaies R. J. Negative regulation of Gcn4 and Msn2 transcription factors by Srb10 cyclin-dependent kinase. Genes Dev. 2001;15:1078–1092. doi: 10.1101/gad.867501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciechanover A. The ubiquitin-proteasome pathway: on protein death and cell life. EMBO J. 1998;15:7151–7160. doi: 10.1093/emboj/17.24.7151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciechanover A., Orian A., Schwartz A. L. Ubiquitin-mediated proteolysis: biological regulation via destruction. Bioessays. 2000;22:442–451. doi: 10.1002/(SICI)1521-1878(200005)22:5<442::AID-BIES6>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- Cox J. S., Chapman R. E., Walter P. The unfolded protein response coordinates the production of endoplasmic reticulum protein and endoplasmic reticulum membrane. Mol. Biol. Cell. 1997;8:1805–1814. doi: 10.1091/mbc.8.9.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J. S., Shamu C. E., Walter P. Transcriptional induction of genes encoding endoplasmic reticulum resident proteins requires a transmembrane protein kinase. Cell. 1993;73:1197–1206. doi: 10.1016/0092-8674(93)90648-a. [DOI] [PubMed] [Google Scholar]
- Cox J. S., Walter P. A novel mechanism for regulating activity of a transcription factor that controls the unfolded protein response. Cell. 1996;87:391–404. doi: 10.1016/s0092-8674(00)81360-4. [DOI] [PubMed] [Google Scholar]
- Deshaies R. J. Phosphorylation and proteolysis: partners in the regulation of cell division in budding yeast. Curr. Opin. Genet. Dev. 1997;7:7–16. doi: 10.1016/s0959-437x(97)80103-7. [DOI] [PubMed] [Google Scholar]
- Deshaies R. J. SCF and Cullin/Ring H2-based ubiquitin ligases. Annu. Rev. Cell Dev. Biol. 1999;15:435–467. doi: 10.1146/annurev.cellbio.15.1.435. [DOI] [PubMed] [Google Scholar]
- Drury L. S., Perkins G., Diffley J. F. The Cdc4/34/53 pathway targets Cdc6p for proteolysis in budding yeast. EMBO J. 1997;16:5966–5976. doi: 10.1093/emboj/16.19.5966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyke C. L. M.Sc. Thesis. Melbourne, Australia: The University of Melbourne; 2003. MAP kinase pathway modulation of the unfolded protein response in Saccharomyces cerevisiae. [Google Scholar]
- Elbein A. D. Inhibitors of the biosynthesis and processing of N-linked oligosaccharide chains. Annu. Rev. Biochem. 1987;56:497–534. doi: 10.1146/annurev.bi.56.070187.002433. [DOI] [PubMed] [Google Scholar]
- Ellgaard L., Helenius A. Quality control in the endoplasmic reticulum. Nat. Rev. Mol. Cell Biol. 2003;3:181–191. doi: 10.1038/nrm1052. [DOI] [PubMed] [Google Scholar]
- Elsasser S., Chi Y., Yang P., Campbell J. L. Phosphorylation controls timing of Cdc6p destruction: a biochemical analysis. Mol. Biol. Cell. 1999;10:3263–3677. doi: 10.1091/mbc.10.10.3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzusoff A., Rothblatt J., Schekman R. Analysis of polypeptide transit through yeast secretory pathway. Methods Enzymol. 1991;194:662–674. doi: 10.1016/0076-6879(91)94048-h. [DOI] [PubMed] [Google Scholar]
- Fong H. K., Hurley J. B., Hopkins R. S., Miake-Lye R., Johnson M. S., Doolittle R. F., Simon M. I. Repetitive segmental structure of the transducin beta subunit: homology with the CDC4 gene and identification of related mRNAs. Proc. Natl. Acad. Sci. USA. 1986;83:2162–2166. doi: 10.1073/pnas.83.7.2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontes M. R., The T., Kobe B. Structural basis of recognition of monopartite and bipartite nuclear localization sequences by mammalian importin-alpha. J. Mol. Biol. 2000;297:1183–1194. doi: 10.1006/jmbi.2000.3642. [DOI] [PubMed] [Google Scholar]
- Gething M. J., McCammon K., Sambrook J. Expression of wild-type and mutant forms of influenza hemagglutinin: the role of folding in intracellular transport. Cell. 1986;46:939–950. doi: 10.1016/0092-8674(86)90076-0. [DOI] [PubMed] [Google Scholar]
- Ghislain M., Udvardy A., Mann C. S. cerevisiae 26S protease mutants arrest cell division in G2/metaphase. Nature. 1993;366:358–362. doi: 10.1038/366358a0. [DOI] [PubMed] [Google Scholar]
- Gilon T., Chomsky O., Kulka R. G. Degradation signals for ubiquitin system proteolysis in Saccharomyces cerevisiae. EMBO J. 1998;17:2759–2766. doi: 10.1093/emboj/17.10.2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guldener U., Heck S., Fielder T., Beinhauer J., Hegemann J. H. A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res. 1996;24:2519–2524. doi: 10.1093/nar/24.13.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustin M. C., Albertyn J., Alexander M., Davenport K. MAP kinase pathways in the yeast Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 1998;62:1264–1300. doi: 10.1128/mmbr.62.4.1264-1300.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinisch J. J., Lorberg A., Schmitz H.-P., Jacoby J. J. The protein kinase C-mediated MAP kinase pathway involved in the maintenance of cellular integrity in Saccharomyces cerevisiae. Mol. Microbiol. 1999;32:671–680. doi: 10.1046/j.1365-2958.1999.01375.x. [DOI] [PubMed] [Google Scholar]
- Henchoz S., Chi Y., Catarin B., Herskowitz I., Deshaies R. J., Peter M. Phosphorylation- and ubiquitin-dependent degradation of the cyclin-dependent kinase inhibitor Far1p in budding yeast. Genes Dev. 1997;11:3046–3060. doi: 10.1101/gad.11.22.3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershko A., Ciechanover A. The ubiquitin system. Annu. Rev. Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- Hinnebusch A. G. Evidence for translational regulation of the activator of general amino acid control in yeast. Proc. Natl. Acad. Sci. USA. 1984;81:6442–6446. doi: 10.1073/pnas.81.20.6442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch A. G., Natarajan K. Gcn4p, a master regulator of gene expression, is controlled at multiple levels by diverse signals of starvation and stress. Eukaryot. Cell. 2002;1:22–32. doi: 10.1128/EC.01.1.22-32.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollenberg S. M., Sternglanz R., Cheng P. F., Weintraub H. Identification of a new family of tissue-specific basic helix-loop-helix proteins with a two-hybrid system. Mol. Cell. Biol. 1995;7:3813–3822. doi: 10.1128/mcb.15.7.3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser P., Flick K., Wittenberg C., Reed S. I. Regulation of transcription by ubiquitination without proteolysis: Cdc34/SCF(Met30)-mediated inactivation of the transcription factor Met4. Cell. 2000;102:303–314. doi: 10.1016/s0092-8674(00)00036-2. [DOI] [PubMed] [Google Scholar]
- Kaiser C., Michaelis S., Mitchell A. Methods in Yeast Genetics: A Cold Spring Harbor Laboratory Course Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1994. [Google Scholar]
- Kaufman R. J. Stress signaling from the lumen of the endoplasmic reticulum: coordination of gene transcriptional and translational controls. Genes Dev. 1999;13:1211–1233. doi: 10.1101/gad.13.10.1211. [DOI] [PubMed] [Google Scholar]
- Kawahara T., Yanagi H., Yura T., Mori K. Endoplasmic reticulum stress-induced mRNA splicing permits synthesis of transcription factor Hac1p/Ern4p that activates the unfolded protein response. Mol. Biol. Cell. 1997;8:1845–1862. doi: 10.1091/mbc.8.10.1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kepes F., Schekman R. The yeast sec53 gene encodes phosphomannomutase. J. Biol. Chem. 1988;263:9155–9161. [PubMed] [Google Scholar]
- Kornitzer D., Raboy B., Kulka R. G., Fink G. R. Regulated degradation of the transcription factor Gcn4. EMBO J. 1994;13:6021–6030. doi: 10.1002/j.1460-2075.1994.tb06948.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozutsumi Y., Segal M., Normington K., Gething M. J., Sambrook J. The presence of malfolded proteins in the endoplasmic reticulum signals the induction of glucose-regulated proteins. Nature. 1988;332:462–464. doi: 10.1038/332462a0. [DOI] [PubMed] [Google Scholar]
- Laney J. D., Hochstrasser M. Substrate targeting in the ubiquitin system. Cell. 1999;94:427–430. doi: 10.1016/s0092-8674(00)80752-7. [DOI] [PubMed] [Google Scholar]
- Leber J. H., Bernales S., Walter P. IRE1-independent gain control of the unfolded protein response. PLoS Biol. 2004;2:E235. doi: 10.1371/journal.pbio.0020235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K. S., Levin D. E. Dominant mutations in a gene encoding a putative protein kinase (BCK1) bypass the requirement for a Saccharomyces cerevisiae protein kinase C homolog. Mol. Cell Biol. 1992;12:172–182. doi: 10.1128/mcb.12.1.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K. S., Irei K., Gotoh Y., Watanabe Y., Araki H., Nishida E., Matsumoto K., Levin E. D. A yeast mitogen-activated protein kinase homolog (Mpk1p) mediates signalling by protein kinase C. Mol. Cell Biol. 1993;13:3067–3075. doi: 10.1128/mcb.13.5.3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin D. E., Bartlett-Heubusch E. Mutants in the S. cerevisiae PKC1 gene display a cell cycle-specific osmotic stability defect. J. Cell Biol. 1992;116:1221–1229. doi: 10.1083/jcb.116.5.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin D. E., Fields F. O., Kunisawa R., Bishop J. M., Thorner J. A candidate protein kinase C gene, PKC1, is required for the S. cerevisiae cell cycle. Cell. 1990;62:213–224. doi: 10.1016/0092-8674(90)90360-q. [DOI] [PubMed] [Google Scholar]
- Lipford J. R., Deshaies R. J. Diverse roles for ubiquitin-dependent proteolysis in transcriptional activation. Nat. Cell Biol. 2003;5:845–850. doi: 10.1038/ncb1003-845. [DOI] [PubMed] [Google Scholar]
- Lipford J. R., Smith G. T., Chi Y., Deshaies R. J. A putative stimulatory role for activator turnover in gene expression. Nature. 2005;438:113–116. doi: 10.1038/nature04098. [DOI] [PubMed] [Google Scholar]
- Longtine M. S., McKenzie A., III, Demarini D. J., Shah N. G., Wach A., Brachat A., et al. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Ma Y., Hendershot L. M. The unfolding tale of the unfolded protein response. Cell. 2001;107:827–830. doi: 10.1016/s0092-8674(01)00623-7. [DOI] [PubMed] [Google Scholar]
- Meimoun A., Holtzman T., Weissman Z., McBride H. J., Stillman D. J., Fink G. R., Kornitzer D. Degradation of the transcription factor Gcn4 requires the kinase Pho85 and the SCF(CDC4) ubiquitin-ligase complex. Mol. Biol. Cell. 2000;11:915–927. doi: 10.1091/mbc.11.3.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori K., Sant A., Kohno K., Normington K., Gething M. J., Sambrook J. F. A 22 bp cis-acting element is necessary and sufficient for the induction of the yeast KAR2 (BiP) gene by unfolded proteins. EMBO J. 1992;11:2583–2593. doi: 10.1002/j.1460-2075.1992.tb05323.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori K., Kawahara T., Yoshida H., Yanagi H., Yura T. Signalling from endoplasmic reticulum to nucleus: transcription factor with a basic-leucine zipper motif is required for the unfolded protein-response pathway. Genes Cells. 1996;1:803–817. doi: 10.1046/j.1365-2443.1996.d01-274.x. [DOI] [PubMed] [Google Scholar]
- Mori K., Ma W., Gething M. J., Sambrook J. A transmembrane protein with a cdc2+/CDC28-related kinase activity is required for signaling from the ER to the nucleus. Cell. 1993;74:743–756. doi: 10.1016/0092-8674(93)90521-q. [DOI] [PubMed] [Google Scholar]
- Mori K., Ogawa N., Kawahara T., Yanagi H., Yura T. Palindrome with spacer of one nucleotide is characteristic of the cis-acting unfolded protein response element in Saccharomyces cerevisiae. J. Biol. Chem. 1998;273:9912–9920. doi: 10.1074/jbc.273.16.9912. [DOI] [PubMed] [Google Scholar]
- Mori K., Ogawa N., Kawahara T., Yanagi H., Yura T. mRNA splicing-mediated C-terminal replacement of transcription factor Hac1p is required for efficient activation of the unfolded protein response. Proc. Natl. Acad. Sci. USA. 2000;97:4660–4665. doi: 10.1073/pnas.050010197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muratani M., Kung C., Shokat K. M., Tansey W. P. The F Box Protein Dsg1/Mdm30 is a transcriptional coactivator that stimulates Gal4 turnover and cotranscriptional mRNA processing. Cell. 2005;120:887–899. doi: 10.1016/j.cell.2004.12.025. [DOI] [PubMed] [Google Scholar]
- Muratani M., Tansey W. P. How the ubiquitin-proteasome system controls transcription. Nat. Rev. Mol. Cell Biol. 2003;4:192–201. doi: 10.1038/nrm1049. [DOI] [PubMed] [Google Scholar]
- Myer V. E., Young R. A. RNA polymerase II holoenzymes and subcomplexes. J. Biol. Chem. 1998;273:27757–27760. doi: 10.1074/jbc.273.43.27757. [DOI] [PubMed] [Google Scholar]
- Nash P., Tang X., Orlicky S., Chen Q., Gertler F. B., Mendenhall M. D., Sicheri F., Pawson T., Tyers M. Multisite phosphorylation of a CDK inhibitor sets a threshold for the onset of DNA replication. Nature. 2001;414:514–521. doi: 10.1038/35107009. [DOI] [PubMed] [Google Scholar]
- Nigg E. A. Cyclin-dependent protein kinases: key regulators of the eukaryotic cell cycle. Bioessays. 1995;17:471–480. doi: 10.1002/bies.950170603. [DOI] [PubMed] [Google Scholar]
- Normington K., Kohno K., Kozutsumi Y., Gething M. J., Sambrook J. S. cerevisiae encodes an essential protein homologous in sequence and function to mammalian BiP. Cell. 1989;57:1223–1236. doi: 10.1016/0092-8674(89)90059-7. [DOI] [PubMed] [Google Scholar]
- Orlicky S., Tang X., Willems A., Tyers M., Sicheri F. Structural basis for phosphodependent substrate selection and orientation by the SCFCdc4 ubiquitin ligase. Cell. 2003;112:243–256. doi: 10.1016/s0092-8674(03)00034-5. [DOI] [PubMed] [Google Scholar]
- Patil C. K., Li H., Walter P. Gcn4p and novel upstream activating sequences regulate targets of the unfolded protein response. PLoS Biol. 2004;2:E246. doi: 10.1371/journal.pbio.0020246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil C. K., Walter P. Intracellular signaling from the endoplasmic reticulum to the nucleus: the unfolded protein response in yeast and mammals. Curr. Opin. Cell Biol. 2001;13:349–355. doi: 10.1016/s0955-0674(00)00219-2. [DOI] [PubMed] [Google Scholar]
- Patton E. E., Willems A. R., Sa D., Kuras L., Thomas D., Craig K. L., Tyers M. Cdc53 is a scaffold protein for multiple Cdc34/Skp1/F-box protein complexes that regulate cell division and methionine biosynthesis in yeast. Genes Dev. 1998;12:692–705. doi: 10.1101/gad.12.5.692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins G., Drury L. S., Diffley J.F.X. Separate SCFCdc4 recognition elements target Cdc6 for proteolysis in S phase and mitosis. EMBO J. 2001;10:231–240. doi: 10.1093/emboj/20.17.4836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petroski M. D., Deshaies R. J. Context of multiubiquitin chain attachment influences the rate of Sic1 degradation. Mol. Cell. 2003;11:1435–1444. doi: 10.1016/s1097-2765(03)00221-1. [DOI] [PubMed] [Google Scholar]
- Petroski M. D., Deshaies R. J. Function and regulation of cullin-ring ubiquitin ligases. Nature Rev. Mol. Cell Biol. 2005;6:9–22. doi: 10.1038/nrm1547. [DOI] [PubMed] [Google Scholar]
- Pickart C. M., Eddins M. J. Ubiquitin: structures, functions, mechanisms. Biochim. Biophys. Acta. 2004;1695:55–72. doi: 10.1016/j.bbamcr.2004.09.019. [DOI] [PubMed] [Google Scholar]
- Pries R., Bomeke K., Irniger S., Grundmann O., Braus G. H. Amino acid-dependent Gcn4p stability regulation occurs exclusively in the yeast nucleus. Eukaryot. Cell. 2002;1:663–672. doi: 10.1128/EC.1.5.663-672.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rechsteiner M., Rogers S. W. PEST sequences and regulation by proteolysis. Trends Biochem. Sci. 1996;21:267–271. [PubMed] [Google Scholar]
- Rogers S., Wells R., Rechsteiner M. Amino acid sequences common to rapidly degraded proteins: the PEST hypothesis. Science. 1986;234:364–368. doi: 10.1126/science.2876518. [DOI] [PubMed] [Google Scholar]
- Rothstein R. Targeting, disruption, replacement, and allele rescue: integrative DNA transformation in yeast. Methods Enzymol. 1991;194:281–301. doi: 10.1016/0076-6879(91)94022-5. [DOI] [PubMed] [Google Scholar]
- Ruegsegger U., Leber J. H., Walter P. Block of HAC1 mRNA translation by long-range base pairing is released by cytoplasmic splicing upon induction of the unfolded protein response. Cell. 2001;107:103–114. doi: 10.1016/s0092-8674(01)00505-0. [DOI] [PubMed] [Google Scholar]
- Salghetti S. E., Muratani M., Wijnen H., Futcher B., Tansey W. T. Functional overlap of sequences that activate transcription and signal ubiquitin-mediated proteolysis. Proc. Natl. Acad. Sci. USA. 2000;97:3118–3123. doi: 10.1073/pnas.050007597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J., Fritsch E. F., Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Schröder M., Kaufman R. J. ER stress and the unfolded protein response. Mutat. Res. 2005;569:29–63. doi: 10.1016/j.mrfmmm.2004.06.056. [DOI] [PubMed] [Google Scholar]
- Shamu C. E., Walter P. Oligomerization and phosphorylation of the Ire1p kinase during intracellular signaling from the endoplasmic reticulum to the nucleus. EMBO J. 1996;15:3028–3039. [PMC free article] [PubMed] [Google Scholar]
- Sidrauski C., Cox J. S., Walter P. tRNA ligase is required for regulated mRNA splicing in the unfolded protein response. Cell. 1996;87:405–413. doi: 10.1016/s0092-8674(00)81361-6. [DOI] [PubMed] [Google Scholar]
- Sidrauski C., Walter P. The transmembrane kinase Ire1p is a site-specific endonuclease that initiates mRNA splicing in the unfolded protein response. Cell. 1997;90:1031–1039. doi: 10.1016/s0092-8674(00)80369-4. [DOI] [PubMed] [Google Scholar]
- Sikorski R. S., Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thrower J. S., Hoffman L., Rechsteiner M., Pickart C. M. Recognition of the polyubiquitin proteolytic signal. EMBO J. 2000;19:94–102. doi: 10.1093/emboj/19.1.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travers K. J., Patil C. K., Wodicka L., Lockhart D. J., Weissman J. S., Walter P. Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell. 2000;101:249–258. doi: 10.1016/s0092-8674(00)80835-1. [DOI] [PubMed] [Google Scholar]
- van Anken E., Braakman I. Versatility of the endoplasmic reticulum protein folding factory. Crit. Rev. Biochem. Mol. Biol. 2005;40:191–228. doi: 10.1080/10409230591008161. [DOI] [PubMed] [Google Scholar]
- Varshavsky A. The ubiquitin system. Trends Biochem. Sci. 1997;22:383–387. doi: 10.1016/s0968-0004(97)01122-5. [DOI] [PubMed] [Google Scholar]
- Verma R., Feldman R. M., Deshaies R. J. SIC1 is ubiquitinated in vitro by a pathway that requires CDC4, CDC34, and cyclin/CDK activities. Mol. Biol. Cell. 1997;8:1427–1437. doi: 10.1091/mbc.8.8.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vodermaier H. C. APC/C and SCF: controlling each other and the cell cycle. Curr. Biol. 2004;14:R787–R796. doi: 10.1016/j.cub.2004.09.020. [DOI] [PubMed] [Google Scholar]
- Wang W., Malcolm B. A. Two-stage PCR protocol allowing introduction of multiple mutations, deletions and insertions using QuikChange site-directed mutagenesis Biotechniques. 1999;26:680–682. doi: 10.2144/99264st03. [DOI] [PubMed] [Google Scholar]
- Weissman A. M. Themes and variations on ubiquitylation. EMBO J. 2001;19:169–178. doi: 10.1038/35056563. [DOI] [PubMed] [Google Scholar]
- Welihinda A. A., Kaufman R. J. The unfolded protein response pathway in Saccharomyces cerevisiae. Oligomerization and trans-phosphorylation of Ire1p (Ern1p) are required for kinase activation. J. Biol. Chem. 1996;271:18181–18187. doi: 10.1074/jbc.271.30.18181. [DOI] [PubMed] [Google Scholar]
- Welcker M., Singer J., Loeb K. R., Grim J., Bloecher A., Gurien-West M., Clurman B. E., Roberts J. M. Multisite phosphorylation by Cdk2 and GSK3 controls cyclin E degradation. Mol Cell. 2003;12:381–392. doi: 10.1016/s1097-2765(03)00287-9. [DOI] [PubMed] [Google Scholar]
- Willems A. R., Goh T., Taylor L., Chernushevich I., Shevchenko A., Tyers M. SCF ubiquitin protein ligases and phosphorylation-dependent proteolysis. Phil Trans. R. Soc. Lond. B. 1999;354:1533–1550. doi: 10.1098/rstb.1999.0497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willems A. R., Schwab M., Tyers M. A hitchhiker's guide to the cullin ubiquitin ligases: SCF and its kin. Biochim. Biophys. Acta. 2004;1695:133–170. doi: 10.1016/j.bbamcr.2004.09.027. [DOI] [PubMed] [Google Scholar]
- Xie K., Lambie E. J., Snyder M. Nuclear dot antigens may specify transcriptional domains in the nucleus. Mol. Cell Biol. 1993;13:6170–6179. doi: 10.1128/mcb.13.10.6170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L., Wei Y., Reboul J., Vaglio P., Shin T. H., Vidal M., Elledge S. J., Harper J. W. BTB proteins are substrate-specific adaptors in an SCF-like modular ubiquitin ligase containing CUL-3. Nature. 2003;425:316–321. doi: 10.1038/nature01985. [DOI] [PubMed] [Google Scholar]
- Yamano H., Tsurumi C., Gannon J., Hunt T. The role of the destruction box and its neighbouring lysine residues in cyclin B for anaphase ubiquitin-dependent proteolysis in fission yeast: defining the D-box receptor. EMBO J. 1998;17:5670–5678. doi: 10.1093/emboj/17.19.5670. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.