Abstract
The cardiac outflow tract develops as a result of a complex interplay among several cell types, including cardiac neural crest cells, endothelial cells, and cardiomyocytes. In both humans and mice, mutations in components of the Notch signaling pathway result in congenital heart disease characterized by cardiac outflow tract defects. However, the specific cell types in which Notch functions during cardiovascular development remain to be defined. In addition, in vitro studies have provided conflicting data regarding the ability of Notch to promote or inhibit smooth muscle differentiation, while the physiological role for Notch in smooth muscle formation during development remains unclear. In this study, we generated mice in which Notch signaling was specifically inactivated in derivatives of the neural crest. These mice exhibited cardiovascular anomalies, including aortic arch patterning defects, pulmonary artery stenosis, and ventricular septal defects. We show that Notch plays a critical, cell-autonomous role in the differentiation of cardiac neural crest precursors into smooth muscle cells both in vitro and in vivo, and we identify specific Notch targets in neural crest that are implicated in this process. These results provide a molecular and cellular framework for understanding the role of Notch signaling in the etiology of congenital heart disease.
Introduction
Mutations in components of the Notch pathway result in cardiovascular defects in both humans and mice, strongly implicating this signaling pathway in the process of cardiac and vascular development. Notch signaling is an evolutionarily conserved pathway that influences cell fate decisions, cell survival, and proliferation and has been implicated in multiple developmental processes (1). Four Notch receptors (Notch1–4) and 5 Notch ligands (Jagged1–2 and Delta-like1, -3, and -4) have been identified in mice and humans. The receptors and ligands are both transmembrane proteins expressed on the cell surface, allowing communication between 2 adjacent cells. Upon ligand binding, the Notch receptor becomes susceptible to proteolytic cleavage mediated by a γ-secretase complex. This cleavage releases the intracellular domain of Notch (NICD), which then translocates to the nucleus, where it is capable of forming an active transcriptional complex with the DNA-binding protein CSL (CBF-1, suppressor of hairless, and Lag-1, also known as RBP-Jκ), mastermind-like (MAML), and other transcriptional coactivators. This complex is responsible for the transcription of Notch target genes, including those of the hairy and enhancer of split (HES) and HES-related transcription factor (HRT; also referred to as Hey, Hesr, HERP, or CHF) families (2, 3).
In humans, the congenital disorder Alagille syndrome has been linked to haploinsufficiency of the Notch ligand Jagged1 (4, 5). One of the hallmarks of this syndrome is congenital heart disease involving the cardiac outflow tract and great vessels, including stenosis of the pulmonary artery and its branches, ventricular septal defects, and tetralogy of Fallot (6). Human mutations in Notch1 have recently been linked to aortic valve defects (7). In mice, combined haploinsufficiency of Jagged1 and Notch2 results in cardiac defects reminiscent of Alagille syndrome (8). In addition, mice deficient in the Notch target gene HRT2 develop ventricular septal defects and pulmonary artery stenosis (9–11). While these models demonstrate the importance of Notch in cardiac outflow tract development, the cellular and molecular mechanisms of Notch action remain largely mysterious.
The cardiac outflow tract forms following a series of complex, poorly understood interactions among multiple different cell types, including endothelial cells, cardiomyocytes, and cardiac neural crest cells. Interestingly, the defects seen in the aforementioned models are reminiscent of those of murine and avian models with defective neural crest cell function. However, there have been no tissue-specific studies to address the role of Notch in the cardiac neural crest or any of the other cell types that contribute to the cardiac outflow tract. The neural crest is a multipotent cell population that develops in the dorsal neural tube and then migrates throughout the embryo, where it is able to differentiate into numerous tissue types. A subpopulation of these cells known as the cardiac neural crest migrates through the pharyngeal arches and into the developing outflow tract. There, these cells contribute to the conotruncal septum that divides the outflow tract into the aorta and pulmonary artery. They also form the vascular smooth muscle layer of the aortic arch arteries (12, 13), a process that is believed to be critical for the proper remodeling of these vessels from their initial state as symmetrically paired arteries into the mature, asymmetric aortic arch (14).
A number of in vitro studies have implicated Notch in multiple aspects of smooth muscle cell biology, including the regulation of smooth muscle cell proliferation and survival (15–18). In addition, Notch has been described as both an inhibitor and a promoter of smooth muscle differentiation in vitro (19–22). However, there have been few studies to address which of these functions of Notch play a significant role in smooth muscle formation in vivo. The fact that cardiac neural crest cells have stereotypical properties of smooth muscle cell precursors makes them an excellent model for studying the process of smooth muscle fate specification. The availability of genetic tools that specifically target the neural crest or smooth muscle cells has allowed us to directly address these questions in vivo.
In this study, we use Cre-lox technology to activate a dominant-negative inhibitor of Notch signaling specifically in the neural crest lineage. The resulting mice displayed cardiac outflow tract defects, including pulmonary artery stenosis, ventricular septal defects, and aortic arch artery patterning defects. Neural crest cell migration was not affected, but the embryos demonstrated localized downregulation of Notch target genes and decreases in expression of smooth muscle markers. We also show that inhibition of Notch prevents the differentiation of cardiac neural crest precursors into smooth muscle cells in vitro. These results demonstrate a critical cell-autonomous role for Notch within neural crest cells during cardiovascular development. Furthermore, we suggest that these cardiac defects can be explained by an effect of Notch on the differentiation of cardiac neural crest cells into vascular smooth muscle. These results suggest a mechanism to explain the cardiovascular manifestations of Alagille syndrome and related disorders. In addition, they provide insight into the physiological role of Notch in vascular smooth muscle development.
Results
Multiple Notch receptors and ligands are expressed in the developing cardiac outflow tract.
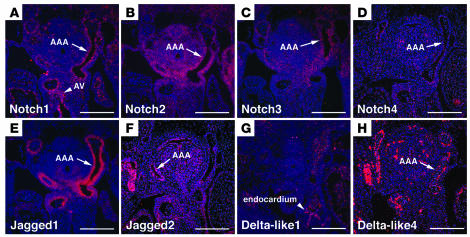
In order to determine which Notch family members are likely to play a role in cardiac outflow tract development, we performed in situ hybridizations to determine the expression patterns of all known Notch receptors and ligands in the developing heart. We examined embryos at E12.5, during a critical time period for remodeling of the outflow tract and the aortic arch arteries. Notch1 and Notch4 were expressed in a thin layer lining the heart and the aortic arch arteries, consistent with expression restricted to the endothelium (Figure 1, A and D). Notch1 was also expressed in the region of the aortic valve (Figure 1A), consistent with its role in development of this valve (7). Notch2 and Notch3 transcripts were detected in the mesenchymal cells surrounding the aortic arch arteries, consistent with expression within neural crest (Figure 1, B and C).
Figure 1. Multiple Notch receptors and ligands are expressed in the developing cardiac outflow tract.
In situ hybridization analysis of cross-sections through the outflow tract and aortic arch arteries of E12.5 embryos is shown. (A) Notch1 is expressed by endothelial cells lining the aortic arch arteries (AAA) and by endocardial cushion tissue of the developing aortic valve (AV). (B) Notch2 is expressed broadly throughout the pharyngeal mesenchyme and notably by the neural crest–derived cells surrounding the aortic arch arteries. (C) Notch3 is expressed by neural crest–derived cells surrounding the aortic arch arteries. (D) Notch4 is expressed by endothelial cells of the aortic arch arteries. (E) Jagged1 is expressed by neural crest–derived cells surrounding the aortic arch arteries. (F) Jagged2 is expressed broadly in the pharyngeal region, including cells surrounding the aortic arch arteries. (G) Delta-like1 is expressed by endocardium but is absent from the aortic arch arteries. (H) Delta-like4 is expressed by endothelium of the aortic arch arteries. Scale bars: 100 μm.
The Notch ligand Jagged1 was expressed in cells surrounding the aortic arch arteries, consistent with expression by neural crest–derived tissues including vascular smooth muscle (Figure 1E). Jagged2 was expressed diffusely throughout the pharyngeal region, including neural crest–derived tissue surrounding the aortic arch arteries (Figure 1F). Delta-like1 transcripts were detectable in the endocardium, but there was no detectable signal in the aortic arch arteries at this stage (Figure 1G). The expression pattern for Delta-like4 was consistent with other reports describing endothelium-specific expression (Figure 1H) (23). Delta-like3 transcripts were not detectable in the developing cardiac outflow tract at this time point (data not shown).
Neural crest–specific expression of dominant-negative MAML results in cardiac outflow tract defects.
The expression of multiple Notch family members suggests possible redundancy among different Notch receptors in the developing cardiac outflow tract. Therefore, we used a dominant-negative approach that would inhibit the activity of all 4 Notch receptors. MAML proteins are important components of the Notch transcriptional complex and are critical for recruitment of other coactivators (24). The dominant-negative version of MAML1 (DNMAML) consists of a 62-amino-acid peptide at the N terminus of MAML1 that has been shown in crystal structures to bind specifically to the Notch-CSL transcriptional complex (25). This construct effectively inhibits canonical (CSL-dependent) signaling by all 4 mammalian Notch receptors (26, 27). We used a mouse model in which DNMAML fused to GFP (DNMAML-GFP) was targeted to the ubiquitously expressed ROSA26 locus. In this model, DNMAML-GFP is placed downstream of a neomycin cassette, which is flanked by loxP sites. In the presence of Cre recombinase, neomycin is excised and DNMAML-GFP comes under control of the ubiquitous ROSA26 promoter. This provides a system in which DNMAML-GFP can be activated in a tissue-specific manner by crossing DNMAML-GFP mice with a mouse line that expresses a tissue-specific Cre recombinase (27).
To inhibit Notch in derivatives of the neural crest, we crossed the DNMAML-GFP mouse with 2 different neural crest–specific Cre recombinase lines. First, we used a mouse in which Cre is knocked into the Pax3 locus (Pax3Cre/+) (28). Pax3 is a paired-type transcription factor that is expressed in premigratory neural crest cells and presomitic mesoderm. Previous fate-mapping studies using the Pax3Cre/+ mouse have shown efficient recombination in derivates of the cardiac neural crest, including the developing aortic arch vessels and the conotruncal septum. In addition to neural crest derivatives, the Pax3Cre/+ mouse also induces recombination in derivatives of the somites, including skeletal muscle of the diaphragm and limb muscles (28).
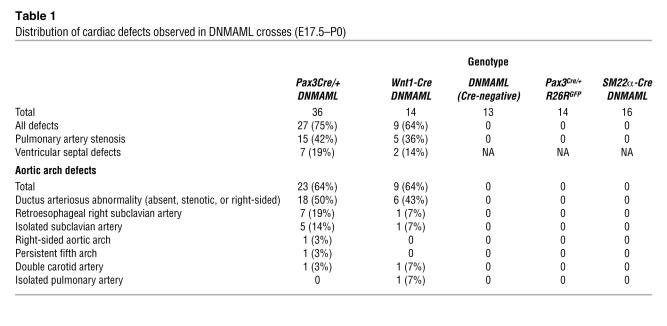
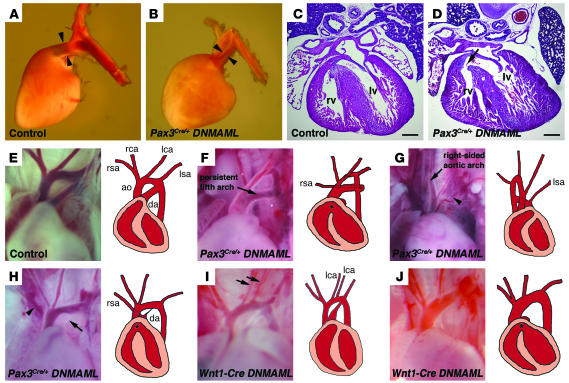
Examination of the hearts of late-gestation Pax3Cre/+ DNMAML embryos revealed multiple significant cardiac defects that mimicked those seen in human congenital heart disease. A detailed list of the defects observed in these mice is presented in Table 1. Twenty-seven of 36 (75%) of the embryos examined at E17.5 or E18.5 showed at least 1 cardiac anomaly. The most common defect observed was stenosis of the pulmonary artery in 15 of 36 (42%) of the embryos (Figure 2, A and B). Some of these embryos also demonstrated membranous ventricular septal defects (Figure 2, C and D) and a malalignment of the outflow tract such that the aorta was located lateral to, rather than posterior to, the pulmonary artery. This suggests a phenotype similar to tetralogy of Fallot in a subset of the embryos.
Table 1 .
Distribution of cardiac defects observed in DNMAML crosses (E17.5–P0)
Figure 2. Neural crest–specific expression of DNMAML results in cardiac outflow tract defects.
(A and B) Lateral view of the left side of hearts removed from E17.5 control (A) and Pax3Cre/+ DNMAML (B) embryos. Compared with the control pulmonary artery, the mutant pulmonary artery and ductus arteriosus are significantly narrowed (arrowheads in A and B). (C and D) Hematoxylin and eosin stained cross-sections through the hearts of E17.5 control (C) or Pax3Cre/+ DNMAML (D) embryos demonstrate a membranous ventricular septal defect in the mutant (arrow in D). (E–J) Photographs and drawings depicting the aortic arch phenotypes in several E17.5 embryos. A control embryo (E) shows the normal branching pattern of the great vessels. (F) Pax3Cre/+ DNMAML mutant showing an abnormal arch structure similar to the human defect known as persistent fifth arch. In addition, the ductus arteriosus is absent, and there is a retroesophageal right subclavian artery. (G) Pax3Cre/+ DNMAML mutant showing a right-sided aortic arch and isolated left subclavian artery arising from the pulmonary artery (arrowhead). (H) Pax3Cre/+ DNMAML mutant with an atretic ductus arteriosus (arrow) and isolated right subclavian artery (arrowhead). (I) Wnt1-Cre DNMAML mutant showing duplication of the left common carotid artery (arrows). (J) Wnt1-Cre DNMAML mutant with an absent ductus arteriosus. The asterisk indicates ventricular septal defects that were observed upon sectioning the hearts. rv, right ventricle; lv, left ventricle; ao, aorta; da, ductus arteriosus; rsa, right subclavian artery; rca, right common carotid artery; lca, left common carotid artery; lsa, left subclavian artery. Scale bars: 200 μm.
Twenty-three of 36 (64%) of these embryos displayed aortic arch patterning defects, with a spectrum of severity (Figure 2, E–H, and Table 1). All of these aortic arch defects could be attributed to improper persistence or regression of the aortic arch arteries during development. Defects in the development of the sixth arch artery were most common, including defects in the pulmonary artery and ductus arteriosus (Figure 2, F–I). However, third and fourth arch artery defects were also apparent. Anomalous origin of the subclavian arteries was observed, either as a retroesophageal right subclavian artery arising from the descending aorta (Figure 2F) or as an isolated subclavian artery arising from the main pulmonary artery (Figure 2, G and H). We also observed other arch anomalies, including right-sided aortic arch (Figure 2G), duplication of the common carotid arteries (Figure 2I), and a defect similar to a rare congenital malformation known as persistent fifth arch (Figure 2F).
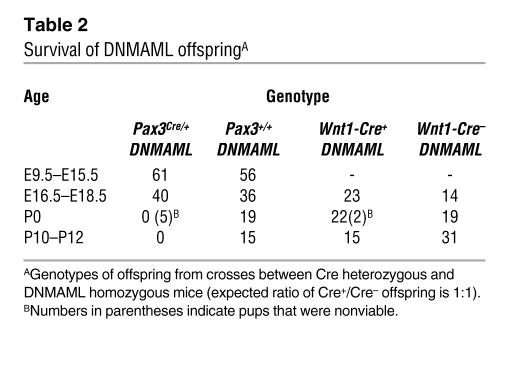
Pax3Cre/+ DNMAML embryos were observed at expected Mendelian ratios throughout gestation.
However, no offspring survived after birth (Table 2). This neonatal mortality could not be explained by cardiac defects alone, as several dead Pax3Cre/+ DNMAML neonates had no obvious cardiac phenotype. On the other hand, all mutant neonates failed to show signs of lung inflation, and examination revealed a very thin diaphragm with severe reduction in skeletal muscle mass (data not shown). Therefore, we conclude that the major cause of death in these mice was respiratory failure. This finding is consistent with the fact that Pax3 is expressed in developing somites and that there is a well-established role for Notch signaling in both somite and skeletal muscle development (29, 30).
Table 2 .
Survival of DNMAML offspringA
To confirm that the cardiac phenotype seen in the Pax3Cre/+ DNMAML mice was specific to the neural crest, we also crossed DNMAML mice with Wnt1-Cre–transgenic mice (13). Wnt1 is also expressed in premigratory neural crest cells, but it is not expressed in skeletal muscle precursors. Wnt1-Cre DNMAML mice also demonstrated cardiac outflow tract and aortic arch abnormalities that were similar to those seen in the Pax3Cre/+ DNMAML mice (Table 1 and Figure 2, I and J). These results confirm a critical cell-autonomous role for Notch signaling in the cardiac neural crest. Unlike the Pax3Cre/+ DNMAML mice, many of the Wnt1-Cre DNMAML mice survived after birth. As Wnt1-Cre does not induce recombination in skeletal muscle precursors, these mice did not display diaphragm defects. However, we did observe significant mortality during the first 2 weeks of life (Table 2). This can be attributed to the cardiac phenotype, as several dead pups had severe cardiac malformations.
Two different control mouse crosses were also generated. First, we examined the Cre-negative littermates from the Pax3Cre/+ DNMAML cross. Second, to control for the presence of Pax3Cre/+, as well as for any possible effects of GFP overexpression, we crossed the Pax3Cre/+ mice with R26RGFP mice (31). The R26RGFP mice contain GFP knocked into the ROSA26 locus, allowing for activation of GFP using Cre recombinase. This acts as an ideal control for the DNMAML-GFP mouse line. Importantly, we did not observe any cardiac defects in either of these control groups (Table 1).
DNMAML is activated by Pax3Cre specifically in derivatives of the neural crest and somites and does not affect neural crest migration.
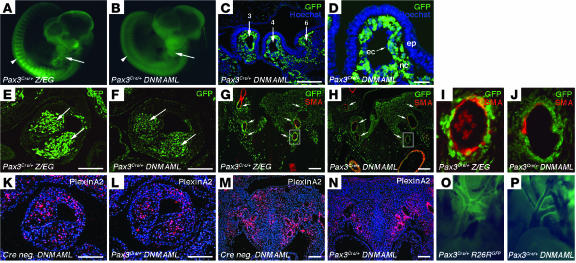
One possible explanation for the cardiac phenotype in these mice is that inhibition of Notch signaling causes a defect in neural crest cell number or migration from the neural tube into the cardiac outflow tract. To follow the fate of neural crest cells in the Pax3Cre/+ DNMAML mice and to ensure that DNMAML was activated specifically within neural crest cells and their derivatives, we made use of the GFP tag on the DNMAML molecule. We compared these embryos with control embryos in which Pax3Cre/+ was crossed with Z/EG mice, which contain a Cre-inducible GFP reporter (32). Pax3Cre/+ DNMAML embryos at E10.5 demonstrated DNMAML-GFP expression as expected in tissues derived from Pax3-expressing precursors, including neural tube, pharyngeal arches, and somites. This pattern was identical to that of Pax3Cre/+ Z/EG controls (Figure 3, A and B).
Figure 3. DNMAML-GFP is activated specifically in neural crest and somites by Pax3-Cre and does not affect neural crest cell number or migration.
(A and B) E10.5 Pax3Cre/+ Z/EG control (A) and Pax3Cre/+ DNMAML mutant (B) embryos demonstrating expression of DNMAML-GFP in pharyngeal arches (arrows) and somites (arrowheads). (C and D) Immunostaining for GFP with Hoechst nuclear counterstain on frontal sections through the pharyngeal arches of E10.5 Pax3Cre/+ DNMAML embryos. (C) Low-magnification view showing GFP-positive cells investing the third, fourth, and sixth aortic arch arteries. (D) Higher magnification showing GFP expression specifically in the neural crest–derived mesenchyme of the pharyngeal arch (nc), but not in pharyngeal epithelium (ep) or endothelial cells (ec). (E and F) Immunostaining for GFP on frontal sections through the conotruncus of E11.5 embryos, showing an equivalent number of GFP-positive cells in the conotruncal cushions (arrows) of control (E) and mutant (F) embryos. (G–J) Immunostaining for GFP and α-SMA on frontal sections through the aortic arch arteries of E11.5 embryos. (G and H) Low-magnification view showing equivalent numbers of GFP-positive cells in the pharyngeal region surrounding the 6 major aortic arch arteries (arrows) in control (G) and mutant (H) embryos. (I and J) High-magnification views of the left sixth aortic arch arteries shown in G and H. (K–N) In situ hybridizations for the neural crest cell marker PlexinA2 on frontal sections through the conotruncus (K and L) and the aortic arch arteries (M and N) of E11.5 embryos, showing equivalent expression in control (K and M) and mutant (L and N) embryos. (O and P) GFP expression in the mature aortic arch of control Pax3Cre/+R26RGFP (O) and mutant Pax3Cre/+ DNMAML (P) mice. Scale bars: 100 μm (C, E–H, and K–N), 20 μm (D).
The developing cardiac outflow tract and aortic arch arteries were observed more closely by sectioning and immunostaining using anti-GFP antibodies. Frontal sections through the pharyngeal arches at E10.5 revealed DNMAML-GFP expression specifically in the neural crest–derived mesenchyme, while it was absent in the pharyngeal endoderm and the endothelium of the aortic arch arteries (Figure 3, C and D). At E11.5, Pax3 derivatives expressing DNMAML-GFP were readily apparent in the cushions of the developing outflow tract and were found in numbers equivalent to those in control Pax3Cre/+ Z/EG embryos (Figure 3, E and F). Sections through the aortic arch arteries at E11.5 also revealed DNMAML-GFP expression throughout the pharyngeal region at levels equivalent to those in controls (Figure 3, G and H). High-magnification views showing costaining for GFP and α-SMA demonstrated that DNMAML-GFP–expressing neural crest cells were present surrounding the aortic arch arteries at the onset of smooth muscle differentiation (Figure 3, I and J).
As an additional method to track these cells, we performed in situ hybridizations for PlexinA2, a marker of migratory and postmigratory cardiac neural crest cells (33). When compared with Cre-negative controls, Pax3Cre/+ DNMAML embryos at E11.5 showed equivalent levels of PlexinA2 in the conotruncal cushions (Figure 3, K and L) and in the mesenchyme surrounding the aortic arch arteries (Figure 3, M and N).
Previous studies have shown that neural crest–derived vascular smooth muscle contributes to the mature aortic arch, ductus arteriosus, and the proximal regions of the major aortic arch branches (13, 28). Pax3Cre/+ DNMAML embryos at late gestation (E17.5–E18.5) showed DNMAML-GFP expression in these regions of the aortic arch, similar to the pattern seen in Pax3Cre/+R26RGFP control embryos (Figure 3, O and P).
Given these results, we conclude that expression of DNMAML within neural crest cells does not affect their overall numbers, their migration into the cardiac outflow tract, or their contribution to the mature aortic arch. This suggests that the cardiac defects in Pax3Cre/+ DNMAML mice are likely due to defective function of postmigratory neural crest cells.
Notch target genes are activated in the developing vascular smooth muscle layer of the aortic arch arteries, and their expression is inhibited by DNMAML.
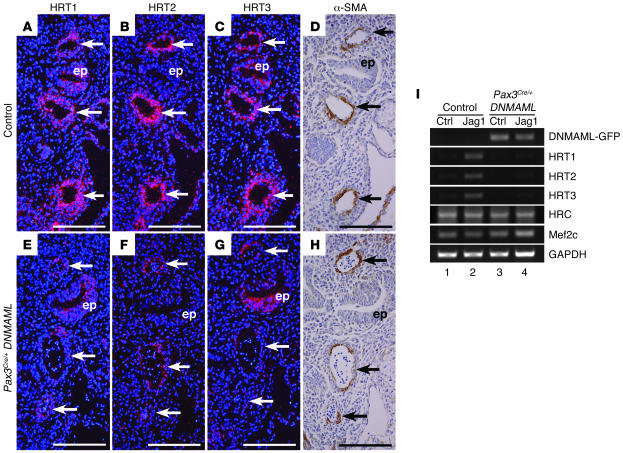
In order to analyze the spatial and temporal patterns of Notch activity in cardiac neural crest cells, we examined the expression of known Notch target genes in both wild-type and Pax3Cre/+ DNMAML embryos. We focused on members of the HRT gene family, as they have been shown to be important targets of Notch in the cardiovascular system (2).
At E10.0, prior to the onset of smooth muscle differentiation, HRT1 was expressed in the mesenchyme of the pharyngeal arches and in a thin layer lining the aortic arch arteries, consistent with endothelium-specific expression (data not shown). Neither HRT2 nor HRT3 showed significant expression around the aortic arch arteries at this time point (data not shown). By E11.5, HRT1, HRT2, and HRT3 showed distinct expression patterns in the cells surrounding the aortic arch arteries in control embryos (Figure 4, A–C). Immunostaining of adjacent sections for α-SMA showed that the activation of HRT genes coincides with the differentiation of neural crest cells into smooth muscle (Figure 4D). Importantly, the expression of all 3 HRT genes was significantly downregulated in the aortic arch arteries of Pax3Cre/+ DNMAML embryos. A small amount of signal was still detected in a thin layer lining the aortic arch arteries. This is consistent with remaining expression of the HRT genes in endothelial cells, where they have also been reported to play a critical role (2). HRT1 and HRT3 expression was also maintained in the non–neural crest–derived pharyngeal epithelium of the Pax3Cre/+ DNMAML mutants (Figure 4, E–G).
Figure 4. Notch target genes are expressed in the developing smooth muscle layer of the aortic arch arteries and are suppressed by DNMAML.
(A–H) Frontal sections through the aortic arch arteries of E11.5 control (A–D) and Pax3Cre/+ DNMAML (E–H) embryos showing the aortic arch arteries (arrows) and pharyngeal epithelium (ep). Adjacent sections were used for in situ hybridizations for the Notch target genes HRT1, HRT2, and HRT3 (A–C and E–G) and immunostaining for α-SMA (D and H). (A–D) In control embryos, HRT1, HRT2, and HRT3 are expressed in the cells surrounding the aortic arch arteries, consistent with expression in the developing smooth muscle layer. (E–H) Pax3Cre/+ DNMAML embryo shows a loss of HRT1, HRT2, and HRT3 expression surrounding the aortic arch arteries, while HRT1 and HRT3 are maintained in the pharyngeal epithelium (ep), and a thin layer of HRT2-positive cells persists in the endothelium of the aortic arch arteries. (I) Semiquantitative RT-PCR from primary smooth muscle cells derived from the aortic arches of late-gestation control and Pax3Cre/+ DNMAML embryos. Cells were stimulated with immobilized control-Fc (ctrl) or Jagged1-Fc (Jag1). Jagged1 stimulation induces HRT1, HRT2, and HRT3 expression in control cells (lane 2) but not in DNMAML-expressing cells (lane 4). In contrast, expression of the Mef2 target genes histidine-rich calcium-binding protein (HRC) and Mef2c is unchanged in cells expressing DNMAML compared with controls. Scale bars: 100 μm.
It was recently reported that, in addition to its role in the Notch transcriptional complex, MAML1 also acts as a coactivator for Mef2c, a member of the MEF2 (myocyte enhancer family 2) family of transcription factors that can influence muscle-specific gene activation (34). We therefore determined whether DNMAML could inhibit Mef2c in addition to Notch. Using luciferase reporter assays, we show that even a low dose of DNMAML robustly inhibited the activity of a Notch-responsive promoter. On the other hand, similar levels of DNMAML had no effect on an artificial promoter containing 4 Mef2 binding sites (Supplemental Figure 1; supplemental material available online with this article; doi:10.1172/JCI30070DS1). Furthermore, a reporter construct driven by an endogenous Mef2c target promoter (histidine-rich calcium-binding protein–luciferase [HRC-luciferase]) was not inhibited over a broad range of both Mef2c and DNMAML doses (Supplemental Figure 2).
The reporter assays suggest that DNMAML acts as a much more potent inhibitor of Notch than Mef2c, but they do not rule out the possibility that DNMAML may inhibit Mef2c in some contexts. Therefore, we also addressed the effect of DNMAML on endogenous Notch and Mef2c targets in our system. We utilized primary smooth muscle cell cultures generated from the aortic arches of Pax3Cre/+ DNMAML and Cre-negative DNMAML (control) embryos. When stimulated with the Notch ligand Jagged 1, control smooth muscle cells demonstrated significant increase in expression of HRT1, HRT2, and HRT3. This increase in expression was completely blocked in DNMAML-expressing smooth muscle cells generated from the Pax3Cre/+ DNMAML embryos (Figure 4I). Importantly, we were unable to detect differences in the expression levels of the Mef2 target genes HRC (35) or Mef2c (Figure 4I). Therefore, in the context of our experiments, DNMAML appears to act as a potent and specific inhibitor of Notch-dependent, but not Mef2c-dependent, transcriptional activation.
Together, these results show that Notch signaling is active in postmigratory neural crest cells, specifically in the developing smooth muscle layer of the aortic arch arteries, and that DNMAML can effectively and specifically block this activity. It also suggests that HRT1, HRT2, and HRT3 are critical Notch targets in neural crest–derived vascular smooth muscle.
Neural crest–specific DNMAML expression is associated with loss of vascular smooth muscle cell markers.
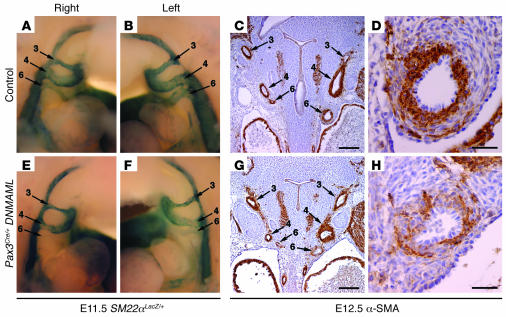
We propose that Notch functions in the postmigratory neural crest to mediate proper development of the cardiac outflow tract and remodeling of the aortic arch arteries. As a major fate of cardiac neural crest cells is to form the vascular smooth muscle cells of the aortic arch, we carefully examined the effect of DNMAML on vascular smooth muscle marker gene expression in vivo. First, we crossed Pax3Cre/+ DNMAML with mice in which LacZ is knocked into the SM22α locus (SM22αLacZ/+ mice) (36). At E11.5, all control SM22αLacZ/+ embryos showed strong LacZ activity in the 6 main aortic arch arteries (left and right third, fourth, and sixth arch arteries) (Figure 5, A and B). However, all Pax3Cre/+ DNMAML, SM22αLacZ/+ embryos showed diminished or absent LacZ activity in 1 or more of the 6 aortic arch arteries (Figure 5, E and F). The left sixth aortic arch artery, the embryonic precursor of the pulmonary artery and ductus arteriosus, was disproportionately affected. However, diminished staining was also observed in the third and fourth aortic arch arteries in some of the embryos.
Figure 5. Neural crest–specific inhibition of Notch results in loss of smooth muscle markers in the developing aortic arch arteries.
(A, B, E, and F) Lateral views of the aortic arch arteries as visualized by whole mount staining for β-galactosidase activity in E11.5 SM22αLacZ/+ embryos. Control embryo shows strong staining in all 3 major aortic arch arteries on both the right (A) and left (B). In contrast, a Pax3Cre/+ DNMAML embryo shows absence of β-galactosidase activity in the right sixth aortic arch artery (E) and diminished β-galactosidase activity in the left sixth aortic arch artery (F). (C, D, G, and H) Immunohistochemistry for α-SMA on frontal sections through the developing aortic arch arteries of E12.5 embryos. At low magnification, the smooth muscle layer surrounding all aortic arch arteries is robust and uniform in control embryos (C). However, a Pax3Cre/+ DNMAML embryo shows diminished α-SMA staining in the sixth aortic arch arteries but normal staining in the other vessels (G). (D and H) High-magnification views of the left sixth aortic arch artery shown in C and G. Compared with control (D), the mutant artery (H) shows diminished α-SMA staining and a general disruption in the architecture of the smooth muscle layer. Scale bars: 100 μm (C and G), 20 μm (D and H).
In addition, we performed immunohistochemistry for α-SMA, another marker of vascular smooth muscle cells. Frontal sections through wild-type embryos at E12.5 showed a thick layer of α-SMA–positive cells surrounding all 6 aortic arch arteries (Figure 5C). However, Pax3Cre/+ DNMAML embryos showed localized downregulation of α-SMA staining in 1 or more of the aortic arch arteries (Figure 5G). As was observed with the SM22αLacZ marker, the sixth aortic arch arteries were disproportionately affected. High-magnification views of sections through the left sixth aortic arch artery of wild-type embryos showed strongly staining smooth muscle cells arranged in an organized, concentric pattern around the vessel (Figure 5D). On the other hand, the left sixth aortic arch arteries of Pax3Cre/+ DNMAML embryos often showed a disorganized smooth muscle cell layer with diminished α-SMA staining (Figure 5H).
These results demonstrate a highly reproducible defect in neural crest–derived vascular smooth muscle cells during the time period critical for aortic arch artery development. The disproportionate effect on smooth muscle markers in the sixth aortic arch artery is consistent with the high frequency of pulmonary artery and ductus arteriosus defects observed in the late-gestation embryos.
Activation of DNMAML after the onset of smooth muscle differentiation has no effect on aortic arch artery remodeling.
A number of in vitro studies indicate that Notch can play a role in the modulation of vascular smooth muscle cell proliferation, apoptosis, migration, and contractility (15–18). One model to explain the aortic arch defects in these mice is that defective vascular smooth muscle cell proliferation or survival may lead to loss of aortic arch artery integrity and subsequent defects in aortic arch remodeling. However, we failed to see any differences in apoptosis or cell division between Pax3Cre/+ DNMAML and control embryos using TUNEL assays and staining for proliferation markers at E11.5 and E12.5 (data not shown).
To directly test the effect of Notch inhibition after the onset of smooth muscle differentiation, we also crossed the DNMAML mice with SM22α-Cre mice (37). Unlike the Pax3Cre/+ DNMAML embryos, which showed strong expression of DNMAML at E10.5, prior to the onset of smooth muscle differentiation, the SM22α-Cre DNMAML embryos did not express DNMAML until after the activation of smooth muscle markers surrounding the aortic arch arteries (Supplemental Figure 3). These mice failed to show defects in the smooth muscle layer of the developing aortic arch arteries (Supplemental Figure 3) and also did not display any cardiac outflow tract or aortic arch defects at later time points (Table 1). These studies indicate that the critical time period for Notch activity in cardiac neural crest–derived tissues is prior to E11.5. Therefore, we do not believe that an effect of Notch on smooth muscle proliferation or survival could account for the defects seen in the neural crest–specific DNMAML mice.
Notch promotes the differentiation of cardiac neural crest precursors into smooth muscle ex vivo.
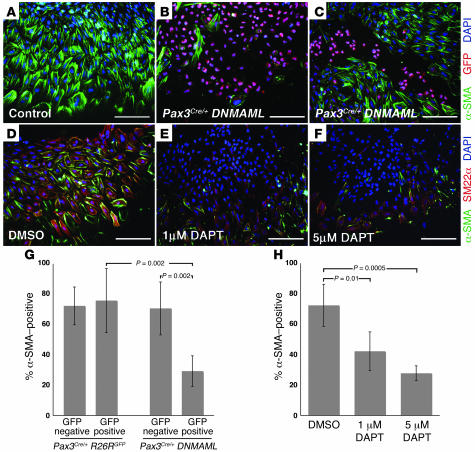
Another explanation for the smooth muscle defects in these mice is the possibility that Notch plays a critical role in the differentiation of neural crest into smooth muscle. We used neural tube explant assays to address this possibility. Neural tubes from the cardiac neural crest region were isolated from E8.5 embryos and grown in culture medium containing serum for 48 hours. Under these conditions, neural crest cells from control embryos readily migrated out of the explanted neural tube and differentiated robustly into smooth muscle cells, as determined by immunofluorescence staining for α-SMA or SM22α (Figure 6, A and D). On the other hand, neural tube explants from Pax3Cre/+ DNMAML embryos showed a dramatic reduction in the number of α-SMA–positive cells (Figure 6B). Some of the explants we examined showed inefficient Cre-mediated recombination, as determined by costaining for GFP. In these explants, the majority of DNMAML-GFP–positive cells were α-SMA negative, while adjacent DNMAML-GFP–negative cells in the same explant were usually α-SMA positive. This demonstrates that the inhibition of smooth muscle differentiation by DNMAML was cell autonomous (Figure 6C).
Figure 6. Inhibition of Notch activity blocks the differentiation of neural crest precursors into smooth muscle ex vivo.
(A–C) Neural tube explants from E8.5 control (A) and Pax3Cre/+ DNMAML (B and C) embryos, immunostained for α-SMA (green) and GFP (red). The majority of cells in the control explant express α-SMA, as do the DNMAML-GFP–negative cells in the Pax3Cre/+ DNMAML explants (B and C). Conversely, most of the DNMAML-GFP–positive cells in B and C are α-SMA negative. (D–F) Wild-type neural tube explants treated with the γ-secretase inhibitor DAPT, immunostained for α-SMA (green) and SM22α (red). Compared with DMSO-treated controls (D), explants treated with 1 μM DAPT (E) or 5 μM DAPT (F) show a decrease in the number of cells expressing smooth muscle markers. (G) Quantification of neural tube explant assays from Pax3Cre/+R26RGFP (control) and Pax3Cre/+ DNMAML embryos. (H) Quantification of assays on neural tube explants treated with the γ-secretase inhibitor DAPT. Error bars indicate 1 SD. P values, indicated by brackets, were determined by Student’s t test. Scale bars: 100 μm.
We quantified this effect by counting cells from the explants. To control for any nonspecific effects of GFP, we also performed neural tube explant assays on Pax3Cre/+R26RGFP embryos. In these control embryos, there was no significant difference in α-SMA expression between GFP-negative and GFP-positive cells (Figure 6G, first and second bars). On the other hand, in Pax3Cre/+ DNMAML explants, an average of 29% of DNMAML-GFP–positive cells coexpressed α-SMA, compared with 71% of DNMAML-GFP–negative cells (Figure 6G, third and fourth bars).
As an additional method of inhibiting Notch, we treated wild-type neural tube explants with the γ-secretase inhibitor DAPT. Both 1 μM and 5 μM DAPT resulted in a statistically significant reduction in the number of cells expressing either α-SMA or SM22α when compared with DMSO-treated controls (Figure 6, D–F and H). Therefore, DAPT mimics the effect of DNMAML, further supporting the specificity of DNMAML as an inhibitor of Notch signaling. Together, these results demonstrate that Notch promotes the differentiation of cardiac neural crest precursors into smooth muscle.
Discussion
Despite genetic evidence from previous studies demonstrating a critical role for Notch signaling in cardiac outflow tract development, the mechanism has remained unclear. In this study, we show that expression of DNMAML in the neural crest lineage is able to reproduce many of the cardiac phenotypes seen in humans and other mouse models in which Notch signaling is compromised, including pulmonary artery stenosis and ventricular septal defects. While we cannot completely rule out the possibility that DNMAML may have effects on other pathways including Mef2c, our results strongly suggest that the phenotype in these mice can be attributed to inhibition of Notch signaling. This provides what we believe to be the first evidence of a cell-autonomous role for Notch in the cardiac neural crest and suggests that some of the defects associated with Alagille syndrome and other Notch mutant mouse models can be attributed to defects in this cell population. In addition, our model appears to be unique with respect to the development of aortic arch patterning abnormalities, which were not reported in association with prior Notch mutants. Therefore, this study is also the first to our knowledge to demonstrate that Notch plays a critical role in remodeling of the aortic arch arteries. This association suggests that components of the Notch signaling pathway are likely to be involved in a broad range of congenital heart abnormalities, including pulmonary artery stenosis, ventricular septal defects, tetralogy of Fallot, and aortic arch abnormalities.
The observation of overlapping expression patterns for Notch receptors and Notch target genes (HRT1–3) in the developing aortic arch arteries suggests the potential for redundancy between these factors, which may explain why aortic arch defects were not reported in Jagged1, Notch2 compound heterozygotes (8) or HRT2 knockouts (9–11). However, it should be noted that in several systems, Notch ligands and receptors do not have strictly overlapping functions. Genetic evidence strongly suggests that Jagged1 is the critical ligand in the process of cardiac outflow tract remodeling despite our observation that several other ligands are expressed. Therefore, future studies will be necessary to determine whether a specific Notch receptor plays a dominant role during cardiac outflow tract development and aortic arch remodeling.
These findings are particularly relevant to cardiovascular defects associated with Alagille syndrome. Pulmonary artery stenosis is the most common cardiac defect in Alagille syndrome and contributes significantly to morbidity and mortality in these patients (6). The high penetrance of pulmonary artery and ductus arteriosus defects in the neural crest–specific DNMAML mice suggests that this is an excellent model for studying the mechanisms underlying Alagille syndrome. These results suggest that Jagged1 haploinsufficiency in Alagille patients would result in defects in neural crest–derived vascular smooth muscle secondary to diminished Notch stimulation. The loss of vascular smooth muscle in the sixth embryonic arch artery would, in turn, be responsible for the development of pulmonary artery stenosis. Jagged1 appears to be expressed in the endothelium and in the neural crest–derived tissue surrounding the aortic arch arteries (Figure 1), suggesting 2 possible models to explain the mechanism of smooth muscle specification. In one model, a ligand expressed on endothelial cells may signal to adjacent neural crest cells to induce their differentiation into smooth muscle. According to a second model, ligand-receptor signaling may take place between neighboring neural crest cells. Further studies will be necessary to understand the precise ligand-receptor interactions that are required for this process.
The identification of Notch as a critical regulator of cardiac neural crest cell development is consistent with a number of studies that implicate Notch in cell fate determination in the neural crest lineage. The most well-characterized role for Notch in neural crest cell fate specification is in developing peripheral nerves, where Notch has been described both as an inhibitor of neurogenesis and as a promoter of glial fate specification (38). Interestingly, under some culture conditions, Notch has also been shown to promote the formation of smooth muscle–like cells from peripheral nerve–derived neural crest stem cells in vitro (39, 40). However, the in vivo relevance of this finding has remained unclear, as the peripheral nervous system does not contain smooth muscle cells that are derived from neural crest. While neural crest cells have been shown to give rise to endoneurial fibroblasts in peripheral nerves, these cells only share some characteristics with smooth muscle cells (40). On the other hand, cardiac neural crest does give rise to bona fide smooth muscle. Therefore, our results provide an important in vivo correlation to explain the role of Notch in determining smooth muscle cell fate in the neural crest lineage.
One of the paradigms of Notch signaling is the observation that the biological response to receptor activation is highly sensitive to dosage, developmental timing, and cellular context (1). There have been several in vitro studies indicating contradictory roles for Notch in smooth muscle cell differentiation. For example, Notch and its downstream target genes have been shown to inhibit myocardin-induced smooth muscle cell differentiation (20, 21). On the other hand, recent reports indicate that Notch can promote smooth muscle cell differentiation and that Notch directly transactivates smooth muscle–specific genes (19, 22). These conflicting data have left unresolved the question of the physiologic role of Notch during smooth muscle cell development. Our results using an in vivo model implicate Notch as a positive regulator of smooth muscle cell differentiation during development. Furthermore, we specify the time period during which Notch activity is critical for neural crest–derived smooth muscle differentiation as between E8.5 and E11.5 (after Wnt1-Cre is activated and before SM22α-Cre is activated).
This study specifically addresses the role of Notch in differentiation of neural crest cells into smooth muscle, a process that is critical in the context of cardiac and aortic arch development. However, these findings are likely to be applicable to a broad range of smooth muscle–related disorders, including those involving non–neural crest–derived smooth muscle. In humans, mutations in Notch3 have been identified in CADASIL syndrome (cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy), an inherited adult neurodegenerative disorder associated with cerebral vascular abnormalities (41). Adult Notch3-knockout mice display defects in the tunica media of the distal muscular arteries, which is attributed to abnormal postnatal maturation of vascular smooth muscle cells (42). Patients with Alagille syndrome are also predisposed to multiple vascular pathologies affecting blood vessels that contain non–neural crest–derived smooth muscle, including stenosis of the peripheral pulmonary vascular tree and intracranial aneurysms (6, 43). Together, these observations demonstrate that Notch signaling plays an important role in multiple regions of the developing and adult vasculature. It will be interesting to address whether any of these findings can be attributed to a parallel function for Notch in the proper differentiation of smooth muscle from non–neural crest progenitors.
This study adds to a growing list of molecules that have been shown to be important in neural crest cells during cardiac outflow tract development and/or neural crest–derived smooth muscle differentiation. Other factors that have been identified as regulators of neural crest–derived smooth muscle formation and aortic arch artery remodeling in vivo include bone morphogenic proteins (BMPs) (39, 44), TGF-β (45, 46), and myocardin-related transcription factor B (MRTF-B) (47, 48). The future challenge will be to elucidate potential links between Notch and these other signaling pathways and to understand how they converge to activate target genes that promote the smooth muscle fate.
Methods
Mice.
DNMAML (27), R26RGFP (31), Pax3Cre (28), Wnt1-Cre (13), SM22α-Cre (37), SM22αLacZ (36), and Z/EG (32) mice were maintained on mixed genetic backgrounds. Genotyping primer sequences are available in Supplemental methods. All animal protocols were approved by the University of Pennsylvania Institutional Animal Care and Use Committee.
In situ hybridization and immunohistochemistry.
The Delta-like4, HRT1, HRT2, and HRT3 probes were generated from mouse EST IMAGE clones 6825525, 478014, 5374813, and 6832398, respectively. The Notch1, Notch2, Notch3, Notch4, Jagged1, Jagged2, and Delta-like1 in situ probes were kindly provided by K. Kusumi (Arizona State University, Tempe, Arizona, USA). Template vectors were linearized with EcoRI (ApaI for Notch1 and BamHI for Notch4) and anti-sense riboprobes were generated with T3 polymerase (T7 polymerase for HRT2 and SP6 polymerase for Notch1). The PlexinA2 probe has been previously described (33). Antibodies used for immunohistochemistry and immunofluorescence microscopy were anti-GFP rabbit polyclonal (Invitrogen) and anti–α-SMA monoclonal 1A4 (Sigma-Aldrich). Radioactive in situ hybridization and immunostaining were performed on paraformaldehyde-fixed, paraffin-embedded sections. Detailed protocols are available in ref. 49.
Primary smooth muscle cell culture.
To specifically isolate neural crest–derived smooth muscle, the aortic arch to the origin of the left subclavian artery and the proximal regions of the major arch vessels were dissected from E17.5 or E18.5 embryos. Primary smooth muscle cell cultures were then performed according to previously described protocols for aortic vascular smooth muscle cells (50).
Jagged1-Fc and control-Fc constructs were kindly provided by T. Kadesch (University of Pennsylvania, Philadelphia, Pennsylvania, USA) and have been described elsewhere (21). Smooth muscle cells were stimulated by plating on Jagged1-Fc– or control-Fc–coated plates at a density of 2.5 × 105 per 10-cm plate. Cells were harvested 24 hours following stimulation, and RNA was extracted using TRI zol reagent (Invitrogen). RT-PCR was performed following standard protocols. Primer sequences are available in Supplemental methods.
Neural tube explants.
E8.5 embryos were dissected in HBSS. The region of the neural tube between the otic placode at the third somite was microdissected from each embryo, along with surrounding mesenchyme. Each sample was incubated with 0.75 mg/ml type I collagenase (Worthington Biochemical Corp.) for 20 minutes at 37°C. The neural tubes were then dissected away from the surrounding mesenchyme, split in half longitudinally, and plated on glass chamber slides precoated with 200 μg/ml fibronectin (Invitrogen). Explants were cultured for 48 hours in DMEM containing 2% horse serum. DAPT (γ-secretase inhibitor IX; Calbiochem) reconstituted in DMSO was added to the culture medium at the time of plating.
For immunofluorescence staining, outgrowths were fixed with 4% paraformaldehyde for 10 minutes at room temperature, permeabilized with PBS plus 0.1% Triton X-100 plus 0.1% deoxycholate, and blocked with 5% normal goat serum (Zymed). Slides were then probed with anti–α-SMA monoclonal 1A4 (1:200; Sigma-Aldrich), anti-GFP rabbit polyclonal (1:200; Invitrogen), and anti-SM22α rabbit polyclonal (1:250; M. Parmacek, as described in ref. 36) antibodies. Secondary antibodies used were Alexa Fluor 488 goat anti-mouse and Alexa Fluor 568 goat anti-rabbit (1:200; Invitrogen). Slides were counterstained for 1 minute with DAPI (Sigma-Aldrich).
Quantification of results was performed by imaging each explant and comparing the number of cells that stained positively for GFP and/or α-SMA with the total number of cells, determined by DAPI nuclear staining. The results of 4–6 explants for each condition were averaged, and statistical significance was determined by 2-tailed Student’s t test. P value of less than 0.05 was considered significant.
Supplementary Material
Acknowledgments
We would like to thank Tom Kadesch, Kenro Kusumi, Brian Black, and Jon Aster for providing reagents. This work was supported by funding from the University of Pennsylvania Medical Scientist Training Program to F.A. High and grants from the NIH to J.A. Epstein, M.S. Parmacek, and W.S. Pear including NIH P01 HL075215 and R01 HL61475 and funds from the WW Smith Endowed Chair for Cardiovascular Research to J.A. Epstein.
Footnotes
Nonstandard abbreviations used: CSL, CBF-1, suppressor of hairless, and Lag-1; DNMAML, dominant-negative MAML1; HRT, HES-related transcription factor; MAML, mastermind-like; Mef2c, myocyte enhancer family 2c.
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J. Clin. Invest. 117:353–363 (2007). doi:10.1172/JCI30070
Maozhen Zhang’s present address is: Department of Cardiology, Xinhua Hospital, Shanghai Jiaotong University School of Medicine, Shanghai, People’s Republic of China.
See the related Commentary beginning on page 299.
References
- 1.Artavanis-Tsakonas S., Rand M.D., Lake R.J. Notch signaling: cell fate control and signal integration in development. Science. 1999;284:770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- 2.Fischer A., Gessler M. Hey genes in cardiovascular development. Trends Cardiovasc. Med. 2003;13:221–226. doi: 10.1016/s1050-1738(03)00082-3. [DOI] [PubMed] [Google Scholar]
- 3.Iso T., Kedes L., Hamamori Y. HES and HERP families: multiple effectors of the notch signaling pathway. J. Cell. Physiol. 2003;194:237–255. doi: 10.1002/jcp.10208. [DOI] [PubMed] [Google Scholar]
- 4.Oda T., et al. Mutations in the human Jagged1 gene are responsible for Alagille syndrome. Nat. Genet. 1997;16:235–242. doi: 10.1038/ng0797-235. [DOI] [PubMed] [Google Scholar]
- 5.Li L., et al. Alagille syndrome is caused by mutations in human Jagged1, which encodes a ligand for Notch1. Nat. Genet. 1997;16:243–251. doi: 10.1038/ng0797-243. [DOI] [PubMed] [Google Scholar]
- 6.McElhinney D.B., et al. Analysis of cardiovascular phenotype and genotype-phenotype correlation in individuals with a JAG1 mutation and/or Alagille syndrome. Circulation. 2002;106:2567–2574. doi: 10.1161/01.cir.0000037221.45902.69. [DOI] [PubMed] [Google Scholar]
- 7.Garg V., et al. Mutations in NOTCH1 cause aortic valve disease. Nature. 2005;437:270–274. doi: 10.1038/nature03940. [DOI] [PubMed] [Google Scholar]
- 8.McCright B., Lozier J., Gridley T.2002. . A mouse model of Alagille syndrome: Notch2 as a genetic modifier of Jag1 haploinsufficiency. Development. 1291075–1082. [DOI] [PubMed] [Google Scholar]
- 9.Gessler M., et al. Mouse gridlock: no aortic coarctation or deficiency, but fatal cardiac defects in Hey2 –/– mice. Curr. Biol. 2002;12:1601–1604. doi: 10.1016/s0960-9822(02)01150-8. [DOI] [PubMed] [Google Scholar]
- 10.Donovan J., Kordylewska A., Jan Y.N., Utset M.F. Tetralogy of Fallot and other congenital heart defects in Hey2 mutant mice. Curr. Biol. 2002;12:1605–1610. doi: 10.1016/s0960-9822(02)01149-1. [DOI] [PubMed] [Google Scholar]
- 11.Sakata Y., et al. Ventricular septal defect and cardiomyopathy in mice lacking the transcription factor CHF1/Hey2. Proc. Natl. Acad. Sci. U. S. A. 2002;99:16197–16202. doi: 10.1073/pnas.252648999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kirby M.L., Gale T.F., Stewart D.E. Neural crest cells contribute to normal aorticopulmonary septation. Science. 1983;220:1059–1061. doi: 10.1126/science.6844926. [DOI] [PubMed] [Google Scholar]
- 13.Jiang X., Rowitch D.H., Soriano P., McMahon A.P., Sucov H.M. Fate of the mammalian cardiac neural crest. Development. 2000;127:1607–1616. doi: 10.1242/dev.127.8.1607. [DOI] [PubMed] [Google Scholar]
- 14.Stoller J.Z., Epstein J.A. Cardiac neural crest. Semin. Cell Dev. Biol. 2005;16:704–715. doi: 10.1016/j.semcdb.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 15.Morrow D., et al. Notch-mediated CBF-1/RBP-Jκ-dependent regulation of human vascular smooth muscle cell phenotype in vitro. Am. J. Physiol. Cell Physiol. 2005;289:C1188–C1196. doi: 10.1152/ajpcell.00198.2005. [DOI] [PubMed] [Google Scholar]
- 16.Wang W., Prince C.Z., Hu X., Pollman M.J. HRT1 modulates vascular smooth muscle cell proliferation and apoptosis. Biochem. Biophys. Res. Commun. 2003;308:596–601. doi: 10.1016/s0006-291x(03)01453-0. [DOI] [PubMed] [Google Scholar]
- 17.Sakata Y., et al. Transcription factor CHF1/Hey2 regulates neointimal formation in vivo and vascular smooth muscle proliferation and migration in vitro. Arterioscler. Thromb. Vasc. Biol. 2004;24:2069–2074. doi: 10.1161/01.ATV.0000143936.77094.a4. [DOI] [PubMed] [Google Scholar]
- 18.Sweeney C., et al. Notch 1 and 3 receptors modulate vascular smooth muscle cell growth, apoptosis and migration via a CBF-1/RBP-Jk dependent pathway. FASEB J. 2004;18:1421–1423. doi: 10.1096/fj.04-1700fje. [DOI] [PubMed] [Google Scholar]
- 19.Doi H., et al. Jagged1-selective Notch signaling induces smooth muscle differentiation via a RBP-Jkappa-dependent pathway. J. Biol. Chem. 2006;281:28555–28564. doi: 10.1074/jbc.M602749200. [DOI] [PubMed] [Google Scholar]
- 20.Doi H., et al. HERP1 inhibits myocardin-induced vascular smooth muscle cell differentiation by interfering with SRF binding to CArG box. Arterioscler. Thromb. Vasc. Biol. 2005;25:2328–2334. doi: 10.1161/01.ATV.0000185829.47163.32. [DOI] [PubMed] [Google Scholar]
- 21.Proweller A., Pear W.S., Parmacek M.S. Notch signaling represses myocardin-induced smooth muscle cell differentiation. J. Biol. Chem. 2005;280:8994–9004. doi: 10.1074/jbc.M413316200. [DOI] [PubMed] [Google Scholar]
- 22.Noseda M., et al. Smooth muscle α-actin is a direct target of Notch/CSL. Circ. Res. 2006;98:1468–1470. doi: 10.1161/01.RES.0000229683.81357.26. [DOI] [PubMed] [Google Scholar]
- 23.Shutter J.R., et al. Dll4, a novel Notch ligand expressed in arterial endothelium. Genes Dev. 2000;14:1313–1318. [PMC free article] [PubMed] [Google Scholar]
- 24.Wu L., Griffin J.D. Modulation of Notch signaling by mastermind-like (MAML) transcriptional co-activators and their involvement in tumorigenesis. Semin. Cancer Biol. 2004;14:348–356. doi: 10.1016/j.semcancer.2004.04.014. [DOI] [PubMed] [Google Scholar]
- 25.Nam Y., Sliz P., Song L., Aster J.C., Blacklow S.C. Structural basis for cooperativity in recruitment of MAML coactivators to Notch transcriptional complexes. Cell. 2006;124:973–983. doi: 10.1016/j.cell.2005.12.037. [DOI] [PubMed] [Google Scholar]
- 26.Maillard I., et al. Mastermind critically regulates Notch-mediated lymphoid cell fate decisions. Blood. 2004;104:1696–1702. doi: 10.1182/blood-2004-02-0514. [DOI] [PubMed] [Google Scholar]
- 27.Tu L., et al. Notch signaling is an important regulator of type 2 immunity. J. Exp. Med. 2005;202:1037–1042. doi: 10.1084/jem.20050923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Engleka K.A., et al. Insertion of Cre into the Pax3 locus creates a new allele of Splotch and identifies unexpected Pax3 derivatives. Dev. Biol. 2005;280:396–406. doi: 10.1016/j.ydbio.2005.02.002. [DOI] [PubMed] [Google Scholar]
- 29.Weinmaster G., Kintner C. Modulation of notch signaling during somitogenesis. Annu. Rev. Cell Dev. Biol. 2003;19:367–395. doi: 10.1146/annurev.cellbio.19.111301.115434. [DOI] [PubMed] [Google Scholar]
- 30.Luo D., Renault V.M., Rando T.A. The regulation of Notch signaling in muscle stem cell activation and postnatal myogenesis. Semin. Cell Dev. Biol. 2005;16:612–622. doi: 10.1016/j.semcdb.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 31.Mao X., Fujiwara Y., Chapdelaine A., Yang H., Orkin S.H. Activation of EGFP expression by Cre-mediated excision in a new ROSA26 reporter mouse strain. Blood. 2001;97:324–326. doi: 10.1182/blood.v97.1.324. [DOI] [PubMed] [Google Scholar]
- 32.Novak A., Guo C., Yang W., Nagy A., Lobe C.G. Z/EG, a double reporter mouse line that expresses enhanced green fluorescent protein upon Cre-mediated excision. Genesis. 2000;28:147–155. [PubMed] [Google Scholar]
- 33.Brown C.B., et al. PlexinA2 and semaphorin signaling during cardiac neural crest development. Development. 2001;128:3071–3080. doi: 10.1242/dev.128.16.3071. [DOI] [PubMed] [Google Scholar]
- 34.Shen H., et al. The Notch coactivator, MAML1, functions as a novel coactivator for MEF2C-mediated transcription and is required for normal myogenesis. Genes Dev. 2006;20:675–688. doi: 10.1101/gad.1383706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Anderson J.P., et al. HRC is a direct transcriptional target of MEF2 during cardiac, skeletal, and arterial smooth muscle development in vivo. Mol. Cell. Biol. 2004;24:3757–3768. doi: 10.1128/MCB.24.9.3757-3768.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang J.C., et al. Analysis of SM22alpha-deficient mice reveals unanticipated insights into smooth muscle cell differentiation and function. Mol. Cell. Biol. 2001;21:1336–1344. doi: 10.1128/MCB.2001.21.4.1336-1344.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lepore J.J., et al. High-efficiency somatic mutagenesis in smooth muscle cells and cardiac myocytes in SM22α-Cre transgenic mice. Genesis. 2005;41:179–184. doi: 10.1002/gene.20112. [DOI] [PubMed] [Google Scholar]
- 38.Cornell R.A., Eisen J.S. Notch in the pathway: the roles of Notch signaling in neural crest development. Semin. Cell Dev. Biol. 2005;16:663–672. doi: 10.1016/j.semcdb.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 39.Morrison S.J., et al. Transient Notch activation initiates an irreversible switch from neurogenesis to gliogenesis by neural crest stem cells. Cell. 2000;101:499–510. doi: 10.1016/s0092-8674(00)80860-0. [DOI] [PubMed] [Google Scholar]
- 40.Joseph N.M., et al. Neural crest stem cells undergo multilineage differentiation in developing peripheral nerves to generate endoneurial fibroblasts in addition to Schwann cells. Development. 2004;131:5599–5612. doi: 10.1242/dev.01429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Joutel A., et al. Notch3 mutations in CADASIL, a hereditary adult-onset condition causing stroke and dementia. Nature. 1996;383:707–710. doi: 10.1038/383707a0. [DOI] [PubMed] [Google Scholar]
- 42.Domenga V., et al. Notch3 is required for arterial identity and maturation of vascular smooth muscle cells. Genes Dev. 2004;18:2730–2735. doi: 10.1101/gad.308904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kamath B.M., et al. Vascular anomalies in Alagille syndrome, a significant cause of morbidity and mortality. Circulation. 2004;109:1354–1358. doi: 10.1161/01.CIR.0000121361.01862.A4. [DOI] [PubMed] [Google Scholar]
- 44.Kaartinen V., et al. Cardiac outflow tract defects in mice lacking ALK2 in neural crest cells. Development. 2004;131:3481–3490. doi: 10.1242/dev.01214. [DOI] [PubMed] [Google Scholar]
- 45.Wurdak H., et al. Inactivation of TGFβ signaling in neural crest stem cells leads to multiple defects reminiscent of DiGeorge syndrome. Genes Dev. 2005;19:530–535. doi: 10.1101/gad.317405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Choudhary B., et al. Cardiovascular malformations with normal smooth muscle differentiation in neural crest-specific type II TGFβ receptor (Tgfbr2) mutant mice. Dev. Biol. 2005;289:420–429. doi: 10.1016/j.ydbio.2005.11.008. [DOI] [PubMed] [Google Scholar]
- 47.Li J., et al. Myocardin-related transcription factor B is required in cardiac neural crest for smooth muscle differentiation and cardiovascular development. Proc. Natl. Acad. Sci. U. S. A. 2005;102:8916–8921. doi: 10.1073/pnas.0503741102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Oh J., Richardson J.A., Olson E.N. Requirement of myocardin-related transcription factor-B for remodeling of branchial arch arteries and smooth muscle differentiation. Proc. Natl. Acad. Sci. U. S. A. 2005;102:15122–15127. doi: 10.1073/pnas.0507346102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wawersik S., Epstein J.A. Gene expression analysis by in situ hybridization. Radioactive probes. Methods Mol. Biol. 2000;137:87–96. doi: 10.1385/1-59259-066-7:87. [DOI] [PubMed] [Google Scholar]
- 50.Lepore J.J., Cappola T.P., Mericko P.A., Morrisey E.E., Parmacek M.S. GATA-6 regulates genes promoting synthetic functions in vascular smooth muscle cells. Arterioscler. Thromb. Vasc. Biol. 2005;25:309–314. doi: 10.1161/01.ATV.0000152725.76020.3c. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.