Abstract
Two analogues of pyruvate, acetylphosphinate and acetylmethylphosphinate were tested as inhibitors of the E1 (pyruvate dehydrogenase) component of the human and Escherichia coli pyruvate dehydrogenase complexes. This is the first instance of such studies on the human enzyme. The acetylphosphinate is a stronger inhibitor of both enzymes (Ki< 1 μM) than acetylmethylphosphinate. Both inhibitors are found to be reversible tight-binding inhibitors. With both inhibitors and with both enzymes, the inhibition apparently takes place by formation of a C2α-phosphinolactylthiamin diphosphate derivative, a covalent adduct of the inhibitor and the coenzyme, mimicking the behavior of substrate and forming a stable analogue of the C2α-lactylthiamin diphosphate. Formation of the intermediate analogue in each case is confirmed by the appearance of a positive circular dichroism signal in the 305-306 nm range, attributed to the 1',4'-iminopyrimidine tautomeric form of the coenzyme. It is further shown that the αHis63 residue of the human E1 has a role in the formation of C2α-lactylthiamin diphosphate since the αHis63Ala variant is only modestly inhibited by either inhibitor, nor did either compound generate the circular dichroism bands assigned to different tautomeric forms of the 4'-aminopyrimidine ring of the coenzyme seen with the wild type enzyme. Interestingly, opposite enantiomers of the carboligase side product acetoin are produced by the human and bacterial enzymes.
Keywords: human pyruvate dehydrogenase complex, Escherichia coli pyruvate dehydrogenase complex, acetylphosphinate, acetylmethylphosphinate, thiamin diphosphate, thiamin 2-thiothiazolone diphosphate, acetylphosphonate methyl ester, circular dichroism, mechanism-based inhibition, acetoin
1. Introduction
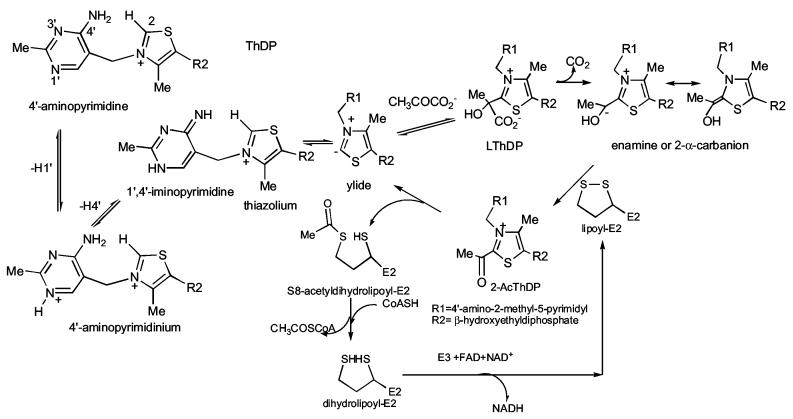
During the past few years, the Rutgers group has shown that the pyruvate analogue methyl acetylphosphonate sodium salt (MAcPho), when added to the thiamin diphosphate(ThDP)-dependent enzymes yeast pyruvate decarboxylase (YPDC) and the E1 subunit (E1ec) of the E. coli pyruvate dehydrogenase complex (PDHc-ec) generates a new UVVIS and a positive circular dichroism (CD) band in the wavelength range of 300-310 nm [1,2]. The same spectrum also results when the preassembled C2α-phosphonolactylthiamin diphosphate (PLThDP, synthesized from thiamin diphosphate and MAcPho) is added to the apo form of E1ec (no ThDP bound) [1]. This electronic transition was assigned to the 1',4'-iminopyrimidine tautomer of the ThDP coenzyme in the bound form on the basis of chemical model studies [3-5]. Concurrently, high-resolution X-ray structure determinations on E1ec with the pre-assembled PLThDP [6], and on pyruvate oxidase from Lactobacillus plantarum with MAcPho added [7] showed that a tetrahedral adduct was being formed between the keto carbon of the MAcPho and the C2 thiazolium atom. In view of these complementary observations, we concluded that the PLThDP when enzyme-bound exists in the 1',4'-iminopyrimidine tautomeric form. The reaction scheme for PDHc is outlined in Scheme 1. Hence, MAcPho can indeed act as a substrate analogue as shown in Scheme 2, but, once the reaction reaches the intermediate state corresponding to LThDP, by virtue of the non-cleavable C-P bond in place of the usual C-C bond, the adduct simply occupies the active center with very long lifetime. The UV and CD results inform us that a consequence of the presence of this stable pre-decarboxylation analogue at the active site is that with the PLThDP in place of ThDP, the 1'.4'-iminopyrimidine tautomer of ThDP is the predominant tautomeric form rather than one of the other two forms on the left-hand side of Scheme 2.
Scheme 1.
Reactions of pyruvate dehydrogenase complexes
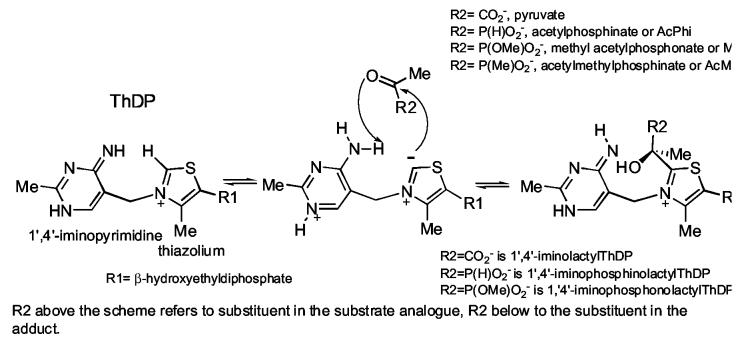
Scheme 2.
Formation of LThDP and its phosphonate and phosphinate analogues
These results encouraged us to synthesize and test two further electrostatic mimics of pyruvate, sodium acetyl phosphinate (AcPhi) and sodium acetyl methylphosphinate (AcMPhi). These compounds possess the advantage over the phosphonate compound used previously in having no ester group present. In this fashion, the –PO2− group becomes a better steric and electrostatic mimic of pyruvate's –CO2− group. Similarly to MAcPho, when bound to ThDP, AcMPhi and AcPhi can not be decarboxylated and form stable analogues of LThDP (Scheme 2). We here report on the kinetics of binding and inactivation of E1ec (homodimer) and the human pyruvate dehydrogenase E1 (α2β2 heterotetramer) (E1h) by AcPhi and AcMPhi. Both substrate analogues behave as tight-binding reversible inhibitors with micromolar values of Kd and Ki. Inactivation of E1ec and E1h resulted on addition of stoichiometric amounts of substrate analogues. The formation of stable analogues of LThDP could be monitored by CD spectroscopy providing strong evidence regarding the mechanism of inhibition. We wish to compare and contrast the inhibition experienced by the human and bacterial PDHc E1.
2. Materials and methods
Methods
2.1. Chemical synthesis
Synthesis of acetyl phosphinate was carried out according to the reported procedure by Baillie et al. [8].
Methyl phosphonousdichloride was purchased from Aldrich and was used without further purification. NaI was dried at 70 °C under 0.1 mm/Hg overnight. Dry acetone was prepared with dry NaI according to literature procedures. NMR spectra were recorded in a 500 MHz Varian INOVA spectrometer NMR instrument.
Synthesis of methyl (1,1-dimethoxyethyl)methylphosphinate
Methyl phosphonousdichloride (5.0 g, 42.7 mmol) was added dropwise, with vigorous stirring to trimethyl orthoacetate (11.67 g, 97.2 mmol) at −30 °C under N2. After the addition was complete the temperature of the reaction mixture was maintained at −30 °C for one h, then the reaction mixture was allowed to warm to room temperature overnight. The excess trimethyl orthoacetate was removed under reduced pressure. The pure product (b.p. 103 °C, 3.2 mm/Hg) was obtained by fractional distillation of the residual reaction mixture. Yield 6.337 g (82%). 1H NMR (500 MHz, CDCl3/TMS): δ 3.801 (d, 3H, J=10.5 Hz,), 3.405 (d, 6H, J= 8 Hz,), 1.490 (d, 3H, J= 3.5 Hz), 1.465 (d, 3H, J =1 Hz). MS (EI) 151, 109, 89, 77, 43.
Synthesis of sodium acetylmethylphosphinate
Methyl (1,1-dimethoxyethyl)methylphosphinate (500 mg, 2.75 mmol) was added to a solution of NaI (8.25 g, 55 mmol) in 30 mL dry acetone under N2 in a sealed tube. The reaction mixture was heated at 70 °C overnight. After cooling to room temperature, the reaction mixture was evaporated to dryness and triturated with ethyl acetate. The crude product was recrystallized from absolute ethanol. Yield 134 mg (37.5 %). 1H NMR (500 MHz, D2O/DSS): δ 2.461 (d, 3H, J=3.5 Hz), 1.406 (d, 3H, J=14 Hz). MS (ESI) 153 (M+ Na) +.
2.2. Protein expression and purification
Bacterial strains and plasmids
E. coli strain JRG 3456 and plasmid pGS878 with aceE1 gene encoding the E1ec subunit of PDHc-ec were from J. Guest (University of Sheffield, UK). A co-expression vector for E1h (pET-28b-PDHA1/PDHB) harboring coding sequences of both human PDHc-E1α and PDHc-E1β subunits was constructed as described previously [9].
The E. coli strain JRG 3456 transformed with pGS878 was used for overexpression of the aceE gene encoding E1ec subunit of PDHc-ec according to reference [10] and purification of E1ec followed the published procedure [11].
The E1h was overexpressed in E. coli BL21 cells harboring pET-28b-PDHA1/PDHB (a coexpression vector with coding sequences of both human E1α and E1β subunits) and purified using Ni-NTA-agarose chromatography as described previously [9].
Recombinant human dihydrolipoamide acetyltransferase (E2h)-dihydrolipoamide dehydrogenase-binding protein (E3BPh) subcomplex and dihydrolipoamide dehydrogenase (E3h) were overexpressed in E. coli and purified as described elsewhere [12].
2.3.a. Activity measurements of E1ec
The E1ec activity was measured in the overall PDHc reaction. First E2ec and E3ec expressed independently were incubated for 1 h at a ratio of 1.0:1.0 (mg/mg) in 20 mM potassium phosphate buffer (pH 7.0) at 4 °C. The E1ec was reconstituted with E2ec-E3ec subcomplex at a ratio of 1:1:1 (mg/mg/mg) with an additional 10 min of incubation. The reaction medium contained (in 1 ml of test volume) 0.10 M Tris HCl (pH 8.0), 1 mM MgCl2, 2 mM sodium pyruvate, 2.5 mM NAD+, 0.20 mM ThDP, and 2.6 mM dithiothreitol at 30 °C. The reaction was initiated by the addition of 0.20 mM CoA and PDHc-ec (1-1.5 μg of protein). Steady-state velocities were taken from the linear portion of the progress curve. One unit of activity is defined as the amount of NADH produced (μmol/min/mg of E1ec).
Activity measurements of E1h
Activity of E1h was determined by two assays: (i) PDHc assay, by the formation of NADH during overall PDHc reaction after reconstitution of E1h with E2h-E3BPh subcomplex and E3h into PDHc; and (ii) by DCPIP assay, by the reduction of 2,6-dichlorophenolindophenol (DCPIP), an artificial electron acceptor, to measure the first partial reaction of E1h, the decarboxylation of pyruvate in the absence of the second substrate, lipoyl moieties of E2h, as described previously [12].
Protein content was measured by the Bradford method using bovine serum albumin as a standard [13].
2.3.b. Kinetic analysis of inhibition of E1h by AcPhi and AcMPhi
To study the inhibition of E1h by AcPhi and AcMPhi, E1h (5 μg) was incubated at room temperature in 50 μl of 50 mM potassium phosphate buffer (pH 7.0) with 2 mM MgCl2, 0.2 mM ThDP and the concentration of substrate analogues indicated. Three-four aliquots (1 μg of E1h each) were withdrawn after 30 min and mixed into a cuvette containing 3 μg of E2h-E3BPh subcomplex, 3 μg of E3h and all reagents necessary to measure PDHc activity except pyruvate. The entire complex was reconstituted for 1 min and the reaction was started by addition of pyruvate and was monitored for 30 sec to record the initial velocity. To determine the inactivation of E1h and its αH63A variant by substrate analogues, 200 μg of E1h or E1h αH63A was incubated at room temperature in 200 μl of 50 mM potassium phosphate buffer (pH 7.0) with 2 mM MgCl2, 0.2 mM ThDP and the indicated concentrations of substrate analogues. Three aliquots (25 μg for wild-type E1h and 50 μg for E1h αH63A) were taken after 30 min to measure the activity of the E1h per se according to the DCPIP assay [12].
2.3.c. Reaction progress curve analysis
E1ec
The progress of the overall PDHc reaction was monitored via the formation of NADH at 340 nm as a function of time (the reaction time was approximately 18 min). The reaction was started by the addition of PDHc after the E1ec was reconstituted with E2-E3 subcomplex (total protein about 0.60 μg) and CoA. A family of progress curves was recorded in the presence of 0.50 mM pyruvate and AcMPhi (0.1-4.5 μM) or AcPhi (0.05 –1..2 μM) and the values of kapp were calculated as reported earlier [10]. The values of Kd were calculated form the plot of kapp vs concentration of substrate analogue according to Kitz and Wilson [14].
E1h
Progress curve analysis was used to determine the Ki for inhibition of wild-type E1h by AcPhi and AcMPhi [10,15]. The PDHc was reconstituted in the cuvette by mixing 0.3 μg of E1h, 1 μg of E2h-E3BP and 1 μg of E3h. The reaction was initiated by addition of pyruvate (300 μM) along with different concentrations of AcPhi (0.4-1.8 μM) or AcMPhi (200-600 μM) as indicated in the figure legends. The progress of the reaction was monitored for 5 min. Less than 5% of pyruvate was consumed under these conditions. The progress curves were analyzed according to equation 1:
| (1) |
where P is the product concentration at time t, vs is the steady-state reaction velocity, vo is the initial velocity and k is the apparent pseudo-first-order rate constant. The dependence of kapp on substrate analogue concentration was linear for both compounds, enabling calculation of k1 and k−1, the rate constants for binding and dissociation of substrate analogue, respectively, were calculated from equation 2:
| (2) |
where I0 is the concentration of substrate analogue, S0 is the concentration of pyruvate used and Km is the value of Km for pyruvate. The values of Ki were calculated from: Ki= k−1/k1. Regression coefficients were > 0.95.
2.4. Circular dichroism spectroscopy
Circular dichroism titration of E1ec with sodium acetylphosphinate and sodium acetylmethylphosphinate
The E1ec (1.50 mg/ml, concentration of active centers = 15.0 μM) in 20 mM potassium phosphate buffer (pH 7.0) containing 2 mM MgCl2 and 0.20 mM ThDP was titrated with 1-100 μM of AcPhi. For experiments with AcMPhi, E1ec (1.63 mg/ml, concentration of active centers = 16.2 μM) in 20 mM potassium phosphate buffer (pH 7.0) containing 0.20 mM ThDP and 2.0 mM MgCl2 was titrated with 1-150 μM of AcMPhi. The difference CD spectra with both substrate analogues were obtained on subtraction of the CD spectrum of E1ec in the presence of ThDP and MgCl2. The Kd for AcPhi or AcMPhi were calculated from a plot of ellipticity at 317 nm vs. concentration of AcPhi or AcMPhi using a quadratic equation [10]. CD spectra were recorded on an Aviv Model 202 CD or an Applied Photophysics Chirascan spectrometer at 30 °C.
Circular dichroism titration of E1h with sodium acetylphosphinate and sodium acetylmethylphosphinate
The E1h (1.50 mg/ml, concentration of active centers = 19.6 μM) in 20 mM potassium phosphate buffer (pH 7.0) containing 1 mM MgCl2 and 0.20 mM ThDP was titrated with 1-150 μM of AcPhi. For experiments with AcMPhi, E1h (1.06 mg/ml, concentration of active centers = 13.8 μM) in 20 mM potassium phosphate buffer (pH 7.0) containing 1 mM MgCl2 and 0.20 mM ThDP was titrated with 1-150 μM of AcMPhi. A plot of ellipticity at 305 nm vs. [AcMPhi] was constructed by subtracting the ellipticity of the enzyme at the same wavelength but recorded in the absence of AcMPhi. The values of Kd were calculated using a quadratic equation [10].
3. Results
Kinetic analysis of the inhibition of E1ec by sodium acetyl phosphinate
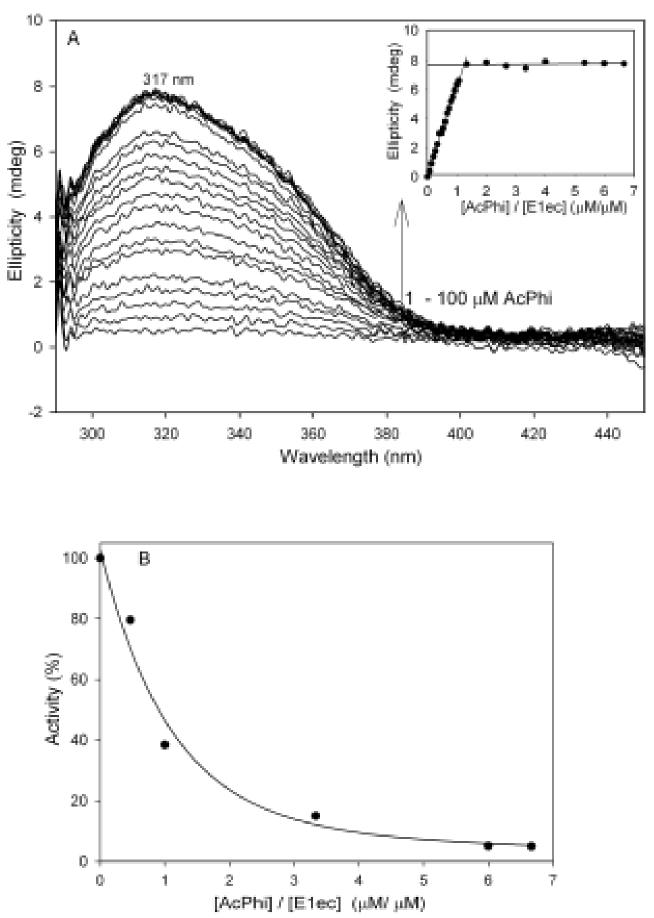
As reported previously, the AcPhi behaves as a tight-binding reversible inhibitor of the PDHc-ec (Ki=0.15μM) and of the E1ec resolved from the complex (Ki=0.21μM) [16]. We studied inhibition of the recombinant E1ec (concentration of active centers = 0.50 μM) by preincubating it with 0.50-1000 μM AcPhi for 20 min at 25 °C and then measuring the activity after reconstitution with its E2ec-E3ec subcomplex. The overall PDHc reaction was triggered by adding CoA and PDHc to the reaction mixture. More than 90 % of E1ec activity was lost at a [AcPhi]/[E1ec] ratio of 1.4/1.0 (data not shown). These kinetic data are in good agreement with a CD titration experiment. On addition of AcPhi to the E1ec, a broad CD band with maximum at 317 nm was observed (Figure 1A). We assigned this band to the 1',4'-iminopyrimidine tautomer of phosphinolactyl-ThDP (iminophosphinolactylThDP), a stable analogue of LThDP, on the basis of similar results obtained with MAcPho (Figure 2) and PLThDP [1]. The CD band at 317 nm reached saturation at a [AcPhi]/[E1ec] ratio of 1.3/1.0 which correlates with the loss of 85% of E1ec activity measured in a parallel experiment (Figure 1B). A Kd=0.062 μM was calculated for AcPhi. Inhibition by AcPhi was reversible according to the following results: (i) approximately 50 % of activity was recovered after overnight dialysis against 20 mM potassium phosphate buffer (pH 7.0) from a sample that had first been inhibited to 95%; (ii) the amplitude of the CD band at 317 nm was reduced by 80% after overnight dialysis (20% of the CD signal was retained on E1ec). These results suggest that AcPhi behaves as a stoichiometric reversible inhibitor of the E1ec. The binding of AcPhi leads to the formation of 1',4'-iminophosphinolactyl-ThDP, a covalent analogue of the LThDP intermediate clearly indicating mechanism-based inhibition.
Figure 1.
CD titration of E1ec by AcPhi: A. Difference spectra of E1ec on addition of AcPhi. Inset: Dependence of the ellipticity at 317 nm on the [AcPhi] / [E1ec active centers in homodimer] ratio. B. E1ec activity remaining with increasing [AcPhi] / [E1ec] ratio. Conditions under Materials and Methods.
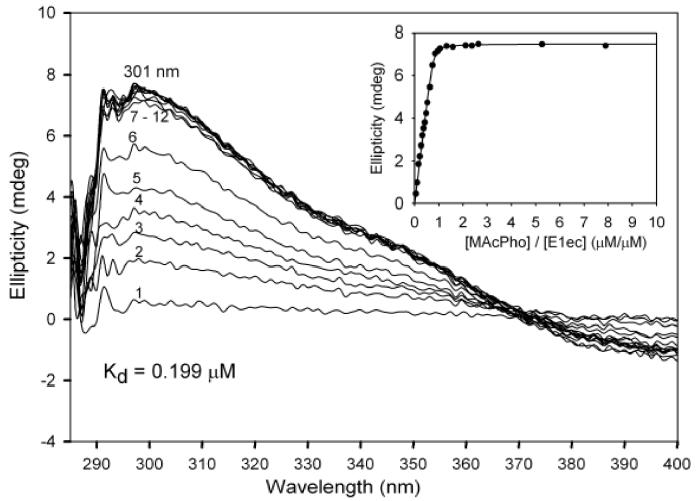
Figure 2.
Difference CD spectra of E1ec on addition of MAcPho. E1ec (1.90 mg/ml, concentration of active centers = 19 μM) in 20 mM potassium phosphate buffer (pH 7.0) containing 1 mM MgCl2 and 0.20 mM ThDP was titrated by MAcPho (μM): spectrum 1 (1.0); 2 (3.0); 3 (5.0); 4 (7.0); 5 (9.0); 6 (12.0); 7 (16.0); 8 (18.0); 9 (20.0); 10 (40.0); 11 (50.0); 12 (150.0). The difference CD spectra were obtained on subtraction of the spectrum of E1ec in the presence of MgCl2 and ThDP. Inset: dependence of the ellipticity at 301 nm on [MAcPho] / [E1ec] ratio.
Circular dichroism analysis of binding of sodium acetylmethylphosphinate to E1ec
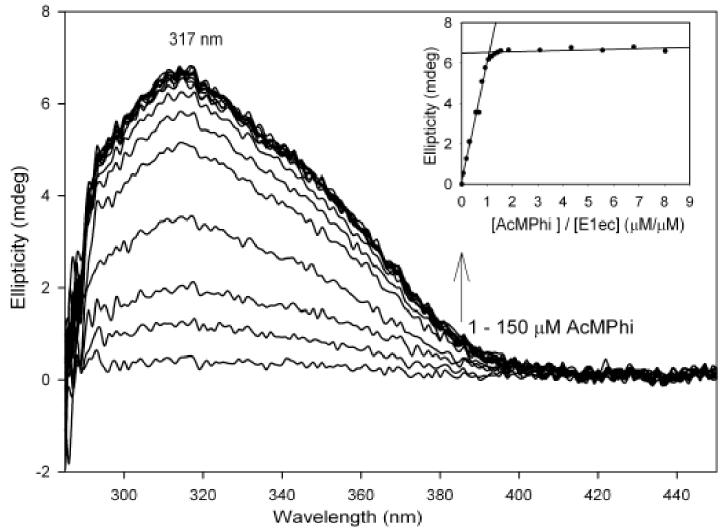
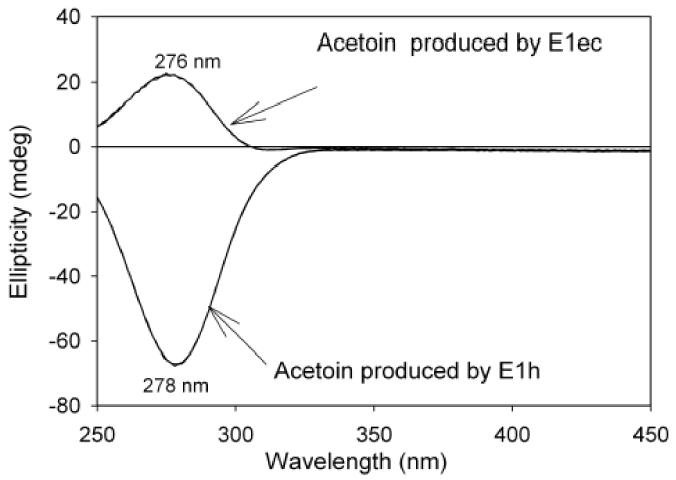
Addition of AcMPhi to the E1ec also resulted in a CD band with maximum at 317 nm suggesting formation of 1',4'-imino,methylphosphinolactyl-ThDP. The maximum at 317 nm was reached at a 1.0/1.0 ratio of [E1ec]/[AcMPhi] indicating stoichiometric binding of AcMPhi (Kd=0.10 μM) (Figure 3). After overnight dialysis approximately 80% of the CD intensity at 317 nm was retained on the enzyme indicating that binding is partially reversible. However, on addition of 2 mM pyruvate, the AcMPhi was displaced from the E1ec, as indicated by the formation of (S)-acetoin according to the CD spectra after removal of protein (Figure 4).
Figure 3.
Difference spectra of E1ec on addition of AcMPhi. Inset: Dependence of the ellipticity at 317 nm on [AcMPhi]/ [E1ec] ratio. Conditions under Materials and Methods.
Figure 4.
Acetoin production by E1ec and E1h. E1ec titrated with AcMPhi (as in Figure 3.) was dialyzed overnight against 20 mM potassium phosphate buffer (pH 7.0). Then 2 mM MgCl2, 0.20 mM ThDP and 2 mM pyruvate were added to the dialyzed E1ec and CD spectra were recorded at different times. After 50 min of incubation of E1ec with 2 mM pyruvate the protein was removed by centrifugation using a Centricon 30 and the spectrum of the reaction mixture was recorded. A similar experiment was conducted with E1h.
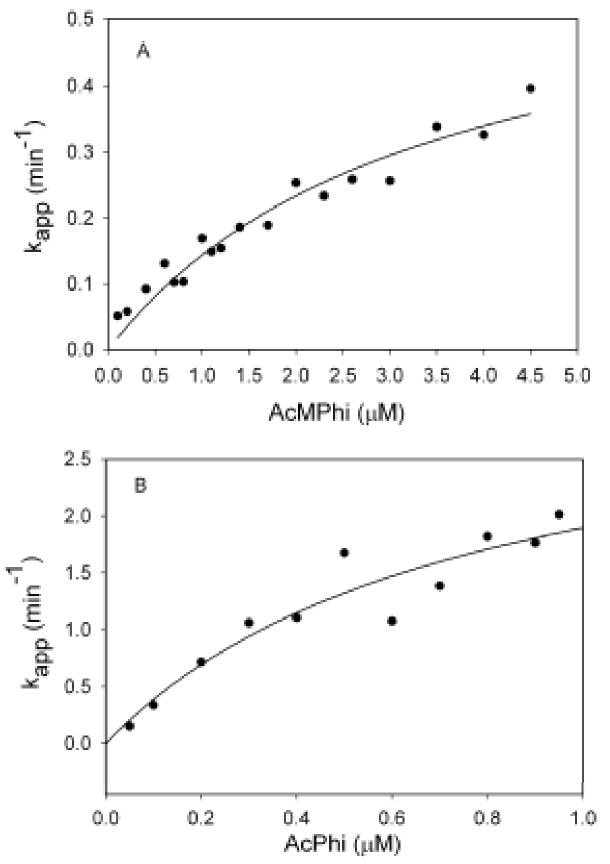
Progress curve analysis of the inhibition of E1ec by sodium acetylphosphinate and sodium acetylmethylphosphinate
The inhibition by AcPhi and AcMPhi was studied using reaction progress curve analysis as we reported earlier [10]. The dependence of the pseudo first-order rate constant (kapp) on the concentration of AcPhi or AcMPhi was hyperbolic, indicating saturation (Figures 5A and 5B) leading to values of Ki of 0.76 μM for AcPhi and 3.33 μM for AcMPhi according to a Kitz-Wilson treatment [ref. 14; Table 1]. The data suggest that inhibition of E1ec by AcPhi or AcMPhi follows the mechanism presented in Scheme 3 for competitive tight-binding inhibitors [17]. The kinetic parameters for inhibition of E1ec by AcPhi and AcMPhi are presented in Table 1.
Figure 5.
Determination of Ki for inhibition of E1ec by AcMPhi and AcPhi from progress curve analysis. Dependence of kapp of inhibition of E1ec by different concentrations of AcMPhi (A) and AcPhi (B). Conditions in Materials and Methods.
Table 1.
Kinetic parameters for the inhibition of E1ec by substrate analogues.
| Compound | Ki (μM) | kinact (min−1) | kinact/Ki (μM min)−1 | Kd (μM) |
|---|---|---|---|---|
| Acetylphosphinate | 0.76 | 3.34 | 4.40 | 0.060a |
| Acetylphosphinateb | 0.12 | 1.30 | 10.83 | 0.005 |
| Acetylphosphinated | 0.33 | 0.55 | 1.67 | |
| Acetylmethylphosphinate | 3.33 | 0.62 | 0.19 | 0.10a |
| Methyl acetylphosphonate | 0.199a | |||
| 2-oxo-3-butynoic acidc | 8.5 | 1.79 | 0.21 | 0.20 |
Ki is the dissociation constant for the initial reversible complex (from kinetic experiment) obtained in the presence of 0.50 mM pyruvate in the incubation mixture.
kinact is the rate constant for conversion of the reversible complex to irreversibly inactivated enzyme
kinact/Ki is the second order rate constant for inactivation
Kd is the dissociation constant obtained from CD analysis.
Determined from kinetic analysis of inhibition for E. coli PDHc from ref. 16.
Determined from kinetic analysis of inhibition for PDHc from bovine heart from ref 19.
Scheme 3.

Kinetic analysis of inhibition of E1h by acetylphosphinate and acetylmethylphosphinate
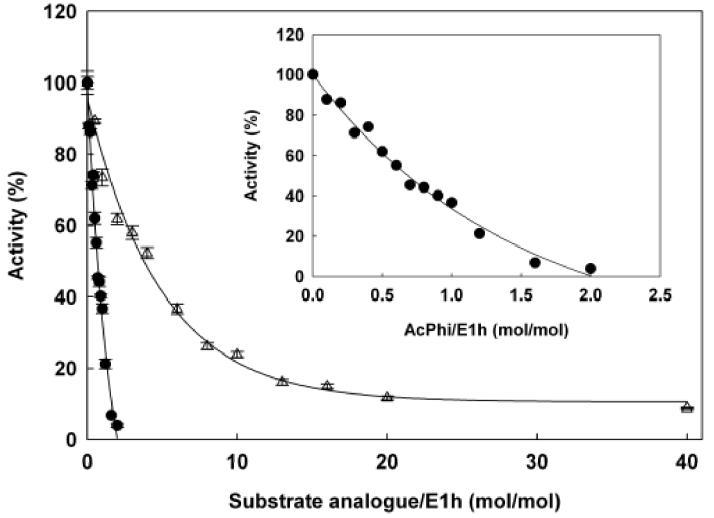
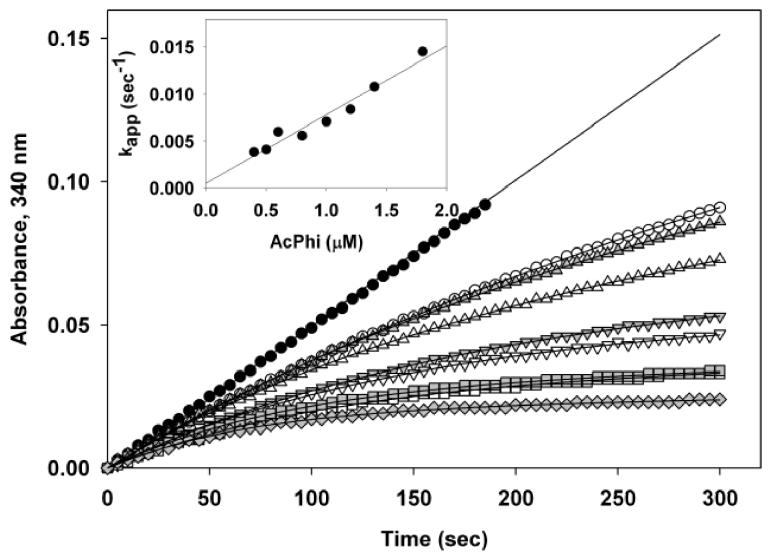
The following results were obtained for inhibition of E1h by AcPhi: (i) full inhibition was achieved with stoichiometric concentrations of AcPhi (Figure 6); (ii) inhibition by AcPhi displayed time dependence, reaching its maximum at approximately 20 min (data not shown); and (iii) inhibition by AcPhi was partially reversible, judging by the partial restoration of activity when an aliquot of inactivated E1h was used in the overall PDHc assay and the reaction was allowed to proceed for longer than 1 min. Kinetic analysis indicated that AcPhi is a tight slow-binding inhibitor of E1h [15]. Analysis of the progress curves of the overall PDHc reaction (E1h was reconstituted with E2h-E3BPh and E3h) in the presence of pyruvate and AcPhi enabled calculation of Ki= 0.014 μM, k−1 = 0.0305 min−1 and k1= 2.18 μM−1min−1 (Figure 7; Table 2).
Figure 6.
Inhibition of E1h by AcPhi ( ) and AcMPhi (
) and AcMPhi ( ). E1h (0.649 μM) was incubated in 50 mM potassium phosphate buffer (pH 7.0), 2 mM MgCl2, 0.2 mM ThDP with different concentrations of AcPhi or AcMPhi for 30 min. Aliquots were taken to determine activity of E1h in the PDHc assay as described in Materials and Methods. 100% of activity was determined in the absence of inhibitors in the incubation mixture and corresponds to 32 U/mg of E1h protein. Data points are means ± SE (n = 3-4). Inset shows inhibition of E1h by AcPhi at lower concentrations.
). E1h (0.649 μM) was incubated in 50 mM potassium phosphate buffer (pH 7.0), 2 mM MgCl2, 0.2 mM ThDP with different concentrations of AcPhi or AcMPhi for 30 min. Aliquots were taken to determine activity of E1h in the PDHc assay as described in Materials and Methods. 100% of activity was determined in the absence of inhibitors in the incubation mixture and corresponds to 32 U/mg of E1h protein. Data points are means ± SE (n = 3-4). Inset shows inhibition of E1h by AcPhi at lower concentrations.
Figure 7.
Progress curve analysis of E1h inhibition by AcPhi. E1h (0.3 μg, 1.95 nM) was reconstituted to PDHc-h with 1 μg of E2h-E3BPh and 1 μg of E3h and the reactions were started by the addition of 300 μM pyruvate and varying concentrations of AcPhi (in μM): 0 ( ); 0.4 (
); 0.4 ( ); 0.5 (
); 0.5 ( ); 0.6 (
); 0.6 ( ); 0.8 (
); 0.8 ( ); 1.0 (
); 1.0 ( ); 1.2 (
); 1.2 ( ); 1.4 (
); 1.4 ( ); 1.8 (
); 1.8 ( ). Lines represent the regression curves generated by SigmaPlot 2001 based on the equation [1] (see Materials and Methods). Inset shows the dependence of kapp on the concentration of APhi.
). Lines represent the regression curves generated by SigmaPlot 2001 based on the equation [1] (see Materials and Methods). Inset shows the dependence of kapp on the concentration of APhi.
Table 2.
Kinetic parameters for the inhibition of E1h by substrate analogue inhibitors.
| Compound | Ki (μM) | k1 (μM−1min−1) | k−1 (min−1) | Kd (μM) |
|---|---|---|---|---|
| Acetylphosphinate | 0.014 | 2.18 | 0.0305 | N/A |
| Acetylmethylphosphinate | 1.46 | 0.0101 | 0.0148 | 0.45 |
Kd is measured from CD titration where experimentally accessible.
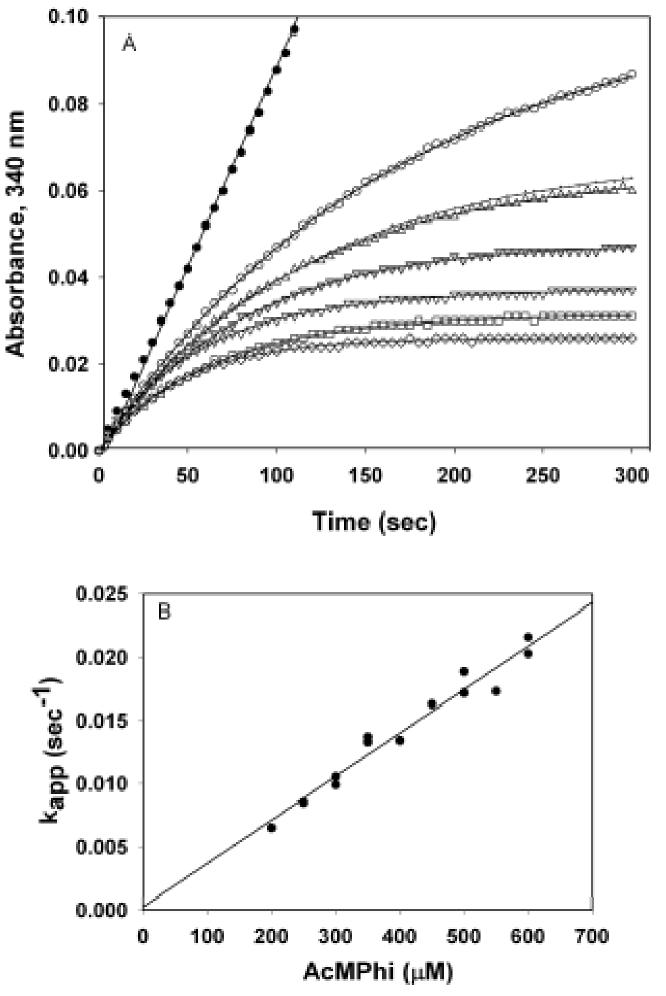
The AcMPhi is a weaker inhibitor than AcPhi of E1h (Figure 6). Progress curve analysis indicated inhibition of E1h at higher concentrations of AcMPhi with Ki = 1.46 μM, k−1 = 0.0148 min−1 and k1 = 0.0101 μM−1 min−1 (Figure 8; Table 2).
Figure 8.
Progress curve analysis of E1h inhibition by AcMPhi. A. E1h (0.3 μg, 1.95 nM) was reconstituted to PDHc-h with 1 μg of E2h-E3BP and 1 μg of E3h and the reactions were started by the addition of 300 μM pyruvate and varying concentrations of AcMPhi (top in μM): 0 ( ); 200 (
); 200 ( ); 250 (
); 250 ( ); 300 (
); 300 ( ); 400 (
); 400 ( ); 450 (
); 450 ( ); 500 (
); 500 ( ); 600 (
); 600 ( ). Lines represent the regression curves generated by SigmaPlot 2001 based on the equation [1] (see Materials and Methods). Several curves used for calculations of kapp are omitted for clarity. B. Dependence of kapp on the concentration of AcMPhi.
). Lines represent the regression curves generated by SigmaPlot 2001 based on the equation [1] (see Materials and Methods). Several curves used for calculations of kapp are omitted for clarity. B. Dependence of kapp on the concentration of AcMPhi.
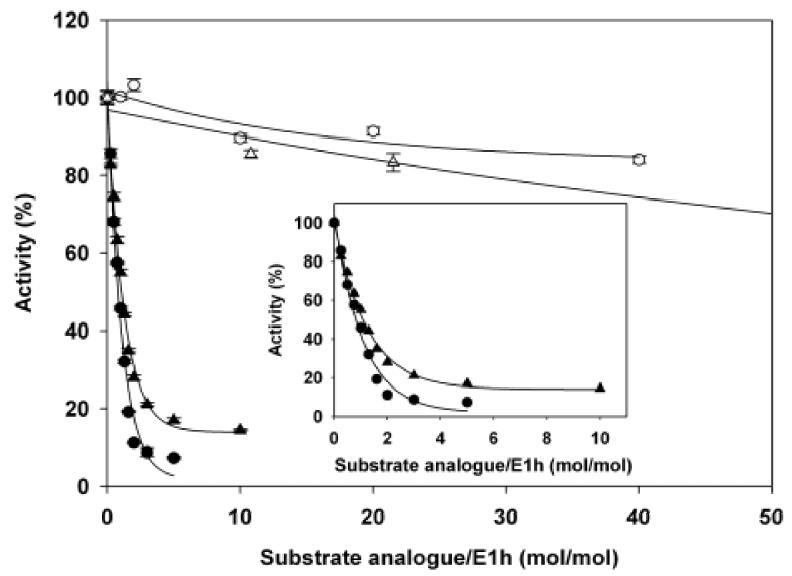
Kinetic analysis of inhibition by AcPhi and AcMPhi according to the DCPIP assay (measuring the rate through the decarboxylation step) revealed similar patterns of inhibition: AcPhi was a stronger inhibitor than AcMPhi (Figure 9). The stoichiometry of inhibition of E1h by AcPhi and AcMPhi when determined with the DCPIP assay was similar to the one determined with the PDHc-h assay (compare Figure 6 to Figure 9 inset).
Figure 9.
Inhibition of wild-type E1h and E1h αH63A by AcPhi and AcMPhi. Symbols are: wild-type E1h inhibited by AcPhi ( ) and AcMPhi (
) and AcMPhi ( ); αH63A E1h inhibited by AcPhi (
); αH63A E1h inhibited by AcPhi ( ) and AcMPhi (
) and AcMPhi ( ). E1h (6.49 μM) or its αH63A variant was incubated in 50 mM KH2PO4 (pH 7.0), 2 mM MgCl2, 0.2 mM ThDP and different concentrations of AcPhi or AcMPhi for 30 min. Aliquots were taken to determine the E1-specific activity according to the DCPIP assay as described in Methods. 100% of the activity was determined in the absence of inhibitors in the incubation mixture and corresponds to 231 mU/mg of E1 protein. Data points are means ± SE (n = 3). Inset shows inhibition of the wild-type E1h by AcPhi (
). E1h (6.49 μM) or its αH63A variant was incubated in 50 mM KH2PO4 (pH 7.0), 2 mM MgCl2, 0.2 mM ThDP and different concentrations of AcPhi or AcMPhi for 30 min. Aliquots were taken to determine the E1-specific activity according to the DCPIP assay as described in Methods. 100% of the activity was determined in the absence of inhibitors in the incubation mixture and corresponds to 231 mU/mg of E1 protein. Data points are means ± SE (n = 3). Inset shows inhibition of the wild-type E1h by AcPhi ( ) and AcMPhi (
) and AcMPhi ( ) at lower concentrations.
) at lower concentrations.
Both AcPhi and AcMPhi are much stronger inhibitors of E1h compared to MAcPho. MAcPho did not result in more than 20% reduction in E1h activity even at 500-fold molar excess (data not shown).
The E1h αH63A variant has undetectable activity when measured in the overall PDHc assay and about 17% when measured with the DCPIP assay. For that reason, inhibition of E1h αH63A could only be studied using the DCPIP assay. Only modest inhibition of E1h αH63A is detected with both AcPhi and AcMPhi (Figure 9), indicating that E1h αH63 is necessary for the interaction with both substrate analogues and is probably involved in the interaction with pyruvate during the decarboxylation reaction.
Circular dichroism titration of E1h with sodium acetylmethylphoshinate
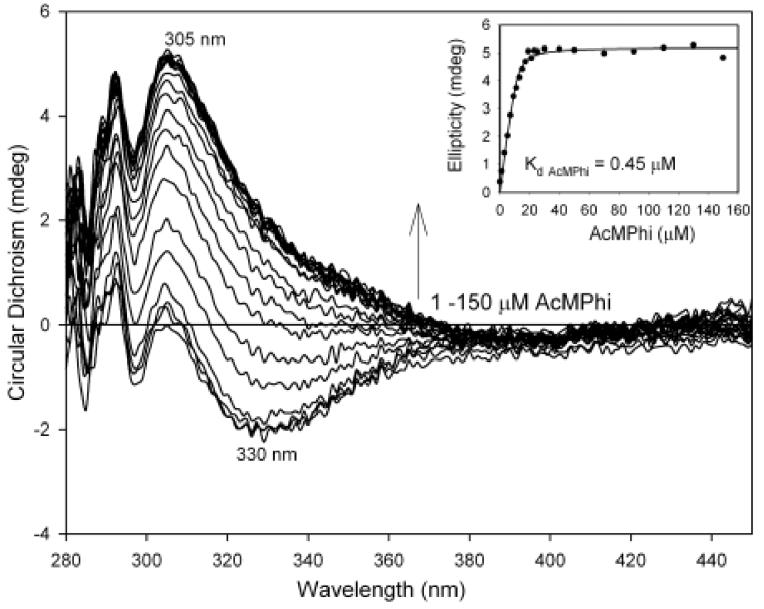
On addition of 1-150 μM of AcMPhi to the E1h the positive band at 305 nm developed and reached the maximum intensity at a molar ratio of [AcMPhi] / [E1h active centers] equal to 2.0/1.0 (Fig. 10). Similar spectra with CD maximum at 306 nm were obtained when AcPhi was used with molar ratio of [AcMPhi ]/ [E1h] equal to 1.0/1.0 (data not shown). A Kd = 0.45 μM was calculated for AcMPhi binding (the Kd for AcPhi is not available). The CD spectra of E1h treated by AcMPhi and AcPhi were not affected by overnight dialysis indicating tight binding. On addition of 5 mM pyruvate to either the E1h.AcPhi or E1h.AcMPhi complex, the CD band at 305-306 nm disappeared suggesting that pyruvate competes with the substrate analogues at the active centers of the E1h, similarly to that seen with E1ec (data not shown). Removal of the enzyme from the incubation mixture revealed the presence of a species with a negative CD band with maximum at 278 nm corresponding to (R)-acetoin. This is in interesting contrast to the (S)-acetoin produced by E1ec (Figure 4) [18]. The data represent the first evidence for the formation of the chiral acetoin by the E1h.
Figure 10.
CD titration of E1h with AcMPhi. The E1h was titrated with the indicated concentrations of AcMPhi, and led to the Kd of 0.45 μM (see inset for ellipticity at 305 nm vs. inhibitor concentration). For conditions see Materials and Methods.
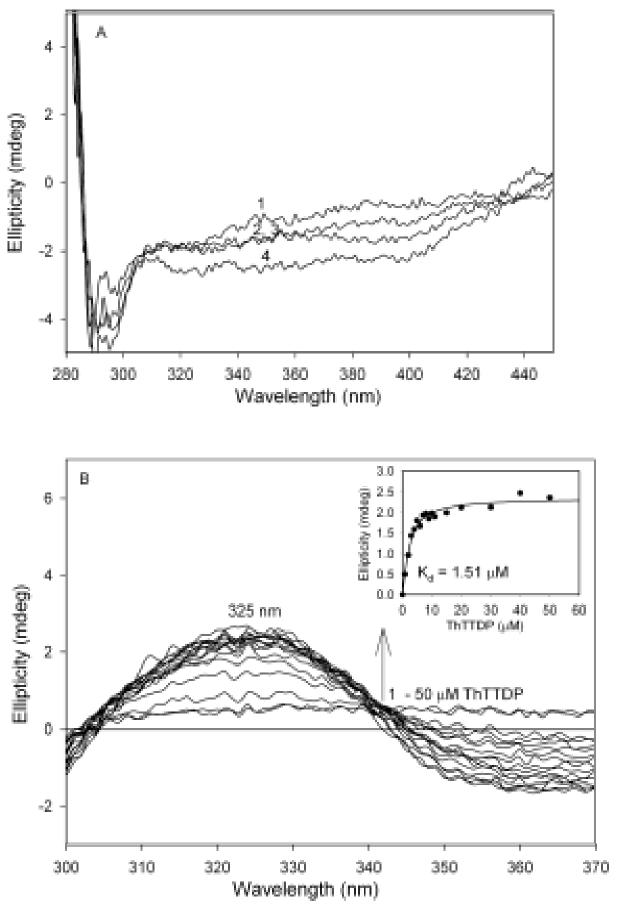
Addition of 1-150 μM of AcPhi to the αH63A E1h variant in the presence of 0.60 mM ThDP did not produce the positive CD band with maximum at 305 nm (Figure 11 A) indicating that E1h αHis63A is important in the reaction(s) leading to formation of LThDP. Significantly, nor did the E1h αH63A variant produce the negative CD band with maximum at 330 nm on addition of 0.20 –0.60 mM ThDP, a signature of ThDP binding (curve 1 in Figure 11A presents the CD spectrum of E1h αH63A in the presence of 0.60 mM ThDP). To confirm that ThDP did bind in the active centers of E1h αH63A, the variant was titrated by thiamin 2-thiothiazolone diphosphate (ThTTDP), a transition state analogue (Figure 11B) that gives a positive CD band near 330 nm, as reported earlier for the E1ec [10]. The data presented in Figure 11B indicate binding of the ThTTDP in the active centers of E1h αHis63A. A Kd of 1.51 μM was obtained for binding ThTTDP to the E1h αH63A, as compared with the value of Kd = 0.59 μM obtained for the wild-type E1h (Korotchkina, unpublished results).
Figure 11.
CD spectra of E1h αH63A titrated by AcPhi. (A) E1h αH63A (concentration of active centers = 16.4 μM) in 20 mM potassium phosphate buffer (pH 7.0) containing 0.60 mM ThDP and 1.0 mM MgCl2 (spectrum 1) was titrated with 5 μM (spectrum 2), 50 μM (spectrum 3) and 150 μM (spectrum 4) of AcPhi. (B) CD spectra of E1h αH63A titrated by thiamin 2-thiothiazolone diphosphate (ThTTDP). E1h αH63A (concentration of active centers = 10.2 μM) in 20 mM potassium phosphate buffer (pH 7.0) was titrated by 1-50 μM ThTTDP. Inset: Dependence of the ellipticity at 325 nm on concentration of ThTTDP.
Discussion
One of our goals in designing and testing inhibitors for ThDP enzymes is to achieve enzyme-selective inhibition. In this paper we wish to compare and contrast inhibition of the bacterial (E1ec) and human (E1h) pyruvate dehydrogenase components by the same substrate analogue-type inhibitor. While inhibition of E1ec by AcPhi and AcMPhi was studied in several laboratories, we here present the first inhibitory analysis of the E1h by such compounds. In an earlier study Laber and Amrhein [19] demonstrated that the compound 1-aminoethylphosphinate [H3C-CH(NH2)-PO2H2] inhibited the PDHc from bovine heart in the presence of alanine aminotransferase and a similar inhibitory effect was observed when synthetic AcPhi was used (see Table 1, ref. 19). Since those early results, inhibition by AcPhi of several ThDP-dependent enzymes was reported, including E1ec [16], pyruvate decarboxylase (YPDC) from brewer's yeast [19], and both YPDC and pyruvate oxidase (POX) from Lactobacillus plantarum [20]. A comparison of kinetic constantswith ours on the recombinant E1ec (Table 1), confirms that AcPhi is a very powerful inhibitor of E1ec, as is to a lesser extent AcMPhi.
The positive broad unsymmetrical band CD band at 317 nm resulting from mixing E1ec and AcPhi signaled formation of 1', 4'-iminophosphinolactyl-ThDP, since this band is a reporter of the tetrahedral intermediates in ThDP enzymes [1,2]. The broad CD band in Figure 1 could be deconvoluted into two bands [21], one of which has a maximum at 309 nm, the other at 343 nm. The CD evidence elucidates the mechanism of inhibition, consistent with data published at Rutgers and elsewhere for interaction of phosphonates and phosphinates with ThDP enzymes (Scheme 2) [1-7].
Regarding the kinetic behavior of E1h with the substrate analogues (Table 2), progress curve analysis revealed that the inhibition involves a single step since there is no evidence for the presence of an E·I* intermediate in Scheme 2 [17]. The analysis yields a second-order rate constant for substrate analogue binding and a first-order ‘off’ rate constant for the dissociation of substrate analogue. A comparison of the second-order rate constants for the human and bacterial enzyme indicates that AcPhi is a strong inhibitor of both E1ec and E1h. Also, the value of kinact/ Ki = 1.67 (μM min)−1 for inhibition of PDHc from bovine heart by AcPhi, is similar to that for E1ec and E1h in Table 1 [19]. For the interaction of YPDC with AcPhi mixed type of inhibition was observed, with Ki = 0.47 mM for competitive inhibition and Ki= 7.14 mM for the non-competitive inhibition [20]. A value of Ki = 50 μM was calculated from the kinetics of inhibition of POX from Lactobacillus plantarum by acetylphosphinate [20].
Among classes of inhibitors studied in our laboratories, the acetylphosphinate appears to be the strongest inhibitor of E1ec and of E1h. This includes the three compounds mentioned in this paper, thiamin 2-thiothiazolone diphosphate (ThTTDP) and thiamin 2-thiazolonediphosphate (ThTDP), and potential Michael acceptor 2-oxo-3-butynoic acid. The 2-oxo-3-butynoic acid is an irreversible inhibitor and activity could not be regained by dilution of inhibitor [22]; its site of reactivity is still not known, but we do know that it does not react with any of the six cysteines of E1ec [11]. The compounds ThTTDP and ThTDP are sometimes referred to as transtion state analogues since the Ki values obtained are much lower than the Kd for ThDP [10, 23]. The nature of inhibition by these compounds has been clarified with the publication of a crystal structure of its complex with E1ec [24].
The phosphonate/phosphinate class mimics the substrate and the cumulative evidence indicates that they inhibit via enzymatic synthesis of LThDP-like adducts (Scheme 2). While these adducts certainly must have significant lifetime, the chemical bond between the keto carbon of the inhibitor and the C2 atom of thiazolium is formed reversibly and excess pyruvate or overnight dialysis (i.e., dilution of the inhibitor concentration) can reverse the inhibition. The koff appears to be slow for both enzymes.
At the same time, we point out that these phosphonate and phosphinate compounds have tuned out to be excellent mechanistic tools since their ThDP-bound adducts (Scheme 2) all give evidence of the positive CD band in the 300-310 nm region (as well as in the UV spectrum) [21].
Another notable finding of our studies is that the two species of pyruvate dehydrogenase studied E1h and E1ec lead to the formation of the opposite enantiomer of acetoin, a carboligase side product of ThDP enzymes (Figure 4). This makes the E1ec truly unusual [produces the (S) enantiomer], since all other ThDP enzymes appear to produce (R)-acetoin as does E1h. As was reasoned elsewhere, this implies that the pyruvate entering the enzyme presents a different face (re or si) to the enamine [18,25].
Finally, the results provide very strong support for the function of residue αH63 in E1h. It is suggested by the absence of the negative 330 CD band for the E1h αH63.ThDP complex, unlike with the wild-type E1h, that the ThDP binding site is perturbed. However, the experiments with ThTTDP indicate that this ThDP analogue can still bind to the active center. The absence of the positive CD band at 305 nm on adding AcPhi to the E1h αH63.ThDP complex and the low level of inhibition of E1h aH63A by AcPhi and AcMPhi confirm that the variant can no longer form the C2α-phosphinolactylThDP (see Scheme 2), nor can it presumably form LThDP. The presence or absence of these CD bands provides a straightforward experimental method to determine whether or not a particular residue is involved in the formation of LThDP, a common intermediate in all ThDP-dependent decarboxylases.
Acknowledgements
Supported at Rutgers by NIH-050380 and at SUNY-Buffalo by NASA Grant NAG8-1924.
Abbreviations
- PDHc
pyruvate dehydrogenase complex
- PDHc-h
human pyruvate dehydrogenase complex
- PDHc-ec
Escherichia coli pyruvate dehydrogenase complex
- E1h, E2h, E3h
refer to the components of the human enzyme
- E3BPh
E3h-binding protein formerly known as protein X
- E1ec, E2ec, E3ec
refer to the subunits of the Escherichia coli enzyme
- ThDP
thiamin diphosphate
- MAcPho
sodium salt of acetylphosphonic acid methyl ester
- AcPhi
sodium salt of acetylphosphinic acid
- AcMPhi
sodium salt of acetylmethylphosphinic acid
- LThDP
C2α-lactylThDP
- PLThDP
C2α-phosphonolactylThDP
- ThTTDP
thiamin 2-thiathiazolonediphosphate
References
- 1.Jordan F, Nemeria NS, Zhang S, Yan Y, Arjunan P, Furey W. J. Am. Chem. Soc. 2003;125:12732–12738. doi: 10.1021/ja0346126. [DOI] [PubMed] [Google Scholar]
- 2.Nemeria N, Baykal A, Joseph E, Zhang S, Yan Y, Furey W, Jordan F. Biochemistry. 2004;43:6565–6575. doi: 10.1021/bi049549r. [DOI] [PubMed] [Google Scholar]
- 3.Jordan F, Zhang Z, Sergienko EA. Bioorg. Chem. 2002;30:188–198. doi: 10.1006/bioo.2002.1249. [DOI] [PubMed] [Google Scholar]
- 4.Baykal AT, Kakalis L, Jordan F. Biochemistry. 2006;45:7522–7528. doi: 10.1021/bi060395k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jordan F, Nemeria NS. Bioorg. Chem. 2005;33:190–215. doi: 10.1016/j.bioorg.2005.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arjunan P, Sax M, Brunskill A, Chandrasekhar K, Nemeria N, Zhang S, Jordan F, Furey W. J. Biol. Chem. 2006;281:15296–15303. doi: 10.1074/jbc.M600656200. [DOI] [PubMed] [Google Scholar]
- 7.Wille G, Meyer D, Steinmetz A, Hinze E, Golbik R, Tittmann K. Nat. Chem. Biol. 2006;6:324–328. doi: 10.1038/nchembio788. [DOI] [PubMed] [Google Scholar]
- 8.Baillie AC, Wright BJ, Wright K. U.S. Patent No 4,339,443. 1982 July 13; granted.
- 9.Korotchkina LG, Sidhu S, Patel MS. J. Biol. Chem. 2006;281:9688–9696. doi: 10.1074/jbc.M511481200. [DOI] [PubMed] [Google Scholar]
- 10.Nemeria N, Yan Y, Zhang Z, Brown AM, Arjunan P, Furey W, Guest JR, Jordan F. J. Biol. Chem. 2001;276:45969–45976. doi: 10.1074/jbc.M104116200. [DOI] [PubMed] [Google Scholar]
- 11.Nemeria N, Volkov A, Brown A, Yi J, Zipper L, Guest JR, Jordan F. Biochemistry. 1998;37:911–922. doi: 10.1021/bi9722251. [DOI] [PubMed] [Google Scholar]
- 12.Korotchkina LG, Patel MS. J. Biol. Chem. 2001;276:5731–5738. doi: 10.1074/jbc.M007558200. [DOI] [PubMed] [Google Scholar]
- 13.Bradford MM. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 14.Kitz R, Wilson IB. J. Biol. Chem. 1962;237:3245–3249. [PubMed] [Google Scholar]
- 15.Morrison JF. Trends Biochem. Sci. 1982;7:102–105. [Google Scholar]
- 16.Schönbrunn-Hanebeck E, Laber B, Amrhein N. Biochemistry. 1990;29:4880–4885. doi: 10.1021/bi00472a018. [DOI] [PubMed] [Google Scholar]
- 17.Bieth JG. Methods Enzymol. 1995;248:59–84. doi: 10.1016/0076-6879(95)48007-2. [DOI] [PubMed] [Google Scholar]
- 18.Nemeria N, Tittmann K, Joseph E, Zhou L, Vazquez-Coll MB, Arjunan P, Hübner G, Furey W, Jordan F. J. Biol. Chem. 2005;280:21473–21482. doi: 10.1074/jbc.M502691200. [DOI] [PubMed] [Google Scholar]
- 19.Laber B, Amrhein N. 1987;248:351–358. doi: 10.1042/bj2480351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Spinka M, Hübner G. In: Biochemistry and Physiology of Thiamin Diphosphate Enzymes. Bisswanger H, Schellenberger A, editors. A. u. C. Intemann, Wissenschaftlicher Verlag; Prien: 1996. pp. 186–194. [Google Scholar]
- 21.Nemeria N, Chakraborty S, Baykal A, Korotchkina L, Patel M, Jordan F. 2006 submitted. [Google Scholar]
- 22.Brown A, Nemeria N, Yi J, Zhang D, Jordan WB, Machado RS, Guest JR, Jordan F. Biochemistry. 1997;36:8071–8081. doi: 10.1021/bi970094y. [DOI] [PubMed] [Google Scholar]
- 23.Gutowski JA, Lienhard GE. J. Biol. Chem. 1976;251:2863–2866. [PubMed] [Google Scholar]
- 24.Arjunan P, Chandrasekhar K, Sax M, Brunskill A, Nemeria N, Jordan F, Furey W. Biochemistry. 2004;43:2405–2411. doi: 10.1021/bi030200y. [DOI] [PubMed] [Google Scholar]
- 25.Baykal A, Chakraborty S, Dodoo A, Jordan F. 2006 doi: 10.1016/j.bioorg.2006.09.006. submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]