Abstract
Background
Members of the family Iridoviridae can cause severe diseases resulting in significant economic and environmental losses. Very little is known about how iridoviruses cause disease in their host. In the present study, we describe the re-analysis of the Iridoviridae family of complex DNA viruses using a variety of comparative genomic tools to yield a greater consensus among the annotated sequences of its members.
Results
A series of genomic sequence comparisons were made among, and between the Ranavirus and Megalocytivirus genera in order to identify novel conserved ORFs. Of these two genera, the Megalocytivirus genomes required the greatest number of altered annotations. Prior to our re-analysis, the Megalocytivirus species orange-spotted grouper iridovirus and rock bream iridovirus shared 99% sequence identity, but only 82 out of 118 potential ORFs were annotated; in contrast, we predict that these species share an identical complement of genes. These annotation changes allowed the redefinition of the group of core genes shared by all iridoviruses. Seven new core genes were identified, bringing the total number to 26.
Conclusion
Our re-analysis of genomes within the Iridoviridae family provides a unifying framework to understand the biology of these viruses. Further re-defining the core set of iridovirus genes will continue to lead us to a better understanding of the phylogenetic relationships between individual iridoviruses as well as giving us a much deeper understanding of iridovirus replication. In addition, this analysis will provide a better framework for characterizing and annotating currently unclassified iridoviruses.
Background
Iridoviruses are large DNA viruses (~120–200 nm in diameter) that replicate in the cytoplasm of infected cells. Iridovirus genomes are circularly permuted and terminally redundant, and range in size from 105 to 212 kbp [1,2]. The family Iridoviridae is currently subdivided into five genera:Chloriridovirus, Iridovirus, Lymphocystivirus, Megalocytivirus, and Ranavirus [3].
Iridoviruses have been found to infect invertebrates and poikilothermic vertebrates, including amphibians, reptiles, and fish [4]. Iridovirus infections produce symptoms that range from subclinical to very severe, which may also result in significant mortality [5-9]. The high pathogenicity associated with some members of the iridovirus family has had a significant impact on modern aquaculture, fish farming, and wildlife conservation. For example, systemic iridovirus infections have been found in economically important freshwater and marine fish species worldwide. In addition, iridovirus infections have been implicated in amphibian population declines, representing a set of emerging infectious diseases whose spread has been accelerated by human activities [10-14].
Despite the economic and ecological significance of iridoviruses, very little is currently known about their molecular biology. One approach towards gaining a deeper understanding of iridoviral pathogenesis is to investigate the core set of essential genes conserved among all members of the family. The genomes of twelve iridoviruses, including at least one from each genus, have been completely sequenced (Table 1). According to the previously published annotations, these genomes contained only 19 core genes associated with a variety of viral activities: transcriptional regulation, DNA metabolism, protein modification, and viral structure. Definition of this core set of genes also highlights those genes that are conserved across some, but not all, genera, and unique genes found within a single species. These non-core genes may be involved in specific virus-host interactions, enhancement of virus replication, and augmented pathogenesis in certain species.
Table 1.
Iridovirus Genomes
| Virus | Abbreviation | Genus | Genome Size (bp) | # ORFs* | GenBank accession # | Ref |
| Frog virus 3 | FV3 | Ranavirus | 105903 | 97 | AY548484 | [27] |
| Tiger frog virus | TFV | Ranavirus | 105057 | 103 | AF389451 | [1] |
| Ambystoma tigrinum virus | ATV | Ranavirus | 106332 | 92 | AY150217 | [30] |
| Grouper iridovirus | GIV | Ranavirus | 139793 | 139 | AY666015 | [21] |
| Singapore grouper iridovirus | SGIV | Ranavirus | 140131 | 139 | AY521625 | [22] |
| Lymphocystis disease virus 1 | LCDV-1 | Lymphocystivirus | 102653 | 108 | L63545 | [34] |
| Lymphocystis disease virus China | LCDV-China | Lymphocystivirus | 186250 | 178 | AY380826 | [24] |
| Infectious spleen and kidney necrosis virus | ISKNV | Megalocytivirus | 111362 | 117 | AF371960 | [20] |
| Rock bream iridovirus | RBIV | Megalocytivirus | 112080 | 116 | AY532606 | [19] |
| Orange-spotted grouper iridovirus | OSGIV | Megalocytivirus | 112636 | 116 | AY894343 | [18] |
| Invertebrate iridescent virus 6 | IIV-6 | Iridovirus | 212482 | 211 | AF303741 | [2] |
| Invertebrate iridescent virus 3 | IIV-3 | Chloriridovirus | 191100 | 126 | DQ643392 | [26] |
*The number of ORFs in each viral genome reflects the results of the analysis done in this paper
Despite the growing number of sequenced iridovirus genomes, no systematic comparative genomic analysis of the family has yet been performed. Thus, annotation of these genomes has been performed without standardization and has so far been guided primarily by the position of start/stop codons rather than the presence of homologous sequences. As a result, some long overlapping potential ORFs have been automatically designated as coding sequences, and smaller homologous ORFs overlooked. In this paper, we have taken a comparative genomics approach to re-examine the annotation of all twelve iridovirus genomes, using the Viral Orthologous Clusters (VOCs) [15] and Viral Genome Organizer (VGO) [16] software. These re-annotated genomes were then analysed further, both to define the core set of iridovirus genes more accurately, and to provide a deeper understanding into the phylogenetic relationship between individual iridovirus species.
Results & discussion
Re-annotation of Iridovirus genomes
One objective of this project was to demonstrate the application of comparative genomics to annotating viral genomes, particularly those that have been poorly characterized experimentally. In an earlier study, we utilized comparative genomics to identify previously unannotated small viral ORFs in the Poxviridae [17]. Here, we focused our analysis on the Iridoviridae family, which represents a challenge in genome annotation since there is little experimental evidence available to confirm gene expression. Another problem is that iridovirus promoter elements have not been well characterized, and thus cannot be used as a reliable criterion for assigning ORFs. These combined factors made previous iridovirus gene annotation a somewhat arbitrary process, resulting in closely related iridovirus species with dramatic differences in their genomic annotations. Therefore, we decided to analyse all members of this family using a standardized comparative genomics approach, using the fact that ORFs that are conserved in more than one divergent species are likely to be functional genes.
Analysis was begun with the Megalocytivirus genus, which contains three sequenced genomes: infectious spleen and kidney necrosis virus (ISKNV), rock bream iridovirus (RBIV), and orange-spotted grouper iridovirus (OSGIV). These three viruses display a co-linear arrangement of genes with an overall DNA sequence identity of greater than 90%. In the analysis of this genus, differences in gene content were examined in detail. Dotplots were used to determine presence of orthologous DNA and a variety of BLAST searches and the VGO genome visualization software were used to determine the reason (frameshifts, extra stop codons) behind the apparent absence of some ORFs.
Using this approach, a substantial number of ORFs were either added to, or deleted from members of the Megalocytivirus genus (Table 2). OSGIV and RBIV share 99% DNA sequence identity, and thus are probably different strains of the same virus; however, previous annotation described only 82 out of 118 total annotated ORFs shared by the two genomes [18,19]. After our re-analysis, the RBIV and OSGIV genomes had an identical complement of annotated genes. Furthermore, this re-annotated ISKNV genome contained 110 ORFs orthologous with both RBIV and OSGIV (compared to 71 in the old annotation.) (Table 2) [18,20].
Table 2.
Re-annotation of the Megalocytivirus genus
| ISKNVa | Start | Stopc | aad | RBIVa | Start | Stopc | aad | OSGIVa | Start | Stopc | aad |
| 1L | 1270 | 134 | 378 | 1L | 1270 | 134 | 378 | 1L | 1270 | 134 | 378 |
| 2R | 1394 | 2044 | 216 | 2Rb | 1394 | 1597/1781 | 67 | 2R | 1394 | 1789 | 131 |
| - | - | - | - | 3R | 1841 | 2056 | 71 | 3R | 1849 | 2064 | 71 |
| 3L | 2634 | 2077 | 185 | 4L | 2605 | 2102 | 167 | 4L | 2613 | 2110 | 167 |
| 4L | 2890 | 2681 | 69 | 5L | 2800 | 2624 | 58 | 5L | 2808 | 2632 | 58 |
| 5L | 3648 | 2893 | 251 | 6L | 3541 | 2876 | 221 | 6L | 3548 | 2883 | 221 |
| 6L | 5155 | 3794 | 453 | 7L | 5147 | 3786 | 453 | 7L | 5154 | 3793 | 453 |
| 7L | 6631 | 5174 | 485 | 8L | 6621 | 5164 | 485 | 8L | 6628 | 5171 | 485 |
| 8R | 6669 | 8246 | 525 | 9R | 6692 | 8239 | 515 | 9R | 6699 | 8246 | 515 |
| 9R | 8342 | 8503 | 53 | 10R | 8335 | 8496 | 53 | 10R | 8342 | 8503 | 53 |
| 10L | 9054 | 8662 | 130 | 11L | 9047 | 8655 | 130 | 11L/12Lb | 9055 | 8849/8662 | 130 |
| 11L | 9311 | 9051 | 86 | 11.5L | 9304 | 9044 | 86 | 13L | 9312 | 9052 | 86 |
| 12R | 9330 | 9659 | 110 | 12R | 9323 | 9655 | 110 | 14R | 9331 | 9663 | 110 |
| 13R | 9669 | 11054 | 461 | 13R | 9662 | 11059 | 465 | 15R | 9670 | 11067 | 465 |
| 14R | 11309 | 12268 | 319 | 14R | 11314 | 12288 | 324 | 16R | 11322 | 12296 | 324 |
| 15R | 12278 | 13069 | 263 | 15R | 12298 | 13089 | 263 | 17R | 12302 | 13093 | 263 |
| 16L | 13716 | 13129 | 195 | 16L | 13733 | 13146 | 195 | 18L | 13738 | 13151 | 195 |
| 17L | 14095 | 13718 | 125 | 17L | 14086 | 13748 | 112 | 19L | 14088 | 13753 | 111 |
| 17.5R | 14089 | 14325 | 78 | 18R | 14171 | 14410 | 79 | 20R | 14094 | 14351 | 85 |
| 18.5Lb | 14563 | 14233 | 109 | 19L | 14648 | 14472 | 58 | 21L | 14607 | 14431 | 58 |
| 19R | 14579 | 17425 | 948 | 20R | 14664 | 17510 | 948 | 22R | 14623 | 17469 | 948 |
| 20L | 17642 | 17454 | 62 | 21L | 17756 | 17574 | 60 | 23L | 17715 | 17533 | 60 |
| 21L | 17900 | 17778 | 40 | 21.5L | 18014 | 17892 | 40 | 24L | 17973 | 17851 | 40 |
| 22L | 19489 | 17990 | 499 | 22L | 19714 | 18104 | 536 | 25L | 19715 | 18063 | 550 |
| 23R | 19562 | 22132 | 856 | 23R | 19787 | 22204 | 805 | 26R | 19788 | 22922 | 1044 |
| 24R | 22300 | 23238 | 312 | 26R | 23035 | 23973 | 312 | 27R | 23207 | 24145 | 312 |
| 25R | 23354 | 23779 | 141 | 27R | 23997 | 24380 | 127 | 28R | 24169 | 24696 | 175 |
| 26L | 24145 | 23822 | 107 | 27.5L | 24697 | 24377 | 106 | 29L | 25013 | 24693 | 106 |
| 27L | 25063 | 24167 | 298 | 28L | 25615 | 24719 | 298 | 30L | 25931 | 25035 | 298 |
| 28L | 28559 | 25080 | 1159 | 29L | 29138 | 25632 | 1168 | 31L | 29454 | 25948 | 1168 |
| 29L | 28814 | 28593 | 73 | 29.5Lb | 29362 | 29087/29145 | 91 | 32L | 29682 | 29461 | 73 |
| 31.5L | 29414 | 28884 | 176 | 30.5L | 29957 | 29430 | 175 | 33.5L | 30277 | 29750 | 175 |
| 32R | 29447 | 30061 | 204 | 31R | 29990 | 30622 | 210 | 34R | 30310 | 30942 | 210 |
| 33L | 31079 | 30138 | 313 | 32L | 31654 | 30713 | 313 | 35L | 31935 | 31033 | 300 |
| 34R | 31144 | 34278 | 1044 | 33R | 31700 | 34861 | 1053 | 36R | 32018 | 35176 | 1052 |
| 35L | 35508 | 34360 | 382 | 34L | 36067 | 34934 | 377 | 37L | 36382 | 35249 | 377 |
| 36R | 35546 | 36601 | 351 | 35.5R | 36061 | 37113 | 350 | 38R | 36376 | 37431 | 351 |
| 37L | 37950 | 36598 | 450 | 37L | 38219 | 37110 | 369 | 39L | 38777 | 37428 | 449 |
| 38L | 39395 | 37959 | 478 | 38L | 39974 | 38469 | 501 | 40L | 40225 | 38786 | 479 |
| 39R | 39439 | 40311 | 290 | 39.5Rb | 40012 | 40506/40857 | 164 | 41R | 40290 | 41168 | 292 |
| 40L | 41443 | 40304 | 379 | 41L | 41995 | 40850 | 381 | 42L | 42306 | 41161 | 381 |
| 41L | 42788 | 41445 | 447 | 42L | 43346 | 41997 | 449 | 43L | 43657 | 42308 | 449 |
| 42R | 42803 | 43396 | 197 | 43R | 43361 | 43959 | 198 | 44R | 43672 | 44271 | 199 |
| 43L | 43842 | 43480 | 120 | 43.5L | 44405 | 43975 | 142 | 45L | 44717 | 44355 | 120 |
| 44L | 44645 | 43845 | 266 | 44L | 45208 | 44408 | 266 | 46L | 45524 | 44724 | 266 |
| 45L | 45564 | 44650 | 304 | 45L | 46127 | 45213 | 304 | 47L | 46443 | 45529 | 304 |
| 46L | 46241 | 45558 | 227 | 46L | 46804 | 46121 | 227 | 48L | 47120 | 46437 | 227 |
| 47R | 46401 | 46664 | 87 | 47R | 47150 | 46887 | 87 | 49R | 47280 | 47543 | 87 |
| 48R | 46661 | 47005 | 114 | 47.5Rb | 47224 | 47433/47588 | 69 | 50R | 47540 | 47893 | 117 |
| 49R | 47021 | 47191 | 56 | 48.5R | 47604 | 47774 | 56 | 51R | 47909 | 48079 | 56 |
| 50L | 47678 | 47250 | 142 | 49L | 48270 | 47842 | 142 | 52L | 48575 | 48147 | 142 |
| 51R | 47733 | 47864 | 43 | - | - | - | - | - | - | - | - |
| 52L | 48403 | 47951 | 150 | 50L | 48999 | 48547 | 150 | 53L | 49306 | 48854 | 150 |
| 53R | 48405 | 48620 | 71 | 51R | 49001 | 49195 | 64 | 54R | 49308 | 49502 | 64 |
| 54L | 49559 | 48633 | 308 | 52L | 50173 | 49229 | 314 | 55L | 50480 | 49536 | 314 |
| 55L | 50508 | 49582 | 308 | 53L | 51137 | 50196 | 313 | 56L | 51444 | 50503 | 313 |
| 56L | 51166 | 50519 | 215 | 54L | 51795 | 51148 | 215 | 57L | 52102 | 51455 | 215 |
| 57L | 51433 | 51173 | 86 | 55L | 52062 | 51802 | 86 | 58L | 52369 | 52109 | 86 |
| 59L | 52414 | 51749 | 221 | 56L | 52839 | 52327 | 170 | 59L | 53146 | 52634 | 170 |
| 61L | 53162 | 52359 | 267 | 57L | 53709 | 52903 | 268 | 60L | 54016 | 53210 | 268 |
| 62L | 56785 | 53159 | 1208 | 58L | 57467 | 53706 | 1253 | 61L/62Lb | 55131 | 54013/53893 | 372 |
| 63L | 59875 | 57227 | 882 | 59L | 60567 | 57919 | 882 | 63L | 60876 | 58228 | 882 |
| 64L | 61393 | 59918 | 491 | 60Lb | 62102 | 60855/60635 | 415 | 64L | 62416 | 60944 | 490 |
| 65L | 61900 | 61439 | 153 | 61L | 62611 | 62144 | 155 | 65L | 62928 | 62458 | 156 |
| 66L | 63025 | 61982 | 347 | 62L | 63744 | 62662 | 360 | 66L | 64061 | 62979 | 360 |
| 67L | 63855 | 63271 | 194 | 63.5L | 64446 | 63865 | 193 | 67.5L | 64763 | 64182 | 193 |
| 68L | 65329 | 63896 | 477 | 64L | 65917 | 64484 | 477 | 69L | 66234 | 64801 | 477 |
| 69L | 66001 | 65336 | 221 | 65Lb | 66661 | 66215/65929 | 148 | 70L | 66977 | 66246 | 243 |
| 70L | 66331 | 66101 | 76 | - | - | - | - | - | - | - | - |
| 71L | 68042 | 66432 | 536 | 68L | 68529 | 67120 | 469 | 71L | 68973 | 67417 | 518 |
| 72R | 68173 | 69177 | 334 | - | - | - | - | - | - | - | - |
| 73R | 69203 | 69622 | 139 | 69Rb | 68717 | 69191/69135 | 157 | 72R | 69078 | 69497 | 139 |
| 74R | 69669 | 70682 | 337 | 70R | 69184 | 70203 | 339 | 73R | 69546 | 70568 | 340 |
| 75L | 71043 | 70777 | 88 | 71L | 70573 | 70304 | 89 | 74L | 70938 | 70669 | 89 |
| 76L | 74017 | 71045 | 990 | 72L | 73541 | 70575 | 988 | 75L | 73912 | 70940 | 990 |
| 77R | 74035 | 75369 | 444 | 73R | 73559 | 74893 | 444 | 76R | 73930 | 75264 | 444 |
| 78R | 75366 | 75830 | 154 | 75R | 74890 | 75354 | 154 | 77R | 75261 | 75725 | 154 |
| 79L | 76053 | 75832 | 73 | 76L | 75580 | 75356 | 74 | 78L | 75951 | 75727 | 74 |
| - | - | - | - | 77R | 75664 | 76137 | 157 | 79R | 76039 | 76512 | 157 |
| 80L | 76368 | 76165 | 67 | - | - | - | - | - | - | - | - |
| 81R | 76367 | 76864 | 165 | 78R | 76150 | 76647 | 165 | 80R | 76525 | 77022 | 165 |
| 82L | 78007 | 76901 | 368 | 79L | 77802 | 76696 | 368 | 81L | 78177 | 77071 | 368 |
| 83R | 78152 | 78418 | 88 | 80R | 77827 | 78225 | 132 | 82R | 78202 | 78600 | 132 |
| 84L | 79881 | 78526 | 451 | 81L | 79556 | 78252 | 434 | 83L | 79931 | 78627 | 434 |
| 85R | 79884 | 80486 | 200 | 82R | 79643 | 80173 | 176 | 84R | 80018 | 80548 | 176 |
| 86R | 80483 | 80947 | 154 | 82.5R | 80170 | 80637 | 155 | 84.5L | 80545 | 81012 | 155 |
| 87R | 80940 | 81710 | 256 | 83R | 80603 | 81400 | 265 | 85R | 80978 | 81775 | 265 |
| 88R | 81717 | 83720 | 667 | 84R | 81503 | 83425 | 640 | 86R/87Rb | 82234 | 83805/82279 | 523 |
| 90.5L | 84663 | 83701 | 320 | 85L | 84457 | 83630 | 275 | 88.5L | 84811 | 83786 | 341 |
| 93L | 85786 | 84860 | 308 | 86L | 85504 | 84578 | 308 | 90L | 85918 | 84992 | 308 |
| 94L | 86296 | 85796 | 166 | 87L | 86014 | 85514 | 166 | 91L | 86428 | 85928 | 166 |
| 95L | 87481 | 86321 | 386 | 88L | 87202 | 86039 | 387 | 92L | 87616 | 86453 | 387 |
| 96L | 88298 | 87489 | 269 | 89.5Lb | 87601 | 87218/87210 | 127 | 93L | 88361 | 87624 | 245 |
| 97.5L | 88723 | 88232 | 163 | 90L | 88443 | 87952 | 163 | 94L | 88857 | 88366 | 163 |
| 99L | 89097 | 88774 | 107 | 91L | 88814 | 88491 | 107 | 95L | 89229 | 88906 | 107 |
| 100L | 89689 | 89144 | 181 | 92L | 89515 | 88868 | 215 | 96L | 90024 | 89377 | 215 |
| 101L | 90251 | 89736 | 171 | 93.5L | 89999 | 89470 | 175 | 97L | 90508 | 89993 | 171 |
| 102R | 90311 | 91753 | 480 | 94R | 90068 | 91513 | 481 | 98R | 90577 | 92022 | 481 |
| 103R | 91760 | 92161 | 133 | 95.5R | 91477 | 91935 | 152 | 99R | 92029 | 92442 | 137 |
| 104R | 92215 | 92991 | 258 | 96R | 91994 | 92773 | 259 | 100R | 92501 | 93280 | 259 |
| 105R | 92993 | 93358 | 121 | 97R | 92775 | 93146 | 123 | 101R | 93282 | 93653 | 123 |
| - | - | - | - | 98R | 93240 | 94127 | 295 | 102R | 93747 | 94634 | 295 |
| 106L | 94501 | 93482 | 339 | 99L | 95042 | 94221 | 273 | 103L | 95548 | 94676 | 290 |
| 108.5L | 95093 | 94494 | 199 | 100.5L | 95699 | 95068 | 209 | 104.5L | 96217 | 95574 | 213 |
| 109L | 97950 | 95185 | 921 | 101L | 98557 | 95792 | 921 | 106L | 99060 | 96298 | 920 |
| 110R | 97997 | 98152 | 51 | 101.5L | 98609 | 98764 | 51 | 107R | 99113 | 99268 | 51 |
| 111L | 99039 | 98149 | 296 | 102L | 99657 | 98761 | 298 | 108Lb | 99885 | 99265/99849 | 206 |
| 112R | 99059 | 99802 | 247 | 103Rb | 99677 | 100426/100422 | 249 | 109R | 100183 | 100926 | 247 |
| 113R | 99937 | 100290 | 117 | 104.5R | 100493 | 100909 | 138 | 110R | 100998 | 101414 | 138 |
| 114L | 103159 | 100334 | 941 | 106Lb | 103615 | 102539/100953 | 358 | 111L | 104041 | 101594 | 815 |
| 115R | 103203 | 104213 | 336 | 108.5R | 104050 | 104781 | 243 | 112R | 104476 | 105486 | 336 |
| 116R | 104219 | 105667 | 482 | 110R | 105060 | 106493 | 477 | 113R | 105547 | 106701 | 384 |
| 117L | 106395 | 105721 | 224 | 111L | 107221 | 106547 | 224 | 114L | 107652 | 106978 | 224 |
| 118L | 108093 | 106723 | 456 | 112L | 108913 | 107549 | 454 | 115L | 109299 | 107986 | 437 |
| 119R | 108105 | 108392 | 95 | 113R | 108931 | 109104 | 57 | 116R | 109369 | 109656 | 95 |
| 120R | 108424 | 108933 | 169 | 114R | 109248 | 109637 | 129 | 117R | 109687 | 110193 | 168 |
| 121L | 109584 | 108934 | 216 | 115L | 110419 | 109756 | 220 | 118L | 110849 | 110214 | 211 |
| 122R | 109594 | 110313 | 239 | 116R | 110429 | 111148 | 239 | 119R | 110859 | 111578 | 239 |
| 123R | 110391 | 110576 | 61 | 117R | 111226 | 111420 | 64 | 120R | 111656 | 111850 | 64 |
| 124L | 111351 | 110665 | 228 | 118L | 112037 | 111576 | 153 | 121L | 112625 | 111939 | 228 |
aORFs that have been added or altered are highlighted in bold. If a previously annotated ORF is not listed in the table, it has been deleted.
bPotentially frameshifted ORF
cWhere an ORF has a potential sequencing error resulting in a frameshift mutation, 2 stop codons are provided in the format X/Y. The first number represents the actual physical stop in the reported sequence. The second number is the proposed stop if a sequencing error occurred.
dLength of ORF in amino acids
In the process of re-examining these genomes, we annotated a number of genes containing apparent frameshift mutations between species. In RBIV we annotated ten genes with potential frameshift mutations, while OSGIV had four such genes (Table 2). All of the genes containing potential frameshift mutations had orthologs in the other two members of the Megalocytivirus genus (Table 2). In some cases, these mutations may be the result of natural mutations within the viruses; however, it is also possible that these apparent frameshift mutations are actually sequencing errors. For both RBIV and OSGIV, PCR primers based on the ISKNV sequence were used to amplify genomic fragments, which were subsequently sequenced [18,19]. It is possible that errors were introduced during the PCR process, leading to apparent frameshifts in the reported sequence. It is interesting to note that the genomic sequence of ISKNV (sequenced using subcloned fragments rather than PCR products) [20], had significantly fewer annotation changes made during our re-analysis. Though we have not experimentally proven that the frameshift mutations in OSGIV and RBIV are the result of sequencing errors, it would be useful to focus future sequencing efforts on these regions, to determine if the reported sequences are indeed correct.
After re-annotating the Megalocytivirus genus, we applied the same comparative genomic analysis to the Ranavirus genus. The genus contains five sequenced members divided into two groups, each with a high degree of sequence conservation and a co-linear arrangement of genes. The first group is comprised of frog virus 3 (FV3), tiger frog virus (TFV), and Ambystoma tigrinum virus (ATV). The second group contains Singapore grouper iridovirus (SGIV) and grouper iridovirus (GIV).
The first step in the re-annotation of the Ranavirus genus was a comparative genomic analysis of FV3, TFV, and ATV. This resulted in an increase in the number of conserved annotated genes from 76 to 87 (Table 3). Subsequent re-analysis of the second Ranavirus group, containing SGIV and GIV, resulted in an increase from 131 to 138 conserved annotated ORFs (Table 4). It should be noted that two of the newly annotated ORFs, SGIV 0.5L and GIV 120.5L, appear to "wrap around", beginning at one end of the genome with the remainder of the ORF located at the opposite end [21,22]. These apparent "split ORFs" are actually the result of the circularly permutated iridovirus genome being represented as a linear genomic sequence, when the arbitrarily chosen start point happens to fall in the middle of an ORF [23].
Table 3.
Re-annotation of FV3, TFV, and ATV of the Ranavirus genus
| FV3a | Start | Stopc | aad | TFVa | Start | Stopc | aad | ATVa | Start | Stopc | aad |
| 1R | 272 | 1042 | 256 | 105R | 103809 | 104576 | 256 | 91R | 104836 | 105606 | 256 |
| 2L | 2611 | 1649 | 320 | 2Lb | 1028 | 315/11 | 237 | 1L | 981 | 70 | 303 |
| 2.5L | 3488 | 2649 | 279 | 2.5L | 1943 | 1065 | 292 | 2L | 1858 | 1019 | 279 |
| 3R | 3418 | 4734 | 438 | 4R | 1937 | 3151 | 404 | 3R | 1892 | 3106 | 404 |
| 4R | 4775 | 4957 | 60 | 5R | 3190 | 3372 | 60 | 4R | 3149 | 3331 | 60 |
| 5R | 5390 | 6004 | 204 | 6R | 3816 | 4418 | 200 | - | - | - | - |
| 6R | 6007 | 6234 | 75 | - | - | - | - | - | - | - | - |
| - | - | - | - | 6.5R | 4411 | 4578 | 55 | - | - | - | - |
| 7.5L | 7025 | 7411 | 128 | 7L | 5452 | 5024 | 142 | 5L | 4416 | 3994 | 140 |
| 8R | 7503 | 11384 | 1293 | 8R | 5531 | 9415 | 1294 | 6R | 4495 | 8379 | 1294 |
| 9L | 14599 | 11753 | 948 | 9L | 12599 | 9753 | 948 | 7L | 11725 | 8879 | 948 |
| 10R | 14615 | 15028 | 137 | 10R | 12615 | 13028 | 137 | 8R | 11741 | 12154 | 137 |
| 11R | 15378 | 15590 | 70 | 11R | 13380 | 13592 | 79 | 88L | 102924 | 102712 | 70 |
| 12L | 16549 | 15656 | 297 | 12L | 14551 | 13658 | 297 | 87R | 101753 | 102646 | 297 |
| - | - | - | - | 13L | 14947 | 14747 | 66 | 86R | 101169 | 101363 | 64 |
| 13R | 17090 | 17296 | 68 | 14.5Rb | 15041 | 15184/15247 | 47 | 85.5L | 101128 | 100871 | 85 |
| 14R | 17311 | 17670 | 119 | 15R | 15261 | 15620 | 119 | 84L | 100856 | 100482 | 124 |
| 15R | 17766 | 18734 | 322 | 16R | 15716 | 16663 | 315 | 83R | 100400 | 99474 | 308 |
| 16R | 19014 | 19841 | 275 | 17R | 16838 | 17665 | 275 | 82.5L | 98809 | 98438 | 123 |
| 17L | 21590 | 20082 | 502 | 18L | 19414 | 17906 | 502 | 81R | 96410 | 97918 | 502 |
| 18L | 21864 | 21628 | 78 | 18.5L | 19687 | 19451 | 78 | 80.5R | 96137 | 96373 | 78 |
| 19R | 21916 | 24471 | 851 | 19R | 19686 | 22271 | 861 | 80L | 96083 | 94086 | 665 |
| 20R | 24519 | 24965 | 148 | 20R | 22319 | 22774 | 151 | 79L | 94038 | 93589 | 149 |
| 21L | 25861 | 25202 | 219 | 21L | 23657 | 23998 | 219 | 78R | 92383 | 93042 | 219 |
| 22R | 25991 | 28912 | 973 | 22R | 23789 | 26716 | 975 | 77L | 92253 | 89326 | 975 |
| 23R | 29290 | 30438 | 382 | 23R | 27093 | 28241 | 382 | 53R | 58082 | 59230 | 382 |
| 24R | 30821 | 31918 | 365 | 24R | 28636 | 29733 | 365 | 54R | 59613 | 60710 | 365 |
| 25R | 32112 | 32900 | 262 | 25R | 29930 | 30709 | 259 | 55R | 62328 | 63335 | 335 |
| - | - | - | - | 26R | 30778 | 30936 | 52 | 56R | 63402 | 63500 | 32 |
| 26Rb | 32967 | 33197 | 76 | 27R | 31033 | 31812 | 259 | 57R | 63659 | 64438 | 259 |
| - | - | - | - | 28L | 32190 | 32002 | 62 | - | - | - | - |
| 27R | 33728 | 36640 | 970 | 29R | 32345 | 35257 | 970 | 58R | 64968 | 67880 | 970 |
| 28R | 36689 | 37177 | 162 | 30R | 35306 | 35794 | 162 | 59R | 67929 | 68417 | 162 |
| 29L | 37652 | 37356 | 98 | 31L | 36122 | 35823 | 99 | - | - | - | - |
| 30R | 37854 | 38006 | 50 | - | - | - | - | - | - | - | - |
| 31R | 38068 | 38487 | 139 | 32R | 36565 | 36984 | 139 | 60R | 68786 | 69205 | 139 |
| 32R | 38537 | 40426 | 629 | 33R | 37098 | 39047 | 649 | 61R | 69255 | 71471 | 738 |
| 33R | 40509 | 40700 | 63 | 34R | 39133 | 39324 | 63 | 62R | 71555 | 71746 | 63 |
| 34R | 40844 | 41164 | 106 | 35.5R | 39467 | 39787 | 106 | 62.5R | 71894 | 72235 | 113 |
| 35L | 41717 | 41256 | 153 | 37L | 40308 | 39772 | 178 | 63L | 72576 | 72220 | 118 |
| 36L | 42353 | 41256 | 365 | 38.5Lb | 40938 | 40543/40367 | 131 | - | - | - | - |
| - | - | - | - | 39R | 41112 | 41246 | 44 | - | - | - | - |
| 37R | 42749 | 43378 | 209 | 40R | 41296 | 41952 | 218 | 64R | 74110 | 74739 | 209 |
| 38R | 43519 | 45216 | 565 | 41R | 42091 | 43788 | 565 | 65R | 74878 | 76575 | 565 |
| 39R | 45322 | 45672 | 116 | 42R | 43899 | 44249 | 116 | 66R | 76682 | 76948 | 88 |
| 40R | 45761 | 46309 | 182 | 43R | 44335 | 44883 | 182 | 67R | 77048 | 77671 | 207 |
| - | - | - | - | 44R | 44973 | 45239 | 88 | 68R | 77899 | 78039 | 46 |
| 41R | 46691 | 50188 | 1165 | 45R | 45270 | 48767 | 1165 | 69R | 78111 | 81608 | 1165 |
| 43.5Lb | 50940 | 51455/5684 | 171 | 46L | 50362 | 49133 | 409 | 70L | 82913 | 82152 | 253 |
| 45L | 52348 | 51938 | 136 | 47L | 50899 | 50489 | 136 | 71L | 83450 | 83040 | 136 |
| 46L | 52968 | 52723 | 81 | 48L | 51411 | 50953 | 152 | 72L | 84331 | 83504 | 275 |
| 47L | 53509 | 53093 | 138 | 49L | 51953 | 51537 | 138 | 73L | 84874 | 84458 | 138 |
| 48L | 53763 | 53512 | 83 | 50L | 52207 | 51956 | 83 | 74L | 85187 | 84804 | 127 |
| 49L | 54621 | 53872 | 249 | 51L | 52899 | 52315 | 194 | 75L | 86776 | 85235 | 513 |
| 50L | 55459 | 54770 | 229 | 52L | 53876 | 53136 | 246 | - | - | - | - |
| 51R | 55539 | 57224 | 561 | 53R | 53956 | 55641 | 561 | 76R | 86858 | 88543 | 561 |
| 52L | 58548 | 57481 | 355 | 54L | 56965 | 55898 | 355 | 52R | 57441 | 57602 | 53 |
| 53R | 58886 | 60454 | 522 | 55R | 57301 | 58869 | 522 | 51L | 57102 | 55522 | 526 |
| 54L | 60899 | 60669 | 76 | - | - | - | - | - | - | - | - |
| 55L | 62232 | 60937 | 431 | 56L | 60615 | 59320 | 431 | 50R | 53770 | 55065 | 431 |
| - | - | - | - | 57L | 60772 | 60623 | 49 | 49R | 53613 | 53762 | 49 |
| 56R | 62320 | 62757 | 145 | 58R | 60809 | 61213 | 134 | 48L | 53576 | 53172 | 134 |
| 57R | 62871 | 64367 | 498 | 59R | 61254 | 62750 | 498 | 47L | 53130 | 51634 | 498 |
| - | - | - | - | 60L | 62888 | 62757 | 43 | - | - | - | - |
| 58.5R | 64819 | 65373 | 184 | 61R | 63264 | 63818 | 184 | 46L | 50770 | 50216 | 184 |
| 59L | 67014 | 65956 | 352 | 62L | 65445 | 64387 | 352 | 45R | 48676 | 49734 | 352 |
| 60R | 67176 | 70217 | 1013 | 63R | 65605 | 68646 | 1013 | 44L | 48512 | 45471 | 1013 |
| 61L | 70408 | 70226 | 60 | - | - | - | - | - | - | - | - |
| - | - | - | - | 64R | 69029 | 69151 | 40 | - | - | - | - |
| 62L | 74516 | 70851 | 1221 | 65L | 72940 | 69281 | 1219 | 43R | 41447 | 45112 | 1221 |
| 62.5R | 74515 | 74778 | 87 | 66.5R | 72927 | 73202 | 91 | 42.5L | 41460 | 41185 | 91 |
| 63R | 74895 | 75389 | 164 | 68R | 73319 | 73813 | 164 | 42L | 41068 | 40631 | 145 |
| 64R | 75529 | 75816 | 95 | 69.5R | 73946 | 74209 | 87 | 40L | 40492 | 40205 | 95 |
| 65L | 76373 | 76209 | 54 | - | - | - | - | - | - | - | - |
| 66L | 76921 | 76370 | 183 | 70.5L | 75301 | 74685 | 204 | 38.5R | 39094 | 36681 | 195 |
| 67L | 78139 | 76976 | 387 | 71L | 76525 | 75362 | 387 | 38R | 37876 | 39039 | 387 |
| 68R | 78422 | 78709 | 95 | 72R | 76785 | 76982 | 65 | 37bL | 37592 | 37416 | 58 |
| - | - | - | - | 73L | 77175 | 77020 | 51 | 36R | 36736 | 36891 | 51 |
| 69R | 78845 | 79111 | 88 | 74R | 77244 | 77507 | 88 | 35L | 36677 | 36411 | 88 |
| 70R | 79129 | 79503 | 124 | 75R | 77507 | 77902 | 131 | 34.5L | 36392 | 36018 | 124 |
| 71R | 79543 | 79776 | 77 | 76R | 77942 | 78175 | 77 | 34L | 35978 | 35742 | 78 |
| 72L | 80549 | 79833 | 238 | 77L | 78948 | 78232 | 238 | 32R | 34970 | 35293 | 107 |
| 73L | 81971 | 80997 | 324 | 78L | 80299 | 79325 | 324 | 31R | 33319 | 34311 | 330 |
| 74L | 83258 | 82146 | 370 | 79L | 81498 | 80506 | 330 | 30R | 31947 | 33128 | 393 |
| 75L | 83544 | 83290 | 84 | 80L | 81809 | 81555 | 84 | 29R | 31637 | 31891 | 84 |
| 76R | 83607 | 83828 | 73 | 81R | 81872 | 82093 | 73 | 28L | 31574 | 31353 | 73 |
| 77L | 84172 | 83825 | 115 | 82L | 82437 | 82090 | 115 | 27R | 31009 | 31356 | 115 |
| 78L | 85395 | 84757 | 212 | 83L | 83568 | 82894 | 224 | - | - | - | - |
| 79R | 85531 | 87249 | 572 | 84R | 83668 | 85386 | 572 | 26L | 30729 | 28999 | 576 |
| 80L | 88987 | 87872 | 371 | 85L | 86988 | 85873 | 371 | 25R | 27224 | 28345 | 373 |
| 81R | 89043 | 89321 | 92 | 86R | 87046 | 87324 | 92 | 24L | 27168 | 26890 | 92 |
| 82R | 89450 | 89923 | 157 | 87R | 87454 | 87927 | 157 | 23L | 26762 | 26289 | 157 |
| - | - | - | - | 88R | 88138 | 88512 | 124 | 22L | 25564 | 25277 | 95 |
| 83R | 90373 | 91017 | 214 | 89R | 88857 | 89501 | 214 | 21L | 24912 | 24268 | 214 |
| 84R | 91389 | 92126 | 245 | 90R | 89903 | 90640 | 245 | 20L | 23923 | 23141 | 260 |
| 85R | 92201 | 92788 | 195 | 91.5R | 90715 | 91302 | 195 | 19L | 23066 | 22479 | 195 |
| 86L | 93363 | 93178 | 61 | 92L | 91943 | 91650 | 97 | 18R | 21742 | 22119 | 125 |
| 87L | 95533 | 93716 | 605 | 93L | 94096 | 92279 | 605 | 17R | 19571 | 21397 | 608 |
| 88R | 95566 | 96018 | 150 | 94R | 94129 | 94581 | 150 | 16L | 19538 | 19086 | 150 |
| 89R | 96086 | 97252 | 388 | 95R | 94649 | 95845 | 398 | 15L | 19018 | 17744 | 424 |
| 90R | 97345 | 98736 | 463 | 96R | 95938 | 97329 | 463 | 14L | 17651 | 16260 | 463 |
| 91R | 98860 | 100047 | 395 | 97R | 97453 | 98640 | 395 | 13L | 16136 | 14949 | 395 |
| 92R | 100398 | 100637 | 79 | 98R | 98927 | 99232 | 101 | - | - | - | - |
| 93L | 100986 | 100819 | 55 | 99L | 99593 | 99426 | 55 | 12R | 14091 | 14246 | 51 |
| 94L | 101563 | 101096 | 155 | 100R | 100169 | 99702 | 155 | 11L | 13512 | 13979 | 155 |
| 95R | 101656 | 102747 | 363 | 101R | 100180 | 101352 | 390 | 10L | 13419 | 12325 | 364 |
| 96R | 103549 | 104220 | 223 | 103R | 102169 | 102840 | 223 | 89R | 103279 | 103965 | 228 |
| 97R | 104303 | 104716 | 137 | 104R | 102923 | 103372 | 149 | 90R | 104031 | 104444 | 137 |
aORFs that have been added or altered are highlighted in bold. If a previously annotated ORF is not listed in the table, it has been deleted.
bPotentially frameshifted ORF
cWhere an ORF has a potential sequencing error resulting in a frameshift mutation, 2 stop codons are provided in the format X/Y. The first number represents the actual physical stop in the reported sequence. The second number is the proposed stop if a sequencing error occurred.
dLength of ORF in amino acids
Table 4.
Re-annotation of SGIV and GIV of the Ranavirus genus
| SGIVa | Start | Stopc | aad | GIVa | Start | Stopc | aad |
| 14L | 12773 | 12348 | 141 | 1.3L | 2020 | 1595 | 141 |
| 15L | 13000 | 12821 | 59 | 1.5L | 2247 | 2068 | 59 |
| 16L | 14289 | 13048 | 413 | 2L | 3536 | 2295 | 413 |
| 18R | 14317 | 15174 | 285 | 3R | 3564 | 4421 | 285 |
| 19R | 15196 | 16224 | 342 | 4R | 4443 | 5399 | 318 |
| 20L | 17246 | 16278 | 322 | 5L | 6421 | 5453 | 322 |
| 21L | 17725 | 17306 | 139 | 6L | 6900 | 6475 | 141 |
| 22L | 18277 | 17777 | 166 | 7L | 7486 | 6950 | 178 |
| 24L | 18774 | 18319 | 151 | 8L | 7983 | 7528 | 151 |
| 25L | 20488 | 18956 | 510 | 9L | 9682 | 8165 | 505 |
| 26R | 20567 | 22267 | 556 | 10R | 9761 | 11461 | 566 |
| 28L | 23363 | 22350 | 337 | 11L | 12559 | 11546 | 337 |
| 29L | 24445 | 23447 | 332 | 12L | 13659 | 12643 | 338 |
| 30L | 25635 | 24610 | 341 | 13L | 14850 | 13816 | 344 |
| 31L | 27160 | 26144 | 338 | 14L | 16384 | 15362 | 340 |
| 32.5L | 28666 | 27609 | 352 | 15L | 17904 | 16846 | 352 |
| 33L | 29760 | 28726 | 344 | 16L | 19010 | 17964 | 348 |
| 34L | 30161 | 29823 | 112 | 16.5L | 19411 | 19073 | 112 |
| 35L | 31388 | 30261 | 375 | 17L | 20638 | 19511 | 375 |
| 36L | 32515 | 31526 | 329 | 18L | 21835 | 20771 | 354 |
| 37L | 33696 | 32668 | 342 | 19L | 23016 | 21988 | 342 |
| 38L | 34236 | 33724 | 170 | 20L | 23556 | 23044 | 170 |
| 39L | 37417 | 34262 | 1051 | 21L | 26738 | 23583 | 1051 |
| 41L | 37978 | 37547 | 143 | 22L | 27296 | 26865 | 143 |
| 42R | 38058 | 38285 | 75 | - | - | - | - |
| 43R | 38285 | 40288 | 667 | 23R | 27608 | 29605 | 665 |
| 45L | 41090 | 40362 | 242 | 24L | 30407 | 29679 | 242 |
| 46L | 41866 | 41120 | 248 | 25L | 31204 | 30467 | 245 |
| 47L | 43063 | 41909 | 384 | 26L | 32401 | 31247 | 384 |
| 48L | 43489 | 43214 | 91 | 27L | 32824 | 32549 | 91 |
| 49L | 44002 | 43535 | 155 | 28L | 33336 | 32857 | 159 |
| 50L | 44695 | 44033 | 220 | 29L | 34029 | 33367 | 220 |
| 51L | 45563 | 44868 | 231 | 30L | 34896 | 34201 | 231 |
| 52L | 37997 | 35097 | 968 | 31L | 37997 | 35097 | 966 |
| 54R | 48777 | 49424 | 215 | 32R | 38100 | 38747 | 215 |
| 55R | 49447 | 50169 | 240 | 33R | 38770 | 39492 | 240 |
| 56R | 50198 | 50938 | 246 | 34R | 39521 | 40261 | 246 |
| 57L | 54510 | 51004 | 1168 | 35L | 43833 | 40327 | 1168 |
| 59L | 55000 | 54560 | 146 | 35.5L | 44323 | 43391 | 146 |
| 60R | 54967 | 57879 | 970 | 36R | 44348 | 47200 | 950 |
| 61R | 57914 | 58528 | 204 | 37R | 47235 | 47849 | 204 |
| 62R | 58593 | 59363 | 256 | 38R | 47914 | 48708 | 264 |
| 64R | 59415 | 61133 | 572 | 39R | 48760 | 50478 | 572 |
| 65R | 61268 | 61510 | 80 | 39.5R | 50614 | 50856 | 80 |
| 66R | 61603 | 61845 | 80 | 39.7R | 50949 | 51191 | 80 |
| 67L | 62482 | 61907 | 191 | 40L | 51829 | 51254 | 191 |
| 68L | 63334 | 62516 | 272 | 41L | 52681 | 51863 | 272 |
| 69L | 64967 | 63321 | 548 | 42L | 54314 | 52668 | 548 |
| 70R | 64994 | 65452 | 152 | 43R | 54341 | 54799 | 152 |
| 71R | 65483 | 66307 | 274 | 44R | 54830 | 55654 | 274 |
| 72R | 66404 | 67795 | 463 | 45R | 55751 | 57142 | 463 |
| 73L | 71185 | 67874 | 1103 | 46L | 60532 | 57221 | 1103 |
| 74R | 68472 | 68738 | 88 | 47R | 57819 | 58085 | 88 |
| 75R | 71239 | 71775 | 178 | 48R | 60586 | 61122 | 178 |
| 76L | 72715 | 71858 | 285 | 49L | 62064 | 61207 | 285 |
| 77L | 73747 | 72839 | 302 | 50L | 63096 | 62188 | 302 |
| - | - | - | - | 51L | 62944 | 62282 | 220 |
| 78L+81La | 76809 | 76246/73855 | 984 | 52L | 66156 | 63202 | 984 |
| 82L | 77592 | 76924 | 222 | 53L | 66939 | 66271 | 222 |
| 83R | 77672 | 79009 | 445 | 54R | 67019 | 68356 | 445 |
| 84L | 80193 | 79066 | 375 | 55L | 69540 | 68413 | 375 |
| 85R | 80251 | 80529 | 92 | 56R | 69598 | 69876 | 92 |
| 86R | 80591 | 81055 | 154 | 57R | 69938 | 70402 | 154 |
| 87R | 81385 | 82032 | 215 | 58R | 70728 | 71375 | 215 |
| 88L | 84187 | 82667 | 506 | 59L | 73355 | 71835 | 506 |
| 89L | 85420 | 84248 | 390 | 60L | 74588 | 73416 | 390 |
| 90L | 86627 | 85506 | 373 | 61L | 75794 | 74673 | 373 |
| 91L | 87886 | 86750 | 378 | 62L | 77051 | 75915 | 378 |
| 92L | 89216 | 88086 | 376 | 63L | 78373 | 77240 | 377 |
| 93L | 90497 | 89280 | 405 | 64L | 79654 | 78437 | 405 |
| 95R | 90635 | 91111 | 158 | 64.5L | 80265 | 79792 | 157 |
| 96R | 91148 | 91618 | 156 | 65R | 80301 | 80771 | 156 |
| 97L | 92774 | 91626 | 382 | 66L | 81926 | 80778 | 382 |
| 98R | 92428 | 93231 | 267 | 67R | 81580 | 82383 | 267 |
| 99R | 93244 | 93492 | 82 | 67.5R | 82380 | 82646 | 88 |
| 101R | 93753 | 94694 | 313 | 68R | 82906 | 83847 | 313 |
| 102L | 95007 | 94774 | 77 | 69L | 84161 | 83928 | 77 |
| 103R | 95092 | 95385 | 97 | 70R | 84246 | 84539 | 97 |
| 104L | 99252 | 95446 | 1268 | 71L | 88406 | 84600 | 1268 |
| 105R | 95498 | 95731 | 77 | 72R | 84652 | 84885 | 77 |
| 107R | 99308 | 100453 | 381 | 73R | 88462 | 89088 | 208 |
| 111R | 100766 | 101533 | 255 | 74R | 89401 | 90168 | 255 |
| 112R | 101588 | 102655 | 355 | 75R | 90223 | 91326 | 367 |
| 114L | 103050 | 102712 | 112 | 77.5L | 91721 | 91383 | 112 |
| 115R | 103122 | 103580 | 152 | 78R | 91793 | 92251 | 152 |
| 116R | 103700 | 104476 | 258 | 79R | 92371 | 93147 | 258 |
| 117L | 104733 | 104575 | 52 | 79.5L | 93401 | 93241 | 52 |
| 118R | 104795 | 105754 | 319 | 80R | 93463 | 94422 | 319 |
| 119R | 105799 | 106050 | 83 | 80.5R | 94467 | 94718 | 83 |
| 120L | 106525 | 106103 | 140 | 81L | 95162 | 94779 | 127 |
| 121R | 106615 | 106869 | 84 | 81.5R | 95291 | 95547 | 84 |
| 122L | 107599 | 106967 | 210 | 82L | 96275 | 95643 | 210 |
| 123L | 108740 | 107652 | 362 | 83L | 97416 | 96328 | 362 |
| 124R | 108863 | 109399 | 178 | 83.5L | 98976 | 97684 | 130 |
| 125R | 109474 | 110028 | 184 | 84R | 98151 | 98705 | 184 |
| 126R | 110101 | 110658 | 185 | 85R | 98692 | 99330 | 212 |
| 127R | 110731 | 111252 | 173 | 86R | 99403 | 99924 | 173 |
| 128R | 112041 | 115070 | 1009 | 87R | 100788 | 103817 | 1009 |
| 129L | 115490 | 115308 | 60 | 87.5L | 104245 | 103884 | 119 |
| 131R | 115749 | 116303 | 184 | 88R | 104499 | 105053 | 184 |
| 132R | 116321 | 117148 | 275 | 89R | 105071 | 105898 | 275 |
| 134L | 118498 | 117527 | 323 | 90L | 107244 | 106273 | 323 |
| 135L | 118885 | 118547 | 112 | 90.5L | 107631 | 107258 | 123 |
| 136R | 118946 | 119260 | 104 | 91R | 107692 | 108006 | 104 |
| 137R | 119282 | 120667 | 461 | 92R | 108028 | 109413 | 461 |
| 138L | 120907 | 120713 | 64 | 92.3L | 109653 | 109457 | 64 |
| 139R | 121013 | 121324 | 103 | 92.5R | 109757 | 110068 | 103 |
| 140R+141Rb | 121397 | 122311/124558 | 93R | 110141 | 113554 | 1137 | |
| 143L | 124882 | 124643 | 79 | 94L | 113878 | 113639 | 79 |
| 144R | 124963 | 125421 | 152 | 95R | 113959 | 114417 | 152 |
| 145R | 125480 | 125977 | 165 | 96R | 114476 | 114973 | 165 |
| 146L | 127052 | 126078 | 324 | 97L | 116050 | 115076 | 324 |
| 147L | 128221 | 127187 | 344 | 98L | 117220 | 116186 | 344 |
| 148R | 128324 | 128803 | 159 | 99R | 117323 | 117802 | 159 |
| 149R | 128843 | 129220 | 125 | 99.5R | 117842 | 118219 | 125 |
| 150L | 130827 | 129301 | 508 | 100L | 119826 | 118300 | 508 |
| 151L | 131435 | 130848 | 195 | 101L | 120434 | 119847 | 195 |
| 152R | 131534 | 132772 | 412 | 102R | 120533 | 121771 | 412 |
| 153L | 132661 | 132089 | 190 | 103L | 121660 | 121088 | 190 |
| 154R | 132788 | 133081 | 97 | 103.5R | 121787 | 122080 | 97 |
| 155R | 133172 | 134899 | 575 | 104R | 122172 | 123896 | 574 |
| 156L | 135860 | 135048 | 270 | 105L | 124852 | 124043 | 269 |
| 157R | 135948 | 136472 | 174 | 106R | 124940 | 125464 | 174 |
| 158L | 136944 | 136528 | 138 | 106.5L | 125936 | 125520 | 138 |
| 159R | 137020 | 137511 | 163 | 106.7R | 126012 | 126503 | 163 |
| 160L | 137996 | 137508 | 162 | 107L | 126988 | 126500 | 162 |
| 161.5L | 138598 | 138309 | 95 | 107.5L | 127561 | 127296 | 87 |
| 162L | 139822 | 138674 | 382 | 108L | 128797 | 127649 | 382 |
| 0.5L | 1029 | 0/140141-140020 | 391 | 109L | 130138 | 128963 | 391 |
| 1L | 1971 | 1057 | 304 | 110L | 131080 | 130166 | 304 |
| 3R | 2018 | 3163 | 381 | 111R | 131127 | 132272 | 381 |
| 4L | 4332 | 3235 | 365 | 113L | 133442 | 132345 | 365 |
| 5L | 5542 | 4400 | 380 | 114L | 134652 | 133510 | 380 |
| 6R | 5570 | 6349 | 259 | 115R | 134680 | 135453 | 257 |
| 7L | 7339 | 6416 | 307 | 116L | 136425 | 135520 | 301 |
| 8L | 7886 | 7194 | 230 | 117L | 136972 | 136280 | 230 |
| 9L | 8444 | 7980 | 154 | 118L | 137530 | 137066 | 154 |
| 10L | 8888 | 8517 | 123 | 118.5L | 137974 | 137603 | 123 |
| 11L | 9132 | 8944 | 62 | 119L | 138218 | 138030 | 62 |
| 12L | 12293 | 9219 | 1024 | 120.5L | 138307/139793 | 1/1540 | 1008 |
aORFs that have been added or altered are highlighted in bold. If a previously annotated ORF is not listed in the table, it has been deleted.
bPotentially frameshifted ORF
cWhere an ORF has a potential sequencing error resulting in a frameshift mutation, 2 stop codons are provided in the format X/Y. The first number represents the actual physical stop in the reported sequence. The second number is the proposed stop if a sequencing error occurred.
dLength of ORF in amino acids
As seen above, our comparative genomic approach was able to identify previously unannotated ORFs, homologous ORFs with potential frameshifts, and ORFs split between the two ends of a circular genome. Although this approach proved extremely successful for the Ranavirus and Megalocytivirus genera, we were unable to use it for the Chloriridovirus, Iridovirus, and Lymphocystivirus genera. This is due to the lack of co-linearity and the highly divergent sets of genes that exist between the members of these genera, as well as the low number of available genome sequences. However, we did modify the annotations of lymphocystis disease virus-China (LCDV-China) and invertebrate iridescent virus-6 (IIV-6). The previous annotations of these genomes of both species had contained a large number of overlapping ORFs [2,24], which we decided to exclude on several grounds. First, LCDV-China and IIV-6 are the only iridoviruses, out of the twelve so far sequenced, in which overlapping ORFs have been annotated. In addition, the original sequencing paper for IIV-6 [2] and a follow-up paper by the same group [25] did not include a number of the overlapping ORFs reported in the database sequence, presumably due to their small size and lack of similarity with other viral and cellular genes. Finally, there is no experimental or bioinformatics evidence to suggest that any of these ORFs encode proteins. Therefore, to improve the overall consistency of the Iridoviridae family annotations, we removed the small overlapping ORF annotations from the LCDV-China and IIV-6 genomic sequences (Table 5, Additional File 1 &2).
Table 5.
Overlapping ORFs deleted from the Iridovirus and Lymphocystivirus genera
| Virus | Deleted |
| LCDV-C | 4L, 8R, 17R, 20L, 21L, 26L, 28L, 30L, 31L, 32L, 35R, 36R, 44R, 46L, 48L, 52R, 55L, 68L, 74R, 76R, 78R, 79R, 81R, 88R, 92R, 94R, 98R, 102R, 103R, 113L, 120R, 130L, 132L, 134L, 138R, 141R, 144L, 152L, 155L, 156L, 163L, 167R, 174L, 183L, 188L, 192L, 193L, 194L, 195L, 198R, 199L, 200R, 204R, 207L, 210L, 213L, 223R, 225R, 232R, 233L, 236L, 238R, 240L |
| IIV-6 | 1R, 2R, 3R, 4R, 5R, 7R, 8R, 11L, 13R, 14R, 15R, 16L, 17R, 18R, 20L, 21R, 23L, 24L, 25R, 26R, 27L, 28L, 31R, 33L, 35L, 36R, 38R, 39R, 40R, 46R, 47R, 48R, 51R 52R, 53R, 54R, 55R, 57L, 58L, 59R, 63R, 64L, 66L, 68L, 70R, 72R, 73R, 74R, 76L, 78R, 79L, 80L, 81L, 86R, 87R, 88L, 89L, 90R, 91R, 92R, 93R, 97L, 99L, 102R, 103R, 105R, 108R, 109R, 112R, 114L, 119R, 124L, 125L, 128L, 129R, 131L, 133R, 134L, 144R, 147L, 150R, 151R, 152R, 153L, 154R, 158R, 163L, 164R, 166L, 167L, 168R, 171R, 173R, 174R, 177L, 178L, 180L, 181L, 182L, 183L, 185L, 186L, 187R, 188L, 189L, 190R, 191L, 194R, 199L, 202L, 204L, 207L, 108L, 210L, 214L, 215R, 217L, 220L, 222R, 223L, 230L, 231R, 233L, 237R, 239R, 243R, 245R, 248R, 252L, 256R, 257R, 258R, 260R, 262R, 263L, 264R, 265L, 266L, 267R, 269R, 270R, 271R, 275R, 276L, 277R, 278L, 279R, 280R, 281R, 282R, 283L, 286L, 288R, 290R, 291R, 292L, 294R, 296R, 297L, 298R, 299R, 303R, 304R, 305L, 310R, 311R, 314L, 316R, 318R, 319L, 320L, 321R, 323L, 324L, 326L, 327R, 328L, 330L, 331R, 333L, 334R, 336R, 338L, 339L, 341R, 344R, 345R, 351R, 352R, 353L, 354L, 355R, 356L, 360R, 362R, 363L, 364L, 365L, 367L, 370R, 371R, 372R, 377R, 379L, 381L, 382R, 383L, 386R, 387R, 390R, 392R, 394R, 397L, 398R, 399R, 402L, 403L, 405L, 406R, 407R, 408R, 409R, 410L, 412L, 416R, 417L, 418R, 419L, 421L, 424R, 425R, 427R, 429R, 430R, 431L, 432R, 433R, 434L, 435R, 440R, 442L, 444R, 445L, 446L, 447L, 448L, 449L, 450L, 452R, 455L, 456R, 459L, 461R, 462R, 464R, 465R |
Defining the conserved genes in Iridoviruses
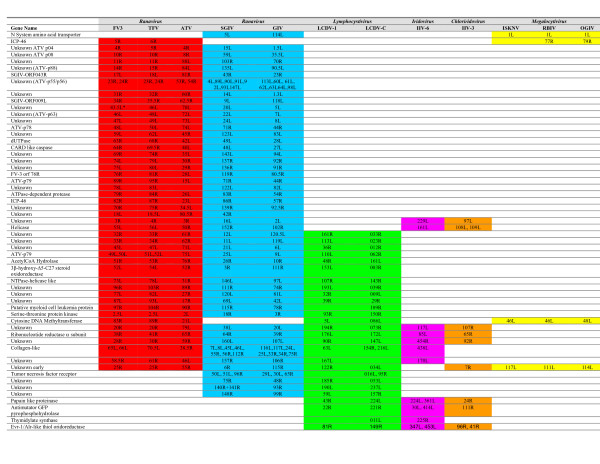
As a result of this re-annotation of the Iridoviridae family, species within each genus now have a much greater consensus among their annotated ORFs. Prior to re-annotation, only 19 ORFs appeared to be conserved across all iridovirus species (Table 6). Although a previous report has suggested that 27 core genes exist within the Iridoviridae family [26], those core genes reported are found in most, but not all published iridoviridal species. In light of our previous results, we re-examined this core set of genes using the VOCs software. We identified seven novel core genes (Table 7), increasing the total number to 26 (Table 6 &7). This increase in the number of core genes was primarily due to the five new genes annotated during the re-analysis of RBIV (Table 7 bold highlighted genes). As expected most of the core genes are predicted to have essential functions, required for transcription, replication, and virus formation. Interestingly, three core genes, the orthologs of FV3 12L, 41R, and 94L, have no predicted functions. As previously stated Delhon et al. [26] identified 27 core genes, one more than we identified after our re-analysis. Delhon et al. [26] report the orthologs of FV3 20L represent a core [26]. However, our analysis shows that orthologs of FV3 20L exist in all genera except the Megalocytivirus (Figure 1) suggesting that FV3 20L is not a core gene. Future research to determine the functions of these genes, which are also likely to be essential, will provide important data for understanding the replication cycle of iridoviruses.
Table 6.
Iridoviridae Core Genes
| Gene Namea | FV3 | TFV | ATV | SGIV | GIV | LCDV-1 | LCDV-C | ISKNV | RBIV | OSGIV | IIV-6 | MIV | |
| 1. | Putative replication factor and/or DNA binding-packing | 1R | 105R | 91R | 116R | 79R | 162L | 181R | 61L | 57L | 60L | 282R | 79L |
| 2. | DNA-dep RNA pol-II Largest subunit | 8R | 8R | 6R | 104L | 71L | 16L | 191R | 28L | 29L | 31L | 176R, 343L | 90L |
| 3. | Putative NTPase I | 9L | 9L | 7L | 60R | 36R | 132L | 075L | 63L | 59L | 63L | 22L | 87L |
| 4. | ATPase-like protein | 15R | 16R | 83R | 134L | 90L | 54R | 114L | 122R | 116R | 119R | 75L | 88R |
| 5. | Helicase family | 21L | 21L | 78R | 54R | 32R | 6L | 7L | 56L | 54L | 57L | 67R | 4R |
| 6. | D5 family NTPase involved in DNA replication | 22R | 22R | 77L | 52L | 31L | 128L | 80L | 109L | 101L | 106L | 184R | 121R |
| 7. | Putative tyrosin kinase/lipopolysaccharide modifying enzyme | 27R | 29R | 58R | 78L+81Lb | 52L | 195R | 173R | 61L, 114L | 57L, 106Lb | 60L, 111L | 179R, 439L | 35R |
| 8. | NIF-NLI interacting factor | 37R | 40R | 64R | 61R | 37R | 82L | 148L | 5L | 6L | 6L | 355R | 104R |
| 9. | Unknown | 41R | 45R | 69R | 57L | 35L | 163R | 235R | 76L | 72L | 75L | 295L | 16R |
| 10. | Myristilated membrane protein | 53R | 55R | 51L | 88L | 59L | 67L | 158R | 7L | 8L | 8L | 118L, 458R | 6R |
| 11. | DNA pol Family B exonuclease | 60R | 63R | 44L | 128R | 87R | 135R | 203L | 19R | 20R | 22R | 37L | 120L |
| 12. | DNA-dep RNA pol-II second largest subunit | 62L | 65L | 43R | 73L | 46L | 25L | 25R | 34R | 33R | 36R | 428L | 9R |
| 13. | Ribonucleotide reductase small subunit | 67L | 71L | 38R | 47L | 26L | 27R | 41L | 24R | 26R | 27R | 376L | 48L |
| 14. | Ribonuclease III | 80L | 85L | 25R | 84L | 55L | 137R | 187R | 87R | 83R | 85R | 142R | 101R |
| 15. | Proliferating cell nuclear antigen | 84R | 90R | 20L | 68L | 41L | 3L | 197L | 112R | 103Rb | 109R | 436L | 60L |
| 16. | Major capsid protein | 90R | 96R | 14L | 72R | 45R | 147L | 43L | 6L | 7L | 7L | 274R | 14L |
| 17. | Putative XPPG-RAD2-type nuclease | 95R | 101R | 10L | 97L | 66L | 191R | 169R | 27L | 28L | 30L | 369L | 76L |
| 18. | Serine-threonine protein kinase | 19R | 19R | 80L | 39L | 21L | 10L | 45R | 55L | 53L | 56L | 380R | 10L |
| 19. | Serine-threonine protein kinase | 57R | 59R | 47L | 150L | 100L | 143L | 178L | 13R | 13R | 15R | 98R | 98L |
The Iridoviridae core genes are shown.
aORFs that have been added or altered are highlighted in bold
bPotentially frameshifted ORF
Table 7.
Additional Iridoviridae Core Genes Identified After Genome Re-analysis
| Newly Characterized Gene Namea | FV3 | TFV | ATV | SGIV | GIV | LCDV-1 | LCDV-C | ISKNV | RBIV | OSGIV | IIV-6 | MIV | |
| 1. | Myristilated membrane protein | 2L | 2Lb | 1L | 19R | 4R | 160L | 38R | 90.5L | 85L | 88.5L | 337L | 47R |
| 2. | Unknown | 12L | 12L | 87R | 118R | 80R | 108L | 100L | 96L | 89.5Lb | 93L | 287R | 56L |
| 3. | Transcription elongation factor TFIIS | 81R | 86R | 24L | 85R | 56R | 171R | 115R | 29L | 29.5Lb | 32L | 349L | 55R |
| 4. | Deoxynucleoside kinase | 85R | 91.5R | 19L | 67L | 40L | 136R | 027R | 32R | 31R | 34R | 143R | 29R |
| 5. | Erv1/Alr family | 88R | 94R | 16L | 70R | 43R | 106L | 142L | 43L | 43.5L | 45L | 347L | 96R |
| 6. | Immediate early protein ICP-46 | 91R | 97R | 13L | 162L | 108L | 47L | 162R | 115R | 108.5R | 112R | 393L | 39R |
| 7. | Hypothetical protein-Clostridium tetani | 94L | 100R | 11L | 98R | 67R | 19R | 153L | 86R | 82.5R | 84.5L | 307L | 33L |
The Iridoviridae core genes are shown.
aORFs that have been added or altered are highlighted in bold
bPotentially frameshifted ORF
Figure 1.
Conserved Iridovirus Genes. Every Iridoviridae gene that has an ortholog in at least 2 Iridoviridae genera are shown. Orthologs share the same row on the table. The genes within each genus are color-coded for easier identification. As long as at least one member of the genus contains an ortholog, the entire genus is highlighted. Where multiple ORFs are listed for a particular gene name, the ORFs represent multiple orthologs of the gene in that viral species. The remainder of the figure showing just the genes conserved between the Iridovirus and Chloriridovirus genera are included in Additional File 3.
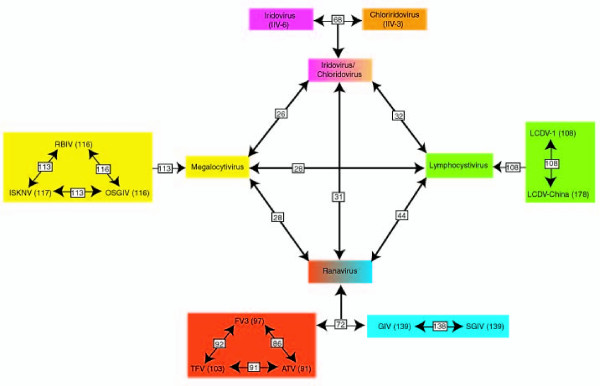
Identifying genes conserved between some, but not all, iridovirus species can give us important information when investigating evolutionary relationships within the family. A number of past phylogenetic analyses of Iridoviridae have used phylogenic trees constructed from aligned protein sequences [1,18-20,22,24,27]. However, there are potential problems with phylogenic analysis based on comparisons of single genes. This type of analysis is rarely consistent due to horizontal gene transfer [28] and variable rates of evolution [29]. Therefore, we decided to take a whole genome comparative phylogenetic analysis to understand the relationship between iridoviruses. Our approach was to identify all the genes conserved between different genera to gain a better understanding of the relationships within the iridovirus family. This approach yields an indication of how similar in gene content 2 genomes are. Our whole-genome comparative analysis, grouped orthologous genes between genera (Figures 1 &2 and Additional File 3), and was consistent with phylogenic trees constructed from single protein sequences. Based on gene conservation, the Ranavirus and Lymphocystivirus genera appear to be most closely related to one another (Figure 2). In addition, the Iridovirus and Chloriridovirus genera are also closely related to one another based on presence of orthologous genes (Figure 2). In contrast, the Megalocytivirus genus and the Iridovirus/Chloriridovirus genera are equally divergent from each other as well as all other Iridoviridae family members (Figure 2).
Figure 2.
Phylogenetic relationships between the five iridovirus genera based on gene content. Individual viral species were compared within a genus to identify the number of orthologous genes. Orthologous genes between viral genera were then determined. The numbers on each line identify the number of orthologous genes shared between viral species or genera including the 26 core genes. The Iridovirus and Chloriridovirus genera have a high degree of gene conservation and a combined genera box (Iridovirus/Chloriridovirus) was used to compare orthologous genes between genera. In addition, two subgroups of the Ranavirus genus are shown. Each subgroup contains a virtually identical complement of genes. However, a comparison between the FV3/TFV/ATV subgroup with the SGIV/GIV subgroup revealed 72 orthologous genes.
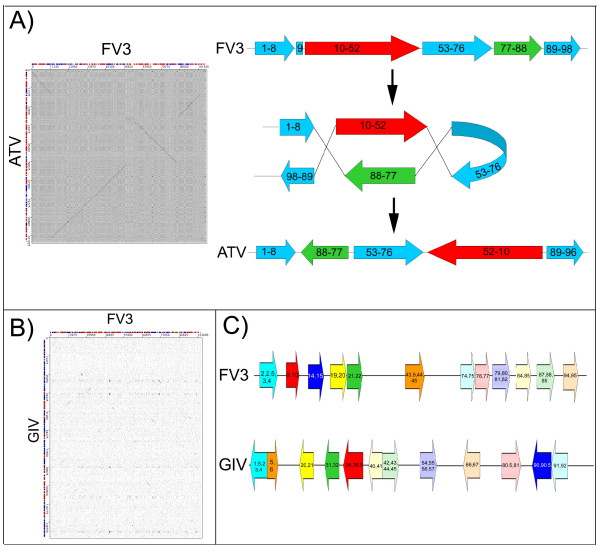
As the list of sequenced iridovirus genomes grows, the non-co-linearity between many of these genomes becomes more apparent. The Megalocytivirus and Ranavirus, but not the Chloriridovirus, Iridovirus, and Lymphocystivirus genera, show a co-linear arrangement of genes within each genus. However, comparisons of genomic sequences from different genera suggest no co-linearity. This trend may be the result of the high recombination rates [30] seen in some iridovirus members [31]. For example, within the Ranavirus genus, ATV has two inversions relative to the FV3 and TFV sequences [30], reducing the co-linearity of these genomes to some degree. Figure 3A shows how two recombination events could convert FV3 to the ATV arrangement of genes. In contrast, a comparison between the more distantly related members within the Ranavirus genus (such as FV3 and GIV) demonstrate a much more dramatic loss of co-linearity. No long stretches of co-linear genes exist between these sequences, although small sections of co-linearity remain as seen through a dotplot analysis between FV3 and GIV (Figure 3B). The dotplot shows small regions of co-linearity scattered throughout the genome of FV3 and GIV as seen by short diagonal lines on the dotplot (Figure 3B). A schematic representation of the co-linearity between FV3 and GIV demonstrates that co-linearity occurs in small clusters of genes often only 2–4 genes in length (Figure 3C).
Figure 3.
Co-linearity found within the Ranavirus genus. (A) FV3 and ATV, both members of the Ranavirus genus possess almost complete co-linearity of orthologous genes as visualized by a dotplot. However, 2 inversions have occurred. The FV3 genes 10–52 and 77–88 have switched genomic locations as shown, potentially through two recombination events. The inversion has also resulted in the loss of the ortholog of FV3 9L in ATV. (B) There is a limited amount of co-linearity found between FV3/TFV/ATV and SGIV/GIV. The co-linearity has been visualized using a dotplot analysis between FV3 (horizontal sequence) and GIV (vertical sequence). Genes are colored either red or blue representing right- or left-ward transcription respectively. (C) The co-linearity between FV3 and GIV is generally composed of stretches of 2 or 3 co-linear orthologous genes. Orthologous genes, in a co-linear arrangement are schematically shown as blocks of the same color on either FV3 or GIV genomic sequence.
Conclusion
The Iridoviridae family can cause severe diseases resulting in significant economic and environmental losses. Very little is known about how iridoviruses cause disease in their host. Our re-analysis of genomes within the Iridoviridae family provides a unifying framework to understand the biology of these viruses. For example, the re-analysis of the Iridoviridae family has increased the consistency of annotated sequences from viruses within the same genus. In addition, the re-analysis has helped create a much greater consensus among Iridoviridae family members and enhanced our understanding of this virus family as a whole. The updated annotations that we have produced for the iridovirus sequences can be found in the additional files to this paper; in addition, the databases and tools to analyse Iridoviridae genomes are available to all researchers [32]. This database will contain genomes from the original GenBank files and also the edited genomes described in this paper. Further re-defining the core set of iridovirus genes will continue to lead us to a better understanding of the phylogenetic relationships between individual iridoviruses as well as giving us a much deeper understanding of iridovirus replication. In addition, this analysis will provide a better framework for characterizing and annotating currently unclassified iridoviruses.
Methods
Re-annotation of the iridoviridae
Annotated sequences for the twelve completely sequenced iridovirus genomes (Table 1) were obtained from GenBank files and imported into the Viral Orthologous Clusters (VOCs) database [15]. Species from the same genus were examined using VOCs to identify all of the orthologous genes. The analysis then focused on the differences found between genomes within the same genus. For those genomes that contained co-linear arrangements of genes (those in the Ranavirus and Megalocytivirus genera), we compared those regions containing annotated ORFs. If more than two sequenced genomes were available for a given genus, and the ORF was present in at least two of the genomes, then we set out to determine if that ORF was also present in the remainder of the genomes. By this method, we were able to re-annotate small segments of each genome without needing to re-analyse the entire genome. The Viral Genome Organizer (VGO) software [16] was used to visualize the annotated ORFs, as well as the start and stop codons found within each genome.
Analysis of orthologous genes
We used a combination of BLAST searches and queries using the VOCs software [32] to define orthologous genes between Iridoviridae genera. VOCs is a JAVA client-server that accesses a sequence query language (SQL) database containing iridovirus genomes. This SQL database permits complex queries to be assembled in an easy to use graphical user interface. VOCs initially groups orthologous genes into families based on BLASTP scores, these can be manually checked and altered if necessary.
Dotplot analysis
Dotplots of FV3 and GIV were done using JDotter [33]. JDotter provides an interactive input window that links JDotter to the VOCs database. The sequences for the FV3 and GIV were obtained through the VOCs database.
Competing interests
The author(s) declare that there are no competing interests.
Authors' contributions
HEE, JM, EP, and CRB carried out the analysis of the Iridoviridae family and generated the tables and figures. VTJ and CU generated the databases and tools to carry out the analysis done in the manuscript. CRB and CU conceived of the study, and participated in its design and coordination and helped to draft the manuscript. All authors read and approved the final manuscript.
Supplementary Material
Revised annotation of the Lymphocystivirus genus. The table highlights the changes made to the Lymphocystivirus genus.
Analysis of the Iridovirus and Chloriridovirus genera. The table highlights the changes made to the Iridovirus and Chloriridovirus genera.
Additional Conserved Genes Between Iridovirus & Chloriridovirus genera. The table is an extension of Figure 1 – showing the genes that are conserved just between the Iridovirus and Chloriridovirus genera.
Acknowledgments
Acknowledgements
We would like to thank Daniel Rock for sharing information about mosquito iridescent virus prior to publication and Cristalle Watson for critically reviewing the manuscript. This work was supported by Discovery Grants (Natural Science and Engineering Research Council (NSERC) of Canada) to C.R.B. and C.U. H.E.E. is the recipient of an NSERC postgraduate scholarship.
Contributor Information
Heather E Eaton, Email: heathereaton@trentu.ca.
Julie Metcalf, Email: juliem@cogeco.ca.
Emily Penny, Email: emilypenny@trentu.ca.
Vasily Tcherepanov, Email: vasilyt@uvic.ca.
Chris Upton, Email: cupton@uvic.ca.
Craig R Brunetti, Email: craigbrunetti@trentu.ca.
References
- He JG, Lu L, Deng M, He HH, Weng SP, Wang XH, Zhou SY, Long QX, Wang XZ, Chan SM. Sequence analysis of the complete genome of an iridovirus isolated from the tiger frog. Virology. 2002;292:185–197. doi: 10.1006/viro.2001.1245. [DOI] [PubMed] [Google Scholar]
- Jakob NJ, Muller K, Bahr U, Darai G. Analysis of the first complete DNA sequence of an invertebrate iridovirus: coding strategy of the genome of Chilo iridescent virus. Virology. 2001;286:182–196. doi: 10.1006/viro.2001.0963. [DOI] [PubMed] [Google Scholar]
- Williams T, Chinchar VG, Darai G, Hyatt A, Kalmakoff J, Seligy V. Iridoviridae. In: van Regenmortel MHV, Bishop DHL, Carstens EB, Estes MK, Lemon SM, Maniloff J, Mayo MA, McGeoch DJ, Pringle CR, Wickner RB, editor. Virus Taxonomy: The Classification and Nomenclature of Viruses. Seventh report of the International Committee on the Taxonomy of Viruses; 2000. pp. 167–182. [Google Scholar]
- Williams T. The iridoviruses. Adv Virus Res. 1996;46:345–412. doi: 10.1016/s0065-3527(08)60076-7. [DOI] [PubMed] [Google Scholar]
- Ahne W, Schlotfeldt HJ, Thomsen I. Fish viruses: isolation of an icosahedral cytoplasmic deoxyribovirus from sheatfish (Silurus glanis) J Vet Med. 1989;36:333–336. doi: 10.1111/j.1439-0450.1989.tb00611.x. [DOI] [PubMed] [Google Scholar]
- Hedrick RP, McDowell TS. Properties of iridoviruses from ornamental fish. Vet Res. 1995;26:423–427. [PubMed] [Google Scholar]
- Hedrick RP, McDowell TS, Ahne W, Torhy C, De Kinkelin P. Properties of three iridovirus-like agents associated with systemic infections of fish. Dis Aquat Org. 1992;13:203–209. [Google Scholar]
- Langdon JS, Humphrey JD, Williams LM, Hyatt AD, Westbury HA. First virus isolation from Australian fish: an iridovirus-like pathogen from redfin perch Perca fluviatilis. J Fish Dis. 1986;9:263–268. doi: 10.1111/j.1365-2761.1986.tb01011.x. [DOI] [Google Scholar]
- Pozet F, Morand M, Moussa A, Torhy C, De Kinkelin P. Isolation and preliminary characterization of a pathogenic icosahedral deoxyribovirus from the catfish Ictalurus melas. Dis Aquat Org. 1992;14:35–42. [Google Scholar]
- Bollinger TK, Mao J, Schock D, Brigham RM, Chinchar VG. Pathology, isolation, and preliminary molecular characterization of a novel iridovirus from tiger salamanders in Saskatchewan. J Wildlife Disease. 1999;35 doi: 10.7589/0090-3558-35.3.413. [DOI] [PubMed] [Google Scholar]
- Jancovich JK, Davidson EW, Seiler A, Jacobs BL, Collins JP. Transmission of the Ambystoma tigrinum virus to alternate hosts. Dis Aquat Organ. 2001;46:159–163. doi: 10.3354/dao046159. [DOI] [PubMed] [Google Scholar]
- Collins JP, Brunner JL, Miera V, Parris MJ, Schock DM, Storfer A. Ecology and evolution of infectious disease. In: Semlitsch R, editor. Amphibian Conservation. Washington , Smithsonian Institution Press; 2003. pp. 137–151. [Google Scholar]
- Daszak P, Cunningham AA, Hyatt AD. Infectious disease and amphibian population declines. Divers Distrib. 2003;9:141–150. doi: 10.1046/j.1472-4642.2003.00016.x. [DOI] [Google Scholar]
- Jancovich JK, Davidson EW, Parameswaran N, Mao J, Chinchar VG, Collins JP, Jacobs BL, Storfer A. Evidence for emergence of an amphibian iridoviral disease because of human-enhanced spread. Molecular Ecology. 2005;14:213–224. doi: 10.1111/j.1365-294X.2004.02387.x. [DOI] [PubMed] [Google Scholar]
- Ehlers A, Osborne J, Slack S, Roper RL, Upton C. Poxirus orthologous clusters (POCs) Bioinformatics. 2002;18:1544–1545. doi: 10.1093/bioinformatics/18.11.1544. [DOI] [PubMed] [Google Scholar]
- Upton C, Hogg D, Perrin D, Boone M, Harris NL. Viral genome organizer: a system for analyzing complete viral genomes. Virus Research. 2000;70:55–64. doi: 10.1016/S0168-1702(00)00210-0. [DOI] [PubMed] [Google Scholar]
- Brunetti CR, Amano H, Ueda Y, Qin J, Miyamura T, Suzuki T, Li X, Barrett JW, McFadden G. Complete genomic sequence and comparative analysis of the tumorigenic poxvirus Yaba monkey tumor virus. Journal of Virology. 2003;77:13335–13347. doi: 10.1128/JVI.77.24.13335-13347.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L, Zhou SY, Chen C, Weng SP, Chan SM, He JG. Complete genome sequence analysis of an iridovirus isolated from the orange-spotted grouper, Epinephelus coicodes. Virology. 2005;339:81–100. doi: 10.1016/j.virol.2005.05.021. [DOI] [PubMed] [Google Scholar]
- Do JW, Moon CH, Kim HJ, Ko MS, Kim SB, Son JH, Kim JS, An EJ, Kim MK, Lee SK, Han MS, Cha SJ, Park MS, Park MA, Kim YC, Kim JW, Park JW. Complete genomic DNA sequence of rock bream iridovirus. Virology. 2004;325:351–363. doi: 10.1016/j.virol.2004.05.008. [DOI] [PubMed] [Google Scholar]
- He JG, Deng M, Weng SP, Li Z, Zhou SY, Long QX, Wang XZ, Chan SM. Complete genome analysis of Mandarin fish infectious spleen and kidney necrosis iridovirus. Virology. 2001;291:126–139. doi: 10.1006/viro.2001.1208. [DOI] [PubMed] [Google Scholar]
- Tsai CT, Ting JW, Wu MH, Wu MF, Guo IC, Chang CY. Complete genomic sequence of the grouper iridovirus and comparison of genomic organization with those of other iridoviruses. Journal of Virology. 2005;79:2010–2023. doi: 10.1128/JVI.79.4.2010-2023.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song WJ, Qin QW, Qiu J, Huang CH, Wang F, C.L. H. Functional genomics analysis of Singapore grouper iridovirus: complete sequence determination and proteomic analysis. Journal of Virology. 2004;78:12576–12590. doi: 10.1128/JVI.78.22.12576-12590.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goorha R, Murti KG. The genome of frog virus 3, an animal DNA virus, is circularly permuted and terminally redundant. Proc Natl Acad Sci USA. 1982;79:248–252. doi: 10.1073/pnas.79.2.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang QY, Xiao F, Xie J, Li ZQ, Gui JF. Complete genome sequence of lymphocystis disease virus isolated from China. Journal of Virology. 2004;78:6982–6994. doi: 10.1128/JVI.78.13.6982-6994.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakob NJ, Kleespies RG, Tidona CA, Muller K, Gelderblom HR, Darai G. Comparative analysis of the genome and host range characteristics of two insect iridoviruses: Chilo iridescent virus and a cricket iridovirus isolate. Journal of General Virology. 2002;83:463–470. doi: 10.1099/0022-1317-83-2-463. [DOI] [PubMed] [Google Scholar]
- Delhon G, Tulman ER, Afonso CL, Lu Z, Becnel JJ, Moser BA, Kutish GF, Rock DL. Genome of invertebrate iridescence virus type 3 (Mosquito iridescent virus) Journal of Virology. 2006;80:8439–8449. doi: 10.1128/JVI.00464-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan WGH, Barkman TJ, Chinchar VG, Essani K. Comparative genomic analyses of frog virus 3, type species of the genus Ranavirus (family Iridoviridae) Virology. 2004;323:70–84. doi: 10.1016/j.virol.2004.02.019. [DOI] [PubMed] [Google Scholar]
- Doolittle WF, Logsdon JM. Archaeal genomics: Do Archaea have a mixed heritage? Current Biology. 1998;8:R209–R211. doi: 10.1016/S0960-9822(98)70127-7. [DOI] [PubMed] [Google Scholar]
- Huynen MA, Bork P. Measuring genome evolution. Proc Natl Acad Sci USA. 1998;95:5849–5856. doi: 10.1073/pnas.95.11.5849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jancovich JK, Mao J, Chinchar VG, Wyatt C, Case ST, Kumar S, Valente G, Subramanian S, Davidson EW, Collins JP, Jacobs BL. Genomic sequence of a ranavirus (family Iridoviridae) associated with salamander mortalities in North America. Virology. 2003;316:90–103. doi: 10.1016/j.virol.2003.08.001. [DOI] [PubMed] [Google Scholar]
- Chinchar VG, Granoff A. Temperature-sensitive mutants of frog virus 3: biochemical and genetic characterization. Journal of Virology. 1986;58:192–202. doi: 10.1128/jvi.58.1.192-202.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viral Bioinformatics Resource Center [www.virology.ca]
- Brodie R, Roper RL, Upton C. JDotter: a Java interface to multiple dotplots generated by dotter. Bioinformatics. 2004;20:279–281. doi: 10.1093/bioinformatics/btg406. [DOI] [PubMed] [Google Scholar]
- Tidona CA, Darai G. The complete DNA sequence of lymphocystis disease virus. Virology. 1997;230:207–216. doi: 10.1006/viro.1997.8456. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Revised annotation of the Lymphocystivirus genus. The table highlights the changes made to the Lymphocystivirus genus.
Analysis of the Iridovirus and Chloriridovirus genera. The table highlights the changes made to the Iridovirus and Chloriridovirus genera.
Additional Conserved Genes Between Iridovirus & Chloriridovirus genera. The table is an extension of Figure 1 – showing the genes that are conserved just between the Iridovirus and Chloriridovirus genera.