Abstract
Ionotropic glutamate receptors (iGluRs), a family of ligand-gated ion channels, are responsible for the majority of fast excitatory neurotransmission in the central nervous system. Within this family, different members serve distinct roles at glutamatergic synapses. Amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) receptors mediate fast depolarization while N-methyl-D-aspartate (NMDA) receptors mediate the slower component of the excitatory postsynaptic potential. These disparate functions suggest alternate modes of regulation. In this work, we show that endogenous regulators of iGluRs have different abilities to bind to specific domains of NMDA NR1-1b and AMPA GluR2 subunits. We have previously shown that the sulfated neurosteroids pregnenolone sulfate and 3α-hydroxy-5β-pregnan-20-one sulfate bind to the extracellular glutamate-binding core (S1S2) of the GluR2 subunit. Here we show that neither neurosteroid binds to the S1S2 domain of the NMDA NR1-1b subunit. This NR1-1b NMDA domain does, however, bind to the endogenous polyamines spermine and spermidine as well as Zn(II). Binding of the polyamines and Zn(II) to the S1S2 domain of the GluR2 subunit was not observed. This binding of Zn(II) and polyamines to the S1S2 domain of the NR1-1b subunit defines a new binding site for each of these modulators.
INTRODUCTION
Ionotropic glutamate receptors (iGluRs) comprise a family of ligand-gated ion channels that include the amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA), kainate, and N-methyl-D-aspartate (NMDA) receptors. These receptors are located in the postsynaptic neural membrane and play important roles in developmental plasticity, learning and memory, sensory transmission and coordination, and control of respiration and blood pressure. Binding of the neurotransmitter glutamate (glycine is a coagonist for the NMDA receptors) to an extracellular binding site on these receptors causes a conformational change which opens a pore, allowing cations to flow into the postsynaptic neural cell.
Each of the iGluRs is believed to be tetrameric in nature. The NMDA receptors are hetero-tetramers composed of two NR1 and two NR2 subunits. There are eight alternatively spliced versions of the NR1 subunit, encoded by a single gene, and four different NR2 subunits (NR2A–D) encoded by separate genes. The non-NMDA receptors are most often homo-tetramers. AMPA receptors are composed of four GluR1, GluR2, GluR3, or GluR4 subunits while kainate receptors are composed of four GluR5, GluR6, GluR7, KA1, or KA2 subunits. Each of these iGluR subunits has a similar modular membrane-spanning topology. The extracellular portion of each subunit has an amino terminal domain (ATD) and an S1 domain that precede the first membrane-spanning region. In addition, there is an extracellular S2 domain that separates the second and third membrane spanning regions, three membrane spanning domains, and a reentrant loop, the latter of which forms the pore region upon association of all four subunits. The extracellular S1 and S2 regions are known to form a clamshell-shaped binding domain for the natural agonist glutamate (in the non-NMDA receptors as well as in the NR2 NMDA subunits) or glycine (in the NR1 NMDA subunits). Soluble extracellular GluR2 (AMPA) and NR1-1b (NMDA) S1S2 domains have been constructed by eliminating all three transmembrane spanning regions plus the reentrant loop and linking the S1 and S2 domains by two amino acids (GT) (1–4). These S1S2 domains have been shown to possess near-native binding affinities for both agonists and antagonists and their structures have been studied by x-ray crystallography (Fig. 1) (1–4).
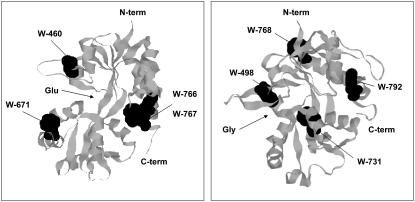
FIGURE 1.
(Left) RASMOL rendering of the apo GluR2 S1S2 domain (PDB: 1FTO) (1). The four tryptophan residues in the S1S2 domain (Trp-460, -671, -766, and -767) are space-filled and are shown in black. In addition, the location of the glutamate (Glu) binding pocket is illustrated. (Right) RASMOL rendering of the glycine bound NMDA NR1-1b S1S2 domain (PDB: 1PB7) (4). The four tryptophan residues in the S1S2 domain (Trp-498, -731, -768, and -792) are space-filled and are shown in black. In addition, the location of the glycine (Gly) binding pocket is illustrated.
As glutamate is the major excitatory neurotransmitter in the central nervous system, the level of activity of iGluRs is tightly controlled. Misregulation has been implicated in the ischemic stroke cascade, schizophrenia, and Alzheimer's, Huntington's, and Parkinson's diseases. A number of endogenous compounds, including Zn(II), polyamines (spermine and spermidine, Fig. 2), and sulfated neurosteroids (pregnenolone sulfate, PS; and 3α-hydroxy-5β-pregnan-20-one sulfate, PregaS, Fig. 2), are known to be involved in regulation.
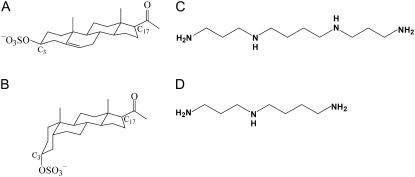
FIGURE 2.
Endogenous ligand structures. (A) Pregnenolone sulfate (PS), (B) 3α-hydroxy-5β-pregnan-20-one sulfate (PregaS), (C) spermine, and (D) spermidine.
PregaS negatively regulates the activity of all iGluRs. PS also negatively regulates the activity of the non-NMDA iGluRs, while differentially regulating the NMDA receptors. It is the most active neurosteroid in its ability to positively regulate NMDA receptors possessing either NR2A or B subunits (5), while it negatively regulates NMDA receptors with either NR2C or D subunits. It has recently been shown that both PS and PregaS bind to the S1S2 domain of the GluR2 subunit of the AMPA receptor (6,7). In addition, it has been demonstrated that PS binds to the NR2B subunit of the NMDA receptor in a region that encompasses portions of the extracellular S2 domain and the final transmembrane spanning region (8).
Zn(II), which is stored and released at many glutamatergic synapses in the brain, regulates the activity of iGluRs by many different pathways, both voltage-dependent and -independent (9,10). This regulation can either potentiate or block activity at iGluRs, depending on the type of iGluR and the concentration of Zn(II) (11). The existence of an NMDA high affinity NR1/NR2A Zn(II) inhibitory site as well as NR1/NR2B, NR1/NR2C, and NR1/NR2D low affinity Zn(II) inhibitory sites have been reported (12–16). The NR2A and NR2B binding sites have been suggested to reside in the extracellular ATD of each subunit (10,17). It has, however, been shown that both the NR1 and NR2 subunits contribute to Zn(II) inhibition (12,15,18), with the presence of exon 5 in the ATD of the NR1 subunit (NR1a forms lack exon 5, while NR1b forms include exon 5) playing a regulatory role.
The endogenous polyamines spermine and spermidine are necessary for a number of cellular functions, including proliferation and differentiation, stabilization of nucleic acids, regulation of protein synthesis, and modulation of ion channel functioning, including that of the iGluRs (19,20). At NMDA receptors, they have been proposed to act by at least four distinct mechanisms, including both glycine and voltage-dependent and -independent mechanisms (19–24). Concentrations of glycine, polyamines, and the NMDA subunit composition determine whether this action is activating or inhibiting (21,25). Control of glycine-independent potentiation has been suggested to occur via residues in both the NR1a and NR2B subunits, including NR1a residues in regions between the third and fourth membrane spanning regions as well as amino acids in the ATD of both subunits (21,26–28).
Thus, while sulfated neurosteroids, polyamines, and Zn(II) do regulate the activities of both NMDA and non-NMDA iGluRs, regulation is achieved in a complicated manner, implicating binding at multiple sites, none of which are yet fully elucidated. The results presented here further describe the binding of each of these three classes of ligands to specific subunit domains from both NMDA (NR1-1b) and non-NMDA (AMPA: GluR2) receptors. We have previously shown that the sulfated neurosteroids PS and PregaS bind to the GluR2 S1S2 domain (6,7). Data is presented here which shows that neither PS nor PregaS bind to the S1S2 domain of the NR1-1b subunit of the NMDA receptor. In addition, it has been found that neither Zn(II) nor the polyamines spermine or spermidine bind to the GluR2 S1S2 domain, however, they do bind to the NMDA NR1-1b S1S2 domain. The latter is significant because it locates a previously unidentified distinct subunit domain to which each of these modulatory ligands bind.
MATERIALS AND METHODS
Protein expression and purification of NMDA NR1-1b S1S2 domain
The cloning plasmid for the NMDA NR1-1b S1S2 domain was obtained from Eric Gouaux (Oregon Health and Science University, Portland, OR) and transformed into chemically competent Escherichia coli Origami B(DE3) cells. The expression system was designed as follows: (His)8-TSG-LVPRG(thrombin cut site)-S1(394-544)-GT-S2(663-800). NMDA NR1-1b S1S2 protein was overexpressed and purified as in Furukawa et al. (4).
Protein expression and purification of wt and mutant AMPA GluR2 S1S2 domains
Mutant AMPA GluR2 S1S2 domains (amino acids: S1:383-524-GT-S2:627-791) were constructed and wild-type and mutant AMPA GluR2 S1S2 proteins were overexpressed and purified as in Stoll et al. (2,3,7).
Fluorescence studies
NMDA
Fluorescence was performed on a Proton Technology International fluorescence spectrophotometer at a cell temperature of 25°C using 4.0-nm excitation and 1.0-nm emission slit widths. All scans were performed from 285 to 450 nm with a 280-nm excitation in a 3-mm pathlength cell. Each sample was equilibrated for 60 s at 25°C in the sample holder before data acquisition.
Blanks were measured by adding the appropriate ligand to buffer 3 (10 mM MES, 25 mM NaCl, 1 mM glycine, pH 6.5). Since all blank signals were <1% of the total fluorescence signal at 325 nm, experimental data was reported without subtraction of a blank. Protein samples for the EC50 and competition binding experiments were at a concentration of 3.75 μM, and for the Stern-Volmer experiments were at a concentration of 3.41 ± 0.16 μM in buffer 3. All ligands were solubilized in water. Spermine (Sigma, St. Louis, MO) and spermidine (Sigma) were diluted to a stock concentration of 100 mM. Zinc acetate (EM Science, Gibbstown, NJ) was diluted to a stock concentration of 150 mM.
In the EC50 experiments (Table 1 and Fig. 2S in the Supplementary Material), spermine was added to a final concentration of 0.300–50.0 mM (79.8:1–13,300:1 molar ratio of ligand/protein), spermidine was added to a final concentration of 0.250–50.0 mM (66.5:1–13,300:1 molar ratio ligand/protein), and zinc acetate was added to a final concentration of 2.0–75.0 mM (320:1–12,000:1 molar ratio ligand/protein). For determination of the EC50, percent binding was defined as [(fluorescence emission in the absence of ligand at 325 nm – fluorescence emission in the presence of a certain concentration of ligand at 325 nm) / (fluorescence emission in the absence of ligand at 325 nm – fluorescence emission in the presence of a saturating concentration of the ligand at 325 nm)] × 100. Saturating concentrations were determined to be 75 mM zinc acetate (20,000:1 molar ratio ligand/protein), 50 mM spermine (13,000:1 molar ratio ligand/protein), and 35 mM spermidine (9300:1 molar ratio ligand/protein).
TABLE 1.
Ligand binding affinity to NMDA NR1-1b and AMPA GluR2 S1S2 domains
| NMDA S1S2, EC50 | GluR2 S1S2, EC50 | |
|---|---|---|
| PS | ND* | 316 μM† |
| PregaS | ND* | 327 μM‡ |
| Spermine | 1649 ± 135 μM | ND* |
| Spermidine | 1052 ± 136 μM | ND* |
| Zinc acetate | 13.5 ± 1.2 mM | ND* |
In each NMDA NR1-1b S1S2 experiment, the fluorescence emission at 325 nm of 3.75 μM NMDA NR1-1b S1S2 in buffer 3 was probed. Ligands were used in the following final concentrations: 2–75.0 mM zinc acetate (320:1–12,000:1 molar ratio ligand/protein), 0.250–50.0 mM spermidine (66.5:1–13,300:1 molar ratio ligand/protein), and 0.300–50.0 mM spermine (79.8:1–13,300:1 molar ratio of ligand/protein). In each GluR2 S1S2 experiment, the fluorescence emission at 340 nm of 7.1 μM GluR2 S1S2 in buffer 9 was probed. Ligands were used in the following final concentrations: 25–1030 μM PS or PregaS (3.5:1–145:1 molar ratio of ligand/protein). EC50 was determined from a plot of % bound versus log [ligand]. Error is reported at the 95% confidence level.
No binding detected.
Error estimated as ±9.1 μM (6).
Error estimated as ±7.0 μM (6).
For competition studies (Table 3) the sample was allowed to equilibrate for 30 s after the addition of the first ligand and an additional 30 s after addition of the second ligand before data was collected. Percent quenching was defined as [(fluorescence emission in the absence of the ligand at 325 nm − fluorescence emission in the presence of the ligand at 325 nm)/fluorescence emission in the absence of the ligand at 325 nm] ×100.
TABLE 3.
Competition binding studies of the NMDA NR1-1b S1S2 domain
| First ligand added | Average (%) quenching | Second ligand added | Total (%) quenching | Quenching (%) due to second ligand |
|---|---|---|---|---|
| Zinc acetate | 41.8 ± 6.6 | Spermidine | 47.3 ± 3.9 | 5.5 ± 7.7 |
| Spermine | 49.1 ± 4.2 | 7.3 ± 7.8 | ||
| Spermidine | 55.0 ± 4.1 | Zinc acetate | 38.3 ± 5.5 | −16.7 ± 6.9 |
| Spermine | 58.4 ± 3.8 | 3.4 ± 5.6 | ||
| Spermine | 34.1 ± 5.5 | Zinc acetate | 44.9 ± 4.9 | 15.8 ± 7.4 |
| Spermidine | 57.5 ± 4.1 | 23.4 ± 6.9 |
The fluorescence emission at 325 nm of 3.75 μM NMDA NR1-1b S1S2 in buffer 3 was probed. Ligands were used in the following final concentrations: 33 mM zinc acetate, 1.3 mM spermidine, and 833 μM spermine. Error is reported at the 95% confidence level.
In the Stern-Volmer experiments (Table 4 and Fig. 4S in the Supplementary Material), potassium iodide (EM Science) ranging in concentration from 25 to 400 mM was added to 3.41 ± 0.16 μM NMDA NR1-1b-S1S2 that had been mixed with ligand at a final concentration of 0.5 mM spermine, 0.8 mM spermidine, or 5 mM Zn (II). Data was analyzed using the Stern-Volmer equation F0/F = 1 + KSV[Q], where the Stern-Volmer constant KSV = kqτ0. The values F0 and F are the fluorescence intensities in the absence and in the presence of quencher, respectively, [Q] is the concentration of quencher, kq is the biomolecular collisional constant, and τ0 is the lifetime of the fluorophore in the absence of quencher. Since it is assumed that the lifetime, τ0, does not change after ligand binding, KSV can be used as a measure of the biomolecular collisional constant and therefore reflects the change in accessibility of the tryptophan residues to quencher upon ligand binding (29,30). Data corresponding to at least six different iodide concentrations were used to construct each Stern-Volmer plot.
TABLE 4.
Stern Volmer constants (Ksv) of iodide accessibility to NMDA NR1-1b S1S2 domain tryptophan residues
| Apo (M−1) | Spermine (M−1) | Spermidine (M−1) | Zinc acetate (M−1) | |
|---|---|---|---|---|
| NMDA S1S2 | 2.52 ± 0.09 | 1.31 ± 0.05 | 1.36 ± 0.05 | 9.33 ± 0.15 |
Iodide ranging in concentration from 25 to 400 mM was added to 3.41 ± 0.16 μM NMDA NR1-1b S1S2 mixed with ligand at a final concentration of 0.5 M spermine, 0.8 M spermidine, or 5 M Zn (II) and the fluorescence emission at 325 nm was probed. Data was analyzed using the Stern-Volmer equation F0/F = 1 + KSV[Q]. Error is reported at the 95% confidence level.
Each data set consists of emission spectra from independent scans of at least four identically prepared samples. The average spectrum of each data set was calculated by averaging the intensity at individual wavelengths between 285 and 450 nm. The standard deviation (SD) was calculated at 325 nm for each data set based on the four data points for each protein-ligand mixture, and the error at the 95% confidence interval was determined. The 95% confidence interval (μ) was defined as (t * SD)/(number of data points)1/2, where t is the Student's t value for the appropriate degrees of freedom at the 95% confidence level.
AMPA
Fluorescence studies were performed at 25°C using 2.5 nm excitation and emission slit widths. All emission scans were performed from 285 to 450 nm with a 280 nm excitation in a 3-mm path length cell. Each sample was equilibrated for 30 s at 25°C in the sample holder before data acquisition. Blank samples of buffer 9 (650 mM arginine-HCl, 400 μM KCl, 10 mM NaCl, 1 mM EDTA, pH 8.5) were analyzed and were shown to contribute <1% to the total fluorescence signal at 340 nm, and therefore, experimental data was reported without subtraction of a blank. Protein samples were at a concentration of 5.2 μM in buffer 9. Neurosteroids (PS, Sigma; and PregaS, Steraloids, Newport, RI) were solubilized in 100% methanol to a stock concentration of 15 mM. To take into account any effects methanol had on the fluorescence emission spectrum of the GluR2-S1S2 domain, fluorescence emission in the absence of quencher was determined in the presence of the final concentration of methanol, thus allowing for a direct comparison between samples with and without neurosteroid. Final methanol concentrations up to 6.0% were determined to be acceptable (6).
To construct a Stern-Volmer plot (Table 2 and Fig. 3S in the Supplementary Material), acrylamide (Sigma) ranging in concentration from 39 to 280 mM was added to 5.2 μM GluR2-S1S2 that had been mixed with ligand at a final concentration of 308 μM PS or 308 μM PregaS. Data was analyzed as described above for NMDA NR1-1b S1S2.
TABLE 2.
Stern Volmer constants (Ksv) of acrylamide accessibility of wt and mutant GluR2 S1S2 tryptophan residues
| Apo (M−1) | PS (M−1) | PregaS (M−1) | |
|---|---|---|---|
| wt | 6.29 ± 0.22 | 6.65 ± 0.19 | 6.67 ± 0.54 |
| W460F | 5.44 ± 0.26 | 5.84 ± 0.28 | 5.28 ± 0.17 |
| W671F | 5.80 ± 0.15 | 6.31 ± 0.29 | 5.92 ± 0.12 |
| W766/767F | 6.36 ± 0.17 | 4.82 ± 0.25 | 4.88 ± 0.19 |
Acrylamide ranging in concentration from 39 to 280 mM was added to 5.2 μM GluR2 S1S2 mixed with ligand that was at a final concentration of 308 μM PS or PregaS, and the fluorescence emission at 340 nm was probed. Data was analyzed using the Stern-Volmer equation F0/F = 1 + KSV[Q]. Error is reported at the 95% confidence level.
Each data set consisted of emission spectra from independent scans of at least three identically prepared samples. The standard deviation (SD) of the intensity at 340 nm was calculated for each data set based on the three data points for each protein-ligand mixture, and the error at the 95% confidence interval was determined as described above.
RESULTS AND DISCUSSION
PS and PregaS bind to the AMPA GluR2 S1S2 domain, but not to the NMDA NR1-1b S1S2 domain
Intrinsic tryptophan fluorescence emission spectroscopy was employed to monitor the binding of PS and PregaS to the GluR2 S1S2 domain (four tryptophans: Trp-460, -671, -766, -767) and the NMDA NR1-1b S1S2 domain (four tryptophans: Trp-498, -731, -768, -792) (Fig. 1S:B-E in the Supplementary Material). We have previously shown (Table 1 and Fig. 2S:A in the Supplementary Material) that the GluR2 S1S2 domain binds with approximate equal affinity to both PS (EC50: 316 μM) and PregaS (EC50: 327 μM) (6,7). Here we report that neither sulfated neurosteroid binds to the NMDA NR1-1b S1S2 domain. This latter result is consistent with the observation that whether PS activates or inhibits the NMDA receptor depends upon the NR2 subunit composition, with NR1-NR2A/NR2B combinations being activated by PS and NR1-NR2C/NR2D combinations being inhibited by PS (31). In addition, it is in agreement with a proposed steroid modulatory domain for PS containing part of the NR2B subunit (8).
Although it has been proposed from electrophysiology studies that PS and PregaS bind to two independent extracellular binding sites on the NMDA receptor, the location of the extracellular binding sites have not been defined, nor has the existence of independent binding sites been shown for other iGluRs (32). To explore the latter, a Stern-Volmer analysis was completed for both the wt and three tryptophan mutants (W460F, W671F, W766/767F) of the GluR2 S1S2 domain (Table 2 and Fig. 3S in the Supplementary Material). These experiments probe the ability of acrylamide to access the tryptophan residues of the GluR2 S1S2 domain and quench their fluorescence emission—with an increase in Ksv corresponding to an increase in the bimolecular collisional constant and an increase in accessibility (29,30). Within the 95% confidence intervals calculated, this Stern-Volmer analysis did not allow for a differentiation of the conformational changes that take place upon binding of each neurosteroid to the GluR2 S1S2 domain.
The polyamines spermine and spermidine bind to the NMDA NR1-1b S1S2 domain, but not to the AMPA GluR2 S1S2 domain
Spermine was found to bind to the glycine-bound NMDA NR1-1b S1S2 domain with an EC50 of 1649 ± 135 μM, while spermidine was found to bind with an EC50 of 1052 ± 136 μM (Table 1 and Fig. 2S:C-D in the Supplementary Material). Neither were found to bind to the GluR2 S1S2 domain (in the presence or absence of glutamate) at concentrations up to 2.1 mM. Competition binding assays (Table 3) show that spermine, spermidine, and Zn(II) all have overlapping binding sites in the NMDA NR1-1b S1S2 domain. For example, upon binding spermidine, the solo quenching of 34.1 ± 5.5% resulting from spermine binding is diminished to 3.4 ± 5.6% and the solo quenching of 41.8 ± 6.6% resulting from Zn(II) binding is diminished to −16.7 ± 6.9%. In an effort to differentiate the nature of the conformational change to the NMDA NR1-1b S1S2 domain upon binding to each of the polyamines, a Stern-Volmer analysis was performed (Table 4 and Fig. 4S:B-C in the Supplementary Material). The Stern-Volmer constant for iodide quenching of the apo S1S2 domain was determined to be 2.52 ± 0.09 M−1, while iodide quenchings of the spermine-bound and spermidine-bound S1S2 domains were determined to be 1.31 ± 0.05 M−1 and 1.36 ± 0.05 M−1, respectively. This indicates that upon binding of either of the polyamines to the NR1-1b S1S2 domain, the conformational change that ensues places the tryptophan residues that are quenched by iodide in less accessible locations. Neither the Ksv values nor the competition binding assays allow differentiation of the binding regions or the induced conformational changes of spermine and spermidine upon binding to the NMDA NR1-1b S1S2 domain.
It has been suggested that the polyamines, whose biosynthesis is under tight and highly regulated control, appear to be well suited for regulation of iGluRs (19). Although not normally found in the synaptic cleft, under certain conditions polyamine synthesis is upregulated and polyamines are released into the synaptic cleft where they can interact with the extracellular surface of iGluRs (33–36). Although the binding affinities determined for these polyamines (EC50s of 1649 and 1052 μM) are not tight, it has been estimated that in proliferating cells as well as mature secretory cells that cytosolic concentrations of spermine and spermidine are at the mM level (33,34). This data is the first report of polyamine binding to the S1S2 domain of an iGluR, and in particular an NMDA NR1b S1S2 domain, extending the list of possible extracellular binding sites.
Zn(II) binds to the NMDA NR1-1b S1S2 domain, but not to the AMPA GluR2 S1S2 domain
Zn(II) was found to bind to the glycine-bound NMDA NR1-1b S1S2 domain with an EC50 of 13.5 ± 1.2 mM (Table 1 and Fig. 2S:B in the Supplementary Material). It was not, however, found to bind to the GluR2 S1S2 domain (in the presence or absence of glutamate) at concentrations up to 21 mM. Competition binding assays (Table 3) show that the Zn(II) binding site on the NR1-1b S1S2 domain overlaps with that of both spermine and spermidine. Upon binding of Zn(II), the solo quenching of 34.1 ± 5.5% resulting from binding of spermine is decreased to 7.3 ± 7.8% and the solo quenching of 55.0 ± 4.1% resulting from binding of spermidine is decreased to 5.5 ± 7.7%. This overlap supports the concept of partial overlapping binding sites or common downstream targets, which have been proposed (9). The results presented here localize at least one possible overlapping binding site to the S1S2 domain of the NMDA NR1-1b subunit. A Stern-Volmer analysis was performed to differentiate the conformational change induced upon Zn(II) binding from that of polyamine binding (Table 4 and Fig. 4S:A in the Supplementary Material). The Stern-Volmer constant for iodide quenching of the apo S1S2 domain was determined to be 2.52 ± 0.09 M−1, while those of spermine-bound, spermidine-bound, and Zn(II)-bound S1S2 domains were determined to be 1.31 ± 0.05 M−1, 1.36 ± 0.05 M−1, and 9.33 ± 0.15 M−1, respectively. This indicates that while binding of either of the polyamines to the NR1-1b S1S2 domain causes a conformational change that places the tryptophan residues that are quenched by iodide in less accessible locations, the conformational change upon Zn(II) binding places them in more accessible regions. Since iodide is an external quencher, this would suggest a more solvent-exposed and more polar location for the tryptophan residues that are quenched by iodide upon Zn(II) binding of the NMDA NR1-1b S1S2 domain. This idea is further supported by the data in Table 5 (and Fig. 1S:A in the Supplementary Material) showing the maximum wavelength of emission of both the apo and ligand bound NMDA NR1-1b S1S2 domains. As can be seen, the maximum emission is red-shifted by ∼8 nm from 326 to 334 nm upon Zn(II) binding, supporting a conformational change that places at least some of the tryptophan residues in a more polar environment.
TABLE 5.
Maximum wavelength of emission of the apo and ligand bound NMDA NR1-1b S1S2 domain
| Ligand | Maximum wavelength (nm) |
|---|---|
| Apo | 326 ± 3 |
| Spermidine | 325 ± 3 |
| Spermine | 324 ± 2 |
| Zinc acetate | 334 ± 2 |
The fluorescence emission at 325 nm of 3.75 μM NMDA NR1-1b S1S2 in buffer 3 was probed. Ligands were used in the following final concentrations: 75 mM zinc acetate, 2 mM spermidine, and 1 mM spermine.
To our knowledge, this is the first report of a low affinity Zn(II) binding site in the S1S2 domain of any iGluR.
Summary
Although the S1S2 domains from the NMDA NR1-1b subunit and the AMPA GluR2 subunit are 33% identical (54% homologous) at the primary amino-acid level and have similar tertiary structures (Fig. 2) (1,4), they have been shown here to bind to different endogenous modulators. It is interesting to speculate that this difference in ability to be modulated correlates with difference in function, as AMPA receptors mediate fast depolarization while NMDA receptors mediate the slower component of the excitatory postsynaptic potential. Of particular significance are two findings. The first is that while PS and PregaS bind to the S1S2 domain of the GluR2 subunit, neither binds to the S1S2 region of the NR1-1b subunit. Since it is known that both PS and PregaS modulate the activity of the NMDA receptor from the extracellular face (32), this leaves the possibility of its binding to either an extracellular region on the NR2 subunit or an extracellular interface between the NR1 and NR2 subunits. The second is that the polyamines spermine and spermidine as well as Zn(II) bind to the S1S2 domain of the NR1-1b subunit, but not to the S1S2 domain of the GluR2 subunit. While the binding site for all three of these modulators overlaps, the conformational change caused by Zn(II) binding is distinct. These findings are important, as they locate previously unidentified distinct subunit domains to which each of these modulatory ligands bind. Understanding specific ligand-receptor binding interactions and the nature of the resultant conformational change has potential for the rational design of therapeutics to be used against diseases caused by iGluR misregulation.
SUPPLEMENTARY MATERIAL
An online supplement to this article can be found by visiting BJ Online at http://www.biophysj.org.
Acknowledgments
NMDA NR1-1b and AMPA GluR2 plasmids were a generous gift from Eric Gouaux, Oregon Health and Science University.
Financial support was from Undergraduate Research and Creative Opportunity Awards to L.S., N.V., A.H., A.M.; Zymogenetics on-campus summer stipend to L.K.; American Chemical Society Petroleum Research Fund grant No. 38815-B4 to L.G.; National Science Foundation CAREER grant No. MCB-0447541 to L.G.; and National Science Foundation Course, Curriculum, and Laboratory Improvement grant No. DUE-0411289 to Steven Emory (fluorimeter).
References
- 1.Armstrong, N., and E. Gouaux. 2000. Mechanism for activation and antagonism of an AMPA-sensitive glutamate receptor: crystal structure of the GluR2 ligand binding core. Neuron. 28:165–181. [DOI] [PubMed] [Google Scholar]
- 2.Chen, G.-Q., Y. Sun, R. Jin, and E. Gouaux. 1998. Probing the ligand binding domain of the GluR2 receptor by proteolysis and deletion mutagenesis defines domain boundaries and yields a crystallizable construct. Protein Sci. 7:2623–2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen, G.-Q., and E. Gouaux. 1997. Overexpression of a glutamate receptor (GluR2) ligand binding domain in Escherichia coli: application of a novel protein folding screen. Proc. Natl. Acad. Sci. USA. 94:13431–13436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Furukawa, H., and E. Gouaux. 2003. Mechanisms of activation, inhibition and specificity: crystal structures of the NMDA receptor NR1 ligand-binding core. EMBO J. 22:2873–2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Irwin, P. R., S. Z. Lin, M. A. Rogawski, R. H. Purdy, and S. M. Paul. 1994. Steroid potentiation and inhibition of N-methyl-D-aspartate receptor-mediated intracellular Ca2+ responses: structure-activity studies. J. Pharmacol. Exp. Ther. 271:677–682. [PubMed] [Google Scholar]
- 6.Spivak, V., A. Lin, P. Beebe, L. Stoll, and L. Gentile. 2004. Identification of a neurosteroid binding site contained within the GluR2–S1S2 domain. Lipids. 39:811–819. [DOI] [PubMed] [Google Scholar]
- 7.Stoll, L., and L. Gentile. 2005. Linking tricyclic antidepressants to ionotropic glutamate receptors. Biochem. Biophys. Res. Commun. 333:622–627. [DOI] [PubMed] [Google Scholar]
- 8.Jang, M.-K., D. F. Mierke, S. J. Russek, and D. H. Farb. 2004. A steroid modulatory domain on NR2B controls N-methyl-D-aspartate receptor proton sensitivity. Proc. Natl. Acad. Sci. USA. 101:8198–8203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dingledine, R., K. Borges, D. Bowie, and S. F. Traynelis. 1999. The glutamate receptor ion channels. Pharmacol. Rev. 51:7–61. [PubMed] [Google Scholar]
- 10.Paoletti, P., F. Perin-Dureau, A. Fayyazuddin, A. Le Goff, I. Callebaut, and J. Neyton. 2000. Molecular organization of a zinc binding N-terminal modulatory domain in a NMDA receptor subunit. Neuron. 28:911–925. [DOI] [PubMed] [Google Scholar]
- 11.Hollmann, M., J. Boulter, C. Maron, L. Beasley, J. Sullivan, G. Pecht, and S. Heinemann. 1993. Zinc potentiates agonist-induced currents at certain splice variants of the NMDA receptor. Neuron. 10:943–954. [DOI] [PubMed] [Google Scholar]
- 12.Amar, M., F. Perin-Dureau, and J. Neyton. 2001. High-affinity Zn block in recombinant NMDA receptors with cysteine substitutions at the Q/R/N site. Biophys. J. 81:107–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Paoletti, P., P. Ascher, and J. Neyton. 1997. High-affinity zinc inhibition of NMDA NR1–NR2A receptors. J. Neurosci. 17:5711–5725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Williams, K. 1996. Separating dual effects of zinc at recombinant N-methyl-D-aspartate receptors. Neurosci. Lett. 215:9–12. [DOI] [PubMed] [Google Scholar]
- 15.Traynelis, S. F., M. F. Burgess, F. Zheng, P. Lyuboslavsky, and J. L. Powers. 1998. Control of voltage-independent zinc inhibition of NMDA receptors by the NR1 subunit. J. Neurosci. 18:6163–6175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rachline, J., F. Perin-Dureau, A. Le Goff, J. Neyton, and P. Paoletti. 2005. The micromolar zinc-binding domain on the NMDA receptor subunit NR2B. J. Neurosci. 25:308–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zheng, F., K. Erreger, C.-M. Low, T. Banke, C. J. Lee, P. J. Conn, and S. F. Traynelis. 2001. Allosteric interaction between the amino terminal domain and the ligand binding domain of NR2A. Nat. Neurosci. 4:894–901. [DOI] [PubMed] [Google Scholar]
- 18.Zheng, X., L. Zhang, G. Durand, M. Bennett, and R. S. Zukin. 1994. Mutagenesis rescues spermine and Zn2+ potentiation of recombinant NMDA receptors. Neuron. 12:811–818. [DOI] [PubMed] [Google Scholar]
- 19.Laube, G., and R. W. Veh. 1997. Astrocytes, not neutrons, show most prominent staining for spermidine/spermine-like immunoreactivity in adult rat brain. Glia. 19:171–179. [DOI] [PubMed] [Google Scholar]
- 20.Igarashi, K., and K. Kashiwagi. 2000. Polyamines: mysterious modulators of cellular function. Biochem. Biophys. Res. Commun. 271:559–564. [DOI] [PubMed] [Google Scholar]
- 21.Turecek, R., K. Vlcek, M. Pterovic, M. Horak, V. Vlachova, and L. Vyklicky. 2004. Intracellular spermine decreases open probability of N-methyl-D-aspartate receptor channels. Neuroscience. 125:879–887. [DOI] [PubMed] [Google Scholar]
- 22.Williams, K., A. M. Zappia, D. B. Pritchett, Y. M. Shen, and P. B. Molinoff. 1994. Sensitivity of the N-methyl-D-aspartate receptor to polyamines is controlled by NR2 subunit. Mol. Pharmacol. 45:803–809. [PubMed] [Google Scholar]
- 23.Durand, G. M., M. V. Bennett, and R. S. Zukin. 1993. Splice variants of the N-methyl-D-aspartate receptor NR1 identify domains involved in regulation by polyamines and protein kinase C. Proc. Natl. Acad. Sci. USA. 90:6731–6735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Williams, K., K. Kashiwagi, J. I. Fukuchi, and K. Igarashi. 1995. An acidic amino acid in the N-methyl-D-aspartate receptor that is important for spermine stimulation. Mol. Pharmacol. 48:1087–1098. [PubMed] [Google Scholar]
- 25.Traynelis, S. F., M. Hartley, and S. F. Heinemann. 1995. Control of proton sensitivity of the NMDA receptor by RNA splicing and polyamines. Science. 268:873–876. [DOI] [PubMed] [Google Scholar]
- 26.Kashiwagi, K., J. Fukuchi, J. Chao, K. Igarashi, and K. Williams. 1996. An aspartate residue in the extracellular loop of the N-methyl-D-aspartate receptor controls sensitivity to spermine and protons. Mol. Pharmacol. 49:1131–1141. [PubMed] [Google Scholar]
- 27.Williams, K. 1995. Polyamines: Regulation and Molecular Interaction. R. A. Casero, editor. R. G. Landes, Austin, TX. 129–170.
- 28.Gallagher, M. J., H. Huang, E. R. Grant, and D. R. Lynch. 1997. The NR2B-specific interactions of polyamines and protons with the N-methyl-D-aspartate receptor. J. Biochem. (Tokyo). 272:24971–24979. [DOI] [PubMed] [Google Scholar]
- 29.Natale, P., T. den Blaauwen, C. van der Does, and A. J. M. Driessen. 2005. Conformational state of the SecYEG-bound SecA probed by single tryptophan fluorescence spectroscopy. Biochemistry. 44:6424–6432. [DOI] [PubMed] [Google Scholar]
- 30.Lakowicz, J. R. 1999. Principles of Fluorescence Spectroscopy, 2nd Ed. Kluwer Academic/Plenum Publishers, New York.
- 31.Malayev, A., T. T. Gibbs, and D. H. Farb. 2002. Inhibition of the NMDA response by pregnenolone sulphate reveals subtype selective modulation of NMDA receptors by sulphated steroids. Br. J. Pharmacol. 135:901–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Park-Chung, M., F. Wu, R. Purdy, A. Malayev, T. Gibbs, and D. H. Farb. 1997. Distinct sites for inverse modulation of N-methyl-D-aspartate receptors by sulfated steroids. Mol. Pharmacol. 52:1113–1123. [DOI] [PubMed] [Google Scholar]
- 33.Williams, K. 1997. Modulation and block of ion channels: a new biology of polyamines. Cell. Signal. 9:1–13. [DOI] [PubMed] [Google Scholar]
- 34.Koh, D. S., N. Burnashev, and P. Jonas. 1995. Block of native Ca2+-permeable AMPA receptors in rat brain by intracellular polyamines generates double rectification. J. Physiol. (Lond.). 486:305–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kamboj, S. K., G. T. Swanson, and S. G. Cull-Candy. 1995. Intracellular spermine confers rectification on rat calcium-permeable AMPA and kainate receptors. J. Physiol. (Lond.). 486:297–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Donevan, S. D., and M. A. Rogawski. 1995. Intracellular polyamines mediate inward rectification of Ca2+-permeable α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptors. Proc. Natl. Acad. Sci. USA. 92:9298–9302. [DOI] [PMC free article] [PubMed] [Google Scholar]