Abstract
Glycerophosphoinositol is produced through deacylation of the essential phospholipid phosphatidylinositol. In Saccharomyces cerevisiae, the glycerophosphoinositol produced is excreted from the cell but is recycled for phosphatidylinositol synthesis when inositol is limiting. To be recycled, glycerophosphoinositol enters the cell through the permease encoded by GIT1. The transport of exogenous glycerophosphoinositol through Git1p is sufficiently robust to support the growth of an inositol auxotroph (ino1Δ). We now report that S. cerevisiae also uses exogenous phosphatidylinositol as an inositol source. Evidence suggests that phosphatidylinositol is deacylated to glycerophosphoinositol extracellularly before being transported across the plasma membrane by Git1p. A genetic screen identified Pho86p, which is required for targeting of the major phosphate transporter (Pho84p) to the plasma membrane, as affecting the utilization of phosphatidylinositol and glycerophosphoinositol. Deletion of PHO86 in an ino1Δ strain resulted in faster growth when either phosphatidylinositol or glycerophosphoinositol was supplied as the sole inositol source. The incorporation of radiolabeled glycerophosphoinositol into an ino1Δ pho86Δ mutant was higher than that into wild-type, ino1Δ, and pho86Δ strains. All strains accumulated the most GIT1 transcript when incubated in media limited for inositol and phosphate in combination. However, the ino1Δ pho86Δ mutant accumulated approximately threefold more GIT1 transcript than did the other strains when incubated in inositol-free media containing either high or low concentrations of Pi. Deletion of PHO4 abolished GIT1 transcription in a wild-type strain. These results indicate that the transport of glycerophosphoinositol by Git1p is regulated by factors affecting both inositol and phosphate availabilities and suggest a regulatory connection between phosphate metabolism and phospholipid metabolism.
Inositol and phosphate are required for the survival of all eukaryotic cells. In the yeast Saccharomyces cerevisiae, each of these nutrients regulates the transcription of a network of genes involved in their metabolism. Starvation for inositol in S. cerevisiae results in the coordinated induction of a set of genes involved in phospholipid biosynthesis. The most highly regulated of these is INO1, the gene encoding inositol-1-phosphate synthase, the rate-limiting enzyme in inositol biosynthesis (7). INO1, as well as the other coregulated genes of phospholipid biosynthesis, contains within its promoter a repeated basic helix-loop-helix (bHLH) consensus sequence (inositol upstream activation sequence [UASINO]) to which the heterodimerized INO2 and INO4 gene products bind to activate transcription when inositol is limiting. Inositol is a precursor of phosphatidylinositol (PI), an essential lipid of eukaryotic cells. PI acts, in turn, as a precursor to sphingolipids, polyphosphoinositides, and glycerophosphoinositol (GroPIns). The GroPIns produced by S. cerevisiae is released into the extracellular milieu and represents a major route of PI catabolism in cultures grown in inositol-containing medium (1). When inositol is limiting, GroPIns is transported into the cell, where it is catabolized and its inositol portion is used for the synthesis of PI (19). The transport of GroPIns is mediated through the permease encoded by the GIT1 gene (20).
Starvation for phosphate (Pi) in S. cerevisiae results in the coordinated induction of a set of phosphatase genes (PHO5, PHO10, and PHO11) and the structural gene for low-Km Pi transport, PHO84 (11, 17). The phosphoinositol (PHO) system is comprised of five regulatory genes: PHO2, PHO4, PHO80, PHO81, and PHO85. The induction of PHO gene transcription during phosphate starvation is mediated by Pho4p, a bHLH transcription factor, and Pho2p, a homeobox DNA binding protein. The transcriptional status of PHO5, a gene whose product supplies more than 90% of the acid phosphatase activity, is typically used as a marker for the entire PHO system. Pho4p binds to two bHLH consensus sequences (CANNTG) in the PHO5 promoter, and Pho2p binds cooperatively with Pho4p (8). In high-Pi medium, Pho80p and Pho85p form a kinase complex, similar to that formed by a cyclin and a cyclin-dependent protein kinase, which hyperphosphorylates and inhibits Pho4p function, thereby blocking PHO5 transcription (16). In low-Pi medium, Pho81p inhibits the Pho80p-Pho85p complex and allows the transcription of PHO5 (25). PHO5 and the other repressible acid phosphatase (rAPase) genes are constitutively expressed in strains bearing mutations in PHO84, the low-Km Pi transport gene. Other genes thought to be involved in Pi transport include pho87, pho88, pho89, pho90, and pho91 (4, 21, 28). Pho86p is an endoplasmic reticulum protein that is required for the transport of Pho84p to the plasma membrane (9). Strains bearing mutations in PHO86 show constitutive rAPase activity and reduced Pi transport.
The transport of exogenously supplied GroPIns through Git1p supports the growth of an inositol auxotroph (ino1Δ), thus defining the Git+ phenotype (20). We now report that an ino1Δ mutant can also grow when PI is supplied as the source of inositol, a phenotype that we have termed Pit+. Genetic and biochemical studies indicate that GIT1 transcription and Git1p transport activity are regulated by factors affecting inositol and phosphate availabilities, revealing a metabolic link between phospholipid metabolism and phosphate metabolism.
MATERIALS AND METHODS
Culture conditions.
The strains used in this study are listed in Table 1. The strains were grown aerobically at 30°C with shaking. Turbidity was monitored by measurement of the A600 with a Beckman DU64 spectrophotometer. Synthetic complete medium was prepared as described previously (19) and contained a high concentration of KH2PO4 (7.4 mM). For the experiments represented in Fig. 1 and 2, synthetic complete medium was supplemented with various amounts of inositol, GroPIns (Sigma), or PI (Sigma). High-Pi and low-Pi media (Table 2) (Fig. 3 to 5) were made by replacing KH2PO4 (1 g/liter) in synthetic complete medium with KCl (1 g/liter) and adding KH2PO4 to 10 mM (high Pi) or 0.2 mM (low Pi). YEPD medium consisted of 20 g of glucose, 10 g of yeast extract, and 20 g of Bacto Peptone per liter.
TABLE 1.
Strains
| Strain | Genotype | Source or reference |
|---|---|---|
| JPV1 | trp1 ura3 leu2 his3 MATα | P. McGraw |
| JPV3 | trp1 ura3 leu2 his3 ino1::HIS3 MATα | P. McGraw |
| JPV22 | trp1 ura3 thr1 ino1::HIS3 MATa | This study |
| JPV89 | trp1 ura3 leu2 his3 git1::HIS3 MATα | 20 |
| JPV90 | trp1 ura3 thr1 his3 ino1::HIS3 pho86::URA3 MATa | This study |
| JPV91 | trp1 ura3 thr1 leu2 his3 MATa | This study |
| JPV93 | trp1 ura3 thr1 his3 pho86::URA3 MATa | This study |
| JPV95 | trp1 ura3 his3 ino1::HIS3 MATα | This study |
| JPV97 | trp1 ura3 his3 git1::HIS3 MATa | This study |
| JPV99 | trp1 ura3 his3 leu2 ino1::HIS3 pho86::URA3 MATα | This study |
| JPV101 | trp1 ura3 his3 ino1::HIS3 git1::his3 MATα | This study |
| JPV103 | trp1 ura3 his pho86::URA3 git1::HIS3 MATα | This study |
| JPV105 | trp1 ura3 his3 thr1 ino1::HIS3 pho86::URA3 git1::HIS3 MATa | This study |
| JPV139 | trp1 ura3 leu2 his3 ino1::HIS3 plb3::kanMX4 MATa | This study |
| JPV142 | trp1 ura3 leu2 his3 ino1::HIS3 plb1::URA3 plb2::kanMX4 MATa | This study |
| JPV148 | trp1 ura3 leu2 his3 ino1::HIS3 plb1::URA3 plb2::kanMX4 MATα | This study |
| JPV145 | trp1 ura3 leu2 his3 ino1::HIS3 MATa | This study |
| JPV146 | trp1 ura3 his3 leu2 plb1::URA3 plb2::kanMX4 plb3::kanMX4 MATa | This study |
| JPV155 | trp1 ura3 leu2 his3 ino1::HIS3 plb2::kanMX4 MATa | This study |
| JPV163 | ura3 his3 ino1::HIS3 plb1::URA3 MATα | This study |
| JPV253 | thr1 MATa | S. Henry |
| MF3 | trp1 ura3 his3 leu2 ade2 plb1::URA3 MATa | F. Paltauf |
| MF11 | trp1 ura3 his3 leu2 ade2 plb2::kanMX4 MATα | F. Paltauf |
| MF30 | trp1 ura3 his3 leu2 plb1::URA3 plb2::kanMX4 plb3::kanMX4 MATa | F. Paltauf |
| JPV203 | ura3 leu2 his3 met15 MATa | Research Genetics |
| JPV296 | ura3 leu2 his3 lys2 pho4::kanMX4 MATa | Research Genetics |
| JPV339 | ura3 leu2 his3 met15 ino2::kanMX4 MATa | Research Genetics |
FIG. 1.
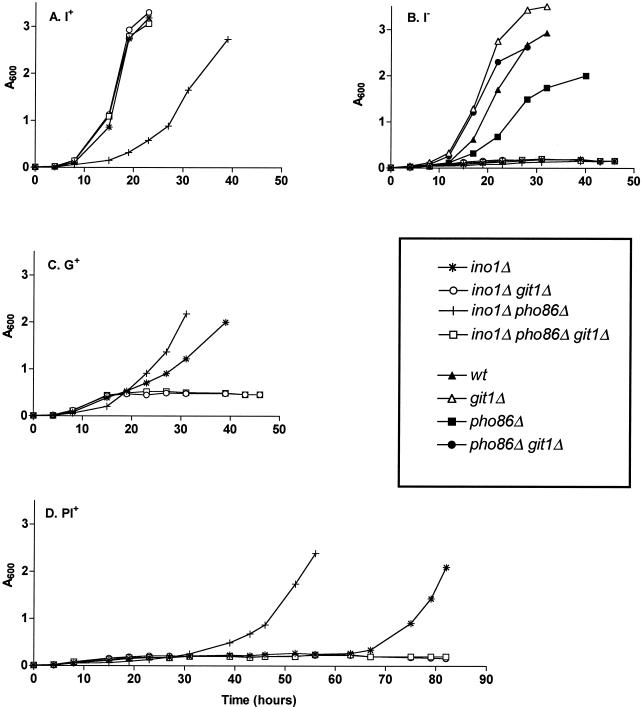
Deletion of PHO86 results in enhanced growth of an ino1Δ strain in GroPIns-containing and PI-containing media. Strains were grown on synthetic media containing I+ (A), no inositol source (I−) (B), 75 mM GroPIns (G+) (C), and 75 mM PI (PI+) (D). At the indicated times, the A600 was measured. Data for wild-type (wt), git1Δ, pho86Δ, and pho86Δ git1Δ strains are shown only in panel B.
FIG. 2.
Growth of an ino1Δ mutant on PI requires one or more PLB gene products. Strains were grown on synthetic media containing 75 mM PI, a source of inositol. At the indicated times, the A600 was measured.
TABLE 2.
Incorporation of radiolabeled GroPInsa
| Expt | Strain | Nmol of GroPIns internalized in medium with the following ingredients:
|
|||
|---|---|---|---|---|---|
| I−, low Pi | I+, low Pi | I−, high Pi | I+, high Pi | ||
| 1 | Wild type | 2.4 | 2.8 | 2.9 | 1.6 |
| ino1Δ | 6.1 | 7.2 | 4.2 | 0.6 | |
| pho86Δ | 4.0 | 7.9 | 3.7 | 1.1 | |
| pho86Δ ino1Δ | 29.4 | 31.2 | 21.4 | 15.5 | |
| 2 | Wild type | 29.2 | 17.6 | 4.8 | 2.1 |
Strains (JPV91, JPV93, JPV95, and JPV99) grown overnight in I+, high-Pi medium were harvested and inoculated to an A600 of 0.05 in 5 ml of each of four separate growth media, each containing 50 μM 3H-GroPIns: low Pi, I−; low Pi, I+; high Pi, I−; and high Pi, I+. Aliquots of each culture were centrifuged, and the resulting pellets and supernatants were subjected to liquid scintillation counting. For experiment 1, cells were harvested following three or four generations of growth. Data are presented as nanomoles of GroPIns internalized/A600 unit. For experiment 2, wild-type cells were harvested at high cell densities (A600, 3 to 4) following 24 h of incubation. The total nanomoles of GroPIns internalized in the stationary phase was determined (data normalized to an A600 of 4). Values represent the average of duplicate determinations. These experiments were repeated, with similar results.
FIG. 3.
GIT1 transcripts accumulate in a wild-type strain limited for phosphate and inositol. A wild-type strain (JPV91) was grown in media containing (I+) or lacking (I−) 75 mM inositol and containing 0.2 mM (low Pi) or 10 mM (high Pi) inorganic phosphate. Cells were harvested in the logarithmic phase, and Northern analysis was performed with digoxigenin-labeled probes for GIT1 and control (SNR17) transcripts as described in Materials and Methods.
FIG. 4.
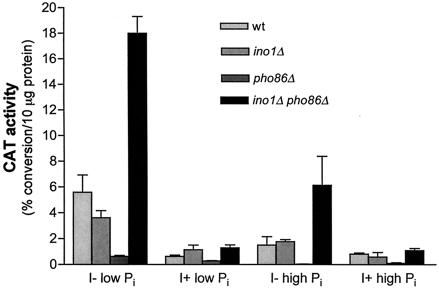
An ino1Δ pho86Δ mutant overexpresses GIT1, as measured by chloramphenicol acetyltransferase (CAT) activity. Cultures transformed with plasmid pCA998 were grown in high-Pi, I+ medium lacking tryptophan and were used to inoculate each of the following media lacking tryptophan: low Pi, I−; low Pi, I+; high Pi, I−; and high Pi, I+. Following three to five generations of growth, lysates obtained by glass bead breakage were assayed for chloramphenicol acetyltransferase activity as described in Materials and Methods. Reactions were carried out for 2.5 h with 5 to 25 μg of protein per assay. Data represent at least three separate determinations and standard errors of the means. wt, wild type.
Strain and plasmid constructions.
JPV3 and JPV253 were crossed, and JPV22 was isolated from the resulting diploids by tetrad dissection. JPV90 and JPV89 were crossed, and JPV91, JPV93, JPV95, JPV99, JPV101, JPV103, and JPV105 were isolated from the resulting diploids by tetrad dissection. Mating, sporulation, and tetrad dissection were performed by standard methods (24). Sporulation medium was supplemented with 100 μM inositol. Strains JPV91, JPV93, JPV95, JPV97, JPV99, JPV101, JPV103, and JPV105 were used in the experiments depicted in Table 2 and Fig. 1, 3, and 4. Strains JPV139, JPV142, JPV145, JPV146, JPV155, and JPV163 were used in the experiment represented in Fig. 2. Strains JPV203, JPV296, and JPV339 were used in the experiment represented in Fig. 5.
FIG. 5.
Pho4p is required for GIT1 expression but not INO1 expression. (A) Strains (JPV203, JPV296, and JPV339) transformed with plasmid pCA999 were grown in high-Pi, I+ medium lacking leucine and were used to inoculate high-Pi, I− and high-Pi, I+ media lacking leucine. Following three to five generations of growth, lysates obtained by glass bead breakage were assayed for chloramphenicol acetyltransferase (CAT) activity as described in Materials and Methods. Reactions were carried out for 2.5 h with 5 to 25 μg of protein per assay. (B) Strains (see above) transformed with plasmid pJH359 (INO1-CYC1-lacZ) were grown in high-Pi, I+ medium lacking uracil and were used to inoculate high-Pi, I− and high-Pi, I+ media lacking uracil. Following three to five generations of growth, cells were assayed for β-galactosidase activity by using the Pierce yeast β-galactosidase assay kit. Data represent at least three separate determinations and standard errors of the means. wt, wild type; OD600, optical density at 600 nm.
The bacterial chloramphenicol acetyltransferase (CAT) gene (cat) under the control of the GIT1 promoter was inserted into vector YCp7-32cat (13) to produce plasmid pCA998. The following PCR primers (Gibco) were used to amplify the GIT1 promoter (nucleotides −7 to −881 upstream of the start codon) and to introduce flanking restriction sites (shown in bold type): SalI forward (5′-TCGCCCATGGGGGTCGACTCGATATCTGCGATAAGG-3′) and BamHI reverse (5′-TCGCCCATGGGGGGATCCTCCTATTCTATTTTTTT-3′). The PCR product and YCp7-32cat were digested with BamHI and SalI and ligated together to form pCA998. To produce a reporter construct with a LEU2 selectable marker, pCA998 was digested with SalI and HindIII to release the 2.7-kb fragment in which the GIT1 promoter is fused to the cat gene. The 2.7-kb fragment was ligated to pRS315 (26) to produce pCA999.
Genetic screen for colonies showing fast growth on PI and isolation of PHO86.
The strain auxotrophic for inositol (JPV3) was transformed to leucine prototrophy with a YEp13-based genomic library. Cells were spread onto plates lacking leucine and incubated at 30°C for 2 days. Colonies were replica plated on synthetic media lacking inositol (I−), containing 75 μM inositol (I+), and lacking inositol but containing 75 μM PI. After 3 to 4 days of incubation at 30°C, colonies showing fast growth on plates containing 75 μM PI but unable to grow on I− plates were selected. Of the 10,000 transformants obtained, 6 displayed increased growth on PI. The plasmids were recovered from yeast cells, amplified in Escherichia coli, and retransformed into the parent strain (JPV3) to verify that the plasmids were responsible for the fast growth phenotype. The six plasmids were assorted into two groups based upon restriction fragment mapping. Representative plasmids from each group were sequenced with an ABI Prism 377 automatic DNA sequencer (University of Pittsburgh Research Support Facilities). The open reading frames contained on the complementing plasmids were analyzed by using the Saccharomyces Genome Database. One set of complementing plasmids contained the GIT1 gene. The GIT1 gene was cloned into multicopy vector pRS424 to produce plasmid pJP104. When transformed with pJP104, strain JPV3 displayed faster growth on PI. The second set of complementing plasmids contained the PHO86 gene. A 2,177-bp SalI-SpeI fragment containing the entire PHO86 gene was cloned into pRS424 to produce plasmid pJP201. When transformed with pJP201, strain JPV3 displayed faster growth on PI.
Construction of PHO86 disruption alleles.
A 1,641-bp HpaI-SphI fragment containing the entire PHO86 gene (−568 to 140 bp after the stop codon) was removed from plasmid pJP201 and replaced with a 1,333-bp SphI-NarI fragment containing the URA3 gene to produce plasmid pJP203. Digestion of pJP203 with SalI and SpeI produced a linear fragment containing the URA3 gene flanked by sequences corresponding to 62 nucleotides at the 5′ end and 474 nucleotides at the 3′ end of PHO86. The SalI-SpeI fragment was used for transformation into strain JPV22 by a one-step gene disruption procedure. Uracil prototrophs were screened by PCR to verify integration at the PHO86 locus, and the resulting strain was named JPV90.
Bacterial and yeast transformations.
Bacterial strains were transformed with plasmid DNA by using calcium chloride (24), and yeast strains were transformed by using lithium acetate (24).
3H-GroPIns incorporation.
Cell cultures (JPV91, JPV93, JPV95, and JPV99) grown overnight in high-Pi, I+ synthetic medium were harvested and used to inoculate 5 ml of each of the following media, all of which contained 50 μM glycerophospho-myo-[2-3H]inositol (3H-GroPIns) (American Radiolabeled Chemicals Inc.): low Pi, I−; low Pi, I+; high Pi, I−; and high Pi, I+. After three to five generations, 0.5-ml aliquots of the cultures were centrifuged, and the resulting pellets and supernatants were subjected to liquid scintillation counting by using a Beckman LS5801 counter with Ecolume liquid scintillation cocktail (ICN Biomedicals). Strains bearing mutations in the INO1 gene grew for approximately three generations in I− media by using their internal reserves of inositol.
Northern analysis.
Wild-type cells (JPV91) pregrown in high-Pi, I+ medium were used to inoculate each of the following media: low Pi, I−; low Pi, I+; high Pi, I−; and high Pi, I+. Cells were harvested in the logarithmic phase, and RNA was extracted by using the hot acid phenol method (6). RNA was separated on a 1% agarose gel, transferred to a positively charged nylon membrane (Roche catalog no. 1-209-299) by blotting, and UV cross-linked to the membrane by using a Stratalinker. Digoxigenin (DIG)-labeled probes for GIT1 and control (SNR17) transcripts were made by using a Roche PCR DIG probe synthesis kit (catalog no. 1-636-090). Prehybridization, hybridization, and detection were performed in accordance with the manufacturer's instructions for a Roche DIG luminescence detection kit (catalog no. 1-363-514).
CAT assays.
Cultures (JPV91, JPV93, JPV95, and JPV99 for Fig. 4; JPV203, JPV296, and JPV339 for Fig. 5) pregrown in high-Pi, I+ medium were harvested and used to inoculate the following media: low Pi, I−; low Pi, I+; high Pi, I−; and high Pi, I+. Following three to five generations of growth, lysates were obtained by glass bead breakage (2). The total protein concentration of each lysate was determined with bicinchoninic acid reagent (27). CAT assays were performed in accordance with the manufacturer's protocol for a FAST CAT green (deoxy) CAT assay kit (Molecular Probes catalog no. F-6616). Reactions were carried out for 2.5 h with 5 to 25 μg of protein per assay. Silica gel-coated thin-layer chromatography plates containing the separated reaction products were analyzed by using Kodak Image Station 440.
β-Galactosidase assays.
Strains (JPV203, JPV296, and JPV339) were transformed to uracil prototrophy with a plasmid (pJH359) bearing an INO1-CYC1-lacZ fusion (12). Cells grown to mid-logarithmic phase in high-Pi, I+ or in high-Pi, I− medium were assayed for β-galactosidase activity by using a Pierce Chemical Company yeast β-galactosidase assay kit.
RESULTS
Deletion of PHO86 accelerates the growth of an ino1Δ mutant in GroPIns- and PI-containing medium.
S. cerevisiae can use exogenous GroPIns as a source of inositol (20). As shown in Fig. 1, S. cerevisiae can also use exogenous PI (the precursor of GroPIns) as a source of inositol, albeit with a lag time even greater than that required for growth on GroPIns. In an attempt to identify factors involved in the breakdown and/or utilization of PI and GroPIns by S. cerevisiae, a gene overexpression scheme was used. A strain auxotrophic for inositol (ino1Δ) was transformed with a high-copy-number YEp13-based genomic library. Transformants displaying an increased growth rate when PI was supplied as the inositol source were chosen. Plasmids bearing two different genes were shown to confer the increased growth rate. Those genes were GIT1 and PHO86.
To discern the role of Pho86p in exogenous PI deacylation and GroPIns utilization, pho86Δ and pho86Δ ino1Δ strains were constructed. The pho86Δ ino1Δ strain grew faster on both GroPIns and PI than the ino1Δ strain, and this growth was, in all instances, dependent upon Git1p (Fig. 1C and D). In fact, the pho86Δ ino1Δ strain (unlike the ino1Δ strain) grew at identical rates when supplied with either inositol or GroPIns as an inositol source (compare Fig. 1A and C). The increased growth rate of the pho86Δ ino1Δ strain on PI was unexpected, since multicopy PHO86 caused an increased growth rate in the original selection. However, others have reported instances in which overexpression and deletion of the PHO86 gene result in equivalent phenotypes. Those phenotypes include reduced Pi uptake (4) and the ability to produce rAPase activity under high-Pi conditions (29). Thus, our findings are consistent with other reports and consistent with the notion that both deletion and overexpression of PHO86 result in a phosphate starvation response. Surprisingly, the pho86Δ ino1Δ strain grew more slowly than the ino1Δ strain when inositol was supplied, but the pho86Δ git1Δ ino1Δ strain grew at the same rate as the ino1Δ strain. This result held true outside of an ino1Δ genetic background: a pho86Δ strain grew more slowly than a wild-type strain, but a pho86Δ git1Δ strain grew at the same rate as a wild-type strain. These complicated growth phenotypes are a further indication of a functional interaction between Git1p and Pho86p. As expected, strains bearing a deletion in INO1 were unable to grow in the absence of an inositol source (Fig. 1B). Strains bearing an intact INO1 gene (git1Δ, pho86Δ, and pho86Δ git1Δ) grew similarly in the absence of inositol (Fig. 1B) as in medium containing PI or GroPIns (data not shown).
PI must be deacylated to GroPIns extracellularly to support the growth of an ino1 mutant.
The finding that an ino1Δ mutant can grow on exogenous PI only when functional Git1p is present suggested that PI is first deacylated to GroPIns before it is transported into the cell. In order to provide further evidence for this hypothesis, we analyzed strains bearing mutations in the three phospholipase B (PLB) genes that have been characterized for S. cerevisiae (10, 14). Plb1p1 and Plb3p are predicted to reside in the plasma membrane and the extracellular space, and Plb2p is predicted to reside in the extracellular space (14). The role of the PLB homologs in exogenous PI deacylation was assessed by monitoring the growth of strains bearing deletions in INO1 in combination with deletions in the PLB homolog genes when PI was supplied as the inositol source (Fig. 2). The ino1Δ plb1Δ, ino1Δ plb2Δ, and ino1Δ plb3Δ strains grew at rates similar to that of the ino1Δ strain in this experiment. However, two different ino1Δ plb1Δ plb2Δ strains and the ino1Δ plb1Δ plb2Δ plb3Δ strain grew more slowly and to a lower density than the ino1Δ strain. These results demonstrate that at least one PLB gene product is necessary for the deacylation of exogenous PI to GroPIns.
An ino1Δ pho86Δ mutant incorporates high levels of exogenous 3H-GroPIns into cells.
The increased growth of the ino1Δ pho86Δ mutant on GroPIns and PI (Fig. 1) indicated that the PHO86 gene product and, possibly, the phosphate concentration might affect GroPIns transport. To measure 3H-GroPIns incorporation, strains (wild type, ino1Δ, pho86Δ, and ino1Δ pho86Δ) pregrown in high-Pi, I+ medium were used to inoculate four separate media, each containing 50 μM 3H-GroPIns (Table 2): low Pi, I−; low Pi, I+; high Pi, I−; and high Pi, I+. These experiments were designed to measure 3H-GroPIns transport activity as a function of inositol and phosphate availabilities prior to the utilization of GroPIns as an inositol source. Thus, the cells were harvested and subjected to scintillation counting after three to five generations of growth (under our experimental conditions, strains carrying ino1Δ will double approximately three times in I− medium by using their internal reserves of inositol).
The ino1Δ pho86Δ strain exhibited greatly increased levels of GroPIns incorporation per cell density compared to the wild-type, ino1Δ, and pho86Δ strains (Table 2, experiment 1). While the levels of incorporation were not vastly different depending upon the media used, all strains showed the lowest level of incorporation in high-Pi, I+ medium. When a wild-type strain was grown to a high density (24 h of incubation), the total incorporation of the label varied according to the growth conditions in the following order (highest to lowest): low Pi, I−; low Pi, I+; high Pi, I−; and high Pi, I+ (Table 2, experiment 2).
GIT1 expression is sensitive to both inositol and phosphate limitations, and an ino1Δ pho86Δ mutant overexpresses GIT1.
Northern analysis of a wild-type strain indicated that the GIT1 transcript was most highly expressed under conditions in which both inositol and phosphate were limiting (Fig. 3). A much lower level of GIT1 expression also occurred in media in which inositol was limiting but phosphate was not and in media in which phosphate was limiting but inositol was not.
In order to facilitate the expression analysis, a plasmid in which the promoter region of GIT1 was fused to the bacterial cat reporter gene in vector yCp7-32cat was constructed (13). CAT activity was determined for wild-type, ino1Δ, pho86Δ, and ino1Δ pho86Δ strains which were grown initially in high-Pi, I+ medium and then transferred to four different media which varied in inositol and phosphate concentrations (Fig. 4). In agreement with the results of the Northern analysis, all strains exhibited the most CAT activity when incubated in media limited for both inositol and phosphate. However, an ino1Δ pho86Δ mutant displayed approximately threefold more CAT activity than the wild-type strain in I− medium containing both high and low levels of phosphate, suggesting that GIT1 transcription was hyperinduced.
GIT1 promoter-driven CAT activity requires Pho4p.
As a first approach to dissecting the regulatory systems involved in controlling GIT1 transcription, GIT1 promoter-driven CAT activity was measured for strains bearing deletion mutations in the transcription factors encoded by PHO4 and INO2 (Fig. 5A). In high-Pi, I+ or high-Pi, I− medium, Pho4p appeared to be required for CAT activity. Under the same growth conditions, strains bearing a deletion in INO2 displayed decreased CAT activity. The requirement of Pho4p for GIT1 transcription was specific, as Pho4p was not required for INO1 promoter-driven β-galactosidase activity (Fig. 5B). As expected, INO1 expression required Ino2p (Fig. 5B).
DISCUSSION
S. cerevisiae scavenges nutrients from exogenous GroPIns and PI. While GroPIns enters the cell intact, several lines of evidence indicate that PI must be deacylated to GroPIns prior to being internalized. To begin with, the growth of an ino1Δ mutant in PI-containing medium requires GIT1 (Fig. 1D), and GIT1 was isolated in the genetic screen as a multicopy suppressor of the slow-growth phenotype of an ino1Δ mutant growing in PI-containing medium. Furthermore, the growth of an ino1Δ mutant in PI-containing medium requires one or more PLB gene products (Fig. 2); the PLB gene products reside in the extracellular space (14) and hydrolyze phospholipids to produce glycerophosphodiesters, such as GroPIns. Finally, inositol auxotrophic strains that display a Git− phenotype (spt7 and ino2) (20) also display a Pit− phenotype (data not shown).
Our prior studies on the regulation of GroPIns transport were performed with media containing high levels of phosphate. Using those conditions, we confirmed that GroPIns transport is regulated by inositol availability (Table 2, compare columns 3 and 4). As an extension of those findings, we now report roles for both inositol and phosphate availabilities in regulating GIT1 transcription (Fig. 3 and 4) and GroPIns transport (Table 2). In a wild-type strain, the upregulation of GIT1 transcription in response to inositol limitation is greatly enhanced when phosphate is limiting, just as the upregulation of GIT1 transcription in response to phosphate limitation is greatly enhanced when inositol is limiting (Fig. 3 and 4). With regard to GroPIns uptake in a wild-type strain, more total 3H-GroPIns is accumulated when cells are grown in low-Pi media as opposed to high-Pi media (Table 2, experiment 2), and that accumulation is increased further by inositol limitation.
Analysis of the ino1Δ pho86Δ mutant has provided more insight into the regulation of GroPIns utilization. Although the ino1Δ pho86Δ strain grows slowly in inositol-containing media (Fig. 1A), it displays virtually the same growth rate when grown in GroPIns-containing media (Fig. 1C). This behavior is in stark contrast to that of the ino1Δ strain, whose lag phase is greatly lengthened in GroPIns-containing media compared to inositol-containing media (Fig. 1A and C). These results suggest that GroPIns transport and/or catabolism are constitutive in the double mutant. Indeed, GroPIns incorporation is constitutively high, although still regulated by inositol and phosphate, in the ino1Δ pho86Δ strain (Table 2). Similarly, GIT1 transcript accumulation in inositol-free media is much higher in the ino1Δ pho86Δ strain than in any other strain tested. The fact that GIT1 transcript accumulation in the ino1Δ pho86Δ strain is similar to that in the wild-type strain grown in I+ media, while 3H-GroPIns accumulation is much higher, suggests that the regulation of GroPIns accumulation does not occur solely at the level of GIT1 transcription. Other possible control points affecting GroPIns accumulation include GIT1 mRNA degradation, Git1p degradation, Git1p transport activity, and the steps involved in the catabolism of GroPIns and its subsequent utilization for de novo PI synthesis. These possibilities are currently under investigation.
Interestingly, both the ino1Δ pho86Δ (Fig. 1) and the pho86Δ (data not shown) strains exhibit a slow-growth phenotype that is alleviated by the deletion of GIT1. Strains lacking Pho86p show constitutive rAPase activity and are unable to efficiently transport Pho84p to the plasma membrane (9). Similarly, pho84Δ mutants show constitutive rAPase activity and have no Pho84p. As an initial attempt to dissect these complex phenotypes, we will examine whether the deletion of GIT1 in a pho84Δ mutant (and in strains carrying mutations in other PHO genes) affects growth, as it does in a pho86Δ background.
In terms of GroPIns incorporation and GIT1 transcript accumulation, the ino1Δ pho86Δ mutant behaves in a manner much different from that of either the ino1Δ or the pho86Δ mutant alone and different from that of the wild-type strain grown in media limited for inositol and phosphate. This finding suggests a synergistic role for inositol and phosphate in regulating the utilization of GroPIns. Furthermore, it suggests that the cell is sensitive to the severity of inositol and phosphate limitation. A wild-type strain growing in I−, low-Pi medium is still capable of making its own inositol and transporting Pi into the cell. An ino1Δ pho86Δ mutant growing in I−, low-Pi medium, on the other hand, has no source of inositol (but grows for a limited time on its internal reserves), has a greatly diminished ability to transport Pi, and shows hyperactivated GIT1 transcription and GroPIns incorporation.
Using genome-wide expression analysis, others have reported that the GIT1 transcript accumulates when phosphate limitation is artificially imposed by deletion of PHO85 or chemical inhibition of Pho85p (5). In another microarray study, GIT1 did not meet the authors' criteria for being a phosphate-regulated gene but did display the induction of transcription in low-Pi versus high-Pi media in one of two wild-type strains tested (15). Those studies were performed with I+ media. In wild-type strain JPV91, GIT1 transcription is significantly affected only by phosphate concentrations in I− media. Thus, the relative contributions of inositol depletion and phosphate depletion in affecting GIT1 transcription appear to be strain dependent. Wykoff and O'Shea (28) reported that the overexpression of GIT1 in high-Pi medium suppresses the nonviability of a strain from which all other known phosphate transporter genes (PHO84, PHO87, PHO89, PHO90, and PHO91) have been deleted. The authors concluded that Git1p is capable of transporting Pi, albeit with a higher Km for Pi than that exhibited by Pho84p. Although differences in strain backgrounds and assay conditions preclude a direct comparison, it is worth noting that the reported apparent Km for Pi transport by Git1p (28) is approximately 10-fold higher than the reported apparent Km for GroPIns transport by Git1p (19).
An initial experiment (Fig. 5) aimed at determining the regulatory systems responsible for controlling GIT1 expression indicates that PHO4 is required for GIT1 transcription under high-phosphate conditions. Future studies will include a thorough analysis of all potential transcription factors (e.g., Ino4p, Ino2p, Pho4p, and Pho2p) and promoter elements involved in controlling GIT1 expression in response to inositol and phosphate. Pho4p, the PHO regulon transcription factor, is a bHLH binding protein whose consensus sequence is CACGTK (17). Five potential Pho4p binding sites and three potential Pho2p binding sites (TAATRA/TAANTAA) exist in the GIT1 promoter region between nucleotides −1 and −700 relative to the ATG start codon. The GIT1 promoter does not contain a copy of the core consensus sequence (CATGTG) for UASINO. However, it does contain a bHLH consensus sequence (CACGTG) to which Ino2p and Ino4p (bHLH proteins) bind, although with less affinity than to UASINO, to activate INO1 transcription (3). Given that mammalian bHLH binding proteins have been shown to form multiple dimer combinations that can act upon diverse sets of genes (23), it is tempting to speculate that Pho4p may heterodimerize with Ino2p or Ino4p to regulate GIT1 transcription in response to inositol and phosphate. In support of this possibility, Ino4p has been shown by a yeast two-hybrid assay and a biochemical assay to interact with Pho4p, as well as other bHLH binding proteins (22). Interestingly, PHO5 expression is partially repressed by inositol availability and the deletion of PHO2 (18). In addition to promoter analysis, future studies will include an analysis of GIT1 mRNA turnover and Git1p turnover as functions of inositol and phosphate availabilities.
Acknowledgments
This work was supported by National Institutes of Health grant GM59817 to J.P.-V.
We thank S. A. Henry for critical reading of the manuscript.
REFERENCES
- 1.Angus, W. W., and R. L. Lester. 1972. Turnover of inositol and phosphorus containing lipids in Saccharomyces cerevisiae; extracellular accumulation of glycerophosphorylinositol derived from phosphatidylinositol. Arch. Biochem. Biophys. 151:483-495. [DOI] [PubMed] [Google Scholar]
- 2.Ashburner, B. P., and J. M. Lopes. 1995. Autoregulated expression of the yeast INO2 and INO4 helix-loop-helix activator genes effects cooperative regulation on their target genes. Mol. Cell. Biol. 15:1709-1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bachhawat, N., Q. Ouyang, and S. A. Henry. 1995. Functional characterization of an inositol-sensitive upstream activation sequence in yeast: a cis-regulatory element responsible for inositol-choline mediated regulation of phospholipid biosynthesis. J. Biol. Chem. 270:25087-25095. [DOI] [PubMed] [Google Scholar]
- 4.Bun-ya, M., K. Shikata, S. Nakade, C. Yompakdee, S. Harashima, and Y. Oshima. 1996. Two new genes, PHO86 and PHO87, involved in inorganic phosphate uptake in Saccharomyces cerevisiae. Curr. Genet. 29:344-351. [PubMed] [Google Scholar]
- 5.Carroll, A. S., A. C. Bishop, J. L. DeRisi, K. M. Shokat, and E. K. O'Shea. 2001. Chemical inhibition of the Pho85 cyclin-dependent kinase reveals a role in the environmental stress response. Proc. Natl. Acad. Sci. USA 98:12578-12583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Elion, E. A., and J. R. Warner. 1984. The major promoter element of rRNA transcription in yeast lies 2 kb upstream. Cell 39:663-673. [DOI] [PubMed] [Google Scholar]
- 7.Henry, S. A., and J. L. Patton-Vogt. 1998. Genetic regulation of phospholipid metabolism: yeast as a model eukaryote. Prog. Nucleic Acid Res. Mol. Biol. 61:133-179. [DOI] [PubMed] [Google Scholar]
- 8.Hirst, K., F. Fisher, P. C. McAndrew, and C. R. Goding. 1994. The transcription factor, the Cdk, its cyclin and their regulator: directing the transcriptional response to a nutritional signal. EMBO J. 13:5410-5420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lau, W. W., R. W. Howson, P. Malkus, R. Schekman, and E. K. O'Shea. 2000. Pho86p, an endoplasmic reticulum (ER) resident protein in saccharomyces cerevisiae, is required for ER exit of the high-affinity phosphate transporter pho84p. Proc. Natl. Acad. Sci. USA 97:1107-1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee, K. S., J. L. Patton, M. Fido, L. K. Hines, S. D. Kohlwein, F. Paltauf, S. A. Henry, and D. E. Levin. 1994. The Saccharomyces cerevisiae PLB1 gene encodes a protein required for lysophospholipase and phospholipase B activity. J. Biol. Chem. 269:19725-19730. [PubMed] [Google Scholar]
- 11.Lenburg, M. E., and E. K. O'Shea. 1996. Signaling phosphate starvation. Trends Biochem. Sci. 21:383-387. [PubMed] [Google Scholar]
- 12.Lopes, J. M., J. P. Hirsch, P. A. Chorgo, K. L. Schulze, and S. A. Henry. 1991. Analysis of sequences in the INO1 promoter that are involved in its regulation by phospholipid precursors. Nucleic Acids Res. 19:1687-1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mannhaupt, G., U. Pilz, and H. Feldmann. 1988. A series of shuttle vectors using chloramphenicol acetyltransferase as a reporter enzyme in yeast. Gene 67:287-294. [DOI] [PubMed] [Google Scholar]
- 14.Merkel, O., M. Fido, J. A. Mayr, H. Prüger, F. Raab, G. Zandonella, S. D. Kohlwein, and F. Paltauf. 1999. Characterization and function in vivo of two novel phospholipases B/lysophospholipases from Saccharomyces cerevisiae. J. Biol. Chem. 274:28121-28127. [DOI] [PubMed] [Google Scholar]
- 15.Ogawa, N., J. DeRisi, and P. O. Brown. 2000. New components of a system for phosphate accumulation and polyphosphate metabolism in Saccharomyces cerevisiae revealed by genomic expression analysis. Mol. Biol. Cell 11:4309-4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.O'Neill, E. M., A. Kaffman, E. R. Jolly, and E. K. O'Shea. 1996. Regulation of PHO4 nuclear localization by the PHO80-PHO85 cyclin-CDK complex. Science 271:209-212. [DOI] [PubMed] [Google Scholar]
- 17.Oshima, Y. 1997. The phosphatase system in Saccharomyces cerevisiae. Genes Genet. Syst. 72:323-334. [DOI] [PubMed] [Google Scholar]
- 18.Paltauf, F., S. D. Kohlwein, and S. A. Henry. 1992. Regulation and compartmentalization of lipid synthesis in yeast, p. 415-500. In E. Jones, J. Pringle, and J. Broach (ed.), The molecular and cellular biology of the yeast Saccharomyces: gene expression, vol. 2. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 19.Patton, J. L., L. Pessoa-Brandao, and S. A. Henry. 1995. Production and reutilization of an extracellular phosphatidylinositol catabolite, glycerophosphoinositol, by Saccharomyces cerevisiae. J. Bacteriol. 177:3379-3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Patton-Vogt, J. L., and S. A. Henry. 1998. GIT1, a gene encoding a novel transporter for glycerophosphoinositol in Saccharomyces cerevisiae. Genetics 149:1707-1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Persson, B. L., A. Berhe, U. Fristedt, P. Martinez, J. Pattison, J. Petersson, and R. Weinander. 1998. Phosphate permeases of Saccharomyces cerevisiae. Biochim. Biophys. Acta 1365:23-30. [DOI] [PubMed] [Google Scholar]
- 22.Robinson, K. A., J. I. Koepke, M. Kharodawala, and J. M. Lopes. 2000. A network of yeast basic helix-loop-helix interactions. Nucleic Acids Res. 28:4460-4466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Robinson, K. A., and J. M. Lopes. 2000. Survey and summary: Saccharomyces cerevisiae basic helix-loop-helix proteins regulate diverse biological processes. Nucleic Acids Res. 28:1499-1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rose, M. D., F. Winston, and P. Hieter. 1990. Methods in yeast genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 25.Schneider, K. R., R. L. Smith, and E. K. O'Shea. 1994. Phosphate-regulated inactivation of the kinase PHO80-PHO85 by the CDK inhibitor PHO81. Science 266:122-126. [DOI] [PubMed] [Google Scholar]
- 26.Sikorski, R. S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smith, P. K., R. I. Krohn, G. T. Hermanson, A. K. Mallia, F. H. Gartner, M. D. Provenzano, E. K. Fujimoto, N. M. Goeke, B. J. Olson, and D. C. Klenk. 1985. Measurement of protein using bicinchoninic acid. Anal. Biochem. 150:76-85. [DOI] [PubMed] [Google Scholar]
- 28.Wykoff, D. D., and E. K. O'Shea. 2001. Phosphate transport and sensing in Saccharomyces cerevisiae. Genetics 159:1491-1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yompakdee, C., N. Ogawa, S. Harashima, and Y. Oshima. 1996. A putative membrane protein, Pho88p, involved in inorganic phosphate transport in Saccharomyces cerevisiae. Mol. Gen. Genet. 251:580-590. [DOI] [PubMed] [Google Scholar]