Abstract
We use the GCN4 oligomerization domain to engineer a covalently linked parallel polyprotein dimer based on the well-studied I27 domain of titin. We use single molecule atomic force microscopy techniques to stretch single polyprotein fibers and verify their mechanical properties. We find that the engineered polyprotein dimers extend in perfect register, doubling the unfolding force and halving the persistence length while keeping the contour length increase unchanged. These experiments directly confirm the mechanical scaling laws proposed for parallel bundles of modular proteins.
Wetherbee and colleagues have reported on intriguing mechanical properties of the adhesive fibers secreted by live diatoms (1,2). These fibers produced traces with the characteristic sawtooth pattern shape of polyproteins, with peak forces up to 800 pN and exceedingly small persistence lengths of ∼0.026 nm. They hypothesized that these sawtooth patterns represented the fingerprint of fibers composed of multiple polyproteins that unfolded simultaneously and in perfect register. Although this is a compelling explanation (3), it has never been demonstrated that a parallel arrangement of polyproteins could produce such in-register high force peaks, together with the low persistence lengths observed in their experiments. Here we report experiments that directly test this hypothesis by examining the mechanical properties of an engineered parallel I27 polyprotein dimer.
The 27th immunoglobulin module from the giant human muscle protein titin (I27), is 89 amino acids long, with 7 antiparallel β-strands (4). In our experience, the I27 protein is a robust platform to engineer a wide variety of polyproteins that can be studied using force spectroscopy (5).
We engineered the parallel polyprotein dimer by making a fusion protein between an (I27)8 polyprotein and a GCN4 oligomerization domains placed at each end of the polyprotein. As demonstrated by Harbury and colleagues, the 33 amino acid α-helical coiled-coil domain GCN4 readily self assembles into dimers (6). Furthermore, the GCN4 oligomerization domain is ideal to bundle together proteins of interest since it forms only parallel coiled-coils, ensuring the directionality of the bundle. The addition of N- and C-terminal cysteines enable the formation of inter\chain S-S bonds, covalently binding the quaternary structure together (see online Supplementary Material).
The unfolding of I27-based polyproteins is now very well characterized using single molecule force spectroscopy. Pulling an (I27)8 polyprotein at ∼400 nm s−1 results in force-extension traces with a characteristic sawtooth pattern shape revealing sequential unfolding events at ∼200 pN (5). The entropic elasticity of the unfolding polyprotein is well characterized by the worm-like-chain model (WLC); fits to our atomic force microscopy force-extension curves describe the restoring force as a result of stretching the polymer, and is given by
 |
where F is the force, x is the extension, p is the persistence length, k is the Boltzmann constant, T is the temperature in degrees Kelvin, and Lc is the contour length of the polymer. The parameters of p, LC are measured by fits to the measured force-extension curves. WLC fits to the force-extension relationship leading up to each unfolding peak of the (I27)8 give values of p ∼ 0.4 nm and contour length increments of ΔLC = 28.4 nm after each unfolding peak (5). Hence, any polyprotein can be uniquely characterized by these three parameters: p, ΔLC, and Fu (peak unfolding force). These same parameters can be used to uniquely identify a parallel polyprotein dimer. We expect that pulling an oligomer made of n perfectly aligned and parallel (I27)8 polyproteins results in a sawtooth-pattern trace where the unfolding force scales with n straightforwardly, as 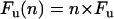 . The observed persistence length scales as
. The observed persistence length scales as 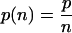 and the contour length, ΔLC, remain unchanged. Although it is generally assumed that these relationships should hold (1,7,8), they have not been verified experimentally for n > 1, because doing so requires knowing exactly how many parallel polyproteins comprise the bundles being measured.
and the contour length, ΔLC, remain unchanged. Although it is generally assumed that these relationships should hold (1,7,8), they have not been verified experimentally for n > 1, because doing so requires knowing exactly how many parallel polyproteins comprise the bundles being measured.
Fig. 1 shows the results of mechanically stretching the polyprotein dimer 2[GCN4-(I27)8-GCN4]. The trace in Fig. 1 A shows the first type of sawtooth pattern observed with this protein sample with unfolding force peaks of Fu ∼ 200 pN, equally spaced by an increase in contour length of ΔLC ∼ 28.4 nm. This is the fingerprint of a single I27 polyprotein, indicating that this trace resulted from a GCN4-(I27)8-GCN4 that failed to form a dimer. Alternatively, since the dimerized molecule can topologically be thought of as a circle, alternative stretching profiles cannot be ruled out. Out of 82 sawtooth patterns recorded from this protein preparation, 53 recordings were of the type shown in Fig. 1 A. In the same protein sample and using the same cantilevers, we also observed sawtooth patterns with similar spacing but with force peaks that were twice as big, Fu ∼ 400 pN, than those shown in Fig. 1 A.
FIGURE 1.
Mechanical fingerprint of a parallel dimeric polyprotein. Probing the 2[GCN4-(I27)8-GCN4] protein preparation, we recorded two types of sawtooth pattern traces. (A) Typical trace obtained from pulling a monomer, GCN4-(I27)8-GCN4 or (B) a self assembled dimer 2[GCN4-(I27)8-GCN4]. Fits of the WLC model to the traces (gray lines for the monomer and green lines for the dimer) measure both the contour length increment after each unfolding event (ΔLc) and the persistence length (p) of the traces. Contour length increment was the same for monomers and dimers (monomers, ΔLc = 28.4 ± 0.3 nm; dimmers, ΔLc = 28.5 ± 0.4 nm); persistence length, p, and the unfolding force, Fu, were significantly different as shown in the histograms in C and D. (C) Histograms of persistence length values measured from monomer traces (gray bars, Gaussian fit; p = 0.37 ± 0.08 nm, n = 53) and dimer traces (green bars, Gaussian fit; p = 0.17 ± 0.04 nm, n = 29). The persistence length was measured only once at the end of each trace, after the final unfolding event. (D) Histogram compares the unfolding force peaks measured from monomer traces (gray bars, Gaussian fit; Fu = 200 ± 25 pN, n = 285, scale on the left) and dimer traces (green bars, Gaussian fit; Fu = 407 ± 65 pN, n = 86, scale on the right). The data show that the dimer proteins have force peaks twice as big as the monomers, with half the persistence length.
Such perfectly in-register sawtooth patterns showing twice the force with the same peak spacing as the wild-type I27 have, to our knowledge, never been observed in recordings obtained from I27 based polyproteins. Thus, the recordings in Fig. 1 B likely resulted from stretching single 2[GCN4-(I27)8-GCN4] polyprotein dimers. Histograms of the peak unfolding forces, Fu, as well as of the persistence length value, p, measured by fitting the WLC traces like those shown in Fig. 1 B, reveal persistence lengths, p = 0.17 ± 0.04 nm (Fig. 1 C), versus similar fits of the WLC to the traces shown in Fig. 1 A, where persistence lengths were twice as large, p = 0.36 ± 0.04 nm. Histograms comparing the peak unfolding forces of these two types of recordings are shown in Fig. 1 D. This figure shows that traces like the one shown in Fig. 1 A have unfolding peaks that average Fu = 200 ± 25 pN, whereas traces like those shown in Fig. 1 B average Fu = 407 ± 65 pN. These results are in good agreement with our expectations regarding the force-extension relationships for stretching either the GCN4-(I27)8-GCN4 polyprotein (Fig. 1 A) or its parallel dimer (2[GCN4-(I27)8-GCN4], Fig. 1 B). As shown, we observe that whereas the unfolding force doubles, the persistence length drops to half, verifying for n = 2 the validity of the simple scaling laws discussed above.
Bundles of parallel polyproteins can indeed unfold in perfect register, directly supporting the hypothesis proposed by Dugdale et al. (2,3). Our observations also demonstrate the manner in which polyprotein fibers might operate in other biological systems as well. For example, quaternary arrangements of modular proteins can be found in the giant muscle protein titin (9). It is likely that the mechanical properties of titin bundles also follow the mechanical scaling laws demonstrated here.
SUPPLEMENTARY MATERIAL
An online supplement to this article can be found by visiting BJ Online at http://www.biophysj.org.
Acknowledgments
This work was supported by funding from the National Institutes of Health (to J.M.F.). A.S. was supported by a fellowship from the Neurosurgery Research and Education Foundation.
Atom Sarkar's present address is Dept. of Neurological Surgery, The Ohio State University, Columbus, OH 43210.
References
- 1.Dugdale, T. M., R. Dagastine, A. Chiovitti, P. Mulvaney, and R. Wetherbee. 2005. Single adhesive nanofibers from a live diatom have the signature fingerprint of modular proteins. Biophys. J. 89:4252–4260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dugdale, T. M., R. Dagastine, A. Chiovitti, and R. Wetherbee. 2006. Diatom adhesive mucilage contains distinct supramolecular assemblies of a single modular protein. Biophys. J. 90:2987–2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fernandez, J. M. 2005. Fingerprinting single molecules in vivo. Biophys. J. 89:3676–3677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Improta, S., A. S. Politou, and A. Pastore. 1996. Immunoglobulin-like modules from titin I-band: Extensible components of muscle elasticity. Structure. 4:323–337. [DOI] [PubMed] [Google Scholar]
- 5.Carrion-Vazquez, M., A. F. Oberhauser, S. B. Fowler, P. E. Marszalek, S. E. Broedel, J. Clarke, and J. M. Fernandez. 1999. Mechanical and chemical unfolding of a single protein: A comparison. Proc. Natl. Acad. Sci. USA. 96:3694–3699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harbury, P. B., T. Zhang, P. S. Kim, and T. Alber. 1993. A switch between 2-stranded, 3-stranded and 4-stranded coiled coils in gcn4 leucine-zipper mutants. Science. 262:1401–1407. [DOI] [PubMed] [Google Scholar]
- 7.Kellermayer, M. S. Z., C. Bustamante, and H. L. Granzier. 2003. Mechanics and structure of titin oligomers explored with atomic force microscopy. Biochimica Et Biophysica Acta-Bioenergetics. 1604:105–114. [DOI] [PubMed] [Google Scholar]
- 8.Li, H. B., A. F. Oberhauser, S. D. Redick, M. Carrion-Vazquez, H. P. Erickson, and J. M. Fernandez. 2001. Multiple conformations of PEVK proteins detected by single-molecule techniques. Proc. Natl. Acad. Sci. USA. 98:10682–10686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liversage, A. D., D. Holmes, P. J. Knight, L. Tskhovrebova, and J. Trinick. 2001. Titin and the sarcomere symmetry paradox. J. Mol. Biol. 305:401–409. [DOI] [PubMed] [Google Scholar]



