Abstract
Hirano bodies are paracrystalline actin filament-containing structures reported to be associated with a variety of neurodegenerative diseases. However, the biological function of Hirano bodies remains poorly understood, since nearly all prior studies of these structures were done with postmortem samples of tissue. In the present study, we generated a full-length form of a Dictyostelium 34-kDa actin cross-linking protein with point mutations in the first putative EF hand, termed 34-kDa ΔEF1. The 34-kDa ΔEF1 protein binds calcium normally but has activated actin binding that is unregulated by calcium. The expression of the 34-kDa ΔEF1 protein in Dictyostelium induces the formation of Hirano bodies, as assessed by both fluorescence microscopy and transmission electron microscopy. Dictyostelium cells bearing Hirano bodies grow normally, indicating that Hirano bodies are not associated with cell death and are not deleterious to cell growth. Moreover, the expression of the 34-kDa ΔEF1 protein rescues the phenotypes of cells lacking the 34-kDa protein and cells lacking both the 34-kDa protein and α-actinin. Finally, the expression of the 34-kDa ΔEF1 protein also initiates the formation of Hirano bodies in cultured mouse fibroblasts. These results show that the failure to regulate the activity and/or affinity of an actin cross-linking protein can provide a signal for the formation of Hirano bodies. More generally, the formation of Hirano bodies is a cellular response to or a consequence of aberrant function of the actin cytoskeleton.
Abnormal protein aggregation results in the formation of distinct types of protein assemblies frequently associated with disease. For example, neurodegenerative diseases are associated with the deposition of peptides derived from β-amyloid precursor protein in senile plaques (54, 63), tau protein in neurofibrillary tangles (33), α-synuclein in Lewy bodies (13, 66), and proteins with polyglutamine repeats in insoluble aggregates (26, 51). Insoluble aggregates containing misfolded proteins in beta-sheet structures underlie a variety of diseases, classified as amyloidoses, that are not specific to the nervous system (2, 4, 29). Furthermore, misfolded proteins collect in structures, termed aggresomes, that can be induced by either the expression of misfolded proteins or inhibition of the proteasome (28, 30).
During the past three decades, Hirano bodies have been reported to be associated with a broad array of conditions, including Alzheimer's disease (21, 44, 45, 60), Parkinson's disease (25), Pick's disease (61), amyotrophic lateral sclerosis (25), ataxic Creutzfeldt-Jakob disease (3), kuru (9), scrapie (10), leukoencephalopathy (23), chronic alcoholism (32), diabetes (64), cancer (12, 20), muscle degeneration (11), and neuronal degeneration associated with abnormal copper homeostasis (1, 46, 53, 71). Hirano bodies are paracrystalline cytoplasmic inclusions that contain actin filaments and actin-associated proteins (19, 22, 39). Hirano bodies have been termed a “non-specific manifestation of neuronal degeneration,” since they have been noted in autopsy examinations of brains from patients with a variety of conditions (62). However, their mechanism of formation, composition, and relationship to disease remain poorly understood.
Maselli et al. recently reported the development of a cultured cell model for studies of Hirano bodies in the cellular slime mold Dictyostelium discoideum (42). Dictyostelium is a lower eucaryote with a well-characterized cytoskeleton and facile methods for protein expression and the creation of mutant strains (41, 48). The 34-kDa protein is one of 11 actin cross-linking proteins present in Dictyostelium (17). In vitro studies have revealed that actin bundling by the purified 34-kDa protein is calcium regulated (5, 6, 8, 34). Experiments with defined segments of recombinant proteins have revealed three actin binding sites located at amino acids 1 to 123, 193 to 254, and 279 to 295 (35). The most active of these sites, located at amino acids 193 to 254, is necessary and sufficient for cosedimentation with F-actin in vitro (35). The carboxy-terminal (CT) fragment, comprising amino acids 124 to 295, lacks the inhibitory domain that is located in the amino terminus and that modulates the activity of the most active actin binding site through an intramolecular interaction (36). The absence of this inhibitory region in the CT fragment results in enhanced binding and cross-linking of actin filaments that are calcium insensitive (36). Maselli et al. found that the expression of low levels of the truncated CT protein in Dictyostelium induces the formation of paracrystalline actin inclusions that resemble Hirano bodies in both ultrastructure and composition, as assessed by immunocytochemistry (42).
The goal of the present work was to investigate the factors leading to Hirano body formation. A form of the 34-kDa protein with a point mutation in the first putative calcium binding EF hand was produced by site-directed mutagenesis. Studies of the calcium binding and actin binding activities of this protein, termed 34-kDa ΔEF1, revealed normal calcium binding but activated actin binding that is calcium insensitive. The 34-kDa ΔEF1 protein induces the formation of Hirano bodies following expression in Dictyostelium and in mammalian cells. Dictyostelium cells with Hirano bodies induced by expression of the 34-kDa ΔEF1 protein grow normally. Moreover, the 34-kDa ΔEF1 protein supplies the function of the 34-kDa protein to null mutants lacking the 34-kDa protein. The results reveal that both loss of calcium sensitivity and activation of actin binding are required to induce the formation of Hirano bodies in cell cultures. Further, Hirano bodies are not necessarily linked to cell death and are not overtly deleterious to cell function.
MATERIALS AND METHODS
Preparation of wild-type and mutant forms of the 34-kDa protein in Escherichia coli.
The wild-type 34-kDa protein was expressed from plasmid pET15b-F18 in E. coli as described previously (34). Mutations in the first putative EF hand of the 34-kDa protein were produced in plasmid pET15b-F18 by using a Chameleon site-directed mutagenesis kit (Stratagene, La Jolla, Calif.) according to the manufacturer's instructions. The notation 34-kDa ΔEF1 designates the protein containing mutations in the first EF hand, as shown in Fig. 1A. Mutagenesis primers were on the basis of the antisense strand of the 34-kDa protein nucleotide sequence with nucleotides numbered as described previously (7). Primer 34kDaΔEF1 has the sequence 5′GAAATAGAGAGCAATAGCGAAAGCCAAAGCGTTACCTTCATC3′, spans nucleotides 306 to 265 in the cDNA sequence, and was used to create the mutations in the first EF hand. Selection primer AccIpET15 has the sequence 5′CGATAGCGGAGTGGTACCTGGCTTAACTATGCG3′ and was used to convert the AccI site in pET15b-F18 to a KpnI site for the selection of mutated plasmids. All primers were synthesized and 5′ phosphorylated at the Molecular Genetics Instrumentation Facility at the University of Georgia. All plasmids were sequenced to confirm that changes in the coding region were restricted to those intentionally introduced by the mutagenesis procedure. Automated sequencing was performed at the Molecular Genetics Instrumentation Facility at the University of Georgia. The wild-type 34-kDa protein was purified from E. coli as described previously (34), and the modified 34-kDa ΔEF1 protein was purified essentially by the same procedure with minor modifications.
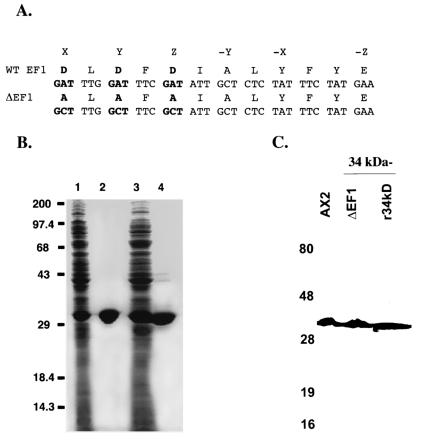
FIG. 1.
Wild-type and calcium-insensitive forms of the 34-kDa protein. (A) Nucleotide and amino acid sequences of the wild-type (WT) 34-kDa and 34-kDa ΔEF1 (modified EF hand) proteins. The altered nucleotides and amino acids are shown in bold type. (B) Expression and purification of the 34-kDa and 34-kDa ΔEF1 proteins. The SDS-polyacrylamide gel was stained with Coomassie brilliant blue. Lanes 1 and 3 were loaded with 100 μg of BL21(DE3) expressing the 34-kDa and 34-kDa ΔEF1 proteins, respectively. Lanes 2 and 4 were loaded with 25 μg of purified 34-kDa and 34-kDa ΔEF1 proteins, respectively. Numbers on the left are in kilodaltons. (C) Western blot showing Dictyostelium AX2 and 34-kDa protein-null (34 kDa−) cells expressing the wild-type 34-kDa protein and the 34-kDa ΔEF1 protein. Proteins from cell lysates were resolved by SDS-polyacrylamide gel electrophoresis, transferred to nitrocellulose, and probed with monoclonal antibody B2C, reactive to the 34-kDa protein. r34kD, recombinant 34-kDa protein.
Analytical methods and antibodies.
Protein concentrations were determined by the bicinchoninic acid (Pierce Chemical Co., St. Louis, Mo.) method (65) with bovine serum albumin (Sigma Chemical Co., St. Louis, Mo.) as a protein standard. Monoclonal anti-34-kDa protein antibody B2C (15) and alkaline phosphatase-conjugated goat anti-mouse antibodies (Promega Corp., Madison, Wis.) were used for Western blot analysis as described previously (70).
Measurement of calcium binding by equilibrium dialysis.
Ultrapure water (Continental Water Systems Corp., San Antonio, Tex.) and buffers were passed over a Chelex 100 (2 by 30 cm; Bio-Rad, Hercules, Calif.) resin to remove any traces of Ca2+. All plasticware was rinsed in Ca2+-free water before use. The proteins were first dialyzed against storage buffer (10 mM Tris [pH 7.0], 50 mM KCl, 0.2 mM dithiothreitol, 0.1 mM EDTA) for 24 h at 4°C. To remove EDTA, the proteins were dialyzed in storage buffer as described above but without the EDTA for 24 h at 4°C with one buffer change. For Ca2+ binding measurements, the proteins were dialyzed in 40 ml of storage buffer without EDTA but containing different concentrations of [45Ca]CaCl2 for 48 h at 25°C. To achieve different concentrations of calcium in the buffer, calcium was added from a 200 μM Ca2+ stock solution that contained approximately 10 μCi of 45Ca (DuPont NEN, Boston, Mass.)/μmol. Protein concentrations in samples were determined after dialysis; 100 μl of buffer from outside the dialysis bag and 100 μl of sample from inside the dialysis bag were sampled in triplicate, and radioactivity was determined by liquid scintillation counting. To determine the amount of calcium bound, the numbers of counts outside were averaged and subtracted from the averaged numbers of counts inside and divided by the specific activity of 45Ca.
Actin isolation.
G-actin was isolated from rabbit skeletal muscle acetone powder and gel filtered as described previously (40, 67). The actin was stored in G-actin buffer (40, 67 ) for not more than 1 week, with fresh buffer changes every day.
Measurement of binding by cosedimentation with actin.
High-speed actin cosedimentation assays were performed as outlined previously (5, 8, 34). G-actin (3 μM) was mixed with 3 μM wild-type 34-kDa protein or 34-kDa ΔEF1 protein in 20 mM piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES) (pH 7.0)-50 mM KCl-50 μM MgCl2-1 mM ATP-0.2 mM dithiothreitol-5 mM EGTA with or without 4.5 mM CaCl2 in a final volume of 130 μl. Supernatant and pellet fractions were analyzed by polyacrylamide gel electrophoresis in the presence of sodium dodecyl sulfate (SDS) (31) and visualized with Coomassie brilliant blue. The intensity of the bands was quantified by scanning densitometry (Molecular Dynamics, Sunnyvale, Calif.) and used to determine the amounts of proteins in the supernatant and pellet fractions. Control experiments verified that none of the 34-kDa proteins showed any significant sedimentation in the absence of actin in either the presence or absence of calcium (data not shown).
Expression of mutant 34-kDa proteins in Dictyostelium.
Wild-type or mutant 34-kDa proteins were expressed in Dictyostelium after subcloning into the BamHI site of vector pBORP (52). The coding sequences were taken from plasmids encoding the full-length 34-kDa protein (pET15b-F18) (34) and the 34-kDa ΔEF1 protein (p34-kDa ΔEF1) by PCR with primers that added terminal BamHI restriction enzyme sites for the purpose of subcloning. The wild-type 34-kDa, 34-kDa ΔEF1, and 34-kDa CT proteins were expressed in previously described Dictyostelium strains lacking the 34-kDa protein and strains lacking both the 34-kDa protein and α-actinin (56, 57). Electroporation and growth of cells were performed as described previously (42). Cells deficient for the 34-kDa protein and α-actinin were cotransformed with pBSK-BSR to impart blasticidin resistance for selection purposes, as the parental cells and pBORP both have G418 resistance cassettes. At 24 h after transformation, the media were removed and replaced with media containing antibiotics required for selection. pBSK-BSR was constructed by subcloning the blasticidin resistance cassette from pBsR479 (55) into the BamHI site of pBluescript SK(−) (Stratagene).
Growth.
Dictyostelium AX2 and the derivatives described below were routinely maintained in axenic cultures with shaking at 150 rpm in HL-5 medium (37) at 20°C. To assess growth rates, cells were inoculated at a starting concentration of 4 × 104 cells/ml into axenic HL-5 culture medium with shaking at 150 or 120 rpm at 20 or 15°C, respectively. Each datum point represents the average for samples obtained daily in duplicate from three flasks and counted on a standard hemocytometer. Growth rates were expressed as doubling times in hours and were calculated from the daily counts from inoculation until the first day on which the stationary phase was approached.
Cell size.
The cells were induced to assume a spherical shape as described previously (56). The cells were then allowed to settle in a Bionique Laboratories (Saranac Lake, N.Y.) chamber, and images were recorded by using a Nikon inverted microscope equipped with a Mighty Max cooled charge-coupled device camera (Princeton, Trenton, N.J.). The major and minor axes of each cell were measured by using IP Lab image analysis software (Scanalytics, Fairfax, Va.). The diameter of each cell was determined by averaging the major and minor axes. Images were recorded by using a Zeiss IM35 microscope equipped with an MTI RC300 cooled charge-coupled device camera (Scion Corp., Frederick, Md.). The perimeter of each cell was determined by using DIAS software (Solltech, Iowa City, Iowa).
Immunofluorescence.
Dictyostelium cells were fixed for 20 min in 3.7% formaldehyde in 17 mM phosphate buffer containing 1 mM CaCl2 (pH 6.1) and permeabilized in acetone at −20°C for 2 min as described previously (5). Cells were stained as described previously (5). Rhodamine-labeled or Oregon green-labeled phalloidin (Molecular Probes, Eugene, Oreg.) was used to localize F-actin, and monoclonal antibody B2C (15) followed by a rhodamine-labeled or a fluorescein-labeled secondary antibody (Sigma) was used to localize the 34-kDa protein.
Electron microscopy.
Dictyostelium cells were fixed for transmission electron microscopy as described previously (49). Cells were embedded in Epon 812 and sectioned by using an RMC 5000 ultramicrotome (RMC Products, Boeckeler Instruments, Inc., Tucson, Ariz.) with a diamond knife. Micrographs were obtained with a Philips 400 transmission electron microscope.
Expression of the 34-kDa ΔEF1 protein in murine L cells.
The sequences encoding the 34-kDa ΔEF1 protein were subcloned into the BamHI site of vector pEGFPN1 (Clontech, Palo Alto, Calif.) following amplification by PCR with custom oligonucleotides. The vector was designed to express the 34-kDa ΔEF1 protein fused to the enhanced green fluorescent protein (EGFP) at the carboxyl terminus. The coding sequence was confirmed by automated DNA sequencing. L-cell fibroblasts were maintained in RPMI 1640 supplemented with 15% bovine serum, 100 U of penicillin-streptomycin/ml, and 2 mM l-glutamine at 37°C in 5% CO2. The cells were plated at 105 cells/22-mm2 coverslip (50 to 60% confluence) and transfected with Lipofectamine Plus (Life Technologies, Rockville, Md.) according to the manufacturer's protocol. The cells were transfected with no DNA (mock) or plasmid p34-kDa ΔEF1-EGFP. The cells were fixed 24 h following replacement of the medium in the transfected cell culture. Coverslips were rinsed in phosphate-buffered saline (PBS), transferred to 3.7% formaldehyde in PBS for 25 min, washed briefly three times in PBS, permeabilized in 0.1% Triton X-100 in PBS for 5 min, washed briefly three times in PBS, blocked in 1% bovine serum albumin in PBS for 1 h, stained with rhodamine-labeled phalloidin for 1 h, washed briefly three times in PBS, and mounted.
RESULTS
Production of a 34-kDa protein with mutations in the first EF hand.
The sequence of the Dictyostelium 34-kDa protein (7) contains two putative calcium binding EF hands, structural motifs involved in the coordination of calcium in a number of calcium binding proteins (68). We produced an altered form of the protein by converting the aspartate residues at the X, Y, and Z positions of the first EF hand (68) to alanines (Fig. 1A) and termed this protein 34-kDa ΔEF1. The wild-type 34-kDa protein and the 34-kDa ΔEF1 protein were expressed and purified from E. coli (Fig. 1B) and then used for studies of calcium binding and actin binding in vitro.
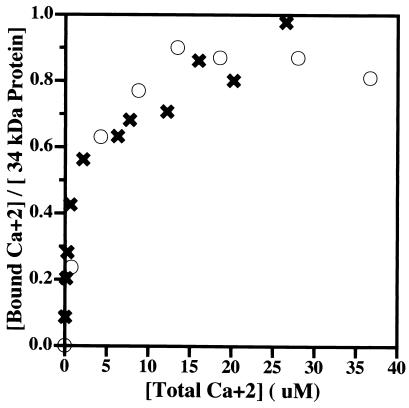
Prior studies of calcium binding to the 34-kDa protein with [45Ca]CaCl2 and equilibrium dialysis to determine the number and affinity of calcium binding sites in the 34-kDa protein revealed that calcium binds saturably to a single site on the 34-kDa protein with an affinity of 2.4 μM (18). Calcium binding to the wild-type 34-kDa protein and that to the 34-kDa ΔEF1 protein were indistinguishable, showing that the first EF hand does not participate in calcium binding to the 34-kDa protein (Fig. 2).
FIG. 2.
Calcium binding to the wild-type 34-kDa and 34-kDa ΔEF1 proteins, as measured by equilibrium dialysis. Approximately 10 μM concentrations of the wild-type 34-kDa and 34-kDa ΔEF1 proteins were dialyzed as a function of the [45Ca]CaCl2 concentration in the buffer as described in Materials and Methods. The radioactive calcium bound is plotted against the free calcium concentration for each protein. Symbols: ○, wild-type 34-kDa protein; ✖, 34-kDa ΔEF1 protein. The 34-kDa protein is reported to bind 1 mol of calcium with a Kd of 2.4 μM (18). The binding of calcium to the wild-type 34-kDa protein and that to the 34-kDa ΔEF1 protein were indistinguishable. The results are representative of independent experiments with two different preparations of the 34-kDa ΔEF1 protein.
The actin binding and cross-linking activities of the purified 34-kDa ΔEF1 protein were compared to those of the wild-type 34-kDa protein by using F-actin cosedimentation assays to assess actin binding. All assays were performed in the presence or absence of micromolar free calcium to investigate calcium regulation of actin binding. The wild-type 34-kDa protein bound to F-actin substoichiometrically and with a moderate affinity in a cosedimentation assay at low calcium ion concentrations (Fig. 3A). In the presence of micromolar calcium, the amount of wild-type 34-kDa protein bound to F-actin was reduced by 70% (Fig. 3B). The wild-type protein formed tight bundles with F-actin in the absence but not in the presence of micromolar calcium (data not shown), in agreement with previous reports of calcium-regulated actin binding by native and recombinant 34-kDa proteins (5, 8, 16, 34).
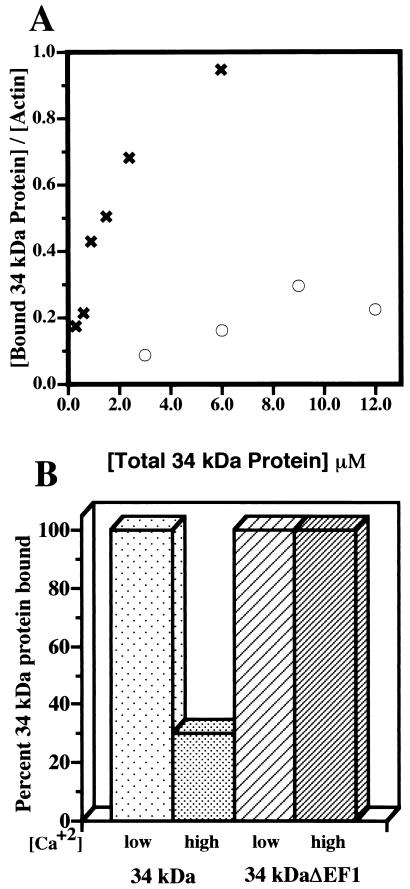
FIG. 3.
F-actin binding to the wild-type 34-kDa and 34-kDa ΔEF1 proteins, as measured by cosedimentation. (A) Moles of 34-kDa protein bound per mole of actin subunits in filaments plotted as a function of the total amount of 34-kDa protein added at a low calcium concentration. Symbols: ○, wild-type 34-kDa protein; ✖, 34-kDa ΔEF1 protein. The 34-kDa ΔEF1 protein binds F-actin more strongly than the wild-type 34-kDa protein. (B) Supernatant and pellet fractions of mixtures of actin and either the wild-type 34-kDa protein or the 34-kDa ΔEF1 protein at high and low calcium concentrations. The binding of the 34-kDa ΔEF1 protein was not inhibited by the presence of micromolar calcium concentrations.
The actin binding activity of the 34-kDa ΔEF1 protein was stronger than that of the wild-type 34-kDa protein at low calcium ion concentrations (Fig. 3A). However, the 34-kDa ΔEF1 protein failed to show calcium-sensitive actin binding, since virtually identical amounts of this protein bound to F-actin in the presence and in the absence of micromolar calcium (Fig. 3B). Thus, this protein shows normal calcium binding, but the calcium binding is no longer coupled to the inhibition of actin binding. Similarly, the 34-kDa ΔEF1 protein induced the formation of large tangled actin bundles in both the presence and the absence of micromolar calcium (data not shown). Thus, actin binding by the 34-kDa ΔEF1 protein was stronger than that of the wild-type 34-kDa protein and was calcium insensitive.
Expression and localization of wild-type and mutant forms of the 34-kDa protein in Dictyostelium.
The wild-type 34-kDa protein and the 34-kDa ΔEF1 protein were expressed in Dictyostelium cells lacking the 34-kDa protein and cells lacking both the 34-kDa protein and α-actinin, and the levels of expression of these proteins were examined by Western blotting with mouse monoclonal antibody B2C to the 34-kDa protein. Expression levels were similar to those observed for the wild-type protein in AX2 cells (Fig. 1C).
The 34-kDa protein-null cells expressing the full-length 34-kDa protein exhibited localization of F-actin and the 34-kDa protein at the leading edge and the cell cortex of vegetative amoebae (Fig. 4). The actin localization pattern in cells expressing the 34-kDa protein was the same as that in wild-type AX2 cells and largely overlapped the pattern of localization of the 34-kDa protein.
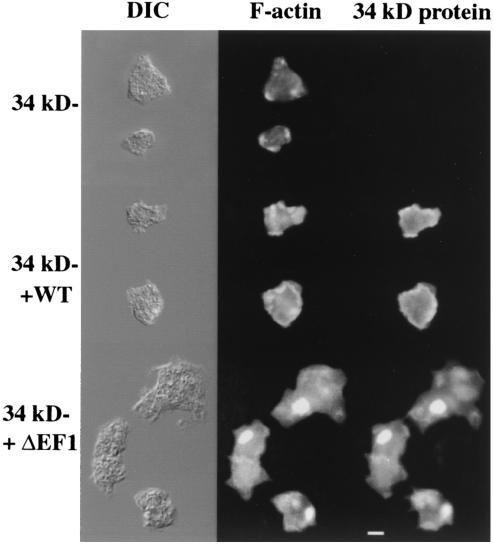
FIG. 4.
Hirano bodies in 34-kDa ΔEF1 protein-expressing cells observed by fluorescence microscopy. Actin and the 34-kDa protein were localized in 34-kDa protein-null cells and in 34-kDa protein-null cells expressing recombinant 34-kDa and 34-kDa ΔEF1 proteins. Cells were observed by differential interference contrast (DIC) microscopy (left column), stained for F-actin with rhodamine-labeled phalloidin (middle column), or stained for the localization of the 34-kDa protein with monoclonal antibody B2C (right column). Cells expressing the 34-kDa ΔEF1 protein exhibited ellipsoidal regions with high concentrations of F-actin and the 34-kDa ΔEF1 protein. WT, wild type. Bar, 10 μM.
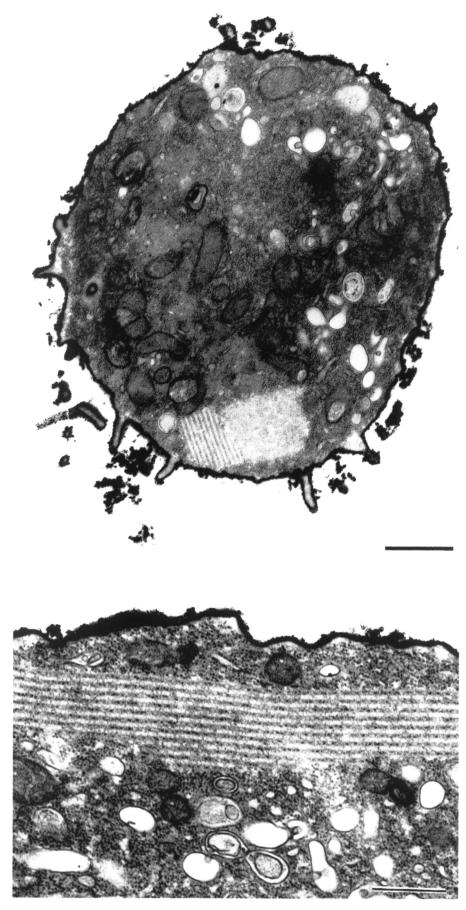
Cells expressing the 34-kDa ΔEF1 protein produced large ellipsoidal inclusions that were labeled strongly with rhodamine-labeled phalloidin and with anti-34-kDa protein monoclonal antibody B2C (Fig. 4). Under optimal conditions, up to 90% of the cells in the cultures expressing the 34-kDa ΔEF1 protein showed the presence of a large actin aggregate, the Hirano body. Examination of these cells by transmission electron microscopy revealed the presence of inclusions not observed in normal Dictyostelium cells. These structures had stacked layers of strata and juxtaposition of ordered and disordered regions, were spatially continuous with the cytosol, lacked a limiting membrane, and excluded membrane-bound organelles (Fig. 5). These features are characteristic of Hirano bodies described previously in studies of autopsy samples of tissue from human and animal disease models (24).
FIG. 5.
Hirano bodies in 34-kDa ΔEF1 protein-expressing cells observed by transmission electron microscopy. The 34-kDa ΔEF1 protein-expressing cells were prepared for electron microscopy as described in Materials and Methods. Note the ordered regions with alternating layers of stacked strata and the presence of disordered regions adjacent to the ordered regions. Bar, 0.5 μm.
Effect of Hirano bodies on the growth and size of Dictyostelium cells.
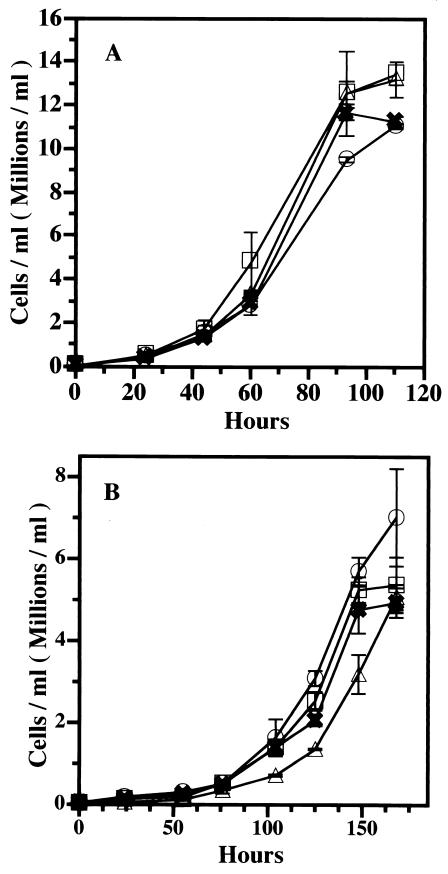
Growth and division are the result of the culmination of a number of processes involving the actin cytoskeleton. To assess directly the significance of the formation of Hirano bodies for cell function, the rates of growth of cells expressing the wild-type 34-kDa protein or the 34-kDa ΔEF1 protein were examined (Fig. 6A). Under standard laboratory growth conditions at 20°C, 34-kDa protein-null cells grew at rates similar to those of wild-type AX2 cells (12.1 h per division), as reported previously (56, 57). The 34-kDa protein-null cells expressing the wild-type and 34-kDa ΔEF1 proteins grew at similar rates (13.3 and 12.6 h per division, respectively). Thus, the presence of Hirano bodies in up to 90% of the cells had no apparent effect on growth rates.
FIG. 6.
Growth curves for 34-kDa protein-null amoebae expressing the wild-type 34-kDa and 34-kDa ΔEF1 proteins. Cells were grown axenically in shaking cultures, and cell density was measured as a function of time after inoculation. Symbols: □, AX2; ▵, 34-kDa protein-null cells; ○, recombinant 34-kDa protein-expressing cells; ✖, recombinant 34-kDa ΔEF1 protein-expressing cells. Error bars indicate standard deviations. (A) The 34-kDa protein-null cells grew as well as the wild-type cells (AX2) at 20°C. The expression of recombinant wild-type 34-kDa or 34-kDa ΔEF1 protein had no effect on growth under these conditions. (B) The 34-kDa protein-null cells grew more slowly than the wild-type cells at 15°C. The expression of recombinant wild-type 34-kDa or 34-kDa ΔEF1 protein restored the normal growth rate to 34-kDa protein-null cells at 15°C.
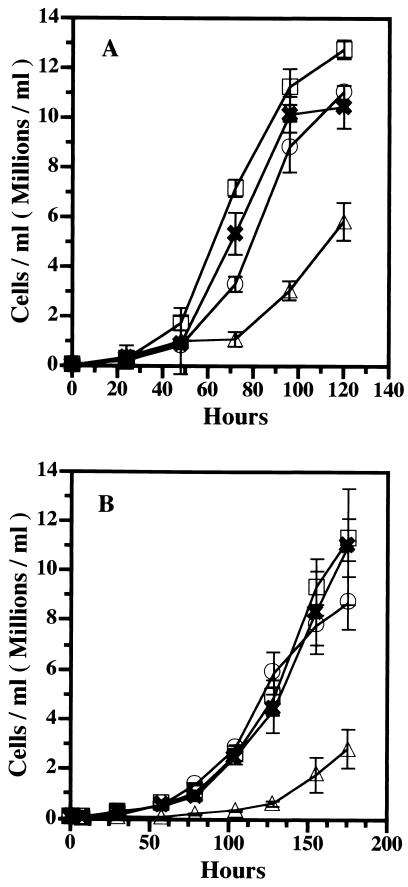
Cells lacking both the 34-kDa protein and α-actinin grew slowly at 20°C compared to AX2 cells (16.5 versus 12.7 h per division), as reported previously (56). The expression of the wild-type 34-kDa protein or the 34-kDa ΔEF1 protein in these cells restored the wild-type growth rate (12.3 or 12.1 h per division, respectively). Thus, the 34-kDa ΔEF1 protein could supply the function of the 34-kDa protein to cells lacking both the 34-kDa protein and α-actinin, even though its expression induced the formation of Hirano bodies in these cells (Fig. 7A).
FIG. 7.
Growth curves for amobae lacking both the 34-kDa protein and α-actinin and rescued with the wild-type 34-kDa and 34-kDa ΔEF1 proteins. Cells were grown axenically in shaking cultures, and cell density was measured as a function of time after inoculation. Symbols: □, AX2; ▵, cells lacking both the 34-kDa protein and α-actinin; ○, recombinant 34-kDa protein-expressing cells; ✖, recombinant 34-kDa ΔEF1 protein-expressing cells. Error bars indicate standard deviations. The cells lacking both the 34-kDa protein and α-actinin grew more slowly than the wild-type cells at both 20°C (A) and 15°C (B). The expression of the wild-type 34-kDa or 34-kDa ΔEF1 protein restored wild-type growth at both 20 and 15°C.
The ability of the 34-kDa ΔEF1 protein to function in vivo was also tested by measurements of cell diameters. Cells lacking both the 34-kDa protein and α-actinin grew to reduced sizes in shaking cultures, attaining an average size (mean and standard deviation) of 9.3 ± 1.9 μm, compared to 12.8 ± 1.9 μm for AX2 cells (significantly different at an α value of 0.01, as determined by the Mann-Whitney-Wilcoxon test) (57). The expression of the wild-type 34-kDa protein restored the cell size to the wild-type level (11.2 ± 2.6 μm; significantly different, at an α level of 0.01, from the value for cells lacking both the 34-kDa protein and α-actinin, but not significantly different, at an α level of 0.05, from the value for AX2 cells). Cells expressing the 34-kDa ΔEF1 protein had an average size of 10.4 ± 2.3 μm (significantly different, at an α level of 0.01, from the value for cells lacking both the 34-kDa protein and α-actinin but not significantly different from that for AX2 cells). Thus, the 34-kDa ΔEF1 protein supplied function to cells lacking both the 34-kDa protein and α-actinin and partially restored wild-type cell size.
Previous studies demonstrated that the CT fragment of the 34-kDa protein also induces the formation of Hirano bodies in either wild-type or 34-kDa protein-null Dictyostelium cells (42). The ability of the CT fragment to provide 34-kDa protein function was tested by expression of the CT fragment in cells lacking both the 34-kDa protein and α-actinin. These cells did exhibit Hirano bodies, as revealed by staining with rhodamine-labeled phalloidin or monoclonal antibody B2C, reactive with the 34-kDa protein (data not shown). Further, expression of the CT fragment did restore normal cell size (13.6 ± 2.0 μm) to cells lacking both the 34-kDa protein and α-actinin.
In its natural environment, Dictyostelium must grow under conditions of various temperatures, osmolarities, and nutrient supplies that are not obtained in standard laboratory culture conditions. To provide an environmental challenge, we grew the cells in shaking cultures at 15°C. The 34-kDa protein-null cells grew slowly (23.2 h per division) (57) compared to wild-type AX2 cells (20.1 h per division) at 15°C. When the wild-type 34-kDa protein was expressed, the wild-type growth rate was nearly restored (21.5 h per division) under these conditions. The expression of the 34-kDa ΔEF1 protein stimulated the rate of growth of the 34-kDa protein-null cells to 22.5 h per division. The cells expressing the 34-kDa ΔEF1 protein grew slightly more slowly than the 34-kDa protein-null cells rescued with the wild-type 34-kDa protein but showed a partial recovery of the function of the 34-kDa protein (Fig. 6B).
The effect of reduced temperature on growth was more pronounced in cells lacking both the 34-kDa protein and α-actinin, which grew extremely slowly at 15°C (28.7 h per division). The expression of the wild-type 34-kDa protein restored the wild-type growth rate (21.4 h per division). The expression of the 34-kDa ΔEF1 protein increased the rate of growth of the mutant lacking both the 34-kDa protein and α-actinin at 15°C to the wild-type level (21.1 h per division) (Fig. 7B).
Formation of Hirano bodies in murine L-cell fibroblasts.
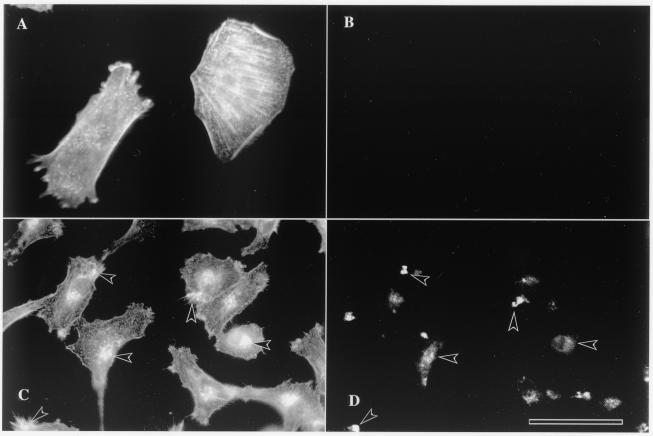
The actin distribution in murine L cells expressing the 34-kDa ΔEF1 protein was investigated to determine whether the results obtained for Dictyostelium might be extended to a mammalian cell culture system. Control (mock-transfected) fibroblasts had a typical F-actin distribution characterized by cortical filaments, filament bundles, and a few punctate foci (Fig. 8A) and no fluorescence from 34-kDa ΔEF1-EGFP (Fig. 8B). Cells expressing 34-kDa ΔEF1-EGFP exhibited a loss of stress fibers and cortical F-actin (Fig. 8C) and accumulated multiple punctate foci and large aggregates of 34-kDa ΔEF1-EGFP (Fig. 8D). Some of the foci and aggregates of 34-kDa ΔEF1-EGFP showed clear enrichment for actin filaments.
FIG. 8.
Formation of Hirano bodies in murine L cells expressing the 34-kDa ΔEF1 protein. The distribution of F-actin was determined by staining with rhodamine-labeled phalloidin (A and C) as described in Materials and Methods. The distribution of the 34-kDa ΔEF1 protein was determined by fluorescence from enhanced green fluorescent protein (EGFP) (B and D). (A) Mock-transfected control cells displayed cortical actin, stress fibers, and a few punctate foci of actin. (B) Mock-transfected cells showed no fluorescence from GFP, as expected. (C) Cells transfected with 34-kDa ΔEF1- EGFP exhibited a loss of stress fibers and an accumulation of punctate foci and large aggregates of F-actin. (D) Cells transfected with 34-kDa ΔEF1- EGFP exhibited punctate foci and large ellipsoidal inclusions that resembled Hirano bodies. Many of the foci and aggregates of 34-kDa ΔEF1- EGFP were also enriched for actin filaments (arrowheads). Scale bar, 10 μm.
DISCUSSION
Hirano bodies have been described in postmortem examination of tissue from patients with a variety of neurodegenerative diseases and other conditions producing persistent injury or stress, including diabetes and chronic alcoholism (24). Since nearly all prior studies of Hirano bodies were performed with fixed tissue and/or autopsy material, cell culture and transgenic models are required for studies of the effects of Hirano bodies on living cells. A truncated form of the Dictyostelium 34-kDa actin bundling protein termed the CT fragment (amino acids 124 to 295) induces the formation of Hirano bodies in Dictyostelium (42). We have used this cell culture model for studies of the formation of Hirano bodies and their effects on cell physiology. Mutations in both of the putative EF hands of the 34-kDa protein were produced to investigate the basis of calcium binding, the nature of the regulation of the activity of the 34-kDa protein, and the requirements for the initiation of the formation of Hirano bodies.
The biochemical properties of the 34-kDa ΔEF1 protein are novel and unexpected compared to those of the wild-type 34-kDa protein, CT fragment, and the 34-kDa ΔEF2 protein (Table 1). Point mutations in the first EF hand do not affect calcium binding (Fig. 2), while the 34-kDa ΔEF2 protein, with point mutations in the second EF hand, exhibits reduced calcium binding (18), consistent with the interpretation that the second EF hand is the site of high-affinity calcium binding to the 34-kDa protein. It is surprising that the 34-kDa ΔEF1 protein shows activated actin binding, even though it is a full-length protein containing the amino-terminal inhibitory domain and a functional calcium binding EF hand (Fig. 3). The truncated CT protein exhibited activated actin binding that was calcium insensitive, an effect that was attributed to the presence of the strong actin binding site at amino acids 193 to 254 and the absence of the amino-terminal inhibitory domain at amino acids 1 to 77 (35, 36). We conjecture that the mutations introduced in the 34-kDa ΔEF1 protein to probe the calcium binding function of the first EF hand actually disrupted the function of the first intramolecular interaction zone (IZ-1), which was mapped to the same region of the protein (36). Thus, it seems most likely that disruption of the intramolecular interaction zone by the mutations in the first EF hand may have activated actin binding, since the inhibitory region is no longer in juxtaposition with the strong actin binding site. A concomitant change is that actin binding is no longer subject to regulation by calcium binding to the intact second EF hand in the 34-kDa ΔEF1 protein (Fig. 3), even though this protein exhibits perfectly normal calcium binding (Fig. 2). Thus, calcium binding is no longer coupled to the inhibition of actin binding in the 34-kDa ΔEF1 protein. This result suggests strongly that calcium regulation of the wild-type 34-kDa protein involves a change in the proximity and/or orientation of the inhibitory region with respect to the strong binding site that is induced by the binding of calcium to the second EF hand. Thus, disruption of the intramolecular interaction zone in the 34-kDa ΔEF1 protein not only activates actin binding but also impedes the regulation of actin binding by calcium.
TABLE 1.
Properties of wild-type and altered forms of the 34-kDa protein in vitro and in vivoa
| Form of 34-kDa protein (amino acids) | Actin binding affinity | Calcium binding | Ca2+-regulated actin binding | Supplies 34-kDa protein function to mutants | Induces Hirano bodies |
|---|---|---|---|---|---|
| Wild type (1-295) | 0.1 μM (35) | 1 mol/mol; Kd, 2.4 μM (18) | Yes (36) | Yes (56, 57) | No (42) |
| CT fragment (124-295) | Activated (36) | None (36) | No (36) | Yes (see text) | Yes (42) |
| 34-kDa ΔEF1 (1-295) | Activated (Fig. 3) | Wild type (Fig. 2) | No (Fig. 3) | Yes (Fig. 6 and 7; see text) | Yes (Fig. 4 and 5) |
| 34-kDa ΔEF2 (1-295) | Wild type (18) | None (18) | No (18) | No (18) | No |
Unless otherwise indicated, items in parentheses indicate reference or sources of the data reported.
The expression of the 34-kDa ΔEF1 protein in Dictyostelium induces formation of Hirano bodies (Fig. 4 and 5). The Hirano bodies induced by the expression of the 34-kDa ΔEF1 protein are similar to the Hirano bodies seen in autopsy samples of brain tissue on the basis of the following criteria: (i) the Hirano bodies are enriched for actin and actin binding proteins (19, 22, 25, 27, 39, 61); (ii) the structures are highly ordered and present elliptical cross-sections that are rod or disk shaped, depending on the overall length (24, 25); (iii) the appearance varies with the plane of section or tilt of the stage due to the paracrystalline order of the structure (62, 69); (iv) the inclusions frequently reveal juxtaposition of ordered and disordered regions (53, 62); and (v) the structures contain strata of intersecting or interwoven parallel filament arrays (62). Hirano bodies are distinct from actin-cofilin rods, which are needle-like structures composed of a single large bundle of longitudinally oriented actin filaments that can form either in the nucleus or in the cytoplasm. These rods have been observed both in Dictyostelium (14, 58, 59) and in mammalian cells (14, 43, 47, 50) but lack the hallmark ultrastructural features of Hirano bodies enumerated above.
Our studies of the effects of the expression of the 34-kDa ΔEF1 protein in cells were also quite informative regarding the signal required to induce the formation of Hirano bodies. First, the 34-kDa ΔEF1 protein induced the formation of Hirano bodies both in Dictyostelium (Fig. 4 and 5) and in mammalian cells (Fig. 8). The 34-kDa ΔEF1 protein and CT fragment, which induce the formation of Hirano bodies, both exhibit activated actin binding that is calcium insensitive. Thus, this combination of properties can perturb the dynamics of the actin cytoskeleton to induce the formation of Hirano bodies. Interestingly, the 34-kDa ΔEF2 protein, which shows normal, albeit calcium-insensitive, actin binding, did not induce the formation of Hirano bodies (Table 1). Thus, loss of calcium regulation alone does not provide a signal for the formation of Hirano bodies.
Finally, our studies of cells expressing the 34-kDa ΔEF1 protein reveal clearly that cells with Hirano bodies are not dying and are quite healthy. The 34-kDa ΔEF1 protein-expressing cells, up to 90% of which contained Hirano bodies, grew identically to wild-type cells in shaking cultures at 20°C. Further, the expression of the 34-kDa ΔEF1 protein or the CT protein, with the concomitant formation of Hirano bodies, did supply function to cells lacking the 34-kDa protein (Fig. 6) and to cells lacking both the 34-kDa protein and α-actinin (Fig. 7 and Table 1; see also Results). In contrast, the 34-kDa ΔEF2 protein did not supply function to cells lacking both the 34-kDa protein and α-actinin and was inhibitory to the growth of cells lacking the 34-kDa protein (18).
The abilities of the 34-kDa ΔEF1 and CT proteins to simultaneously induce the formation of Hirano bodies in which they are highly enriched and provide the function of the 34-kDa protein to mutant cells pose an apparent paradox. The results can be explained by proposing that the formation of Hirano bodies effectively removes the mutant form of the 34-kDa protein from the dynamic cortical cytoskeleton. Thus, the 34-kDa ΔEF2 protein, which remains dispersed throughout the actin cytoskeleton, is inhibitory, revealing that calcium regulation of actin filament cross-linking is required in vivo (18). Moreover, the expression of high levels of the CT fragment of the 34-kDa protein is apparently toxic to Dictyostelium (42). Thus, the levels of 34-kDa proteins that have activated actin binding and that are not sequestered in Hirano bodies must be carefully regulated. The 34-kDa ΔEF1 protein is largely sequestered in Hirano bodies (Fig. 4) and so is not inhibitory to the function of the cytoskeleton. However, since the 34-kDa ΔEF1 protein has activated actin binding, a small amount of this protein may rescue the phenotypes of 34-kDa protein-null cells.
In summary, our results show that the failure to regulate the activity and/or affinity of an actin cross-linking protein can provide a signal for the formation of Hirano bodies. More generally, the formation of Hirano bodies is a general response of eucaryotic cells to or a consequence of aberrant function of the actin cytoskeleton. Hirano bodies cannot be regarded as “non-specific manifestations of neuronal degeneration” (62) or a “non-specific finding largely devoid of cytopathologic significance” (20). The formation of Hirano bodies may sequester actin, actin-associated proteins, and other components that contribute to morphogenesis and to the plasticity of synaptic connections essential to neuronal function (38). In addition, Hirano bodies may modulate the formation of actin-cofilin rods, which may contribute to the loss of synapses in Alzheimer's disease by impeding axonal transport (43). Assessing the potential role of Hirano body formation as either an adaptive response or a step in the progression of disease presents an exciting challenge for future investigations.
Acknowledgments
This work was supported by grants from the National Science Foundation (MCB 98-08748) and the Alzheimer's Association.
REFERENCES
- 1.Anzil, A., H. Herrlinger, K. Blinzinger, and A. Heldrich. 1974. Ultrastructure of brain and nerve biopsy tissue in Wilson's disease. Arch. Neurol. 31:94-100. [DOI] [PubMed] [Google Scholar]
- 2.Buxbaum, J. N., and C. E. Tagoe. 2000. The genetics of the amyloidoses. Annu. Rev. Med. 51:543-569. [DOI] [PubMed] [Google Scholar]
- 3.Cartier, L., S. Galvez, and D. C. Gajdusek. 1985. Familial clustering of the ataxic form of Creutzfeldt-Jakob disease with Hirano bodies. J. Neurol. Neurosurg. Psychiatry 48:234-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dobson, C. M. 2001. Protein folding and its links with human disease. Biochem. Soc. Symp. 68:1-26. [PubMed] [Google Scholar]
- 5.Fechheimer, M. 1987. The Dictyostelium discoideum 30,000-dalton protein is an actin filament-bundling protein that is selectively present in filopodia. J. Cell Biol. 104:1539-1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fechheimer, M., and R. Furukawa. 1993. A 27,000 dalton core of the Dictyostelium 34,000 dalton protein retains Ca+2-regulated actin cross-linking but lacks bundling activity. J. Cell Biol. 120:1169-1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fechheimer, M., D. Murdock, M. Carney, and C. V. C. Glover. 1991. Isolation and sequencing of cDNA clones encoding the Dictyostelium discoideum 30,000 dalton actin bundling protein. J. Biol. Chem. 266:2883-2889. [PubMed] [Google Scholar]
- 8.Fechheimer, M., and D. L. Taylor. 1984. Isolation and characterization of a 30,000-dalton calcium-sensitive actin cross-linking protein from Dictyostelium discoideum. J. Biol. Chem. 259:4514-4520. [PubMed] [Google Scholar]
- 9.Field, E. J., J. D. Mathews, and C. S. Raine. 1969. Electron microscopic observations on the cerebellar cortex in kuru. J. Neurol. Sci. 8:209-224. [DOI] [PubMed] [Google Scholar]
- 10.Field, E. J., and H. K. Narang. 1972. An electron-microscopic study of scrapie in the rat: further observations on “inclusion bodies”and virus-like particles. J. Neurol. Sci. 17:347-364. [DOI] [PubMed] [Google Scholar]
- 11.Fisher, E. R., A. R. Gonzalez, R. C. Khurana, and T. S. Danowski. 1972. Unique, concentrically laminated, membranous inclusions in myofibers. Am. J. Clin. Pathol. 58:239-244. [DOI] [PubMed] [Google Scholar]
- 12.Fu, Y., J. Ward, and H. F. Young. 1975. Unusual, rod-shaped cytoplasmic inclusions (Hirano bodies) in a cerebellar hemangioblastoma. Acta Neuropathol. 31:129-135. [DOI] [PubMed] [Google Scholar]
- 13.Fujiwara, H., M. Hasegawa, N. Dohmae, A. Kawashima, E. Masliah, M. S. Goldberg, J. Shen, K. Takio, and T. Iwatsubo. 2002. α-Synuclein is phosphorylated in synucleinopathy lesions. Nat. Cell Biol. 4:160-164. [DOI] [PubMed] [Google Scholar]
- 14.Fukui, Y., and H. Katsumaru. 1979. Nuclear actin bundles in Amoeba, Dictyostelium, and human HeLa cells induced by dimethyl sulfoxide. Exp. Cell Res. 120:451-455. [DOI] [PubMed] [Google Scholar]
- 15.Furukawa, R., S. Butz, E. Fleischmann, and M. Fechheimer. 1992. The Dictyostelium discoideum 30,000 dalton protein contributes to phagocytosis. Protoplasma 169:18-27. [Google Scholar]
- 16.Furukawa, R., and M. Fechheimer. 1996. Role of the Dictyostelium 30 kDa protein in actin bundle formation. Biochemistry 35:7224-7232. [DOI] [PubMed] [Google Scholar]
- 17.Furukawa, R., and M. Fechheimer. 1997. The structure, function, and assembly of actin filament bundles. Int. Rev. Cytol. 175:29-90. [DOI] [PubMed] [Google Scholar]
- 18.Furukawa, R., A. G. Maselli, S. A. M. Thomson, R. W.-L. Lim, J. V. Stokes, and M. Fechheimer. 2003. Calcium regulation of actin cross-linking is important for function of the actin cytoskeleton in Dictyostelium. J. Cell Sci. 116:187-196. [DOI] [PubMed] [Google Scholar]
- 19.Galloway, P. G., G. Perry, and P. Gambetti. 1987. Hirano body filaments contain actin and actin-associated proteins. J. Neuropathol. Exp. Neurol. 46:185-199. [DOI] [PubMed] [Google Scholar]
- 20.Gessaga, E. C., and A. P. Anzil. 1975. Rod-shaped filamentous inclusions and other ultrastructural features in a cerebellar astrocytoma. Acta Neuropathol. 33:119-127. [DOI] [PubMed] [Google Scholar]
- 21.Gibson, P. H., and B. E. Tomlinson. 1977. Numbers of Hirano bodies in the hippocampus of normal and demented people with Alzheimer's disease. J. Neurol. Sci. 33:199-206. [DOI] [PubMed] [Google Scholar]
- 22.Goldman, J. E. 1983. The association of actin with Hirano bodies. J. Neuropathol. Exp. Neurol. 42:146-152. [DOI] [PubMed] [Google Scholar]
- 23.Hadfield, M. G., A. J. Martinez, and R. C. Gilmartin. 1974. Progressive multifocal leukoencephalopathy with paramyxovirus-like structures, Hirano bodies and neurogibrillary tangles. Acta Neuropathol. (Berlin) 27:277-288. [DOI] [PubMed] [Google Scholar]
- 24.Hirano, A. 1994. Hirano bodies and related neuronal inclusions. Neuropathol. Appl. Neurobiol. 20:3-11. [DOI] [PubMed] [Google Scholar]
- 25.Hirano, A., H. M. Dembitzer, L. T. Kurland, and H. M. Zimmerman. 1968. The fine structure of some intraganglionic alterations. J. Neuropathol. Exp. Neurol. 27:167-182. [PubMed] [Google Scholar]
- 26.Hughes, R. E., and J. M. Olson. 2001. Therapeutic opportunities in polyglutamine disease. Nat. Med. 7:419-423. [DOI] [PubMed] [Google Scholar]
- 27.Izumiyama, N., K. Ohtsubo, T. Tachikawa, and H. Nakamura. 1991. Elucidation of three-dimensional ultrastructure of Hirano bodies by the quick-freeze, deep-etch and replica method. Acta Neuropathol. 81:248-254. [DOI] [PubMed] [Google Scholar]
- 28.Johnston, J. A., C. L. Ward, and R. R. Kopito. 1998. Aggresomes: a cellular response to misfolded proteins. J. Cell Biol. 143:1883-1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Koo, E. H., P. T. J. Lansbury, and J. W. Kelly. 1999. Amyloid diseases: abnormal protein aggregation in neurodegeneration. Proc. Natl. Acad. Sci. USA 96:9989-9990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kopito, R. R. 2000. Aggresomes, inclusion bodies, and protein aggregation. Trends Cell Biol. 10:524-530. [DOI] [PubMed] [Google Scholar]
- 31.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 227:680-685. [DOI] [PubMed] [Google Scholar]
- 32.Lass, R., and C. Hagel. 1994. Hirano bodies and chronic alcoholism. Neuropathol. Appl. Neurobiol. 20:12-21. [DOI] [PubMed] [Google Scholar]
- 33.Lee, V. M., M. Goedert, and J. Q. Trojanowski. 2001. Neurodegenerative tauopathies. Annu. Rev. Neurosci. 24:1121-1159. [DOI] [PubMed] [Google Scholar]
- 34.Lim, R. W. L., and M. Fechheimer. 1997. Overexpression, purification, and characterization of recombinant Dictyostelium discoideum calcium regulated 34,000 dalton F-actin bundling protein from Escherichia coli. Protein Expr. Purif. 9:182-190. [DOI] [PubMed] [Google Scholar]
- 35.Lim, R. W. L., R. Furukawa, S. Eagle, R. C. Cartwright, and M. Fechheimer. 1999. Three distinct F-actin binding sites in the Dictyostelium discoideum 34,000 dalton actin bundling protein. Biochemistry 38:800-812. [DOI] [PubMed] [Google Scholar]
- 36.Lim, R. W. L., R. Furukawa, and M. Fechheimer. 1999. Evidence of intramolecular regulation of the Dictyostelium discoideum 34,000 dalton F-actin bundling protein. Biochemistry 38:16323-16332. [DOI] [PubMed] [Google Scholar]
- 37.Loomis, W. F. 1971. Sensitivity of Dictyostelium discoideum to nucleic acid analogues. Exp. Cell Res. 64:484-486. [DOI] [PubMed] [Google Scholar]
- 38.Luo, L. 2002. Actin cytoskeleton regulation in neuronal morphogenesis and structural plasticity. Annu. Rev. Cell Dev. Biol. 18:601-635. [DOI] [PubMed] [Google Scholar]
- 39.Maciver, S. K., and C. R. Harrington. 1995. Two actin binding proteins, actin depolymerizing factor and cofilin, are associated with Hirano bodies. Neuroreport 6:1985-1988. [DOI] [PubMed] [Google Scholar]
- 40.MacLean-Fletcher, S. D., and T. D. Pollard. 1980. Identification of a factor in conventional muscle actin preparations which inhibits actin filament self-association. Biochem. Biophys. Res. Commun. 96:18-27. [DOI] [PubMed] [Google Scholar]
- 41.Mann, S. K. O., P. N. Devreotes, S. Eliott, K. Jermyn, A. Kuspa, M. Fechheimer, R. Furukawa, C. A. Parent, J. Segall, G. Shaulsky, P. H. Vardy, J. Williams, K. Williams, and R. A. Firtel. 1998. Cell biological, molecular genetic, and biochemical methods to examine Dictyostelium, p. 431-465. In J. E. Celis (ed.), Cell biology: a laboratory handbook, vol. 1. Academic Press, Inc., San Diego, Calif.
- 42.Maselli, A. G., R. Davis, R. Furukawa, and M. Fechheimer. 2002. Formation of Hirano bodies in Dictyostelium and mammalian cells induced by expression of a modified form of an actin cross-linking protein. J. Cell Sci. 115:1939-1952. [DOI] [PubMed] [Google Scholar]
- 43.Minamide, L. S., A. M. Striegl, J. A. Boyle, P. J. Meberg, and J. R. Bamburg. 2000. Neurodegenerative stimuli induce persistent ADF/cofilin-actin rods that disrupt distal neurite function. Nat. Cell Biol. 2:628-636. [DOI] [PubMed] [Google Scholar]
- 44.Mitake, S., K. Ojika, and A. Hirano. 1997. Hirano bodies and Alzheimer's disease. Kao-Hsiung I Hsueh K'o Hsueh Tsa Chih 13:10-18. [PubMed] [Google Scholar]
- 45.Mori, H., M. Tomonaga, N. Baba, and K. Kanaya. 1986. The structure analysis of Hirano bodies by digital processing on electron micrographs. Acta Neuropathol. 71:32-37. [DOI] [PubMed] [Google Scholar]
- 46.Nagara, H., K. Yajima, and K. Suzuki. 1980. An ultrastructural study on the cerebellum of the brindled mouse. Acta Neuropathol. (Berlin) 52:41-50. [DOI] [PubMed] [Google Scholar]
- 47.Nishida, E., K. Iida, and N. Yonezawa. 1987. Cofilin is a component of intranuclear and cytoplasmic actin rods induced in cultured cells. Proc. Natl. Acad. Sci. USA 84:5262-5266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Noegel, A. A., and M. Schleicher. 2000. The actin cytoskeleton of Dictyostelium: a story told by mutants. J. Cell Sci. 113:759-766. [DOI] [PubMed] [Google Scholar]
- 49.Novak, K. D., M. D. Peterson, M. C. Reedy, and M. A. Titus. 1995. Dictyostelium myosin I double mutants exhibit conditional defects in pinocytosis. J. Cell Biol. 131:1205-1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ono, S., H. Abe, R. Nagaoka, and T. Obinata. 1993. Colocalization of ADF and cofilin in intranuclear rods of cultured muscle cells. J. Muscle Res. Cell Motil. 14:195-204. [DOI] [PubMed] [Google Scholar]
- 51.Orr, H. T. 2001. Beyond the Q's in the polyglutamine diseases. Genes Dev. 15:925-932. [DOI] [PubMed] [Google Scholar]
- 52.Ostrow, B. D., P. X. Chen, and R. L. Chisholm. 1994. Expression of a myosin regulatory light-chain phosphorylation site mutant complements the cytokinesis and developmental defects of Dictyostelium RMLC null cells. J. Cell Biol. 127:1945-1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Peterson, C., K. Suzuki, Y. Kress, and J. E. Goldman. 1986. Abnormalities of dendritic actin organization in the brindled mouse. Brain Res. 382:205-212. [DOI] [PubMed] [Google Scholar]
- 54.Price, D. L., and S. S. Sisodia. 1998. Mutant genes in familial Alzheimer's disease and transgenic models. Annu. Rev. Neurosci. 21:479-505. [DOI] [PubMed] [Google Scholar]
- 55.Putka, F., and C. Zeng. 1998. Blasticidin resistance cassette in symmetrical polylinkers for insertional inactivation of genes in Dictyostelium. Folia Biol. 44:185-188. [PubMed] [Google Scholar]
- 56.Rivero, F., R. Furukawa, A. A. Noegel, and M. Fechheimer. 1996. Dictyostelium discoideum cells lacking the 34,000 dalton actin binding protein can grow, locomote, and develop, but exhibit defects in regulation of cell structure and movement: a case of partial redundancy. J. Cell Biol. 135:965-980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rivero, F., R. Furukawa, A. A. Noegel, and M. Fechheimer. 1999. Dictyostelium mutants lacking the 34 kD bundling protein and alpha-actinin or gelation factor. J. Cell Sci. 112:2737-2751. [DOI] [PubMed] [Google Scholar]
- 58.Sameshima, M., Y. Chijiiwa, Y. Kishi, and Y. Hashimoto. 1994. Novel actin rods appeared in spores of Dictyostelium discoideum. Cell Struct. Funct. 19:189-194. [DOI] [PubMed] [Google Scholar]
- 59.Sameshima, M., Y. Kishi, M. Osumi, D. Mahadeo, and D. A. Cotter. 2000. Novel actin cytoskeleton: actin tubules. Cell Struct. Funct. 25:291-295. [DOI] [PubMed] [Google Scholar]
- 60.Schmidt, M. L., V. M. Lee, and J. Q. Trojanowski. 1989. Analysis of epitopes shared by Hirano bodies and neurofilament proteins in normal and Alzheimer's disease hippocampus. Lab. Investig. 60:513-522. [PubMed] [Google Scholar]
- 61.Schochet, S. S., Jr., P. W. Lampert, and R. Lindenberg. 1968. Fine structure of the Pick and Hirano bodies in a case of Pick's disease. Acta Neuropathol. (Berlin) 11:330-337. [DOI] [PubMed] [Google Scholar]
- 62.Schochet, S. S., Jr., and W. F. McCormick. 1972. Ultrastructure of Hirano bodies. Acta Neuropathol. 21:50-60. [DOI] [PubMed] [Google Scholar]
- 63.Selkoe, D. J. 1998. The cell biology of β-amyloid precursor protein and presenilin in Alzheimer's disease. Trends Cell Biol. 8:447-453. [DOI] [PubMed] [Google Scholar]
- 64.Sima, A. A., and D. Hinton. 1983. Hirano-bodies in the distal symmetric polyneuropathy of the spontaneously diabetic BB-Wistar rat. Acta Neurol. Scand. 68:107-112. [DOI] [PubMed] [Google Scholar]
- 65.Smith, P. K., R. I. Krohn, G. T. Hermanson, A. K. Mallia, F. H. Gartner, M. D. Provenzano, E. K. Fujimoto, N. M. Goeke, B. J. Olson, and D. C. Klenk. 1985. Measurement of protein using bicinchoninic acid. Anal. Biochem. 150:76-85. [DOI] [PubMed] [Google Scholar]
- 66.Spillantini, M. G., M. L. Schmidt, V. M.-Y. Lee, J. Q. Trojanowski, R. Jakes, and M. Goedert. 1997. α-Synuclein in Lewy bodies. Nature 388:839-840. [DOI] [PubMed] [Google Scholar]
- 67.Spudich, J. A., and S. Watt. 1971. The regulation of rabbit skeletal muscle contraction. J. Biol. Chem. 246:4866-4871. [PubMed] [Google Scholar]
- 68.Strynadka, N. C. J., and M. N. G. James. 1989. Crystal structures of the helix-loop-helix calcium binding proteins. Annu. Rev. Biochem. 58:951-998. [DOI] [PubMed] [Google Scholar]
- 69.Tomonaga, M. 1974. Ultrastructure of Hirano bodies. Acta Neuropathol. 28:365-366. [DOI] [PubMed] [Google Scholar]
- 70.Towbin, H., T. Staehelin, and J. Gordon. 1979. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl. Acad. Sci. USA 76:4350-4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Waggoner, D. J., T. B. Bartnikas, and J. D. Gitlin. 1999. The role of copper in neurodegenerative disease. Neurobiol. Dis. 6:221-230. [DOI] [PubMed] [Google Scholar]