Abstract
The effects of cholesterol (Chol) on phospholipid bilayers include ordering of the fatty acyl chains, condensing of the lipids in the bilayer plane, and promotion of the liquid-ordered phase. These effects depend on the type of phospholipids in the bilayer and are determined by the nature of the underlying molecular interactions. As for Chol, it has been shown to interact more favorably with sphingomyelin than with most phosphatidylcholines, which in given circumstances leads to formation of lateral domains. However, the exact origin and nature of Chol-phospholipid interactions have recently been subjects of speculation. We examine interactions between Chol, palmitoylsphingomyelin (PSM) and palmitoyl-oleoyl-phosphatidylcholine (POPC) in hydrated lipid bilayers by extensive atom-scale molecular dynamics simulations. We employ a tailored lipid configuration: Individual PSM and Chol monomers, as well as PSM-Chol dimers, are embedded in a POPC lipid bilayer in the liquid crystalline phase. Such a setup allows direct comparison of dimeric and monomeric PSMs and Chol, which ultimately shows how the small differences in PSM and POPC structure can lead to profoundly different interactions with Chol. Our analysis shows that direct hydrogen bonding between PSM and Chol does not provide an adequate explanation for their putative specific interaction. Rather, a combination of charge-pairing, hydrophobic, and van der Waals interactions leads to a lower tilt in PSM neighboring Chol than in Chol with only POPC neighbors. This implies improved Chol-induced ordering of PSM's chains over POPC's chains. These findings are discussed in the context of the hydrophobic mismatch concept suggested recently.
INTRODUCTION
The lipid bilayer is responsible for some remarkable physical properties of cellular membranes, for they are as tight and robust as they are thin and flexible (1,2). Membrane proteins in turn are responsible for specific membrane functions such as signaling, channeling, or cell recognition (1,2). Based on these fundamental roles of lipids and proteins in biological membranes, our views on their detailed molecular organization have been changing for the past decade.
The classical Singer-Nicolson model of membrane structure of 1972 (3) has proven to be highly useful but incomplete. It describes the lipid bilayer part of cell membranes as a uniform fluid phase, in which all membrane proteins dissolve and diffuse evenly (3). Yet, studies of model membranes show that bilayer mixtures of already a few different physiological lipids exhibit rather complex phase behavior (1,4). In particular, sphingolipids and other phospholipids with mostly saturated fatty acid residues can form a liquid-ordered (lo) phase that may coexist in the bilayer with a conformationally more disordered (ld) phase (1,5).
The formation of the lo phase is greatly facilitated by the presence of cholesterol (Chol), which partitions rather into an lo than an ld lipid environment (6–8). The coexistence of lo and ld phases inflict lateral fine structure on a lipid bilayer: Separate lo domains are known to form in the ld matrix of model membranes that mimic physiological conditions (6–8). Understandably, the phase behavior of real cell membrane lipid bilayers, which contain hundreds of different lipids (2,9), is even more complex and domain formation in them is not fully established (10,11).
In the case of lipid domains in cell membranes, as so often in biology, structural aspects are closely related to function: Their possible physiological consequences were first described in the lipid raft model, introduced by Simons and Ikonen in 1997 (12). In this model, certain membrane proteins were suggested to segregate in lo lipid domains, or rafts, while others are excluded from them (2,8,9,12–14). The earliest functions connected to lipid rafts were protein trafficking and cell signaling (12). Later on, raft lipids have been associated also with viral budding, prion diseases, and cancer, though clearcut evidence is lacking (11,13,14). Nevertheless, it is nowadays largely accepted that the functioning of proteins in membranes depends on their local membrane composition, which highlights the role of lipid membranes in the understanding of various cellular functions. For example, insulin receptor activity is greatly inhibited in kidney cells grown with desmosterol instead of Chol (15).
Full understanding of lipid domains and in particular raftlike ordered patches in cell membranes requires detailed knowledge of their properties in model membranes (6). Yet, the exact mechanisms of lo domain formation are largely unknown (6,7,14). While the presence of Chol is generally reckoned to be a necessary requirement for lo phase formation, Chol interactions with phospholipids have not been resolved in satisfying detail. In particular, Chol is thought to interact preferentially with sphingolipids such as sphingomyelin (SM)—a major component of lipid rafts—rather than with comparable glycerophospholipids. For instance, Chol fractions of 0.3 or more have been shown to reduce the bilayer lateral elasticity by up to 25% more in palmitoyl (16:0) or stearoyl (18:0) SM than in their chain-matched PC analogs myristoyl (14:0)-palmitoyl PC and myristoyl-stearoyl PC (16). The same study showed similar observations also in a monounsaturated case. What is more, the rate of Chol desorption from SM monolayers into a water-β-cyclodextrin solution is significantly slower than that from dipalmitoyl PC (17,18). In addition, several partitioning experiments indicate a preference of Chol for SM over acyl-chain matching PC (7).
The origin of this putative specific Chol-SM interaction has remained unresolved despite different attempts of explanation, including direct hydrogen bonding, hydrophobic mismatch, and lipid packing. SM features two potential hydrogen bond (H-bond) donors in the bilayer interfacial region, while PC features none. Consequently, the formation of direct H-bonds between SM donors and the Chol oxygen is a commonly proposed mechanism of SM–Chol interaction (8,16,19,20). An infrared spectroscopy study hinted that, after inclusion of Chol, SM H-bonding patterns change more than those of PC. These changes included a change in H-bonding of the SM amide group with water (21), possibly involving direct SM–Chol H-bonds. Other experiments suggest that Chol association with phospholipids is driven by minimization of hydrophobic mismatch (22,23) instead of any specific interactions. In that line of thought, Chol gathers at the interfaces of lipid bilayer regions of different hydrophobic thickness, regardless of the involved phospholipids. In this way, Chol would promote the formation of ordered patches by significantly reducing the associated line tension between the thicker (more ordered) and thinner (less ordered) bilayer regions. Another idea based on hydrophobic interactions is that phospholipid headgroups may help to shield bilayer Chols from unwanted water contact (the so-called umbrella model) (24). An experimental NMR comparison of Chol interactions with synthetic DPPC and natural brain SM (two phospholipid mixtures with similar main-chain transition temperatures) found only minor differences between both two-component systems (25). This lead the authors to the conclusion that, in actual cellular membranes, Chol preference for SM probably arises mainly from the higher saturation levels of SM compared to other phospholipids (25).
The case of Chol–SM interactions highlights why a thorough understanding of lipid-lipid interactions is crucial to the understanding of the basic principles of membrane function. In the past 10 years simulation methods have developed far enough to help tackle lipid membrane problems. Especially the atomic-level picture provided by molecular dynamics (MD) has become an important complementary means to the understanding of soft-matter systems (26–30). MD simulations should be well suited to shed light on some open questions about lo phases and lipid rafts, particularly regarding the atom-level mechanisms of phospholipid-Chol interactions and their implications on domain formation. First steps have already been taken in that direction (31–38).
This study continues to develop this idea: Atomic level MD simulations are analyzed for specific interactions of Chol, palmitoyl sphingomyelin (PSM, a typical raft lipid), and palmitoyl-oleoyl phosphatidylcholine (POPC). A special lipid setup was chosen to address the problem from a novel perspective: the dilute limit of only few SM and Chol molecules in a matrix of monosaturated glycerophospholipids. The analysis presented in this work focuses on specific, atomic level mechanisms of the interactions between the involved lipids. Therefore, it provides a foundation for further studies on actual lipid raft formation.
SIMULATION DETAILS
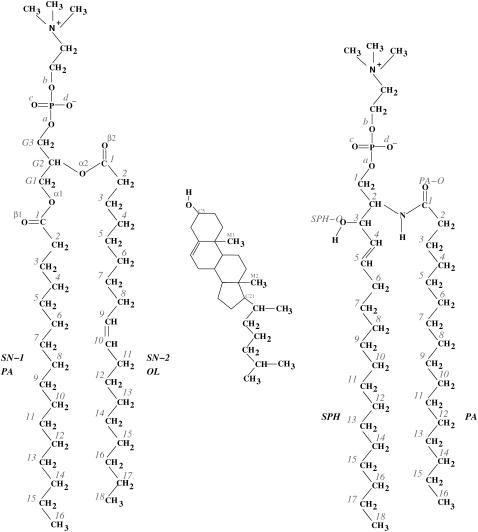
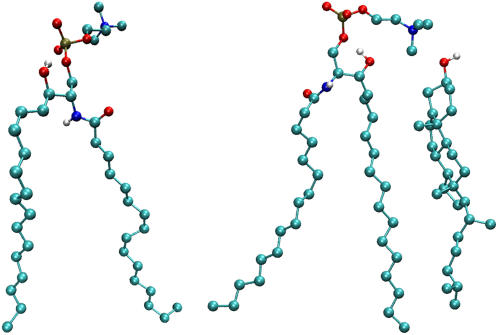
We have simulated a three-component lipid bilayer comprised of palmitoyl-sphingomyelin (PSM), palmitoyl-oleoyl-phosphatidylcholine (POPC), and cholesterol (Chol) in explicit water using the GROMACS package (39,40). Chemical structures of the involved lipids are shown in Fig. 1.
FIGURE 1.
Structures of palmitoyl-oleoyl-phosphatidylcholine (POPC, left), cholesterol (Chol, middle), and palmitoyl-sphingomyelin (PSM, right). Despite their broad structural similarities, POPC and PSM interact differently with Chol.
We employ a special lipid configuration tailored to study the problem of specific interactions: a matrix of 992 POPC lipids (496 per monolayer) embedded with PSM and Chol monomers (four each per monolayer) as well as PSM-Chol dimers (four dimers per monolayer). Thus, in total, this system features 1024 lipids in molar fractions of POPC/PSM/Chol 62:1:1. A snapshot of one monolayer of this system is shown in Fig. 2.
FIGURE 2.
Snapshot of one monolayer of the simulated system. For clarity, PSM (red) and Chol (blue) are rendered with a space-filling model, while the POPC matrix is rendered with sticks. In both monolayers, the system features four PSM monomers, four Chol monomers, and four PSM-Chol dimers.
Starting coordinates for the system were obtained by expanding a previously equilibrated POPC bilayer (41) to a total number of 1024 lipids. We replaced 32 selected POPC molecules to result in a POPC matrix with eight Chol–PSM dimers and 16 monomers that are as far away as possible from each other. The force-field parameters for POPC (42), PSM (35), and Chol (43) were obtained from previously published works. The system was fully hydrated with 27.8 SPC water molecules per lipid (44), resulting in a total of 138,147 atoms. Using GROMACS (39,40) for integrating the equations of motion with a 2 fs time step, the system was initially equilibrated with a Langevin thermostat in NVT-ensemble (for 50 ps) and then in NpT ensemble (for 500 ps). The first 5 ns of the actual simulation were run in NpT ensemble (T = 310 K, p = 1 atm) using Berendsen thermostat and barostat (45), after which we switched to the Nosé-Hoover thermostat (46,47) and Parrinello-Rahman (48,49) barostat to produce the correct ensemble. The chosen temperature of 310 K is well above the main phase transition temperature of POPC (Tm = 268 K (50)), the lipid that forms the bulk of the simulated bilayer. The pressure coupling was applied in a semi-isotropic way to result in zero surface tension. Long-range electrostatic interactions were accounted for by the reaction-field technique (with rc = 2.0 nm and εr = 80), whereas 1.0 nm cutoff was used for the Lennard-Jones interactions. Reaction-field has been shown to be a reliable method for simulating noncharged lipid bilayers, giving results that are comparable to those of the particle-mesh Ewald method (51). The total simulation time was 50 ns. For the analysis, we have considered the last 40 ns of the trajectory.
RESULTS
Structural and dynamic features
Before going into detailed analysis, we collect some overall properties of the system. Calculating them separately for PSMs and POPCs with and without Chol neighbor reveals first differences in phospholipid-Chol interactions.
Equilibration and area per molecule
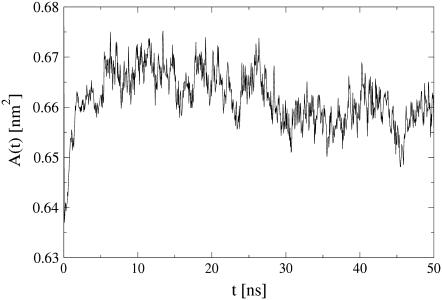
The area per molecule is a central structural quantity for lipid bilayers. In simulations, it is often used as a measure of system equilibration. Fig. 3 shows this quantity, calculated as the quotient of simulation box size over number of lipids per monolayer. Since the simulated composition is very close to the all-POPC system used as the starting structure, the system reached a dynamic equilibrium rather quickly, within 10 ns. After equilibration, the mean area per molecule attains a value of A = 0.66 ± 0.01 nm2. The original all-POPC structure featured an area per lipid of A = 0.69 nm2, a value that decreased quickly during the initial 50 ps stochastic simulation. A recent experimental study of pure POPC bilayers resulted in an area per lipid of A = 0.683 ± 0.015 nm2 at a temperature of 303 K (52). Thus, the POPC force field we use in this simulation reproduces the experimental area per lipid very well.
FIGURE 3.
Area per lipid versus time for the simulated bilayer.
Density profile
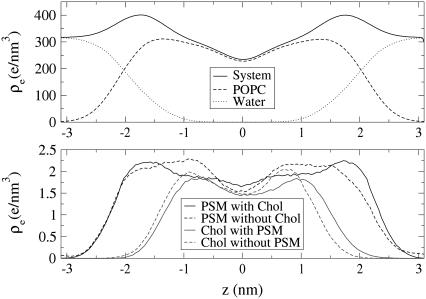
The electron density profile across the bilayer is shown in Fig. 4 separately for the different lipid species with or without specific nearest neighbors. More specifically, the density graphs of PSM and Chol in PSM–Chol pairs and those of PSM and Chol monomers are shown separately.
FIGURE 4.
Electron densities of different lipids with and without specific nearest neighbors.
The width of the bilayer, measured as the separation of the total electron density peaks, is 3.5 nm. To facilitate comparison to experiments of pure POPC bilayers, we also calculated the peak-to-peak distance of the combined electron density of only POPC and water, which resulted in a value of 3.64 nm (graph not shown). This is just slightly less than previous results for pure POPC bilayers, such as the 3.9 nm and 3.7 nm measured by x-ray scattering (52,53), or the result of 3.7 nm found in a MD simulation of a pure POPC bilayer (with a slightly different force field than used here) (W. Zhao, T. Róg, A. A. Gurtovenko, I. Vattulainen, and M. Karttunen, unpublished; S. Ollila, M. T. Hyvönen, and I. Vattulainen, unpublished).
Comparing the profiles of PSM with and without Chol neighbor hints of an important Chol effect: In the region, where the Chol density is highest, the PSM density is reduced. Chol seems to flatten the SM density and push it farther up in the bilayer, a clear sign of Chol-induced lipid ordering. The peak-to-peak distances of electron densities can be used as a measure of the hydrophobic thickness of individual lipid components. For POPC and non-Chol neighboring PSM this gives a similar measure (∼2.7 nm). Already one Chol neighbor raises this measure in PSM to ∼3.3 nm, a significantly higher value, but still far from that seen in one-component PSM bilayers (∼4.2 nm (35)).
Order parameters
The conformational ordering of lipid acyl chains is described by the deuterium order parameter,
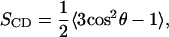 |
(1) |
where θ is the angle between a selected C–H vector and the reference direction (bilayer normal). In a united-atom simulation, SCD can be derived from the carbon chain positions, by assuming the hydrogens at their equilibrium bond angles. This derivation starts from the general order parameter tensor S for the carbon atoms,
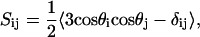 |
(2) |
in which θi is the angle between the ith molecular axis and the bilayer normal (56). At any nonterminal carbon position Cn along the chain, the molecular axis system can be defined using neighboring carbon atoms of the chain: The z axis is taken to be the (normalized) vector 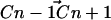 , the x axis is the unit vector perpendicular to
, the x axis is the unit vector perpendicular to 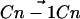 and
and 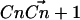 , and the y axis is the cross product of the z and x axes. Now, for hydrogens in a sp3 hybridized CH2 group, this reduces to
, and the y axis is the cross product of the z and x axes. Now, for hydrogens in a sp3 hybridized CH2 group, this reduces to
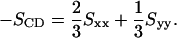 |
(3) |
For a sp2 hybridized CH group with an in-plane hydrogen at angle 120° we get
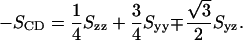 |
(4) |
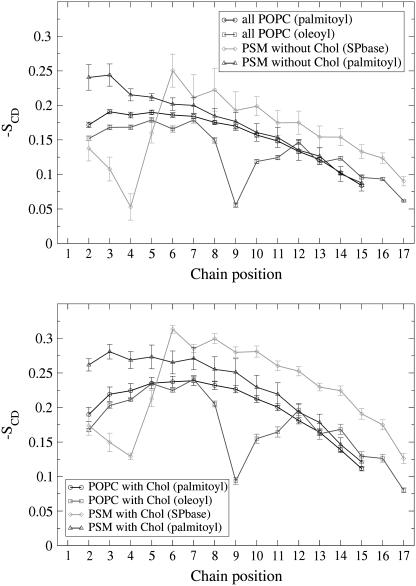
The SCD order parameters for the individual acyl chains are shown in Fig. 5 separately for those chains that interact with Chol and for those that do not. More specifically, the whole POPC matrix represents POPC without Chol contact, the monomeric PSM represents PSM without Chol contact, and the PSM in PSM–Chol pairs represents PSM with Chol contact. To establish POPC lipids with Chol contact, the order profile was collected for those POPCs, whose center-of-mass is the closest neighbor of a Chol center-of-mass, in terms of distance in the two dimensions of the bilayer plane (i.e., in the x,y plane).
FIGURE 5.
SCD order parameters of the acyl chains of phospholipids without contact to a Chol molecule (upper graph) and of those that have a Chol as a nearest neighbor (lower graph). SPbase refers to the sphingosine chain of PSM.
Except for the unsaturated carbon positions, POPC and PSM without Chol neighbor are about equally ordered. Since the PSM ordering falls short of that found in pure PSM-bilayers (35,57), it seems reasonable to state that the PSM monomers adopt the (lesser) ordering of the POPC matrix. Interaction with Chol changes this: Being nearest neighbor to a Chol increases the order of PSM chains more than it increases the order of POPC chains. While the mean POPC order grows by 0.041 for palmitoyl (to 0.201) and by 0.042 for oleoyl (to 0.173), the mean PSM order rises by 0.067 for the sphingosine base (to 0.224) and 0.066 for the palmitoyl residue (to 0.230). These numbers underline two general properties of Chol also seen in experiments: its ordering effect, and a preference for SM over a monounsaturated PC.
A recent 2H-NMR study of a pure POPC bilayer at 321 K found a mean order parameter value of SCD = 0.146 ± 0.003 for the whole palmitoyl chain (57), and a mean value of SCD = 0.185 ± 0.004 for the plateau region of the palmitoyl chain. The plateau region chosen in (57) corresponds to chain positions 3–5 in Fig. 5. This experiment allows for a comparison with our simulated system, which consists mostly of POPC. In the simulation, the mean order of POPC-palmitoyl without contact to neither a Chol nor a PSM is SCD = 0.152 ± 0.005 for the whole chain and SCD = 0.184 ± 0.005 for carbon positions 3–5. The agreement with the experimental values is very good.
Cholesterol tilt and ordering
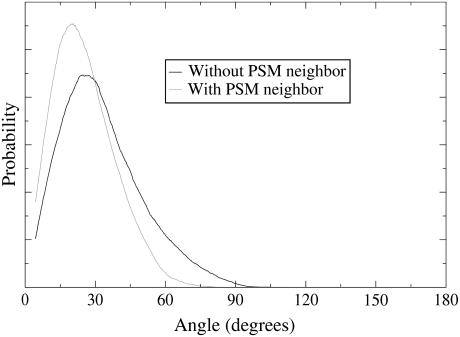
We define the Chol tilt as the angle that the vector across the steroid nucleus (from C21 to C5 in Fig. 1) forms with the bilayer normal. Distributions of this tilt are shown in Fig. 6 separately for the Chol in PSM-Chol pairs and the monomeric Chol. The averaged tilt angles are 25° for Chol with PSM neighbor and 33° for Chol without.
FIGURE 6.
Distribution of Chol tilt angles for those Chols with a PSM neighbor and those without.
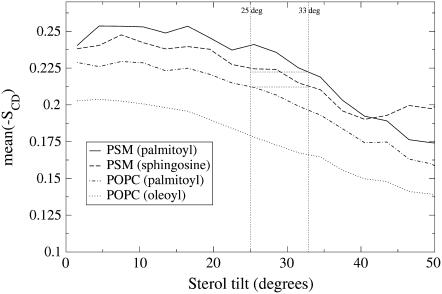
Recently, we established a relationship of a sterol's tilt angle to its local ordering effect: A higher sterol tilt weakens the ordering effect on the sterol's nearest neighbors (58). The same relationship applies in this system as well: The mean order parameters of the acyl chains of Chol nearest neighbors as a function of Chol's tilt are shown in Fig. 7. The clear downward slopes indicate a weakened ordering with higher Chol tilt. In a more detailed analysis (data not shown), we found that the observed effect splits into two contributions. First, an increased tilt of cholesterol decreases the trans/gauche fraction of the neighboring acyl chain, which is reflected as a decrease in the order parameter values. Second, a more tilted cholesterol also increases the overall tilt of the neighboring acyl chains, causing an additional decrease in the order parameter values.
FIGURE 7.
Order parameter of Chol neighboring chains (mean SCD averaged over all carbons in a chain) plotted against that Chol's tilt angle. The added lines indicate the average tilts of PSM neighboring Chol (25°) and monomeric Chol (33°). Neighborship to cholesterol was established using a 0.7 nm cutoff for the center-of-mass distances in the bilayer plane (the x,y plane).
PSM and POPC both carry palmitoyl residues, which can be compared as suggested by the lines in Fig. 7. It seems as if the different tilts of PSM neighboring and non-PSM neighboring Chol explain a major part of the observation, that Chol orders PSM better than POPC. Now, having established the importance of the Chol tilt, we should put special emphasis on the effects of different interaction mechanisms on the Chol tilt.
Headgroup orientations
A revealing quantity for studying phosphocholine headgroup structure and interactions is the headgroup orientation distribution, as measured using the angle of the P-N vector (from the phosphate to the nitrogen) with the bilayer normal.
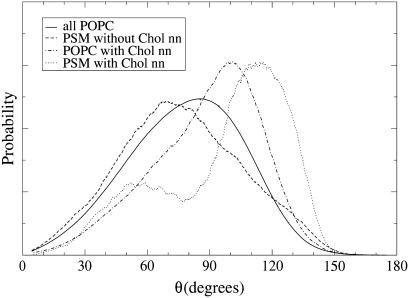
Such distributions are plotted in Fig. 8. Neighborship to Chol was determined similarly as in the calculation of the order profiles (i.e., using the POPC matrix, the monomeric PSM, the paired PSM, and POPCs that are closest neighbors to Chol). The headgroup orientations of non-Chol neighboring PSMs and that of all (i.e., mostly non-Chol neighboring) POPCs have similar shapes, with the PSM distribution peaking at a lower angle than the POPC distribution (70° vs. 85°). As a reference for pure POPC, a simulation of DPPC (with a similar force field than used here for POPC) resulted in a monomodal P-N angle distribution peaking at 90° (35). Comparing that result to Fig. 8 shows that POPC headgroup orientation is very similar to that of DPPC. Since a simulation of a hydrated PSM-only bilayer with the same force field showed a bimodal P-N vector angular distribution, with peaks at 55° and 105° (35), the appearance of a monomodal distribution in non-Chol neighboring PSM suggests that the POPC headgroups inflict their preferred orientation also onto the headgroups of the PSM monomers.
FIGURE 8.
Orientation distributions of choline headgroups, measured as the angle θPN of the P-N vector with the outward bilayer normal. Of all POPCs, only a small fraction is in contact with Chol. SM without chol refers to the monomeric non-Chol neighboring PSMs. POPC/PSM with chol nn contains only those lipids that actually have a Chol as nearest neighbor.
Interactions with Chol bring remarkable changes to the headgroup orientations. The angular distribution of the headgroups of those PSMs that are in contact with Chol becomes bimodal, with maxima at 60° and 115°. The POPC distribution remains monomodal but with a narrower maximum at an increased angle of 100°. Thus, when a PSM has just one Chol as nearest neighbor, the neighboring POPC headgroups no longer inflict their orientation on its headgroup. Presumably the PSM choline moves into the free space above the Chol, which has hardly any headgroup of its own. In this way, the headgroup-free space of Chol allows a neighboring PSM to adopt headgroup orientations similar to those in the pure PSM bilayer. To visualize different possible headgroup orientations, snapshots of a PSM molecule with and without Chol neighbor are shown in Fig. 9.
FIGURE 9.
Simulation snapshots representing typical PSM orientations, for a PSM without Chol neighbor (left) and a PSM with Chol neighbor (right). The PSM and Chol in the right image form a charge pair between the headgroup positive charge and the Chol oxygen (see Charge-Pairing).
Other factors that cause shifts toward higher angles for both POPC and PSM headgroups are charge-pair interactions between the choline nitrogen moiety and Chol oxygens, as well as hydrophobic interactions in the sense of Chol water shielding. These aspects will be revisited later.
Rotational motions
Along with lateral translation, rotation around a molecular axis is a major degree of freedom of bilayer lipids. In a liquid-ordered (lo) phase, rotational motion should be restricted just as acyl chain conformational changes are. Thus, Chol—an efficient promoter of the lo phase—should affect rotational motions of neighboring lipids. We study rotational motions using second rank reorientational autocorrelation functions C2(t),
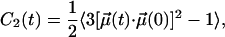 |
(5) |
where 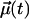 is a unit vector that defines the chosen rotational mode. We examine two rotational modes, one in the headgroup and one in the interfacial region, separately for the POPC matrix and the Chol paired and non-Chol paired PSM. The headgroup mode was defined as the vector from phosphate to nitrogen (P–N vector), while the interfacial vector was taken from carbons G1 to G3 in POPC and SPH-3 to SPH-1 in SM (see Fig. 1).
is a unit vector that defines the chosen rotational mode. We examine two rotational modes, one in the headgroup and one in the interfacial region, separately for the POPC matrix and the Chol paired and non-Chol paired PSM. The headgroup mode was defined as the vector from phosphate to nitrogen (P–N vector), while the interfacial vector was taken from carbons G1 to G3 in POPC and SPH-3 to SPH-1 in SM (see Fig. 1).
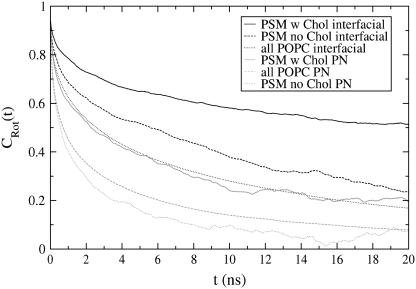
The resulting autocorrelation functions are shown in Fig. 10. Since the motions of the examined lipid parts are limited, the autocorrelation functions do not decay to zero. Instead, their plateau values represent inherent ordering of the studied vectors (59,60). It seems that our simulation is not long enough for the autocorrelation functions to reach their plateau values. Yet, Fig. 10 contains interesting information about the mobility of the studied lipid parts in different local lipid compositions.
FIGURE 10.
C2 reorientational autocorrelation functions of vectors in the headgroup (P-N) and in the interfacial region for POPC/PSM molecules with (w) or without (no) Chol contact.
The POPC matrix is mostly without Chol contact, so its rotations should be compared to those of PSMs without a Chol neighbor. But, since ∼10% of POPCs have a Chol neighbor, the rotations of non-Chol neighboring POPC are probably a bit faster than indicated in Fig. 10. Keeping that in mind, headgroup reorientations in POPC and PSM without Chol contact seem about equally fast (a bit slower in POPC), whereas interfacial reorientations are slower in (non-Chol neighboring) PSM than in POPC. The latter finding is probably due to PSM intermolecular hydrogen bonding (see next section).
Interaction with Chol slows down PSM rotations remarkably, both in the headgroup as well as the interfacial regions. In both regions, the C2(t) decay half-times grow approximately fourfold. A less stringent analysis of reorientations of Chol-neighboring POPCs shows that Chol contact slows POPC rotations, but far less than it does in PSM (data not shown): Decay half-times in the interfacial region roughly double, while those in the headgroup region grow only slightly.
Hydrogen bonding
Although classical MD simulations fail to include quantum effects (such as proton sharing) entirely, many such simulations have been able to predict the correct qualitative static and dynamic features of water and other hydrogen-bonding liquids (61). In this work, we employ geometric criteria to define a hydrogen bond (H-bond): the acceptor-hydrogen distance dAH < 0.25 nm and the donor-hydrogen-acceptor angle θDHA < 90°. Geometric conditions like these are a commonly used operational definition in the field (35,62,63). The average hydrogen-bond numbers for the different lipids are gathered in Table 1. In the following sections, we will analyze them in more detail.
TABLE 1.
Average numbers of hydrogen bonds per corresponding pair for different molecules
| POPC | PSM | Water | |
|---|---|---|---|
| POPC | — | 0.93 | 6.99 |
| PSM without Chol | 0.93 | 1.08* | 6.39 |
| PSM with Chol | 0.93 | 1.12* | 6.2 |
| Chol without PSM | 0.88 | — | 0.54 |
| Chol with PSM | 0.82 | 0.08 | 0.44 |
PSM intramolecular H-bonds, including both OH⋯OPa and OH⋯OPb contacts.
PSM H-bonding
In the simulation of the dilute system, hardly any direct H-bonds form between PSM and Chol (0.08 ± 0.02 H-bonds on average per Chol-PSM pair). Thus, direct H-bonding of PSM and Chol is too rare to be considered of importance for the lipid-lipid interactions. Yet, the simulations show a change in the PSM amide group's H-bonding to water as a consequence of Chol interactions: The H–N–C=O entity of a PSM without a Chol neighbor forms on average 0.45 ± 0.03 H-bonds to water, while that of a PSM with a Chol neighbor forms only 0.34 ± 0.12 of them. Therefore, just as in the IR spectroscopy experiment (21), interactions with Chol change the SM amide group's H-bonding to water. However, at least under the conditions of this simulation, this is not due to direct SM–Chol H-bonds.
Instead of binding to Chol, the PSM donors are occupied with forming H-bonds to POPC (through the N–H group) and intramolecular H-bonds (the O–H group). The intramolecular PSM H-bond forms between the hydroxyl group and oxygens of the phosphate group, mostly oxygen atom OPa (see Fig. 1), and is virtually always present in every PSM molecule. This intramolecular H-bond has been found to be very stable and popular also in other simulations of all-PSM bilayers, using both the same PSM force field as here (35) as well as an all-atom CHARMM force field in an earlier study (64). The four oxygens and net negative charge of the phosphate group probably stabilize this H-bond. PSM intramolecular H-bonding is increased in those PSMs with Chol contact: While the number of O–H⋯ OPa contacts remain virtually one, the number of O–H⋯OPb H-bond-like contacts increases from 0.08 ± 0.01 per non-Chol neighboring PSM to 0.12 ± 0.02 per Chol-neighboring PSM. The resulting case of a proton being shared between three oxygens is probably unphysical. Nonetheless, the increase in OH⋯OPb H-bonds is indicative of a strengthening interaction. This increase in intramolecular H-bonding is related to a change in headgroup orientations (see below).
It is important to remember that the studied three-component system features merely pairs of Chol and PSM. It does not allow conclusions about PSM-Chol H-bonding in a two-component (or mostly two-component) phase. A simulation study of SM-Chol bilayers found diverse hydrogen bonding between these lipids (33). Thus, the virtually complete absence of direct SM-Chol H-bonds probably results from the dilute PSM and Chol concentrations, so that PSM has to compete with POPC about the popularity of H-bonds to Chol. Chol prefers H-bonds to POPC, while PSM prefers intramolecular H-bonds (O–H group) and those to POPC (N–H group).
PSM and POPC form, on average, 0.93 H-bonds per PSM molecule through the N–H donor of PSM. All POPC ester-bond oxygens participate as acceptors. H-bonding of the SM amide group to neighboring phospholipids has been established in other studies as well (35,65). Such H-bonds probably stabilize the PSM molecule orientation with respects to its neighbors. It seems reasonable to assume that PSM intermolecular H-bonding explains its slower reorientations in the interfacial region (see Rotational Motions).
POPC-Chol
POPC and Chol form on average 0.85 H-bonds per Chol, i.e., far more than Chol forms with PSM. In these H-bonds, the Chol hydroxyl group acts as donor while POPC ester bond oxygens (in 90% of bonds) and phosphate oxygen OPb (in 10% of bonds) act as acceptors. Notably, oxygen Oβ2 of the oleoyl chain is the by far most common acceptor of POPC-Chol H-bonds, acting as acceptor in two-thirds of the bonds. Thus, Oβ2 is better suited to accept Chol H-bonds than the other POPC oxygens, including those of the saturated fatty acid residue.
H-bonding to POPC has interesting effects on the Chol tilt. Chol without a PSM neighbor has an average tilt of 34° when H-bonded to POPC, but only 28° when not H-bonded to POPC. The equivalent numbers for Chol with PSM neighbor are 25° (with H-bond to POPC) and 24° (without H-bond to POPC). H-bonding to oxygens in the interfacial region might force Chol to keep rather low in the bilayer, which in turn would cause a higher Chol tilt. In addition, PSM neighboring Chol has significantly lower tilts when H-bonded to POPC oxygens Oα2 and Oβ2 (∼25°) than when H-bonded to POPC oxygens Oα1 and Oβ1 (∼30°).
Direct H-bonding is a rather favorable interaction. It is especially pronounced between Chol and POPC. Yet, such H-bonds enforce a high tilt on the Chol, especially if that Chol is not neighboring a PSM. The higher Chol tilt in turn weakens the ordering and condensing effects of Chol, as well as other forms of interactions, which are analyzed next.
Charge-pairing
Both phospholipids in the simulation feature a positive charge in the amide moiety of their choline headgroups. Thus, they interact favorably with oxygens, all of which carry negative partial charges. In the context of this study, the most relevant oxygen for a choline group to pair with is the cholesterol oxygen.
The fundamental nature of such N+(CH3)3⋯O interactions is ambiguous: Some consider them to be hydrogen bonds, with the partially charged methyl groups acting as donors (37). Others prefer to talk simply about charge-pair interactions (62). Quantum chemical calculations indeed suggest that a carbon with an electronegative substituent can donate a proton to an oxygen, forming a H-bond-like interaction (66). Yet, this interaction was found to be energetically weaker than a classical OH⋯O H-bond, and to react less sensitively to changes in donor-hydrogen-acceptor angles and distances (66). The CH⋯O interaction decays more slowly with increasing H⋯O distance, so, at certain distances, it can be effectively stronger than a OH⋯O H-bond (66).
In the united atom descriptions of the simulations at hand, no choline group hydrogens are included explicitly. This forbids a similar geometric analysis of the N+(CH3)3⋯O interaction than was used above for OH⋯O and NH⋯O H-bonds. As an alternative, we employ an approach based on groupwise Coulomb energies to analyze these interactions. After all, the CH⋯O interaction is mainly of electrostatic nature (66). In this work, the N+(CH3)3⋯O interaction is referred to as a charge-pair interaction, because, as outlined above, it is different from “classical” H-bonds. This choice also distinguishes it from the OH⋯O and NH⋯O H-bonds in the interfacial region of the bilayer, and reminds the reader that, in this analysis, N+(CH3)3⋯O interactions are defined energetically, not geometrically.
Charge-pairing criterion
To study charge-pairing of choline groups to the polar atoms of Chol, the Coulomb energy between the involved groups (the nitrogen and the three adjacent methyl groups for the phospholipid, and the C–O–H moiety for the cholesterol) is calculated as a sum of atom-pairwise interactions. A charge-pair binding mode is identified as a local maximum in the negative tail of the interaction energy histograms (graphs not shown). These choline-cholesterol-COH histograms have local minima at −2.8 kcal/mol (POPC) and −2.5 kcal/mol (PSM), with local maxima at −3.5 kcal/mol (POPC) and −4.0 kcal/mol (PSM). Thus, energy cutoffs of −2.8 kcal/mol (POPC) and −2.5 kcal/mol (PSM) give an operational definition for charge-pair interactions. In a similar simulation, Pandit et al. found a cutoff energy of −2.8 kcal/mol for the interaction of the DPPC choline group and the Chol polar atoms (37), which is exactly the same as observed here for the POPC-Chol charge pairing. For comparison, the histogram of the Chol OH to POPC Oβ2 hydrogen bond has a maximum at −11 kcal/mol (graph not shown). Thus, in this simulation, the charge-pair interactions have roughly one-third of the nominal strength of actual hydrogen bonds.
Charge-pairing occurrence
On average, there are 0.17 ± 0.02 charge-pair bonds to a PSM choline group per (PSM neighboring) Chol. To POPC choline groups, there are on average 0.36 ± 0.05 charge pairs per PSM neighboring Chol and 0.44 ± 0.06 charge pairs per non-PSM neighboring Chol. Thus, between Chol and PSM, charge pairs are more frequent than conventional H-bonds. To put these numbers into proper relation, the different configuration numbers should be noted: Chol has typically six closest POPC neighbors, but only at most one PSM neighbor. Thus, if Chol formed charge pairs to both phospholipids with equal probability, we should see 5–6 times more Chol-POPC charge pairs than Chol-PSM charge pairs. This is obviously not the case, since Chol seems rather eager to charge-pair to PSM instead of POPC.
To bind to a Chol oxygen, the phospholipid headgroup has to bend low, deep into the interfacial region of the bilayer. This can be clearly seen in the P-N vector angular distributions. Headgroups that are charge-pair-bonded have almost exclusively P-N vector angles >90°, with maxima at 110° (for POPC) and 125° (for PSM) (graphs not shown). Thus, headgroup pairing with Chol oxygens helps to explain Chol-induced shifts in P-N angle histogram maxima (see Fig. 8).
PSM structure and charge-pairing
PSM's preference for higher headgroup angles and Chol's preference for charge pairs to PSM are probably caused by certain structural differences in PSM and POPC. The intramolecular H-bonds between PSM hydroxyl groups and phosphate oxygens—without match in POPC—seem to pull the headgroups down and stabilize their higher angles. Indeed, as mentioned earlier, the prevalence of PSM intramolecular OH⋯OPb H-bonds increases from 0.08 in Chol without a PSM neighbor to 0.12 in Chol with a PSM neighbor, an increase that is mostly explained by an increase to 0.18 in those PSM with a charge-pair bond to Chol. In a simulation of pure SM and SM-Chol bilayers (with other force fields than those used here), the frequency of the SM OH··OPa intramolecular bond rose remarkably in the bilayer system with Chol (33). It seems reasonable to assume that this increase is connected to SM choline charge-pairing to Chol oxygens.
The PSM choline also forms intramolecular charge pairs with the carbonyl oxygen OPA of the same molecule. The occurrence of this interaction is 0.14 charge pairs per PSM in those PSM without a Chol neighbor, and 0.24 in those PSM with a Chol neighbor. Again, it seems reasonable to believe that this interaction stabilizes headgroup interactions with Chol. In POPC, no charge pairs form between the choline group and the corresponding carbonyl oxygen Oβ2.
Other forms of interaction
Hydrophobic interactions
Hydrophobic interactions are the main factor that drives structural lipids into bilayer form. Being of such overall importance, they might be involved in lipid-lipid interactions as well. After all, Chol is mostly hydrophobic, with only a small polar headgroup.
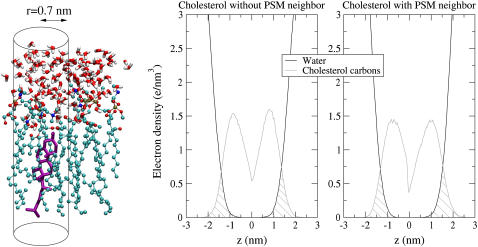
Unfortunately, hydrophobic interactions are difficult to quantify energetically in molecular dynamics simulations. Water contact may be easily derived from a simulation, but the free energies related to it are hard to establish. Therefore, in this analysis, hydrophobic interactions are examined through changes in Chol-water contact. To this end, density profiles of water and the Chol nonpolar parts are calculated over small cylinders centered around Chol molecules. In other words, only atoms that are, in the x,y (bilayer) plane, within 0.7 nm of the Chol center of mass are included in the density profile. The overlap of the densities of water and Chol nonpolar carbons indicates unfavorable water contacts (see Fig. 11).
FIGURE 11.
Hydrophobic interaction calculation. Densities of water and Chol are calculated inside small cylinders (of radius 0.7 nm), that are centered on Chol molecules (illustrated above on the purple-colored cholesterol molecule). Density overlap of water and Chol nonpolar carbons (highlighted by stripes) indicates unfavorable water contacts. The left profile is of non-PSM-paired Chol and the right one is of Chol that is paired to a PSM.
Upon visual inspection, no differences are visible in the graphs of Fig. 11. As a measure for the water contact, the area overlap of the density profiles of water and Chol carbons (i.e., the striped area of the minimum of the two curves) can be related to the total Chol carbon density area. This dimensionless fraction ξ is then a measure for the unfavorable hydration of the Chol nonpolar part,
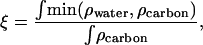 |
(6) |
where ρ denotes electron density of the water or the cholesterol carbon molecules.
For Chol with no PSM neighbor, this fraction is ξ = 0.17 ± 0.03. For those with a PSM neighbor, this fraction is ξ = 0.15 ± 0.03. Keeping in mind the bad statistics implied by the dilute Chol (and PSM) concentration, these numbers could be interpreted to point toward a better Chol water shielding by PSM.
A more interesting point is the effect on the water contact of choline headgroup charge-pairing to Chol. When the cylindrical density calculations are restricted only to those Chols, whose oxygen forms a charge pair to a PSM choline group, the density overlap fraction drops to ξ = 0.11 ± 0.01. However, for those non-PSM neighboring Chols that form the same bond to a POPC choline group, this fraction decreases merely to ξ = 0.16 ± 0.03. Thus, in these simulations, choline group charge-pairing to the Chol oxygen (forming on average 0.17 ± 0.02 charge pairs per Chol) is responsible for a great part of the PSM-caused reduction in Chol water contact.
When forming a charge pair with Chol oxygen, the PSM headgroup folds down, after which it should be able to accommodate the Chol underneath itself and protect it from unwanted water contact (a snapshot of a charge-paired PSM-Chol pair is shown in Fig. 9). The POPC headgroup does not shield a neighboring Chol from water as well as the PSM headgroup. This is probably related to the Chol tilt, which is significantly higher for Chols without PSM neighbor. A more tilted Chol should be more accessible to water.
The differences in steric shielding by the choline headgroup might explain the observed variation in Chol efflux rates from SM and PC monolayers to cyclodextrin subphase (accelerating efflux with decreasing SM content (23)), and the reduced accessibility of Chol oxidase to Chol in SM monolayers than in PC monolayers (7,23).
Van der Waals interactions and lipid packing
Direct calculation of van der Waals energies is out of scope of classical MD force fields. Those few lipid bilayer simulation articles that analyze van der Waals interactions usually examine only order parameters or atom packing, and relate those directly to van der Waals interactions (e.g., in (67)). In that spirit, since this work found Chol to order PSM more than POPC (in the sense of the SCD order parameter), van der Waals interactions between PSM and Chol can be seen as more favorable than those between POPC and Chol. This line of thought also means that a lower Chol tilt improves van der Waals interactions and lipid packing.
Steric hindrance issues affect Chol-phospholipid orientations. To study lipid arrangements with reference to the Chol α (smooth) and β (two protruding methyl groups) faces, center-of-mass trajectories were produced for the Chols, the protruding Chol methyl groups, and the individual phospholipid chains. In such a trajectory, the two-dimensional angle (angle in the x,y-plane) formed by the centers of mass of the phospholipid chain, the Chol, and the Chol methyl groups, indicates on which face the chain is located (angle > 90°: α-face; angle < 90°: β-face). This analysis shows a clear preference (of ∼60:40) of PSM to reside on a neighboring Chol's smooth α-face, while POPC shows no face preference. In analogous simulations, Pandit et al. found similar preferences of PSM and DOPC for the Chol faces (38).
The Chol face affects acyl-chain ordering: The saturated acyl chains (both PSM chains and the palmitoyl in POPC) are more ordered when next to a Chol α-face than when next to a Chol β-face. For the POPC-oleoyl chain, there is no significant change in ordering between the Chol faces. The differences in chain ordering between Chol α- and β-faces are greater in PSM chains (with changes in the mean SCD order of ∼0.020) than in POPC-palmitoyl (changes of ∼0.010). This difference is probably related to the differences in the tilt angles of Chols neighboring these phospholipids.
The changes in order on the two Chol faces are minor when compared to the overall order increment of having a Chol neighbor at all (which increases the mean SCD order by 0.030 – 0.070).
DISCUSSION
Cholesterol favors PSM over POPC
As established in the density and order profiles, already single Chol molecules order and condense the bilayer locally. Thus, no large phospholipid-Chol complexes or networks are needed to initiate these ultimately macroscopic effects of Chol. Moreover, just one Chol neighbor is enough to promote significant changes in PSM: Its hydrophobic thickness increases, its acyl-chain order increases, its headgroup orientation becomes bimodal and resembles more closely that in an all-PSM bilayer, and its rotational motions become slower. Most of these changes are also visible in Chol neighboring POPC, but with significantly smaller magnitudes. In addition, having just one PSM neighbor significantly reduces the tilt of Chol. Taken together, these pieces of evidence convincingly assert a preference, in the simulations, of Chol for saturated SM over monounsaturated PC. The existence of such a preferential interaction is in line with many experiments.
Yet, the simulations do not support one of the main lines of speculation on the origin of the SM-Chol interaction specificity: direct H-bonding of SM donors to the Chol oxygen. Instead, in the simulations, the PSM hydroxyl group is constantly H-bonded to a phosphate oxygen of the same molecule, while the PSM amide group forms H-bonds almost exclusively to neighboring POPCs. Chol forms hydrogen bonds through its hydroxyl group mainly to POPC oxygens in the interfacial region, a consequence of the higher number, greater partial charge, and greater flexibility of the POPC acceptors over the SM acceptors in the interfacial region.
Since the simulated system includes only monomers and dimers of PSM and Chol, no conclusions should be drawn from it on the PSM-Chol hydrogen bonding in a phase rich in PSM and Chol. Still, the unique approach of this study, with its dilute PSM and Chol concentration, shows that direct PSM-Chol hydrogen bonds are of little importance in the pairing of these molecules. Consequently, the initial formation of PSM-Chol enriched phases must be driven by alternative factors.
Alternatives to hydrogen bonding
Alternatives to hydrogen bonding include charge-pairing of the phospholipid headgroup nitrogen moieties with Chol oxygens, hydrophobic interactions, and van der Waals interactions. The differences in PSM and POPC van der Waals interactions with Chol are manifested in a greater Chol-induced increase in PSM ordering than in POPC ordering, as well as the clear preference of PSM for the smooth α-face of Chol, which is not seen for POPC. Some evidence suggests that charge pairs of the Chol polar group to the PSM choline group are more stable than those to the POPC choline group. This seems to originate from features related to structural differences of POPC and PSM, namely the formation of the PSM intramolecular hydrogen bonds and PSM choline group intramolecular charge pairs to the PSM carbonyl oxygen. What is more, the headgroup-free space above Chol allows the PSM headgroup to adopt a bimodal orientation distribution, which is also seen in one-component PSM bilayers. Analysis of the water contact of the nonpolar parts of Chol shows a small decrease in water contact for SM-neighboring Chol versus Chol with only POPC neighbors. To a great part, this decrease is explained by the PSM choline charge-pairing to Chol, which provides clearly better water shielding than similar charge-pairing of POPC choline.
Of these mechanisms, at least van der Waals and hydrophobic interactions should benefit from small Chol tilts. The ordering capability of Chol clearly decreases with increasing tilt, and a more tilted Chol seemingly exposes more of its nonpolar carbons to water. H-bonding of Chol to POPC ester bond oxygens was found to increase the Chol tilt in comparison to Chol without such H-bonds. This raises the idea that Chol H-bonding to POPC, an energetically very favorable bond, competes with the other interactions mentioned above. PSM has fewer and (at least in our force field) weaker H-bond acceptors in the interfacial region, and its peptide bond renders the PSM less flexible in the interfacial region. Therefore, between PSM and Chol, H-bonding is weakened and the other interactions become stronger, which leads to a lower Chol tilt and improved ordering of surrounding lipids.
A central role of hydrophobicity in phospholipid-Chol interactions has been suggested in connection with the concept of hydrophobic mismatch (22). In that concept, Chol positions itself preferentially at boundaries of more and less ordered patches not because of specific interactions but to smooth the mismatch in hydrophobic thickness between the regions. In our system, the monomeric (non-Chol neighboring) PSM features a hydrophobic thickness similar to that of the POPC matrix, but already a single Chol neighbor raises the hydrophobic thickness of a PSM molecule significantly. Thus, a Chol molecule in a lipid bilayer allows for differences in hydrophobic thickness to form. What is more, due to differences in Chol interaction mechanisms with PSM and POPC, Chol not only allows but also promotes differences in PSM and POPC hydrophobic thickness. The change in PSM hydrophobic thickness is accompanied by a lowering of the tilt angle of the neighboring Chol. It seems that Chol can adapt to different hydrophobic environments by adjusting its tilt angle.
In conclusion, this work suggests that the initial phases of raft formation are not driven by direct H-bonding between PSM and Chol. Rather, the “specific” nature of the interaction between these molecules is more subtle and comprises a shift in interactions away from H-bonding toward electrostatic (charge-pair) interactions between PSM headgroups and Chol oxygens, together with improved van der Waals interactions and better water-shielding of Chol. Unlike direct H-bonding, these latter interactions benefit from a lower Chol tilt, which in turn promotes higher ordering of hydrocarbon chains. In addition, the concept of hydrophobic mismatch seems to hold, in the sense that Chol smoothens a difference in hydrophobic thickness that is itself created in the first place. In a bilayer of Chol and phospholipids with different acyl-chain lengths, the role of hydrophobic mismatch is probably more pronounced.
Acknowledgments
We thank Juha M. Holopainen for discussions. We acknowledge the Finnish IT Center for Science and the HorseShoe (DCSC) supercluster computing facility at the University of Southern Denmark for computer resources.
This work has, in part, been supported by the Academy of Finland (to I.V., M.T.H., P.S.N., and M.K.), the Academy of Finland Center of Excellence Program (to P.S.N. and I.V.), the Jenny and Antti Wihuri Foundation (to M.T.H.), the Finnish Academy of Science and Letters (to P.S.N.), the Emil Aaltonen foundation (M.K.), and the Natural Sciences and Engineering Council (NSERC) of Canada (to M.K.).
References
- 1.Bloom, M., E. Evans, and O. G. Mouritsen. 1991. Physical properties of the fluid lipid-bilayer component of cell membranes: a perspective. Q. Rev. Biophys. 24:293–397. [DOI] [PubMed] [Google Scholar]
- 2.Nelson, D. L., and M. M. Cox. 2004. Lehninger Principles of Biochemistry, 4th Ed. W.H. Freeman and Company, New York.
- 3.Singer, S. J., and G. L. Nicolson. 1972. The fluid mosaic model of the structure of cell membranes. Science. 175:720–731. [DOI] [PubMed] [Google Scholar]
- 4.Vist, M. R., and J. H. Davis. 1990. Phase equilibria of cholesterol/dipalmitoylphosphatidylcholine mixtures: 2H nuclear magnetic resonance and differential scanning calorimetry. Biochemistry. 29:451–464. [DOI] [PubMed] [Google Scholar]
- 5.Ipsen, J. H., G. Karlström, O. G. Mouritsen, H. Wennerström, and M. J. Zuckermann. 1987. Phase equilibria in the phosphatidylcholine-cholesterol system. Biochim. Biophys. Acta. 905:162–172. [DOI] [PubMed] [Google Scholar]
- 6.London, E. 2002. Insights into lipid raft structure and formation from experiments in model membranes. Curr. Opin. Struct. Biol. 12:480–486. [DOI] [PubMed] [Google Scholar]
- 7.Silvius, J. R. 2003. Role of cholesterol in lipid raft formation: lessons from lipid model systems. Biochim. Biophys. Acta. 1610:174–183. [DOI] [PubMed] [Google Scholar]
- 8.Simons, K., and W. L. C. Vaz. 2004. Model systems, lipid rafts, and cell membranes. Annu. Rev. Biophys. Biomol. Struct. 33:269–295. [DOI] [PubMed] [Google Scholar]
- 9.Alberts, B., A. Johnston, J. Lewis, M. Raff, K. Roberts, and P. Walter. 2002. Molecular Biology of the Cell, 4th Ed. Garland Science, New York.
- 10.Cottingham, K. 2004. Do you believe in lipid rafts? Anal. Chem. 76:403A–406A. [PubMed] [Google Scholar]
- 11.Munro, S. 2003. Lipid rafts: elusive or illusive? Cell. 115:377–388. [DOI] [PubMed] [Google Scholar]
- 12.Simons, K., and E. Ikonen. 1997. Functional rafts in cell membranes. Nature. 387:569–572. [DOI] [PubMed] [Google Scholar]
- 13.Brown, D. A., and E. London. 1998. Functions of lipid rafts in biological membranes. Annu. Rev. Cell Dev. Biol. 14:111–136. [DOI] [PubMed] [Google Scholar]
- 14.Edidin, M. 2003. The state of lipid rafts: from model membranes to cells. Annu. Rev. Biophys. Biomol. Struct. 32:257–283. [DOI] [PubMed] [Google Scholar]
- 15.Vainio, S., M. Jansen, M. Koivusalo, T. Róg, M. Karttunen, I. Vattulainen, and E. Ikonen. 2006. Significance of sterol structural specificity: desmosterol cannot replace cholesterol in lipid rafts. J. Biol. Chem. 281:348–355. [DOI] [PubMed] [Google Scholar]
- 16.Li, X.-M., M. M. Momsen, J. M. Smaby, H. L. Brockman, and R. E. Brown. 2001. Cholesterol decreases the interfacial elasticity and detergent solubility of sphingomyelins. Biochemistry. 40:5954–5963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ohvo, H., and J. P. Slotte. 1996. Cyclodextrin-mediated removal of sterols from monolayers: effects of sterol structure and phospholipids on desorption rate. Biochemistry. 35:8018–8024. [DOI] [PubMed] [Google Scholar]
- 18.Ramstedt, B., and J. P. Slotte. 1999. Interaction of cholesterol with sphingomyelins and acyl-chain-matched phosphatidylcholines: a comparative study of the effect of the chain length. Biophys. J. 76:908–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bittman, R., C. R. Kasireddy, P. Mattjus, and J. P. Slotte. 1994. Interaction of cholesterol with sphingomyelin in monolayers and vesicles. Biochemistry. 33:11776–11781. [DOI] [PubMed] [Google Scholar]
- 20.Sankaram, M. B., and T. E. Thompson. 1990. Interaction of cholesterol with various glycerophospholipids and sphingomyelin. Biochemistry. 29:10670–10675. [DOI] [PubMed] [Google Scholar]
- 21.Veiga, M. P., J. L. R. Arrondo, F. M. Goni, A. Alonso, and D. Marsh. 2001. Interaction of cholesterol with sphingomyelin in mixed membranes containing phosphatidylcholine, studied by spin-label ESR and IR spectroscopies. A possible stabilization of gel-phase sphingolipid domains by cholesterol. Biochemistry. 40:2614–2622. [DOI] [PubMed] [Google Scholar]
- 22.Holopainen, J. M., A. J. Metso, J.-P. Mattila, A. Jutila, and P. K. J. Kinnunen. 2004. Evidence for the lack of a specific interaction between cholesterol and sphingomyelin. Biophys. J. 86:1510–1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Slotte, J. P. 1999. Sphingomyelin-cholesterol interactions in biological and model membranes. Chem. Phys. Lipids. 102:13–27. [DOI] [PubMed] [Google Scholar]
- 24.Huang, J., and G. W. Feigenson. 1999. A microscopic interaction model of maximum solubility of cholesterol in lipid bilayers. Biophys. J. 76:2142–2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guo, W., V. Kurze, T. Huber, N. H. Afdhal, K. Beyer, and J. A. Hamilton. 2002. A solid-state NMR study of phospholipid-cholesterol interactions: sphingomyelin-cholesterol binary systems. Biophys. J. 83:1465–1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Feller, S. E. 2000. Molecular dynamics simulations of lipid bilayers. Curr. Op. Coll. Interface Sci. 5:217–223. [Google Scholar]
- 27.Saiz, L., and M. L. Klein. 2002. Computer simulation studies of model biological membranes. Acc. Chem. Res. 35:482–489. [DOI] [PubMed] [Google Scholar]
- 28.Scott, H. L. 2002. Modeling the lipid component of membranes. Curr. Opin. Struct. Biol. 12:495–502. [DOI] [PubMed] [Google Scholar]
- 29.Tieleman, D. P., S. J. Marrink, and H. J. C. Berendsen. 1997. A computer perspective of membranes: molecular dynamics studies of lipid bilayer systems. Biochim. Biophys. Acta. 1331:235–270. [DOI] [PubMed] [Google Scholar]
- 30.Vattulainen, I., and M. Karttunen. 2004. Modeling of biologically motivated soft matter systems. In Computational Nanotechnology. M. Rieth and W. Schommers, editors. American Scientific Publishers, Stevenson Ranch, California.
- 31.Falck, E., M. Patra, M. Karttunen, M. T. Hyvönen, and I. Vattulainen. 2004. Impact of cholesterol on voids in phospholipid membranes. J. Chem. Phys. 121:12676–12689. [DOI] [PubMed] [Google Scholar]
- 32.Falck, E., M. Patra, M. Karttunen, M. T. Hyvonen, and I. Vattulainen. 2004. Lessons of slicing membranes: interplay of packing, free area, and lateral diffusion in phospholipid/cholesterol bilayers. Biophys. J. 87:1076–1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Khelashvili, G. A., and H. L. Scott. 2004. Combined Monte Carlo and molecular dynamics simulation of hydrated 18:0 sphingomyelin-cholesterol lipid bilayers. J. Chem. Phys. 120:9841–9847. [DOI] [PubMed] [Google Scholar]
- 34.Kupiainen, M., E. Falck, S. Ollila, P. Niemela, A. A. Gurtovenko, M. T. Hyvonen, M. Patra, M. Karttunen, and I. Vattulainen. 2005. Free volume properties of sphingomyelin, DMPC, DPPC, and PLPC bilayers. J. Comput. Theor. Nanosci. 2:401–413. [Google Scholar]
- 35.Niemelä, P., M. T. Hyvönen, and I. Vattulainen. 2004. Structure and dynamics of sphingomyelin bilayer: insight gained through systematic comparison to phosphatidylcholine. Biophys. J. 87:2976–2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Niemelä, P. S., M. T. Hyvönen, and I. Vattulainen. 2006. Influence of chain length and unsaturation on sphingomyelin bilayers. Biophys. J. 90:851–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pandit, S. A., D. Bostick, and M. L. Berkowitz. 2004. Complexation of phosphatidylcholine lipids with cholesterol. Biophys. J. 86:1345–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pandit, S. A., E. Jakobsson, and H. L. Scott. 2004. Simulation of the early stages of nano-domain formation in mixed bilayers of sphingomyelin, cholesterol, and dioleoylphosphatidylcholine. Biophys. J. 87:3312–3322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Berendsen, H. J. C., D. van der Spoel, and R. van Drunen. 1995. GROMACS: a message-passing parallel molecular dynamics implementation. Comput. Phys. Comm. 91:43–56. [Google Scholar]
- 40.Lindahl, E., B. Hess, and D. van der Spoel. 2001. GROMACS 3.0: a package for molecular simulation and trajectory analysis. J. Mol. Model. (Online). 7:306–317. [Google Scholar]
- 41.Patra, M., E. Salonen, E. Terama, I. Vattulainen, R. Faller, B. W. Lee, J. Holopainen, and M. Karttunen. 2006. Under the influence of alcohol: the effect of ethanol and methanol on lipid bilayers. Biophys. J. 90:1121–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tieleman, D. P., and H. J. C. Berendsen. 1998. A molecular dynamics study of the pores formed by Escherichia coli OmpF porin in a fully hydrated palmitoyloleoylphosphatidylcholine bilayer. Biophys. J. 74:2786–2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Höltje, M., T. Förster, B. Brandt, T. Engels, W. von Rybinski, and H.-D. Höltje. 2001. Molecular dynamics simulations of stratum corneum lipid models: fatty acids and cholesterol. Biochim. Biophys. Acta. 1511:156–167. [DOI] [PubMed] [Google Scholar]
- 44.Berendsen, H. J. C., J. P. M. Postma, W. F. van Gunsteren, and J. Hermans. 1981. Interaction models for water in relation to protein hydration. In Intermolecular Forces. B. Pullman, editor. Reidel, Dordrecht, The Netherlands.
- 45.Berendsen, H. J. C., J. P. M. Postma, W. F. van Gunsteren, A. DiNola, and J. R. Haak. 1984. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 81:3684–3690. [Google Scholar]
- 46.Hoover, W. G. 1985. Canonical dynamics: equilibrium phase-space distributions. Phys. Rev. A. 31:1695–1697. [DOI] [PubMed] [Google Scholar]
- 47.Nosé, S. 1984. A molecular dynamics method for simulation in the canonical ensemble. Mol. Phys. 52:255–268. [Google Scholar]
- 48.Nosé, S., and M. L. Klein. 1983. Constant pressure molecular dynamics for molecular systems. Mol. Phys. 50:1055–1076. [Google Scholar]
- 49.Parrinello, M., and A. Rahman. 1981. Polymorphic transitions in single crystals: a new molecular dynamics method. J. Appl. Phys. 52:7182–7190. [Google Scholar]
- 50.Seelig, J., and N. Waespe-Šarčević. 1978. Molecular order in cis and trans unsaturated phospholipid bilayers. Biochemistry. 17:3310–3315. [DOI] [PubMed] [Google Scholar]
- 51.Patra, M., M. Hyvönen, E. Falck, M. Sabouri-Ghomi, I. Vattulainen, and M. Karttunen. 2004. Long-range interactions and parallel scalability in molecular simulations. Comput. Phys. Commun. 176:14–22. [Google Scholar]
- 52.Kucerka, N., S. Tristram-Nagle, and J. F. Nagle. 2005. Structure of fully hydrated fluid phase lipid bilayers with monounsaturated chains. J. Membr. Biol. 208:193–202. [DOI] [PubMed] [Google Scholar]
- 53.Vogel, M., C. Münster, W. Fenzl, and T. Salditt. 2000. Thermal unbinding of highly oriented phospholipid membranes. Phys. Rev. Lett. 84:390–393. [DOI] [PubMed] [Google Scholar]
- 54.Reference deleted in proof.
- 55.Reference deleted in proof.
- 56.Douliez, J.-P., A. Leonard, and E. J. Dufourc. 1995. Restatement of order parameters in biomembranes: calculation of C–C bond order parameters from C–D quadrupolar splittings. Biophys. J. 68:1727–1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mehnert, T., K. Jacob, R. Bittman, and K. Beyer. 2006. Structure and lipid interaction of N-palmitoylsphingomyelin in bilayer membranes as revealed by 2H-NMR spectroscopy. Biophys. J. 90:939–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Aittoniemi, J., T. Rog, P. Niemela, M. Pasenkiewicz-Gierula, M. Karttunen, and I. Vattulainen. 2006. Tilt: major factor in sterols' ordering capability in membranes. J. Phys. Chem. B. 110:25562–25564. [DOI] [PubMed] [Google Scholar]
- 59.Lindahl, E., and O. Edholm. 2001. Molecular dynamics simulation of NMR relaxation rates and slow dynamics in lipid bilayers. J. Chem. Phys. 115:4938–4950. [Google Scholar]
- 60.Pastor, R. W., R. M. Venable, and S. E. Feller. 2002. Lipid bilayers, NMR relaxation and computer simulations. Acc. Chem. Res. 35:438–446. [DOI] [PubMed] [Google Scholar]
- 61.Ladanyi, B. M., and M. S. Skaf. 1993. Computer simulation of hydrogen-bonding liquids. Annu. Rev. Phys. Chem. 44:335–368. [Google Scholar]
- 62.Pasenkiewicz-Gierula, M., T. Róg, K. Kitamura, and A. Kusumi. 2000. Cholesterol effects on the phosphatidylcholine bilayer polar region: A molecular simulation study. Biophys. J. 78:1376–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.van der Spoel, D., E. Lindahl, B. Hess, A. R. van Buuren, E. Apol, P. J. Meulenhoff, D. P. Tieleman, A. L. T. M. Sijbers, K. A. Feenstra, R. van Drunen, and H. J. C. Berendsen. 2004. GROMACS User Manual, Ver. 3.2. www.gromacs.org.
- 64.Hyvönen, M. T., and P. T. Kovanen. 2003. Molecular dynamics simulation of sphingomyelin bilayer. J. Phys. Chem. B. 107:9102–9108. [Google Scholar]
- 65.Chiu, S. W., S. Vasudevan, E. Jacobsson, R. J. Mashl, and H. L. Scott. 2003. Structure of sphingomyelin bilayers: a simulation study. Biophys. J. 85:3624–3635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gu, Y., T. Kar, and S. Scheiner. 1999. Fundamental properties of the CH⋯O interaction: is it a true hydrogen bond? J. Am. Chem. Soc. 121:9411–9422. [Google Scholar]
- 67.Róg, T., and M. Pasenkiewicz-Gierula. 2004. Nonpolar interactions between cholesterol and phospholipids: a molecular dynamics simulation study. Biophys. Chem. 107:151–164. [DOI] [PubMed] [Google Scholar]