Abstract
The enzymes and genes of the lignin biosynthetic pathway have been studied for several decades, but the gene encoding ferulate 5-hydroxylase (F5H) was cloned only 3 years ago by T-DNA tagging in Arabidopsis. To characterize the enzyme in detail, we have expressed F5H in yeast. According to current models of the phenylpropanoid pathway, F5H catalyzes the hydroxylation of ferulate to 5-hydroxyferulate; however, our studies indicate that the enzyme also uses coniferaldehyde and coniferyl alcohol as substrates. Unexpectedly, the Km values measured for the latter two substrates are three orders of magnitude lower than that measured for ferulic acid, suggesting that in lignifying tissues, syringyl monomers may be derived from their guaiacyl counterparts by hydroxylation and subsequent methylation. Thus, F5H may function later in the lignin biosynthetic pathway than was originally proposed. To further test this model, recombinant F5H was incubated together with ferulic acid, coniferaldehyde, or coniferyl alcohol in the presence of native or recombinant Arabidopsis caffeic acid/5-hydroxyferulic acid O-methyltransferase and [14C]S-adenosylmethionine. In all cases, the corresponding radiolabeled sinapyl derivatives were synthesized, indicating that the necessary enzymes required for this pathway are present in Arabidopsis. Taken together, these data suggest that the previously accepted pathway for lignin biosynthesis is likely to be incorrect.
Lignin is a cross-linked phenylpropanoid polymer that is a major sink for fixed carbon in plants. It is essential for the rigidification of tracheary-element cell walls that enables them to resist the tension generated during transpiration (1). Lignin is responsible for the decay resistance that enables woody plants to live for centuries in an environment teeming with parasitic and saprophytic organisms capable of degrading nonlignified plant cell walls. It is also elaborated in nonwoody plants in response to pathogen attack (2). Lignin is important from a human perspective because it constrains our use of lignocellulosic materials in agriculture and forestry (3). Lignin decreases the nutritional value of forages by limiting the access of digestive hydrolytic enzymes to cell-wall polysaccharides (4). In the costly and polluting pulping process, lignin is removed from wood to produce cellulose for the manufacture of paper products. The potential economic benefits of modifying lignin through biotechnology has inspired many efforts to engineer lignin quantity and quality (5).
The phenylpropanoid pathway, which generates the coniferyl and sinapyl alcohol monomers (monolignols) required for lignin biosynthesis, has been the subject of investigation for many decades. The conversion of ferulic acid to sinapic acid via 5-hydroxyferulate has been accepted as the route to syringyl lignin monomers since it was proposed over thirty years ago (6). The activity of the enzyme ferulate 5-hydroxylase (F5H, CYP84) was first detected in extracts of lignifying poplar xylem where it was shown to catalyze the hydroxylation of ferulate to 5-hydroxyferulate (7). The NADPH dependence and the characteristic light-reversible carbon monoxide inhibition of this activity gave the first indication that this enzyme is a cytochrome P450-dependent monooxygenase (P450). Unfortunately, F5H activity was lost on solubilization, thus preventing the purification of the enzyme. The identification of the F5H-deficient fah1 mutant of Arabidopsis enabled the isolation of the F5H gene by T-DNA tagging (8, 9). Overexpression of F5H in Arabidopsis abolished the tissue specificity of syringyl monomer deposition and led to the accumulation of lignin that was almost solely derived from syringyl units (10). These data indicate that F5H plays a key regulatory role in syringyl lignin biosynthesis.
To address the biochemical properties of F5H, we have expressed the protein in yeast, thus providing sufficient enzyme for detailed kinetic analyses. Here we report that in addition to ferulate, F5H uses coniferaldehyde and coniferyl alcohol as substrates, and that the Km values for the latter two substrates are substantially lower than that for ferulate. Furthermore, we have demonstrated that the O-methyltransferase (OMT) activity required for conversion of the 5-hydroxyguaiacyl-substituted products to their corresponding syringyl derivatives is present in Arabidopsis and that this reaction is catalyzed by caffeic acid/5-hydoxyferulic acid O-methyltransferase (COMT). These data indicate that, instead of involving sinapic acid as an intermediate, the biosynthesis of syringyl lignin monomers most likely proceeds by hydroxylation and methylation of coniferaldehyde and/or coniferyl alcohol and that a further revision of the lignin biosynthetic pathway is necessary.
EXPERIMENTAL PROCEDURES
Yeast Strains.
The construction of the Saccharomyces cerevisiae strain WAT11, a derivative of the W303-B strain (MAT a; ade2–1; his3–11,-15; leu2–3,-112; ura3–1; canR; cyr+) expressing the ATR1 Arabidopsis NADPH–P450 reductase has been described (11, 12).
Constructs.
For the construction of the pYeDP60-F5H expression construct, the F5H ORF was amplified by PCR using Takara Extaq (Panvera, Madison WI) with primers 5′-aaggatccatggagtcttctatatcacaaa-3′ and 5′-aagaattcacttaaagagcacagatgaggc-3′. These primers correspond to the 5′ and 3′ ends of the ORF and introduce a BamHI site upstream of the start codon and an EcoRI site downstream of the stop codon, respectively. The resulting 1.5-kilobase PCR product was subcloned, sequenced, and ligated into BamHI–EcoRI-digested pYeDP60 (13) to yield the plasmid pYeDP60-F5H. WAT11 cells were transformed with pYeDP60 and pYeDP60-F5H as described (14).
Yeast Growth Conditions and Preparation of Yeast Extracts.
WAT11 cells carrying pYeDP60 or pYeDP60-F5H were grown, induced with galactose, and extracted, and microsomes were prepared as described (15). The microsomal pellet was resuspended in assay buffer [50 mM sodium Pipes (pH 7.0) containing 20% glycerol and 4 mM disodium EDTA] and stored at −70°C.
Triton Phase Partitioning of Microsomal Proteins.
To isolate membrane-associated proteins from the polyethylene glycol-precipitated microsomal preparation, Triton X-114 phase partitioning was used (16). Microsomal and Triton phase-partitioned samples were precipitated with trichloroacetic acid before SDS/PAGE analysis on 12% acrylamide gels (17).
Spectroscopy.
The carbon monoxide difference spectrum of F5H [200 μl of the Triton X-114 phase-partitioned protein sample, 40 μl of Triton X-100, 10 μl of 10% CHAPS (wt/vol), 1.75 ml of assay buffer] was performed as described (18).
Plant Material and Preparation of Plant Extracts.
Arabidopsis thaliana was grown under a 16 h light/8 h dark photoperiod at 100 microeinstein⋅m−2⋅s−1 at 22°C, cultivated in ProMix potting mixture (Premier Horticulture, Red Hill, PA). Extracts for assay of OMT activity were prepared from wild-type Arabidopsis rachis (flowering stem) tissue as described (7) by using F5H assay buffer. For fractionation on FPLC, extracts were desalted into 50 mM Tris⋅HCl and chromatographed on a Mono Q HR5/5 column (solvent A, 50 mM Tris⋅HCl, pH 7.8; solvent B, 50 mM Tris⋅HCl, pH 7.8, containing 1.0 M NaCl; 0% B for 15 min, 0 to 50% B in 30 min, 50 to 100% B in 1 min, 100% B for 10 min; flow rate 1.0 ml⋅min−1). For both plant and yeast extracts, protein content was determined with the bicinchonic acid assay procedure (Pierce) with BSA as a standard.
Bacterial Strains and Preparation of Bacterial Extracts.
Clone 154J19T7 harboring an Arabidopsis cDNA insert identical to a putative COMT homologue (19) fused in-frame with the plasmid β-galactosidase gene was obtained from the Ohio State University Arabidopsis Stock Center and transformed into DH5α. Logarithmic-phase cells carrying the COMT clone were induced with isopropyl-β-d-thiogalactoside (IPTG) for 30 min, pelleted by centrifugation, washed once in assay buffer, and resuspended in assay buffer. Cells were disrupted in a French press, and the extract was clarified by centrifugation and desalted into assay buffer berore use.
Enzyme Assays. For F5H assays, an NADPH regenerating system consisting of 1 mM NADP+/10 mM glucose 6-phosphate/1 unit of glucose-6-phosphate dehydrogenase was preincubated at 30°C for 5 min to permit the generation of NADPH in the presence of ferulic acid, coniferaldehyde, or coniferyl alcohol in a final volume of 450 μl of assay buffer. Assays were initiated by the addition of 50 μl of microsomes and were allowed to incubate for 20 min at 30°C before being terminated by the addition of 100 μl of glacial acetic acid. Assays conducted with ferulic acid or coniferaldehyde were extracted with 600 μl of ethyl acetate, which was reduced to dryness in vacuo and dissolved in 50 μl of methanol for analysis. Assays conducted with coniferyl alcohol were clarified by centrifugation for 5 min at 13,000 × g and were analyzed directly. The products of F5H assays were analyzed by HPLC on a Microsorb-MV C-18 column (Rainin Instruments) by using method 1 (solvent A, 1.5% acetic acid in water; solvent B, acetonitrile; 0 to 11% B in 2 min, 11% B isocratic for 6 min, 11 to 18% B in 1 min, 18 to 35% B in 11 min; flow rate 1 ml⋅min−1) and UV detection (ferulic acid, 322 nm; coniferaldehyde, 340 nm; coniferyl alcohol, 265 nm). Assays conducted with microsomes isolated from yeast transformed with pYeDP60 served as negative controls.
For F5H–OMT coupled assays, F5H assays as described above were incubated for 2 h, at which time 100 μl of Arabidopsis rachis extract or COMT (from Arabidopsis COMT expressed in Escherichia coli) preparation and 125 nCi (1 Ci = 37 GBq) S-adenosyl-l-[methyl-14C]-methionine (final concentration 3.7 μM) was added. After a further 2-h incubation period, assays were terminated by the addition of 100 μl of glacial acetic acid and extracted with 600 μl of ethyl acetate. The organic phase was dried in a stream of nitrogen gas, dissolved in methanol, and spiked with 5 μg of the unlabeled expected product. The entire sample was then applied to the origin of a silica gel TLC plate, which was developed by using a mobile phase of toluene/acetic acid (2:1) saturated with water. Products were visualized under UV light with a Packard Instant Imager. Radiolabeled products were scraped from the TLC plate, eluted in methanol, and analyzed by HPLC using method 2 (solvent A, 5% acetic acid in water; solvent B, 20% acetic acid, 25% acetonitrile in water; 5 to 55% B in 20 min; flow rate 1 ml⋅min−1) and UV detection. One-milliliter fractions were collected and analyzed by liquid scintillation counting.
Product Identification.
For the large scale production of 5-hydroxyferulic acid and 5-hydroxyconiferyl alcohol for mass spectral identification, 400 μl of 250 mM ferulic acid or coniferyl alcohol was added to 100 ml of galactose-induced yeast cultures at an OD600 of 8. Cultures were incubated for 3 h, after which cells were pelleted by centrifugation for 10 min at 10,000 × g. The supernatant was acidified by the addition of 20 ml of glacial acetic acid and extracted with 100 ml of ethyl acetate. The ethyl acetate phase was removed and concentrated to dryness in vacuo, and the residue was derivatized in 100 μl of N,O-bis(trimethylsilyl) trifluoroacetamide (Pierce) before analysis by GC–electron impact MS. For the identification of 5-hydroxyconiferaldehyde, a large-scale in vitro F5H assay was conducted in a final volume of 16 ml in the presence of 10 μM coniferaldehyde. After quenching the reaction with 2.4 ml of glacial acetic acid, the reaction was extracted with 20 ml of ethyl acetate. The ethyl acetate phase was dried in vacuo, redissolved in methanol, and subjected to analysis by HPLC–chemical ionization MS.
RESULTS
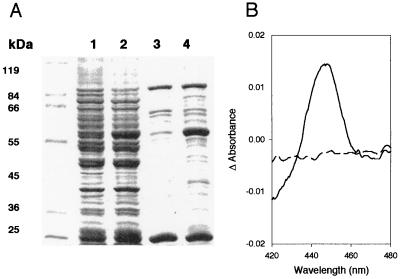
Expression of F5H in Yeast. After induction in the presence of galactose, yeast cells harboring pYeDP60 (control yeast) and pYeDP60-F5H were harvested and used for the isolation of microsomal proteins. SDS/PAGE analysis of total microsomal proteins (Fig. 1a, lanes 1 and 2) suggested that microsomes of yeast carrying pYeDP60-F5H contained an abundant protein with a molecular mass of ≈58 kDa, very close to the expected mass of 58,727 Da for the inferred translation product of the F5H cDNA. This protein was absent in samples prepared from control yeast. The microsomal preparations were then further fractionated by using a Triton X-114 phase-partition procedure in which integral membrane proteins are extracted into a detergent-rich phase in the presence of high concentrations of glycerol (16). When analyzed by using SDS/PAGE (Fig. 1a, lanes 3 and 4), the Triton phase prepared from microsomes of control yeast contains a number of bands, whereas similar preparations from yeast expressing F5H contain an additional prominent protein with a molecular mass of ≈58 kDa.
Figure 1.
Analysis of F5H expressed in yeast. (A) SDS/PAGE analysis of microsomal and membrane proteins from yeast harboring pYeDP60 (lanes 1 and 3) and the F5H expression vector pYeDP60-F5H (lanes 2 and 4). Lanes 1 and 2, yeast microsomal proteins; lanes 3 and 4, membrane protein-enriched detergent phase from Triton X-114 phase-partitioning of yeast microsomal proteins. (B) Carbon monoxide difference spectrum of dithionite-reduced microsomes from yeast expressing F5H. Dotted line, baseline before CO treatment; solid line, difference spectrum after CO treatment.
To characterize the recombinant F5H protein in greater detail, it was analyzed spectroscopically. The reduced carbon monoxide difference spectrum of the Triton phase exhibited a peak at 450 nm, characteristic of the Soret band shift seen with most P450s (Fig. 1b; ref. 20). No such peak was observed when similar analyses were performed on the Triton phase prepared from control yeast. These data indicate that in yeast carrying the pYeDP60-F5H vector, F5H is expressed and folded into a near-native, and possibly catalytically active, form.
Kinetic Analysis of F5H Substrate Specificity. Yeast cultures and microsomal protein preparations were assayed for F5H activity to explore the kinetics and substrate specificity of F5H catalysis. The WAT11 yeast strain is the system of choice for F5H expression because this strain has been engineered to express the ArabidopsisP450 reductase gene ATR1 under the control of a galactose-inducible promoter (12). Our initial characterization of F5H was based on the conventional view that the enzyme catalyzes the conversion of ferulate to 5-hydroxyferulate. The activity of F5H was first investigated by using an in vivo assay for F5H modeled after that described for cinnamate 4-hydroxylase (15). Two hours after the addition of ferulate to the medium of yeast expressing F5H, a novel UV-absorbent compound could be extracted from the medium with an HPLC retention time identical to that of authentic 5-hydroxyferulate (data not shown). No such compound was identified in the medium of control yeast. GC-electron impact MS analysis of the N,O-bis(trimethylsilyl) trifluoroacetamide-derivatized ethyl acetate extract of the acidified medium revealed the presence of a compound that co-chromatographed with the trimethylsilylated derivative of authentic 5-hydroxyferulate and exhibited a parental ion of the expected m/z ratio (M+ 426). These data demonstrated that the yeast harboring pYeDP60-F5H were expressing a catalytically active form of F5H.
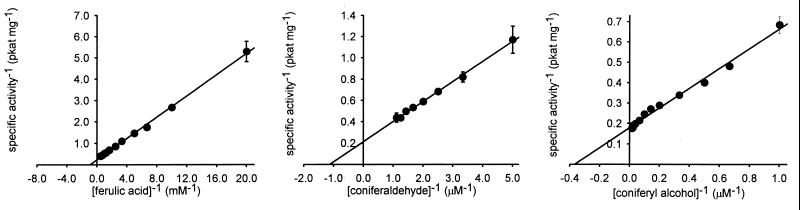
To examine the kinetics of F5H in vitro, we assayed for the conversion of ferulate to 5-hydroxyferulate by HPLC. F5H demonstrated Michaelis–Menten kinetics with regard to ferulate hydroxylation with a Km of 1 mM and a Vmax of 4 pkat⋅mg−1 protein (Fig. 2). This Km was unexpectedly high considering that cinnamate 4-hydroxylase, a P450 earlier in the pathway, exhibits a 4 μM Km for its substrate (15). This inconsistency led us to test the hypothesis that phenylpropanoid pathway intermediates other than ferulate might be better substrates for F5H. Assays conducted with coniferaldehyde demonstrated that the Km and Vmax of F5H for this substrate were 1 μM and 5 pkat⋅mg−1 respectively, and the corresponding values for coniferyl alcohol were 3 μM and 6 pkat⋅mg−1 (Fig. 2). The identity of the product derived from coniferyl alcohol was confirmed to be 5-hydroxyconiferyl alcohol by using GC-electron impact MS (m/z M+ 412) after its isolation from an in vivo assay similar to that described above for 5-hydroxyferulate. The identity of the 5-hydroxyconiferaldehyde produced from coniferaldehyde in a large-scale in vitro assay was confirmed by HPLC-chemical ionization MS (m/z M + H+ 195). In both cases, these compounds were not produced by control yeast or control yeast microsomes. No data were obtained indicating that feruloyl CoA is an F5H substrate (data not shown). These data indicate that coniferaldehyde and coniferyl alcohol are the preferred substrates for F5H in vitro.
Figure 2.
Kinetic analysis of F5H-catalyzed substrate 5-hydroxylation with ferulate, coniferaldehyde, and coniferyl alcohol as substrates. Error bars represent 1 SD for triplicate assays.
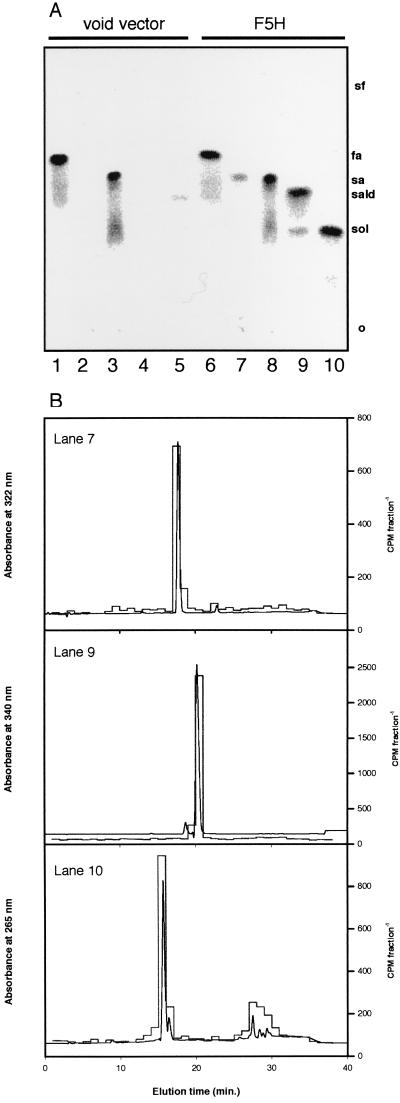
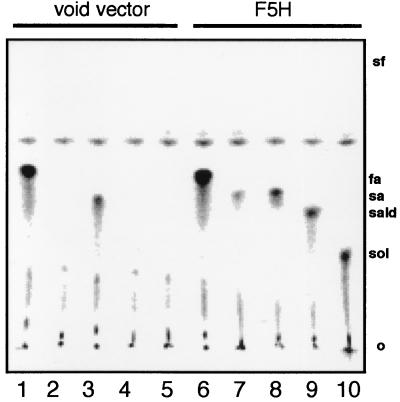
O-Methylation of 5-Hydroxyconiferaldehyde and 5-Hydroxyconiferyl Alcohol by Arabidopsis OMT Activity. The F5H-catalyzed conversion of coniferaldehyde and coniferyl alcohol to their corresponding 5-hydroxy derivatives suggested that these reactions may be relevant for the production of syringyl-substituted monolignols. This pathway would require the existence of one or more OMT activities capable of acting on 5-hydroxyconiferaldehyde and/or 5-hydroxyconiferyl alcohol. To determine whether this hypothetical pathway could exist in Arabidopsis, rachis extracts were prepared and tested for OMT activity in assays coupled to the F5H-catalyzed production of these putative substrates. When ArabidopsisOMT extracts were incubated with control yeast microsomes, products labeled from S-adenosyl-l-[methyl-14C]-methionine were found only in assays in which caffeate or 5-hydroxyferulate were used as substrates, consistent with the presence of COMT activity in the Arabidopsis extracts (Fig. 3a). Identical experiments carried out with microsomes from yeast expressing F5H gave similar results with these two substrates, but also gave radiolabeled products in assays in which ferulate, coniferaldehyde, and coniferyl alcohol were used as substrates in the coupled assay (Fig. 3a). Although the identity of these compounds was strongly suggested by co-chromatography of the radioactive products on TLC with authentic sinapic acid, sinapaldehyde, and sinapyl alcohol, their identity was confirmed by subsequent co-chromatography on HPLC after elution of the compounds from the TLC stationary phase (Fig. 3b).
Figure 3.
TLC/HPLC analysis of products from F5H assays coupled to OMT activity from Arabidopsis rachis extracts. (A) Ethyl acetate-soluble assay products were separated by TLC, and radioactive products labeled from S-adenosyl-l-[methyl-14C]methionine were visualized on a Packard Instant Imager. The substrates employed were caffeic acid (lanes 1 and 6), ferulic acid (lanes 2 and 7), 5-hydroxy- ferulic acid (lanes 3 and 8), coniferaldehyde (lanes 4 and 9), and coniferyl alcohol (lanes 5 and 10). Lanes 1–5, control yeast microsomes; lanes 6–10, microsomes from yeast expressing F5H. The relative mobilities of the putative products, ferulic acid (fa), sinapic acid (sa), sinapaldehyde (sald), and sinapyl alcohol (sol), were determined by observation of nonradioactive standards under UV light. (B) The identity of the products was confirmed by elution of the radiolabeled compounds from the TLC stationary phase and co-chromatography of the radiolabel with authentic standards on HPLC. UV absorbance is shown as a continuous line; radioactivity is shown as a histogram.
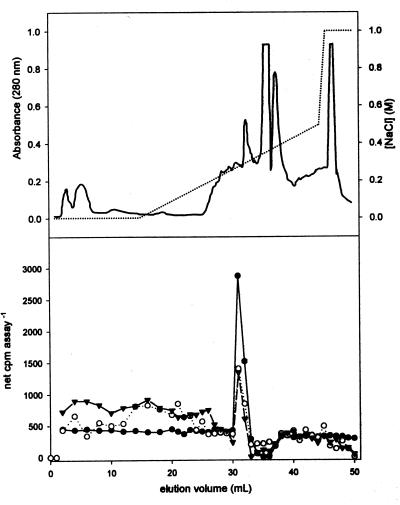
To determine whether the enzyme(s) that catalyzes the 5-O-methylation of 5-hydroxyconiferaldehyde and 5-hydroxyconiferyl alcohol could be distinguished from COMT by anion exchange chromatography, a crude Arabidopsis extract was fractionated on a Mono Q FPLC column. Aliquots of individual fractions were tested for activity with caffeic acid as a substrate for COMT and by using the coupled F5H–OMT assay described above with coniferaldehyde and coniferyl alcohol as substrates. Assays for COMT revealed a single peak of activity that eluted in the middle of the NaCl gradient (Fig. 4). The 5-hydroxyconiferaldehyde and 5-hydroxyconiferyl alcohol O-methylation activity coeluted with the peak of COMT activity.
Figure 4.
Separation of OMT activities from Arabidopsis rachis extracts by anion-exchange chromatography. Proteins were eluted from the column by using a salt gradient (dotted line), and monitored by the UV absorbance of the eluate (solid line) (Upper). Aliquots of individual fractions were used in coupled assays with F5H. Ethyl acetate-soluble assay products labeled from S-adenosyl-l-[methyl-14C]methionine were analyzed by liquid scintillation counting (Lower). The substrates used were caffeic acid (●), coniferaldehyde (○), and coniferyl alcohol (▴).
O-Methylation of 5-Hydroxyconiferaldehyde and 5-Hydroxyconiferyl Alcohol by Arabidopsis COMT Expressed in E. coli.
Co-chromatography of the OMT activities on FPLC suggested that all three of these reactions may be catalyzed by the same enzyme. To determine whether the Arabidopsis COMT can catalyze 5-O-methylation of 5-hydroxyconiferaldehyde and 5-hydroxyconiferyl alcohol, the protein was expressed in E. coli and used in an F5H–OMT-coupled assay. The results obtained by using the heterologously expressed protein were identical to those obtained when Arabidopsis rachis tissue was used as the source of OMT activity. Caffeate and 5-hydroxyferulate were the only substrates methylated in control reactions. When F5H was used to produce 5-hydroxyferulate, 5-hydroxyconiferaldehyde, and 5-hydroxyconiferyl alcohol, these compounds were all used as COMT substrates (Fig. 5 and data not shown).
Figure 5.
TLC analysis of products from F5H assays coupled to OMT activity from the Arabidopsis COMT expressed in E. coli. Ethyl acetate-soluble assay products were separated by TLC (A), and radioactive products labeled from S-adenosyl-l-[methyl-14C]-methionine were visualized on a Packard Instant Imager. The substrates used were caffeic acid (lanes 1 and 6), ferulic acid (lanes 2 and 7), 5-hydroxyferulic acid (lanes 3 and 8), coniferaldehyde (lanes 4 and 9), and coniferyl alcohol (lanes 5 and 10). Lanes 1–5, control yeast microsomes; lanes 6–10, microsomes from yeast expressing F5H. The relative mobilities of the putative products, ferulic acid (fa), sinapic acid (sa), sinapaldehyde (sald), and sinapyl alcohol (sol), were determined by observation of nonradioactive standards under UV light.
DISCUSSION
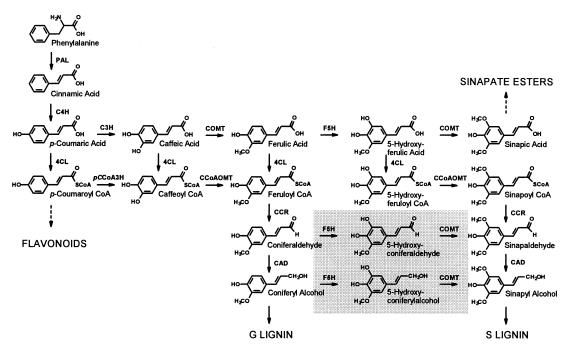
The potential success or failure of metabolic engineering efforts hinge on a thorough understanding of the target pathway. Recent experiments have demonstrated that our understanding of lignin biosynthesis is far from complete, and as a result, our ability to modify the pathway may be limited. Monolignol biosynthesis involves a series of hydroxylations and methylations of the cinnamic acid-derived phenylpropane skeleton (Fig. 6). These reactions have long been thought to occur at the level of the corresponding free acids, including the conversion of ferulate to 5-hydroxyferulate catalyzed by F5H. From the results reported here, it is apparent that the previous model of lignin biosynthesis is likely to be incorrect, at least with respect to F5H and COMT. Although F5H is capable of catalyzing the conversion of ferulate to 5-hydroxyferulate, later intermediates in the pathway, coniferaldehyde and coniferyl alcohol, are more effective substrates. Whereas it is difficult to determine or predict the concentrations of each of these substrates in vivo, the very large difference in Km between ferulate and coniferaldehyde as well as coniferyl alcohol is consistent with the hypothesis that the latter compounds are the more relevant substrates in lignifying tissues.
Figure 6.
Revised pathway of phenylpropanoid biosynthesis in Arabidopsis including the reactions now known to be catalyzed by F5H and COMT (boxed in gray). The enzymes and their abbreviations are caffeoyl CoA O-methyltransferase (CCoAOMT), cinnamate 4-hydroxylase (C4H), cinnamoyl alcohol dehydrogenase (CAD), cinnamoyl CoA reductase (CCR), p-coumarate 3-hydroxylase (C3H), 4-(hydroxy)cinnamoyl CoA ligase (4CL), ferulate 5-hydroxylase (F5H), p-coumaroyl CoA 3-hydroxylase (pCCoA3H), phenylalanine ammonia-lyase (PAL), caffeic acid/5-hydroxyferulic acid O-methyltransferase (COMT).
This new route to syringyl monolignols may explain enigmatic results obtained through enzymologic studies and the generation of transgenic plants in which the expression of lignin biosynthetic pathway genes was either up- or down-regulated. First, the long-accepted pathway for syringyl monomer biosynthesis, in which sinapic acid is activated to its corresponding CoA ester by 4-(hydroxy)cinnamoyl CoA ligase (4CL), has recently been called into question because 4CL in Arabidopsis and other plants has no activity toward sinapic acid, suggesting that a 4CL-independent pathway to syringyl lignin monomers must exist (21–24). If syringyl lignin monomers are instead synthesized from coniferaldehyde and/or coniferyl alcohol, a sinapoyl CoA ligase activity is not necessary. Second, an alternative pathway to guaiacyl subunits has recently been elucidated in which p-coumaroyl CoA is hydroxylated and subsequently methylated to give feruloyl CoA (25, 26). The importance of this route of lignin monomer synthesis has been demonstrated by antisense suppression of caffeoyl CoA O-methyltransferase expression in tobacco, which led to an overall decrease in lignin accumulation (27). In the context of this alternative pathway, it was difficult to reconcile the results of transgenic experiments in Arabidopsis in which F5H overexpression led to lignin that was almost solely derived from syringyl units (10). According to the classical model of the phenylpropanoid pathway, the flow of carbon through the alternative pathway would provide a route by which guaiacyl lignin precursors could escape hydroxylation by F5H. As a result, the effect of F5H overexpression should have been limited by the ratio of flux through the free-acid pathway and the alternative pathway. The demonstration that F5H is capable of hydroxylating coniferaldehyde and coniferyl alcohol places its activity downstream of the alternative pathway and explains how F5H overexpression is capable of diverting virtually all monolignol precursors into syringyl lignin biosynthesis. Finally, the finding that antisense suppression of COMT gene expression in poplar and tobacco preferentially suppresses syringyl monomer synthesis raised questions concerning how this enzyme fits into the lignin biosynthetic pathway as well (28, 29). These questions have been at least partially answered in this study by the demonstration that COMT is also capable of catalyzing the conversion of 5-hydroxyconiferaldehyde and 5-hydroxyconiferyl alcohol to their syringyl-substituted counterparts. Thus, down-regulation of COMT expression would be expected to impact the synthesis of syringyl monomers late in the lignin biosynthetic pathway, whereas the alternative pathway would provide a COMT-independent route for the synthesis of guaiacyl monomers.
Recent heavy-isotope feeding studies have provided evidence for a pathway involving coniferyl alcohol and/or coniferaldehyde hydroxylation and subsequent O-methylation (30). Our results are consistent with these data and identify the enzymes involved (Fig. 6). F5H is an obligatory catalyst in syringyl lignin monomer synthesis (8) that is likely to act primarily on coniferaldehyde and coniferyl alcohol in vivo. The 5-O-methylation step is almost certainly catalyzed by COMT, although we cannot currently rule out the existence of other OMTs with assay requirements that were not met in our F5H–OMT-coupled system.
Although most of the animal P450s that have been studied have broad substrate specificity and function in the catabolism of xenobiotics, the plant P450s that have been studied to date have anabolic roles and are highly substrate-specific (31). F5H is unusual in that it is a multifunctional plant P450 with three physiologically relevant substrates. The Km values for coniferaldehyde and coniferyl alcohol are consistent with the Km values of cinnamate 4-hydroxylase for cinnamate (15). The high Km for ferulate demonstrated by F5H raises the question of whether the compound is an actual substrate in vivo. The presence of soluble sinapic acid-derived secondary metabolites in Arabidopsis and some other plants suggests that ferulate can be converted, at least in some plants, to sinapate by F5H and COMT. Alternatively, these two enzymes might act to convert coniferaldehyde to sinapaldehyde, which could subsequently be oxidized to sinapic acid by an aldehyde oxidase.
Acknowledgments
We thank Drs. Philip Urban and Denis Pompon for the generous donation of pYeDP60 and the WAT11 yeast strain, Dr. Don Gray (Bioanalytical Systems, West Lafayette, IN) for the HPLC-MS analysis, and Joanne Cusumano for careful review of the manuscript. This work was supported by a grant from the Division of Energy Biosciences, United States Department of Energy to C.C. This is journal paper number 16043 of the Purdue University Agricultural Experiment Station.
ABBREVIATIONS
- 4CL
4-(hydroxy)cinnamoyl CoA ligase
- COMT
caffeic acid/5-hydroxyferulic acid O-methyltransferase
- F5H
ferulate 5-hydroxylase
- OMT
O-methyltransferase
- P450
cytochrome P450-dependent monooxygenase
References
- 1.Lewis N G, Yamamoto E. Annu Rev Plant Physiol Plant Mol Biol. 1990;41:455–496. doi: 10.1146/annurev.pp.41.060190.002323. [DOI] [PubMed] [Google Scholar]
- 2.Nicholson R L, Hammerschmidt R. Annu Rev Phytopathol. 1992;30:369–389. [Google Scholar]
- 3.Whetten R, Sederoff R. Forest Ecol Management. 1991;43:301–316. [Google Scholar]
- 4.Jung H G, Deetz D A. In: Forage Cell Wall Structure and Digestibility. Jung H G, Buxton D R, Hatfield R D, Ralph J, editors. Madison, WI: Am. Soc. Agron./Crop Sci. Soc. Am./Soil Sci. Soc. Am. Press; 1993. pp. 315–346. [Google Scholar]
- 5.Campbell M M, Sederoff R R. Plant Physiol. 1996;110:3–13. doi: 10.1104/pp.110.1.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Higuchi T, Brown S A. Can J Biochem Physiol. 1963;41:613–620. [PubMed] [Google Scholar]
- 7.Grand C. FEBS Lett. 1984;169:7–11. [Google Scholar]
- 8.Chapple C C S, Vogt T, Ellis B E, Somerville C R. Plant Cell. 1992;4:1413–1424. doi: 10.1105/tpc.4.11.1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meyer K, Cusumano J C, Somerville C, Chapple C C S. Proc Natl Acad Sci USA. 1996;93:6869–6874. doi: 10.1073/pnas.93.14.6869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meyer K, Shirley A M, Cusumano J C, Bell-Lelong D A, Chapple C. Proc Natl Acad Sci USA. 1998;95:6619–6623. doi: 10.1073/pnas.95.12.6619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Truan G, Cullin C, Reisdorf P, Urban P, Pompon D. Gene. 1993;125:49–55. doi: 10.1016/0378-1119(93)90744-n. [DOI] [PubMed] [Google Scholar]
- 12.Pompon D, Louerat B, Bronine A, Urban P. Methods Enzymol. 1996;272:51–64. doi: 10.1016/s0076-6879(96)72008-6. [DOI] [PubMed] [Google Scholar]
- 13.Urban P, Cullin C, Pompon D. Biochimie. 1990;72:463–472. doi: 10.1016/0300-9084(90)90070-w. [DOI] [PubMed] [Google Scholar]
- 14.Geitz R D, St. Jean A, Woods R A, Schiestl R H. Nucleic Acids Res. 1992;20:1425. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Urban P, Werck-Reichhart D, Teutsch H, Durst F, Regnier S, Kazmaier M, Pompon D. Eur J Biochem. 1994;222:843–850. doi: 10.1111/j.1432-1033.1994.tb18931.x. [DOI] [PubMed] [Google Scholar]
- 16.Werck-Reichhart D, Benveniste I, Teutsch H, Durst F, Gabriac B. Anal Biochem. 1991;197:125–131. doi: 10.1016/0003-2697(91)90367-3. [DOI] [PubMed] [Google Scholar]
- 17.Laemmli U K. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 18.Estabrook R W, Werringloer J. Methods Enzymol. 1978;52:212–220. doi: 10.1016/s0076-6879(78)52024-7. [DOI] [PubMed] [Google Scholar]
- 19.Zhang H, Wang J, Goodman H M. Biochim Biophys Acta. 1997;1353:199–202. doi: 10.1016/s0167-4781(97)00096-1. [DOI] [PubMed] [Google Scholar]
- 20.Loew G H, Rohmer M-M. J Am Chem Soc. 1980;102:3655–3657. [Google Scholar]
- 21.Lee D, Douglas C J. Plant Physiol. 1996;112:193–205. doi: 10.1104/pp.112.1.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee D, Meyer K, Chapple C, Douglas C J. Plant Cell. 1997;9:1985–1998. doi: 10.1105/tpc.9.11.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meng H, Campbell W H. Phytochemistry. 1997;44:605–608. [Google Scholar]
- 24.Allina S M, Pri-Hadash A, Theilmann D A, Ellis B E, Douglas C J. Plant Physiol. 1998;116:743–754. doi: 10.1104/pp.116.2.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ye Z-H, Kneusel R E, Matern U, Varner J E. Plant Cell. 1994;6:1427–1439. doi: 10.1105/tpc.6.10.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ye Z-H, Varner J E. Plant Physiol. 1995;108:459–467. doi: 10.1104/pp.108.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhong R, Morrison W H, Negrel J, Ye Z-H. Plant Cell. 1998;10:2033–2045. doi: 10.1105/tpc.10.12.2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Atanassova R, Favet N, Martz F, Chabbert B, Tollier M-T, Monties B, Fritig B, Legrand M. Plant J. 1995;8:465–477. [Google Scholar]
- 29.Van Doorsselaere J, Baucher M, Chognot E, Chabbert B, Tollier M-T, Petit-Conil M, Leplé J-C, Pilate G, Cornu D, Monties B, et al. Plant J. 1995;8:855–864. [Google Scholar]
- 30.Chen F, Yasuda S, Fukushima K. Planta. 1999;207:597–603. [Google Scholar]
- 31.Chapple C. Annu Rev Plant Physiol Plant Mol Biol. 1998;49:311–343. doi: 10.1146/annurev.arplant.49.1.311. [DOI] [PubMed] [Google Scholar]