Abstract
Herbivorous insect species are constantly challenged with reactive oxygen species (ROS) generated from endogenous and exogenous sources. ROS produced within insects because of stress and prooxidant allelochemicals produced by host plants in response to herbivory require a complex mode of antioxidant defense during insect/plant interactions. Some insect herbivores have a midgut-based defense against the suite of ROS encountered. Because the Hessian fly (Mayetiola destructor) is the major insect pest of wheat worldwide, and an emerging model for all gall midges, we investigated its antioxidant responses during interaction with its host plant. Quantitative data for two phospholipid glutathione peroxidases (MdesPHGPX-1 and MdesPHGPX-2), two catalases (MdesCAT-1 and MdesCAT-2), and two superoxide dismutases (MdesSOD-1 and MdesSOD-2) revealed high levels of all of the mRNAs in the midgut of larvae on susceptible wheat (compatible interaction). During development of the Hessian fly on susceptible wheat, a differential expression pattern was observed for all six genes. Analysis of larvae on resistant wheat (incompatible interaction) compared with larvae on susceptible wheat showed increased levels of mRNAs in larvae on resistant wheat for all of the antioxidant genes except MdesSOD-1 and MdesSOD-2. We postulate that the increased mRNA levels of MdesPHGPX-1, MdesPHGPX-2, MdesCAT-1, and MdesCAT-2 reflect responses to ROS encountered by larvae while feeding on resistant wheat seedlings and/or ROS generated endogenously in larvae because of stress/starvation. These results provide an opportunity to understand the cooperative antioxidant defense responses in the Hessian fly/wheat interaction and may be applicable to other insect/plant interactions.
Keywords: Hessian fly, insect/plant interaction, reactive oxygen species, wheat
Reactive oxygen species (ROS) such as superoxide radicals (O2−), hydroxyl radical (OH−), H2O2, and hydroperoxides (ROOH) are generated by exogenous and endogenous sources (1). Exogenous sources, including prooxidant allelochemicals, pose a serious challenge to herbivorous insect species during host interactions, whereas ROS generated because of stress/starvation are an important endogenous source. However, insects have evolved a complex antioxidant mechanism to overcome the toxic effects of ROS. The antioxidant defense is primarily constituted by the enzymatic actions of glutathione peroxidase (GPX), catalase (CAT), superoxide dismutase (SOD), and ascorbate peroxidase (2).
GPXs reduce H2O2 and hydroperoxides, thereby scavenging oxidative radicals in tissues and cell membranes (3). According to Behne and Kyriakopoulos (4), the mammalian GPX enzymes, which have selenium associated with the active cysteine (selenium-dependent), can be grouped into five forms: the classical or cytosolic GPX, gastrointestinal GPX, plasma GPX, phospholipid hydroperoxide GPX (PHGPX), and sperm nuclei GPX. However, PHGPXs reported in nematodes (5), endoparasitoids (6), insects (7), and plants (8) encode selenium-independent forms. In particular, the PHGPX forms reduce phospholipid and cholesterol hydroperoxides and thereby play an important role in protecting biological membranes against oxygen toxicity. SODs are characterized by the presence of metal prosthetic groups and can be classified into two major families in Drosophila melanogaster: Cu/Zn-SOD (Sod1), located mainly in the cytosol; and Mn-SOD (Sod2), found in mitochondria (9). SOD converts O2− to molecular O2 and H2O2 (10). H2O2 is subsequently scavenged by CAT, resulting in the production of water and molecular oxygen. Ascorbate peroxidase also scavenges H2O2, but activity is probably limited to the H2O2 not scavenged by CAT (11).
Because of its agricultural importance as the major pest of wheat worldwide, more knowledge about the Hessian fly (Mayetiola destructor) and its interaction with its host at the molecular level would be useful. Additionally, the Hessian fly is emerging as a general model for members of the Cecidomyiidae (gall midges), the sixth largest family of the Diptera. The life cycle of the Hessian fly consists of three larval instars, pupa, and adult. Duration of the first stadium is 6 days, and that of the second stadium is 5–6 days (12). The third instar is a nonfeeding stadium contained within a puparium and under field conditions normally diapauses over the winter or summer. However, when the insect completes its development continuously under favorable temperature conditions the duration of the third stadium is 6–7 days (13). Damage to wheat is entirely due to feeding first and second larval instars. On seedling wheat (fall infestation), larval infestation causes stunting and development of a dark green color in infested shoots or tillers and can lead to the death of seedling plants (14). However, on jointing wheat (spring infestation), larval feeding prevents normal elongation of the stem and transport of nutrients to the developing grain (15). To date, the most effective means of control for the Hessian fly has been via genetic resistance in the host plant (16), with 32 Hessian fly resistance genes identified so far (17). This resistance is expressed as larval antibiosis and is controlled mostly by single plant genes that are partially to completely dominant (18).
There are two types of the Hessian fly/wheat interactions. First, compatible interactions allow first-instar larvae to survive on susceptible wheat plants. In these interactions, larvae establish a sustained feeding site, develop normally, and complete their life cycle. However, the susceptible wheat seedlings are severely affected (19). Second, incompatible interactions inhibit survival of first-instar larvae on resistant wheat plants. These interactions are characterized by larvae that fail to establish a sustained feeding site or develop normally and usually die within a period of 5–6 days after hatching (20). Resistant wheat undergoes little or no physiological stress during Hessian fly attack (21) and yields normally. Furthermore, resistant wheat in response to attack by larval Hessian fly has been shown to produce greater levels of mRNAs for a number of putative defense genes including lipoxygenase (LOX), an ROS-generating enzyme (22).
The presence of glutathione-S-transferases (GSTs) (23) and cytochrome P450s (24) has been documented in the Hessian fly. These studies suggest a plausible role for two delta GSTs (MdesGST-1 and MdesGST-3) and a CYP6 cytochrome P450 (CYP6AZ1) in detoxifying wheat allelochemicals during feeding. Furthermore, a sigma GST (MdesGST-2) and another CYP6 cytochrome P450 (CYP6BA1) were speculated to have general functions during development (23, 24). In this study we report the transcription profiles of two PHGPXs (MdesPHGPX-1 and MdesPHGPX-2), two CATs (MdesCAT-1 and MdesCAT-2), and two SODs (MdesSOD-1 and MdesSOD-2) in larval tissues during development and in larvae participating in compatible and incompatible interactions. Results are discussed in the context of protection against possible peroxide-induced damage in feeding Hessian fly larvae and during development.
Results
Characterization of the Hessian Fly Antioxidant Genes.
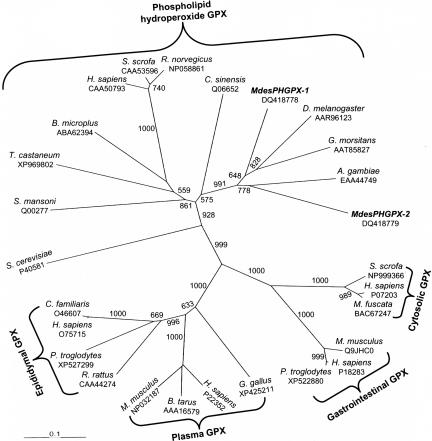
Compared with vertebrates little is known about PHGPX genes in insects (6). The MdesPHGPX-1 deduced amino acid sequence revealed 70% similarity (3e-67 threshold) with a Glossina morsitans (AAT85827) PHGPX. MdesPHGPX-2 showed greatest amino acid similarity (66%, 9e-57) with an Anopheles gambiae (EAA44749) PHGPX. The deduced protein sequences for both the Hessian fly PHGPXs revealed the presence of a conserved catalytic triad cysteine (C), glutamine (Q), and tryptophan (W) (3, 6). The deduced catalytic triads for MdesPHGPX-1 and MdesPHGPX-2 were C47-Q78-W137 and C44-Q75-W134, respectively. Phylogenetic analyses using maximum parsimony and distance/neighbor-joining criteria both yielded dendrograms with the same topology supporting identity of the Hessian fly PHGPXs by grouping them specifically with PHGPXs from other Diptera, whereas other classes of GPXs (cytosolic, gastrointestinal, plasma, and epididymal) grouped in the dendrograms separate from the PHGPXs (Fig. 1). The deduced protein sequences for both Hessian fly CATs and SODs also revealed a high level of homology with other members of Diptera.
Fig. 1.
Dendrogram of the GPX families calculated from aligned amino acid sequences. The topology and branch lengths of the radial phylogram were produced by the distance/neighbor-joining criteria. Numbers at the branches correspond to bootstrap support >50%. M. destructor phosopholipid hydroperoxide GPXs (PHGPXs) MdesPHGPX-1 and MdesPHGPX-2 group with PHGPXs from other Diptera. Taxa and GenBank accession numbers included are as follows: Saccharomyces cerevisiae, P40581; Schistosoma mansoni, QO0277; Tribolium castaneum, XP_969802; Boophilus microplus, ABA62394; Homo sapiens, CAA50793; Sus scrofa, CAA53596; Rattus norvegicus, NP058861; Citrus sinensis, Q06652; M. destructor, DQ418778; D. melanogaster, AAR96123; G. morsitans, AAT85827; A. gambiae, EAA44749; M. destructor, DQ418779; Sus scrofa, NP999366; H. sapiens, P07203; Macaca fuscata, BAC67247; Mus musculus, Q9JHC0; H. sapiens, P18283; Pan troglodytes, XP_522880; Gallus gallus, XP_425211; H. sapiens, P22352; Bos taurus, AAA16579; Mus musculus, NP032187; Rattus rattus, CAA44274; P. troglodytes, XP_527299; H. sapiens, O75715; Canis familiaris, O46607.
Transcriptional Patterns of the Hessian Fly Antioxidant Genes in Larval Tissues.
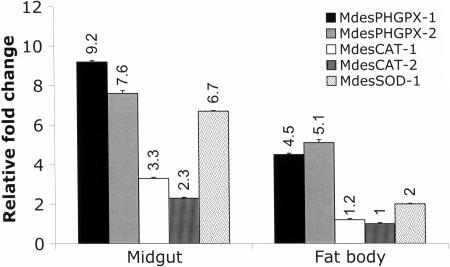
The mRNA level in all larval tissues was assessed in larvae that were reared on susceptible wheat. All of the antioxidant genes had the highest expression in the midgut and the lowest in salivary glands with fat body producing intermediate mRNA levels (Fig. 2). Hence, the quantitation of antioxidant gene mRNA levels in the midgut and fat body is presented relative to the salivary glands, which was taken as the calibrator sample. Of the genes assayed, MdesPHGPX-1 and Mdes-PHGPX-2 showed the greatest mRNA levels in all tissues, whereas MdesCAT-2 showed the least. Furthermore, a significant difference (P < 0.05) between the mRNA levels in fat body and salivary glands was revealed for all genes assayed except MdesCAT-2 (P > 0.05). The transcript of MdesSOD-2 was detected at a very low level in the midgut and was below detection in the fat body and salivary glands (data not shown).
Fig. 2.
Temporal gene expression of the Hessian fly antioxidant genes in larval tissues. Gene expression was studied in midgut, salivary glands, and fat body. Expression in the salivary glands was taken as the calibrator, and the expression in midgut and fat body samples was calculated relative to the expression in the salivary glands to reveal the fold changes. The standard error is represented by the error bars for three technical replicates.
Transcriptional Patterns of the Hessian Fly Antioxidant Genes During Development.
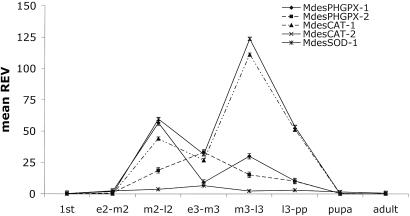
The mRNA level in all of the life stages of the Hessian fly was assessed in compatible interactions because there is no developmental progress in incompatible interactions. For all of the antioxidant genes, mRNA levels increased from first larval instar to third larval instar and thereafter declined in the pupal and adult stages (Fig. 3). Therefore, the lowest levels of mRNA for the six antioxidant genes were calculated in the pupal and adult stages. Compared with the first larval instar, a significant (P < 0.05) fold increase in MdesPHGPX-1 and MdesPHGPX-2 mRNA levels was observed for second and third larval instars but not for pupae and adults (P > 0.05). Both the CATs (MdesCAT-1 and MdesCAT-2) showed similar expression patterns throughout development with two distinct peaks. The first peak in the mRNA level was observed in mid to late second larval instars, and the second peak was observed in mid to late third larval instars (Fig. 3). The mRNA levels for MdesCAT-1 and MdesCAT-2 were calculated to be significant (P < 0.05) between any two given stages. The mRNA level for MdesSOD-1 was significantly different only between the first larval instar and the later second and third larval instars. MdesSOD-2 mRNA, although detected at very low levels, also showed an expression profile similar to that of MdesSOD-1 (data not shown).
Fig. 3.
Temporal gene expression of the Hessian fly antioxidant genes during development. Gene expression was studied for all of the developmental stages including first, second, and third larval instars, pupae, and adults. REV for all of the genes was calculated by using an endogenous Hessian fly ubiquitin gene. The standard error is represented by the error bars for three technical replicates. e2, early second instar; m2, mid second instar; l2, late second instar; e3, early third instar; m3, mid third instar; l3, late third instar.
Differential mRNA Levels of the Hessian Fly Antioxidant Genes During Interactions with Wheat.
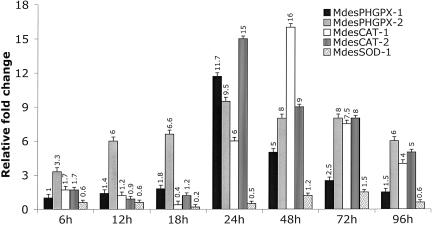
The expression patterns of the Hessian fly antioxidant genes were assessed in larvae on resistant and susceptible wheat representing incompatible and compatible interactions, respectively. We observed the mRNA levels for all six Hessian fly antioxidant genes to be greater in larvae during incompatible interactions (Fig. 4). In the initial phase of the interaction (6–18 h after hatching), mRNA levels for only MdesGPX-2 were high in avirulent larvae. However, in the later phase of the interaction (24–96 h after hatching), except for MdesSOD-1 there were significant increases in antioxidant gene mRNA levels (P < 0.05) in larvae during incompatible interactions compared with similar-aged larvae during compatible interactions (Fig. 4). MdesGPX-2 and MdesCAT-2 transcripts were the most abundant during these interactions. The highest level of these transcripts was observed 24 and 48 h after hatching and thereafter declined in the later time points. The average fold increase in mRNA levels for all genes at 6, 12, 18, 24, 48, 72, and 96 h after hatching was 1.6, 2.0, 2.0, 8.5, 7.8, 5.5, and 3.4, respectively. No significant difference (P > 0.05) for MdesSOD-1 was detected for mRNA levels in larvae during incompatible/compatible interactions. The expression pattern for MdesSOD-2 during these interactions was not assessed because of the very low level of transcript detected.
Fig. 4.
Temporal gene expression patterns of the Hessian fly antioxidant genes during interactions with wheat. Gene expression was studied in compatible and incompatible interactions. Relative fold change for all of the genes was determined by dividing the REV calculated for Biotype L larvae on resistant Iris wheat (incompatible interaction) by the REV calculated for Biotype L larvae on susceptible Newton wheat (compatible interaction). The standard error is represented by the error bars for two biological replicates (two technical replicates within each).
Discussion
We report the transcriptional expression patterns for six antioxidant genes in the Hessian fly. The classification of these Hessian fly antioxidant genes was primarily based on the identity shared at the amino acid level with other known insect antioxidant enzymes. The deduced amino acid sequences for the Hessian fly antioxidant enzymes were in agreement in length and contained conserved residues that are characteristic of similar enzymes. In particular, the deduced amino acid sequences of both Hessian fly PHGPX genes revealed the presence of a nonselenium cysteine residue in the active pocket of the protein, thus classifying these enzymes as selenium-independent forms of PHGPX, or cys-PHGPX. The C47 of MdesPHGPX-1 and C44 of MdesPHGPX-2 are assumed to be the active catalytic residue in all cys-PHGPX enzymes reported thus far (5–8, 25). Furthermore, phylogenetic analyses grouped the PHGPXs, including the Hessian fly PHGPXs, separate from other forms of GPXs. Topology of the dendrograms in our analyses grouped the various classes of GPXs in agreement with previous phylogenetic analysis of GPXs (26).
To date, the exact source(s) of oxidative stress in the Hessian fly is unknown. Results obtained in this study support ROS generation due to both exogenous and endogenous sources. Several of these possibilities include oxidative stress from plant-generated ROS, starvation/stress, or even the reduced availability of ingested low-molecular-weight antioxidants such as reduced ascorbate and glutathione. Thus, changes in expression for the Hessian fly antioxidant mRNAs cannot be singly attributed to either endogenous or exogenous ROS.
Tissue-specific analysis in the current study revealed the highest mRNA levels of the six Hessian fly antioxidant genes in the midgut compared with the levels in fat body and salivary glands. This is similar to the patterns described for Spodoptera littoralis (27), G. morsitans (7), and Aulocara ellioti (2). In S. littoralis and A. ellioti high levels of SOD and CAT activity occur in the midgut contents and/or midgut tissues (2, 27), whereas the G. morsitans GPX-like gene (GTP0092) showed greatest expression in the midgut compared with fat body and flight muscle tissues (7). Generally, high midgut expression of antioxidant genes in herbivorous insects is hypothesized to be a protective response to ROS ingested during feeding or generated during food processing (27).
The developmental expression patterns for all of the antioxidant genes were assessed only in compatible Hessian fly/wheat interactions because in incompatible interactions the first-instar larvae are dead within 5–6 days after hatching (20). Large changes in expression of antioxidant genes occurred within the Hessian fly development. The highest antioxidant mRNA levels during development occurred between mid second larval instars and late third larval instars. These developmental stages represent both feeding and nonfeeding larva. The mRNA peaks observed for MdesPHGPX-1, MdesCAT-1, and MdesCAT-2 support the basis for their role in the midgut of feeding second larval instars against ROS generated because of ingested wheat allelochemicals. Additionally, the high mRNA levels observed in late second and early third larval instars provide clues to the processing of ROS during postfeeding digestion.
The digestive physiology of most insects excluding members of Lepidoptera is poorly studied. From observations on the Hessian fly it is clear that the gut of early to mid third larval instars contains material that is thought to be plant sap ingested by the preceding larval instars (R.H.S., unpublished observation). The larval Hessian fly could be atypical in that food material from previous instars is carried to the next. However, food in the gut of wandering larvae of other flies has been reported to be continuously processed until it has reached a critical size for metamorphosis (28).
The peak expression levels observed for MdesCAT-1 and MdesCAT-2 in nonfeeding (mid to late) third larval instars imply their function against ROS generated endogenously. It is thus plausible that the products of MdesCAT-1 and MdesCAT-2 are important in quenching ROS produced during stages of rapid development and differentiation, which are usually associated with high rates of metabolic activity (7). Data presented in the current study with respect to both the CATs are in agreement with studies of CAT expression during development in D. melanogaster (29, 30) and the housefly, Musca domestica (31). It was reported that peak levels for a CAT mRNA coincided with pulses of ecdysteroid synthesis in late third larval instar as well as in prepupal (larval–pupal transition) stages of D. melanogaster (29). Also, as observed in M. domestica (31), a tremendous increase in H2O2 due to alterations in substrate catabolism before pupation could result in the concurrent increase in mRNA levels of both the Hessian fly CATs.
In several insect species the developmental processes are regulated by ecdysteroid titer. Indeed, the expression patterns of antioxidant genes, especially CATs, are thought to be under such hormonal influence in D. melanogaster (29, 30). Thus, in the present study, the second peak observed with respect to both the Hessian fly CATs in late third larval instars could address this cooperative function of ecdysteroids and CATs. These results are in corroboration with the hypothesis that during development the cellular environment gradually becomes more prone to the process of oxidation (31).
The clearest evidence for effects of food on antioxidant gene expression is from comparison of compatible and incompatible interactions. Larvae in incompatible interactions displayed higher mRNA levels of antioxidant genes than larvae in compatible interactions. Because larvae in incompatible interactions fail to establish a feeding site, the ingested plant material may be quantitatively and qualitatively different from material ingested in compatible interactions. The higher antioxidant mRNA levels suggest that larvae in incompatible interactions experience higher oxidative stress. We hypothesize that this higher oxidative stress is due to ROS produced either exogenously in the resistant plants and/or endogenously in larvae on resistant plants (because of stress and failure to establish a sustained feeding site).
It is implicated that elevated H2O2 is a primary plant response to herbivorous insect attack (32). H2O2 can cause lipid peroxidation, which severely damages insect cells and thereby retards development (33). Recent evidence suggests that resistant wheat plants increase LOX in response to feeding Hessian fly larvae (22). LOX produces ROS including LOOH and H2O2 as a by-product (32, 34). Two resistant wheat lines, P19346A1-2-5-5-2 and Iris (the line used in this study), carrying the Hessian fly resistance genes H13 and H9 increase expression of a LOX mRNA (WCI-2) in response to feeding first larval instar. WCI-2 is initially up-regulated in Iris wheat (2.5-fold compared with uninfested wheat plants) 6 h after hatching of Hessian fly larvae (22). Peak expression of WCI-2 (>30-fold) occurs 24 h after hatching in both resistant wheats participating in incompatible interactions.
The timing of up-regulation in WCI-2 mRNA levels is consistent with the hypothesis that resistant plants can produce increased ROS during attack by Hessian fly larvae. The timing of increased antioxidant gene mRNA levels in Hessian fly larvae in incompatible interactions is also consistent with the timing of increased LOX expression in the plant. Indeed, a cumulative peak expression of the Hessian fly antioxidant genes is observed in larvae on resistant plants 24 h after hatching. Furthermore, the mRNA levels exhibited by these larvae remain high through the remaining time points examined (48–96 h after hatching), suggesting their continued stress state even as larvae fail to thrive. The higher expression of MdesPHGPX-1, MdesPHGPX-2, MdesCAT-1, and MdesCAT-2 could be an adaptive response to increased ingestion of LOOH and H2O2. The lack of higher expression of MdesSOD-1 in larvae on resistant plants may indicate that superoxide concentrations are no higher in incompatible than compatible interactions and that the primary source of H2O2 is from the host plant as a result of the resistance reaction.
Conclusions
Based on the data presented in this article and recent information on gene expression in incompatible Hessian fly/wheat interactions, we propose the following oxidative stress model. In the compatible interaction, feeding is established by the first larval instar under conditions of relative low oxidative stress and continues through the second larval instar. However, in the third larval instar, high levels of endogenous ROS resulting from high metabolic activity and postfeeding digestion could lead to a concurrent increase in the antioxidant mRNA levels, specifically CATs. On the other hand, the incompatible interaction is characterized by higher mRNA levels of antioxidant genes in larvae on resistant plants. This finding suggests that larvae in incompatible interactions experience significantly greater oxidative stress. This may be due to increased ROS in the resistant wheat plants attacked by Hessian fly larvae as suggested by studies of LOX expression in resistant wheat. Alternatively, the increase in mRNA levels of antioxidant genes in larvae on resistant plants can also be due to endogenous sources resulting from the incompatible interaction. These models can be tested by further studies of Hessian fly/wheat interactions and by interactions of other gall-forming flies with their host plants.
Materials and Methods
Insect and Plant Material.
Biotype L of the Hessian fly was used in this study. The laboratory culture of Biotype L was selected from a field collection made from Posey County, Indiana, in 1986 and maintained (13). Biotype L is defined as virulent to the wheat genes H3, H5, H6, and H7H8. For compatible interactions Biotype L was reared on the wheat line Newton (which carries no resistance gene), and for incompatible interactions Biotype L was reared on the wheat line Iris (which carries the resistance gene H9).
Phylogenetic Analysis.
Amino acid sequences for the GPXs were aligned by using ClustalX (1.81) software (35). Phylogenetic analyses for the GPXs were conducted according to both the maximum parsimony and distance/neighbor-joining criteria using the software package PAUP* 4.0b10 (36). For the parsimony analysis, starting tree(s) were obtained via stepwise addition with the tree bisection–reconnection branch-swapping algorithm. The distance/neighbor-joining analysis used the total number of pairwise character differences (TOTAL) as the distance setting. Gaps were treated as missing data. All analyses were performed with the PHGPX from Saccharomyces cerevisiae as an outgroup. Confidence values for groupings in the trees were assessed by bootstrap resampling (37) with 1,000 repetitions.
Larval Dissections, RNA Extraction, and cDNA Library Construction.
Larval tissues including midgut, salivary glands, and fat body were dissected as described earlier (24). RNA was isolated from the larval tissues and different stages of development (first, second, and early third larval instars, pupae, and adults) using the RNAqueous-4PCR kit from Ambion (Austin, TX). RNA extracted from 200 midguts was used to construct a cDNA library using a Smart cDNA library construction kit from Clontech (Mountain View, CA) as described earlier (24).
To assess the midgut contents of nonfeeding third larval instars, dissections were performed (24) and direct visual observations were made. These observations included comparison of the midgut contents of early (first and second) larval instars with the midgut contents of third-instar larvae. Furthermore, characteristics of the midgut cuticle were also noted.
Transcription Patterns of the Hessian Fly Antioxidant Genes.
RNA extracted from each pool of the isolated tissues was used to determine the transcription pattern for each gene. Similarly, to assess the transcription patterns during development, RNA extracted from all of the developmental stages was used as the template. To study the transcription patterns of the Hessian fly antioxidant genes in larvae during compatible and incompatible interactions, RNA was extracted from 6- to 96-h posthatching larvae in both interactions. Larvae from a compatible interaction between Biotype L and Newton and larvae from an incompatible interaction between Biotype L and Iris were obtained for this analysis. Quantitative real-time PCR was performed to reveal the antioxidant gene mRNA levels in tissues during development and in larvae participating in compatible and incompatible interactions.
Quantitative Real-Time PCR.
Quantitative real-time PCR was performed by using total RNA extracted as described above. The software Primer Express from Applied Biosystems (Foster City, CA) was used to design real-time primers used in this study. The relative expression analysis was performed by using a Hessian fly ubiquitin as an internal reference. Quantification of mRNA levels, displayed as relative expression value (REV), was based on the Relative Standard Curve method (Applied Biosystems User Bulletin No. 2 for the ABI Prism 7700 Sequence Detection System). In brief, to calculate the REV, first the target quantities were calculated by using serial dilutions of a cDNA sample containing the target sequence. The threshold cycle value for each dilution was plotted against the log of its concentration, and threshold cycle values for the experimental samples were plotted onto this dilution series standard curve. Target quantities were calculated from separate standard curves generated for each experiment. REVs were then determined by dividing the target quantities of the gene of interest with the target quantity obtained for ubiquitin. PCR cycling parameters included 50°C for 2 min, 95°C for 10 min, and 40 cycles of 95°C for 15 sec, and 60°C for 1 min.
Statistical Analysis.
For calculations of significance, the logs of the REVs for each gene were analyzed by ANOVA using the PROC MIXED procedure of SAS (SAS/STAT User's Guide, Version 9.1; SAS Institute, Cary, NC). For expression analysis in tissues and developmental stages, the statistical model included treatment and interaction between treatments, whereas for the analysis of expression in different interactions (compatible and incompatible), the statistical model included treatment, time points, and interaction between treatments and time points as fixed effects. Biological replicates were included as a random effect in the analysis model. Treatment differences at each time point were evaluated by using orthogonal contrasts and were considered statistically significant if the P value associated with the contrast was <0.05.
Relative fold change in tissues was determined by taking the sample (REV) that showed the lowest level of expression known as the calibrator sample (38). Hence, the fold changes in the midgut and fat body tissues for all of the genes assessed were calculated relative to the salivary gland tissue, which showed the lowest level of expression for all of the transcripts. During development, the mean REV of three technical replicates was plotted against each developmental stage. The fold change during development was calculated by taking the expression level of the first larval instar as the calibrator. The fold change in antioxidant gene mRNA levels during compatible and incompatible interactions was assessed by dividing the REV for larvae on resistant plants by the REV for larvae on susceptible plants for all of the seven times points examined (6, 12, 18, 24, 48, 72, and 96 h after hatching). The standard error represented the variance in three technical replicates for the tissue/development expression analysis and two biological replicates (two technical replicates within each) for the interaction study.
Acknowledgments
Technical support provided by John Shukle is greatly appreciated. This is a joint contribution of the U.S. Department of Agriculture/Agricultural Research Service and Purdue University. This work was supported through U.S. Department of Agriculture Current Research Information System No. 3602-22000-014D.
Abbreviations
- ROS
reactive oxygen species
- CAT
catalase
- SOD
superoxide dismutase
- LOX
lipoxygenase
- REV
relative expression value
- GPX
glutathione peroxidase
- PHGPX
phospholipid hydroperoxide GPX
- GST
glutathione-S-transferase.
Footnotes
References
- 1.Ahmad S, Pardini RS. Free Radical Biol Med. 1990;8:401–413. doi: 10.1016/0891-5849(90)90107-t. [DOI] [PubMed] [Google Scholar]
- 2.Barbehenn RV. J Chem Ecol. 2002;28:1329–1347. doi: 10.1023/a:1016288201110. [DOI] [PubMed] [Google Scholar]
- 3.Maiorino M, Scapin M, Ursini F, Biasolo M, Bosello V, Flohe L. J Biol Chem. 2003;278:34286–34290. doi: 10.1074/jbc.M305327200. [DOI] [PubMed] [Google Scholar]
- 4.Behne D, Kyriakopoulos A. Annu Rev Nutr. 2001;21:453–473. doi: 10.1146/annurev.nutr.21.1.453. [DOI] [PubMed] [Google Scholar]
- 5.Tang L, Gounaris K, Griffiths C, Selkirk E. J Biol Chem. 1995;270:18313–18318. doi: 10.1074/jbc.270.31.18313. [DOI] [PubMed] [Google Scholar]
- 6.Li D, Blasevich F, Theopold U, Schmidt O. J Insect Physiol. 2003;49:1–9. doi: 10.1016/s0022-1910(02)00189-0. [DOI] [PubMed] [Google Scholar]
- 7.Munks RJL, Sant'Anna MRV, Grail W, Gibson W, Igglesden T, Yoshiyama M, Lehane SM, Lehane M. Insect Mol Biol. 2005;14:483–491. doi: 10.1111/j.1365-2583.2005.00579.x. [DOI] [PubMed] [Google Scholar]
- 8.Eshdat Y, Holland D, Paltin Z, Ben-Hayyim G. Physiol Plant. 1997;100:234–240. [Google Scholar]
- 9.Landis GN, Tower J. Mech Ageing Dev. 2006;126:907–908. doi: 10.1016/j.mad.2004.08.012. [DOI] [PubMed] [Google Scholar]
- 10.Fridovich I. Science. 1978;201:875–880. doi: 10.1126/science.210504. [DOI] [PubMed] [Google Scholar]
- 11.Mathews MC, Summers CB, Felton GW. Arch Insect Biochem Physiol. 1997;34:57–68. [Google Scholar]
- 12.Gagne RJ, Hatchett JH. Ann Entomol Soc Am. 1989;82:73–79. [Google Scholar]
- 13.Sosa O, Gallun RL. Ann Entomol Soc Am. 1973;66:1065–1070. [Google Scholar]
- 14.Byers RA, Gallun RL. J Econ Entomol. 1972;65:955–958. [Google Scholar]
- 15.Buntin GD. J Econ Entomol. 1999;92:1190–1197. [Google Scholar]
- 16.El Bouhssini M, Hatchett JH, Cox TS, Wilde GE. Bull Entomol Res. 2001;91:327–331. doi: 10.1079/ber2001115. [DOI] [PubMed] [Google Scholar]
- 17.Sardesai N, Nemacheck JA, Subramanyan S, Williams CE. Theor Appl Genet. 2005;111:1167–1173. doi: 10.1007/s00122-005-0048-6. [DOI] [PubMed] [Google Scholar]
- 18.Gallun RL. Ann NY Acad Sci. 1977;287:223–229. [Google Scholar]
- 19.Shukle RH, Grover PB, Jr, Mocelin G. Environ Entomol. 1992;21:845–853. [Google Scholar]
- 20.Painter RH. J Econ Entomol. 1930;23:322–326. [Google Scholar]
- 21.Williams CE, Collier CC, Nemacheck JA, Liang C, Cambron SE. J Chem Ecol. 2002;28:1411–1428. doi: 10.1023/a:1016200619766. [DOI] [PubMed] [Google Scholar]
- 22.Sardesai N, Subramanyam S, Nemacheck JA, Williams CE. J Plant Interact. 2005;1:39–50. [Google Scholar]
- 23.Yoshiyama M, Shukle RH. Ann Entomol Soc Am. 2004;97:1285–1293. [Google Scholar]
- 24.Mittapalli O, Neal JJ, Shukle RH. Insect Biochem Mol Biol. 2005;35:981–989. doi: 10.1016/j.ibmb.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 25.Jovanovic-Galovic A, Blagojevic DP, Grubor-Lajsic G, Worland R, Spasic MB. Arch Insect Biochem Physiol. 2004;55:79–89. doi: 10.1002/arch.10126. [DOI] [PubMed] [Google Scholar]
- 26.Ursini F, Maiorino M, Brigelus-Flohi R, Aumann KD, Roveri A, Schomburg D, Flohe L. Methods Enzymol. 1995;252:38–53. doi: 10.1016/0076-6879(95)52007-4. [DOI] [PubMed] [Google Scholar]
- 27.Krishnan N, Kodrík D. J Insect Physiol. 2006;52:11–20. doi: 10.1016/j.jinsphys.2005.08.009. [DOI] [PubMed] [Google Scholar]
- 28.Denlinger DL, Zaarek J. Annu Rev Entomol. 1994;39:243–266. doi: 10.1146/annurev.en.39.010194.001331. [DOI] [PubMed] [Google Scholar]
- 29.Radyuk SN, Klichko VI, Orr W. Arch Insect Biochem Physiol. 2000;45:79–93. doi: 10.1002/1520-6327(200010)45:2<79::AID-ARCH4>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 30.Klichko VI, Radyuk SN, Orr W. Arch Insect Biochem Physiol. 2004;56:34–50. doi: 10.1002/arch.10142. [DOI] [PubMed] [Google Scholar]
- 31.Allen RG, Oberley LW, Elwell JH, Sohal RS. J Cell Physiol. 1991;146:270–276. doi: 10.1002/jcp.1041460212. [DOI] [PubMed] [Google Scholar]
- 32.Bi JL, Felton GW. J Chem Ecol. 1995;21:1511–1530. doi: 10.1007/BF02035149. [DOI] [PubMed] [Google Scholar]
- 33.Downer RGH. In: Comprehensive Insect Physiology, Biochemistry, and Pharmacology. Kerkut GA, Gilbert LI, editors. Oxford: Pergamon; 1985. pp. 77–113. [Google Scholar]
- 34.Kanofsky JR, Axelrod B. J Biol Chem. 1986;261:1099–1104. [PubMed] [Google Scholar]
- 35.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. Nucleic Acids Res. 1997;22:4673–4680. [Google Scholar]
- 36.Swofford DL. PAUP*: Phylogenetic Analysis Using Parsimony (*and Other Methods) Sunderland, MA: Sinauer; 2002. Version 4. [Google Scholar]
- 37.Felsenstein J. Evolution (Lawrence, Kans) 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 38.Pfaffl WM. Nucleic Acids Res. 2001;29:2002–2007. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]