Abstract
Nuclei of the mammalian thalamus are aggregations of neurons with unique architectures and input–output connections, yet the molecular determinants of their organizational specificity remain unknown. By comparing expression profiles of thalamus and cerebral cortex in adult rhesus monkeys, we identified transcripts that are unique to dorsal thalamus or to individual nuclei within it. Real-time quantitative PCR and in situ hybridization analyses confirmed the findings. Expression profiling of individual nuclei microdissected from the dorsal thalamus revealed additional subsets of nucleus-specific genes. Functional annotation using Gene Ontology (GO) vocabulary and Ingenuity Pathways Analysis revealed overrepresentation of GO categories related to development, morphogenesis, cell–cell interactions, and extracellular matrix within the thalamus- and nucleus-specific genes, many involved in the Wnt signaling pathway. Examples included the transcription factor TCF7L2, localized exclusively to excitatory neurons; a calmodulin-binding protein PCP4; the bone extracellular matrix molecules SPP1 and SPARC; and other genes involved in axon outgrowth and cell matrix interactions. Other nucleus-specific genes such as CBLN1 are involved in synaptogenesis. The genes identified likely underlie nuclear specification, cell phenotype, and connectivity during development and their maintenance in the adult thalamus.
Keywords: development, excitatory neurons, inhibitory neurons, thalamocortical, Wnt signaling pathway
The mammalian thalamus is made up of groupings of neurons that reflect its evolutionary and developmental history, its function as a sensory relay, and its involvement in forebrain activities that underlie states of consciousness (1–4). The three major subdivisions of thalamus (epithalamus, dorsal thalamus, and ventral thalamus) emerge during embryogenesis from the wall of the diencephalic alar plate (5–7). Aggregation of fate-determined postmitotic neurons leads to the formation of multiple subnuclei within these divisions characterized by different chemo-, cyto-, and myeloarchitectures and differing patterns of connections.
The establishment of architecture and connections in all brain regions is modulated by molecular cues that govern cell aggregation, neurotransmitter phenotype, axon guidance, and synaptogenesis (8, 9). The unique architecture, connectivity, and transmitter/receptor characteristics of each thalamic nucleus are unlikely to be established or maintained in the absence of molecular genetic guidance. Clues to the nature of underlying mechanisms can be found by identifying genes that give a molecular identity to thalamic nuclei. Sets of regulatory genes distinguish the three major thalamic subdivisions in the developing and adult rodent and primate (10, 11), but expression occurs across multiple nuclei, in regional rather than nucleus-specific patterns. Some examples of nucleus-specific expression, however, do occur, e.g., Id-2 in the primate centre médian nucleus (CM; ref. 10). Expression of neurotransmitter- or receptor-related genes in the thalamus also tends to be regional rather than nucleus-specific, but there are exceptions to this rule as well (12, 13).
To determine the extent to which subnuclei of the major thalamic divisions are molecularly distinct, we used high-density oligonucleotide arrays to identify thalamus-specific genes in adult monkeys. By comparing expression profiles of thalamus and cerebral cortex, we identified genes not hitherto known to be expressed in thalamus. Further profiling of microdissected nuclei identified more comprehensive sets of genes with nucleus-specific expression. Confirmation of gene expression by RT-PCR, in situ hybridization histochemistry, and/or immunohistochemistry revealed remarkable cell- and nucleus-specific patterns of expression. Functionally, the genes are related to development and cell–cell interactions, implying their involvement in the establishment and maintenance of thalamic nuclear and cellular specificity.
Results
Genes specific to thalamus were identified by comparing expression patterns in posterior thalamus (PT) with those in visual cortex (VC) and frontal cortex (FC). The vast majority of genes called “present” were equally distributed between cortex and thalamus. Of 1,978 genes called “present” in PT, VC, and FC, 89 (4.4%) were enriched in PT by ≥1.2-fold. A more stringent cutoff of ≥3-fold reduced this number to 31 (1.5%) thalamus-specific genes [ supporting information (SI) Table 3].
A significant overrepresentation of thalamus-enriched probe sets was found for Gene Ontology (GO) terms related to development, morphogenesis, and cell signaling. The terms structural molecule activity (six genes, P < 0.001), cell differentiation (six genes, P < 0.001), morphogenesis (13 genes, P < 0.01), and extracellular matrix (ECM) (three genes, P < 0.01) were all overrepresented. Of the 17 most overrepresented genes (SI Table 4), 12 were associated with cellular growth and development (FGFR2, HSPA2, ILGFBP7, MOG, NDRG1, NHLH2, NTNG1, PCP4, PRDX1, SPARC, SPP1, and WWP1). Four genes associated with cell–cell interactions (NTNG1, RLN, SPARC, and SPP1) were localized to the ECM.
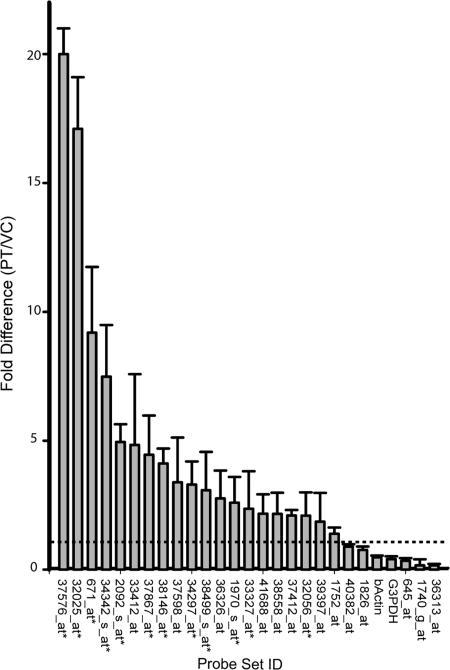
Validation of the microarray results by RT-PCR was limited to the 31 genes enriched in PT by ≥3-fold. Twenty-five (81%) were successfully amplified (Figs. 1 and 2, Table 1). Twenty (65%) displayed elevated expression in PT vs. VC.
Fig. 1.
RT-PCR measures of regional transcript expression levels confirm microarray results. Bar graph indicates relative expression between PT and VC for transcripts overrepresented in thalamus by ≥3-fold in microarray analysis. Dotted line indicates a ratio of 1, denoting no change in expression. Error bars indicate standard deviation of the mean. Glyceraldehyde-3-phosphate dehydrogenase (G3PDH) and β-actin genes were included as internal controls. Asterisks indicate probe sets analyzed by in situ hybridization histochemistry.
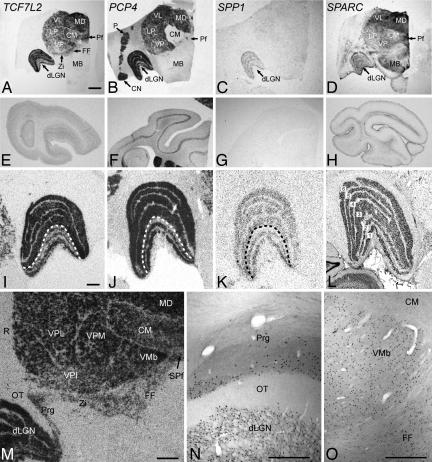
Fig. 2.
Autoradiograms and immunocytochemical preporations from monkey thalamus and cortex. Images from film audioradiograms show abundance of transcripts of TCF7L2 (A, E, I, and M), PCP4 (B, F, and J), SPP1 (C, G, and K), and SPARC (D, H, and L) in thalamus (A–D and I–M) relative to cortex (E–H). Expression of TCF7L2 and SPP1 is restricted to thalamus, whereas PCP4 and SPARC are expressed in other subcortical regions as well. Higher-magnification images of the dorsal lateral geniculate nucleus (dLGN) illustrate the differential expression of TCF7L2, PCP4, and SPP1 in dLGN (I–L). Dashed line indicates separation between magnocellular (1 and 2) and parvocellular (3–6) layers of the dLGN, as marked in adjacent Nissl section (L). In situ hybridization for TCF7L2 mRNA (M) and immunoreactivity for TCF7L2 (N and O) reveal expression in posterior division of the zona incerta (M), the deep lamina of the pregeniculate nucleus (N), and the field of Forel (O). (Scale bars: A–H, 3 mm; M, 1 mm; I–L, N, and O, 500 μm.) CN, caudate nucleus; FF, field of Forel; LP, lateral posterior nucleus; MB, midbrain; MD, mediodorsal nucleus; OT, optic tract; P, putamen; Prg, pregeniculate nucleus; R, reticular nucleus; SPf, sub-Pf; VL, ventral lateral nucleus; VMb, basal ventral medial nucleus; VP, ventral posterior nuclei; VPI, ventral posterior inferior nucleus; VPL, ventral posterior lateral nucleus; VPM, ventral posterior medial nucleus; ZI, zona incerta.
Table 1.
Candidate genes validated by in situ hybridization
| Expression pattern | Gene symbol |
|---|---|
| Restricted to thalamus | SPP1 |
| TCF7L2 | |
| Enriched in thalamus | SPARC |
| SEPT4 | |
| PCP4 | |
| Equal in cortex and thalamus | CALR |
| FGFR2 | |
| ST18 | |
| Associated with fiber tracts | MAG |
| MOBP | |
| MOG | |
| Cortex specific or enriched | NONE |
| None detected | GPR37 |
| C11ORF9 |
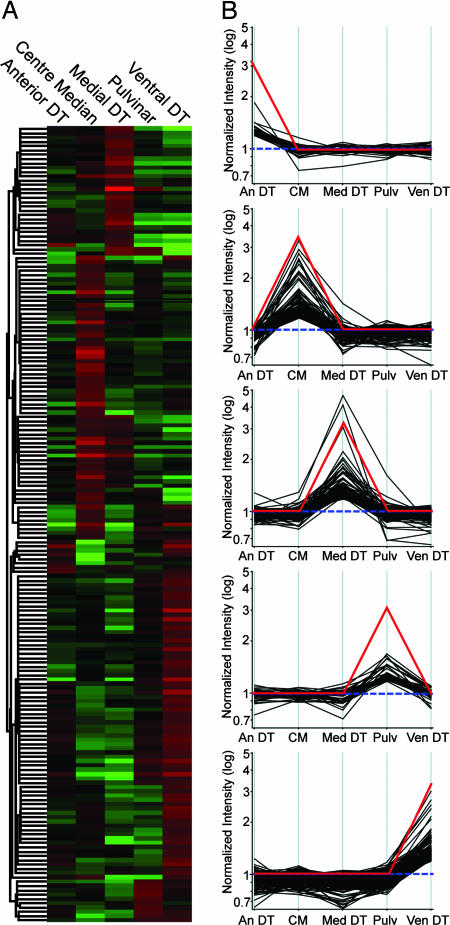
Transcriptional profiling of samples microdissected from anterior nuclei of dorsal thalamus, CM, mediodorsal nucleus, pulvinar, and ventral posterior nuclei confirmed and extended the thalamus-specific results. Unsupervised hierarchical clustering broadly parsed thalamic nuclei into groups displaying similar gene expression profiles (Fig. 3). When further analyzed by the Drawn Gene function in GeneSpring software (Agilent Technologies, Foster City, CA) (Fig. 3), >550 genes expressed in a nucleus-specific manner were identified within the dorsal thalamus (SI Dataset 1).
Fig. 3.
GeneSpring analysis of five thalamic nuclei. Nuclei of the dorsal thalamus display unique gene expression profiles. (A) Genes with significant expression (P < 0.05) in at least one of the five regions examined were hierarchically clustered by similarity in expression profile. The resulting heat map of the dendrogram tree reveals groups of genes with high (red) expression levels in one thalamic nucleus and moderate (black) or low (green) levels in the other four nuclei. (B) Genes with thalamic nucleus-specific expression profiles were identified by using the Drawn Gene function in the GeneSpring analysis program. For each region examined, sets of genes whose expression profiles significantly correlated (R2 > 0.95) to a template pattern (red) were identified.
Ingenuity Pathways Analysis of the nucleus-enriched genes also showed significant overrepresentation of functions related to development and cell signaling (Fig. 4). Analysis of candidate genes for potential interactions revealed putative protein networks involving development and cell signaling as top functional categories (Table 2). Other top network functions included cell cycle, cancer, and gene expression. Strikingly, members of the canonical Wnt/β-catenin signaling cascade featured prominently in four of the five top networks (Table 2).
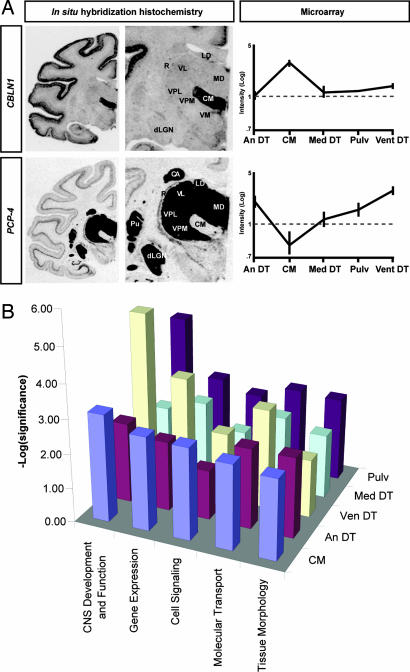
Fig. 4.
Microarray results confirmed by in situ hybridization histochemistry (A) and functional analysis of thalamus-enriched genes (B). In situ hybridization and functional annotation validate thalamic nucleus-specific gene expression. (A) Images from film autoradiograms show differential expression of CBLN1 (Upper) and PCP4 (Lower) in monkey dorsal thalamus, corroborating the microarray results (Right). (B) The Ingenuity Pathways Analysis program enabled biological/functional annotations of candidate genes identified by the Drawn Gene Function in GeneSpring analysis. The top five functions are compared across all regions. Abbreviations are as in Fig. 2.
Table 2.
Top gene networks identified in Ingenuity Pathways analysis
| Region | Score | Focus genes | Wnt/β-catenin canonical pathway genes | Top biological functions |
|---|---|---|---|---|
| Ven DT | 24 | 18 | 0 | Cell-to-cell signaling and interaction, cellular assembly and organization, cellular function and maintenance |
| CM | 23 | 15 | 3 | Skeletal and muscular system development and function, cancer, cell cycle |
| Med DT | 33 | 21 | 2 | Embryonic development, cellular assembly and organization, cellular development |
| Pulv | 18 | 10 | 3 | Cell cycle, carbohydrate metabolism, small molecule biochemistry |
| An DT | 33 | 16 | 3 | Gene expression, cancer, cellular growth and proliferation |
Genes displaying region-specific expression were mapped to corresponding objects in the Ingenuity Pathways Knowledge Base. A score based on the number of mapped “focus” genes and network size is used for ranking purposes. The canonical Wnt/β-catenin signaling pathway was well represented among the top networks. An DT, anterior dorsal thalamus; CM, centre médian nucleus; Med DT, medial dorsal thalamus; Pulv, pulvinar; Ven DT, ventral dorsal thalamus.
The distribution of genes validated by RT-PCR and of certain of those defined by nucleus-specific assay was examined by in situ hybridization. Four patterns of expression were observed (Table 1): (i) restricted to thalamus, (ii) enriched in thalamus, (iii) equal in thalamus and cortex, and (iv) associated with fiber tracts. For example, expression of SPP1, TCF7L2, and CBLN1 was restricted to thalamus. Expression of SPARC, PCP4, and SEPT4 was enriched in thalamic nuclei. Expression of CALR, FGFR2, and ST18 was equal in cortex and thalamus. Expression of genes associated with myelin production MAG, MOG, and MOBP was restricted to fiber tracts.
Expression of TCF7L2, PCP4, CBLN1, and SPP1 was delimited by the borders of thalamic nuclei. TCF7L2 expression was restricted to nuclei of the dorsal thalamus (Fig. 2), except for some aspects of the ventral thalamus (the deep lamina of the pregeniculate nucleus, the posterior division of the zona incerta, and among scattered cells of the field of Forel), as well as in a small focus in the medial habenular nucleus of the epithalamus (Fig. 2). PCP4 was expressed throughout dorsal thalamus, with the notable exception of the CM and parafascicular nucleus (Pf); CBLN1, by contrast, was expressed only in CM and Pf (Fig. 4). PCP4 was also expressed in the striatum (caudate nucleus and putamen) and in layer V of the cerebral cortex. SPP1 and PCP4 were expressed only in the parvo- and magnocellular layers of the dorsal lateral geniculate nucleus; TCF7L2 was expressed only in the parvocellular, s layers, and interlaminar layers (Fig. 2).
Immunocytochemistry for TCF7L2 protein showed enrichment in the dorsal thalamus (SI Fig. 5). In double-labeling experiments with markers of excitatory and inhibitory neurons, TCF7L2 immunostaining was absent from GABA neurons (SI Fig. 5) in dorsal thalamus, reticular nucleus, zona incerta, or pregeniculate nucleus. TCF7L2 was coexpressed in dorsal thalamic neurons immunostained for α type II calcium/calmodulin-dependent protein kinase, a marker for excitatory neurons (14–16) (SI Fig. 5). SPP1 and PCP4 immunoreactive cells could be costained for neuronal markers, whereas SPARC immunoreactive cells could not be costained for neuronal, astrocytic, or oligodendrocytic markers, implying they may be microglial cells (data not shown).
Discussion
Thalamic nuclei, classically defined by cyto-, myelo-, and chemoarchitecture and by connectional and physiological characteristics, are molecularly distinct. Expression profiling revealed a subset of genes that were enriched or exclusively expressed in nuclei of adult thalamus. Most prominent were genes associated with development, cell signaling, morphogenesis, the ECM, and the Wnt signaling pathway. Cell signaling molecules encoded by several candidate genes are localized to ECM, suggesting that molecules within the extracellular environs are important determinants of how thalamic cells aggregate into nuclei.
The results demonstrate that human oligonucleotide arrays can be successfully used for large-scale transcriptional profiling of other Old World primate brains, as demonstrated by the delineation of novel thalamic markers, e.g., TCF7L2, PCP4, and CBLN1, whose expression is not only limited to specific thalamic nuclei, but in the case of TCF7L2, restricted to excitatory neurons. Two members of the bone matricellular family, SPP1 (Osteopontin) and SPARC (Osteonectin), with cell type- and nucleus-specific patterns of expression, were also detected. Although from the same protein family, SPP1 and SPARC are expressed in neurons and nonneuronal cells, respectively, suggesting they may contribute to nuclear and cellular identities in unique ways.
The thalamus-specific genes identified, whether highly restricted (e.g., SPP1 and CBLN1) or broad (e.g., TCF7L2 and PCP4) in their expression, were invariably delimited by classical nuclear boundaries, and none redefined thalamic boundaries. Similar adherence of expression patterns to cytoarchitecture occurs in mouse hippocampus (17, 18) and amygdala (19), often providing less equivocal delineations of borders than cytoarchitecture alone (20). Establishment of nuclear identity from homogenous masses of cells in the developing thalamus is associated with determination of cell size, aggregation, packing density, and establishment of connections; the GO associations of the majority of the genes identified implicate them in these activities.
Expression of two genes, PCP4 and SPP1, was restricted to cells of dorsal thalamus, but none to ventral thalamus, although TCF7L2 marked subpopulations of non-GABA cells in the pregeniculate nucleus, zona incerta, and field of Forel. In ferrets, Kawasaki et al. (21) demonstrated molecular distinctions between the dLGN, a component of dorsal thalamus and the perigeniculate nucleus, a part of the reticular nucleus. In their study, PCP4 was more heavily expressed in larger putative Y cells of the dLGN than in smaller putative X cells. We observed PCP4 expression in both magno- and parvocellular laminae of monkey dLGN and in most other dorsal thalamic nuclei. However, PCP4 expression was absent from the CM and Pf nuclei that form the caudal group of intralaminar nuclei. CM, absent in rodents and many other mammals, is particularly elaborated in primates and displays a unique expression profile compared with other thalamic nuclei (Fig. 3 and SI Dataset 1). CM and Pf also exclusively express CBLN1 and the transcription factor Id-2 (10) and are characterized by patterns of GABA and glutamate receptor expression that are very different from those in other dorsal thalamic nuclei (12, 13). CBLN1, a glycoprotein that is structurally related to the C1q and tumor necrosis factor families of proteins, in the cerebellum is associated with synaptogenesis in Purkinje cells (22).
The transcription factor, TCF7L2, was highly expressed and restricted to excitatory neurons in dorsal thalamus and to non-GABA neurons in the ventral thalamus. TCF7L2 is specific for thalamus, unlike other markers of excitatory thalamic neurons, such as α type II calcium/calmodulin-dependent protein kinase, which are expressed in other forebrain regions as well (13, 14). TCF7L2 is a member of the high mobility group box proteins of the T cell factor/lymphoid enhancer factor subfamily that mediate Wnt protein signaling through the canonical β-catenin pathway (23–25). Wnts direct the emergence of functionally distinct regions along the developing neuraxis and have consequently been implicated in the formation of neuronal connectivity (26–30). The Wnt pathway also influences axonal remodeling through interactions with the cytoskeleton (31, 32), and interfering with Wnt signaling disrupts synapse formation (31, 33). The Wnt/β-catenin signaling cascade featured prominently in the top functional networks (Table 2). Hence, Wnt/β-catenin signaling is a candidate for involvement not only in formation and maintenance of dorsal thalamic connectivity but also in the plasticity of connections in the adult. This notion is supported by the observation that mice null for the low-density lipoprotein receptor-related protein 6, a required signaling coreceptor for the Wnt/β-catenin pathway, exhibit profound disruptions in the development of dorsal thalamus and epithalamus (34).
The present observations illustrate that nuclei of the dorsal thalamus are comprised of groups of cells sharing common molecular phenotypes. The variety of cellular and molecular mechanisms likely to be required for establishing and maintaining the specific architectural, connectional, and functional signatures of each thalamic nucleus is reflected in the range of functions associated with the thalamus-enriched genes.
Materials and Methods
Animals.
Brains from eight adult male rhesus monkeys (Macaca mulatta) were used. All procedures were carried out by using protocols approved by the Institutional Animal Care and Use Committee.
Microarray Probe Generation.
Brains were processed according to protocols developed for postmortem human brain tissue (35). Pieces were excised or micropunched from frozen coronal slices to give samples from PT, VC, or FC or from different nuclear regions of the thalamus. Total RNA was extracted by TRIzol reagent (Invitrogen, Carlsbad, CA) followed by cleanup by using an RNeasy Lipid Mini Extraction Kit (Qiagen, Valencia, CA). RNA quality was monitored by agarose gel electrophoresis, spectrophotometry, and an Agilent Bioanalyzer (Agilent, Foster City, CA). Total RNA was processed for Affymetrix GeneChip analysis according to the manufacturer's protocol (Affymetrix, Santa Clara, CA), using human HuU95Av2 or U133A GeneChips, as described (36, 37). Criteria used to assess quality of chip hybridization were percent “present” calls, scaling factor, background noise, and mean average difference value.
Microarray Data Analysis.
To identify genes enriched in thalamus, mean average difference values for PT were compared with those for VC and independently for FC. Only genes called “present” in all three samples and with the same direction of fold-change in both comparisons (PT-VC and PT-FC) were included. A list of genes in rank order of fold difference was generated (SI Table 3). To identify genes displaying regional expression in dorsal thalamic nuclei, Affymetrix Cel files were preprocessed for robust multiarray analysis (RMA) in GeneSpring GX (Ver. 7.3.1) software (Agilent Technologies) followed by per-gene and per-chip median polishing. Hierarchical clustering and GeneSpring Drawn Gene analyses were performed on genes filtered by confidence (P < 0.05) and normalized expression value (>1.2). Gene profiles highly correlated with a drawn template (R2 > 0.95) were included in further analyses.
Gene sets defined by transcriptional profiling on samples micropunched from thalamic nuclei were divided into groups displaying similar gene expression profiles by unsupervised hierarchical clustering and by the Drawn Gene function in GeneSpring.
Genes that were expressed in a region- or nucleus-specific manner were analyzed for association with biological functions and/or diseases using Onto-Express (http://vortex.cs.wayne.edu/ontoexpress) (38–40) and Ingenuity Pathways Analysis (Ingenuity Systems, Redwood City, CA). Fischer's exact test was used to determine P values. To interrogate potential functional interactions among candidate genes, each gene identifier was mapped to its corresponding gene object, and these “Focus Genes” were overlaid onto a global molecular network developed from information in the Ingenuity Pathways Knowledge Base. Networks of the “Focus Genes,” based on their connectivity, were then algorithmically generated.
Real-Time RT-PCR.
Total RNA was isolated from VC or PT, and a cDNA template was prepared from 1 μg by using oligo(dT)18 primer and Moloney murine leukemia virus reverse transcriptase (Clontech, Palo Alto, CA). PCRs were performed by using 4% of the cDNA product as starting material and measured in real time as a function of SYBR green I incorporation using the Bio-Rad iCycler (BioRad, Hercules, CA) (41). After PCR, a first derivative melting-curve analysis was performed to confirm the specificity of the PCR. PCR products were electrophoresed, purified, and sequenced to verify amplicon identity. The relative fold difference in mRNA between samples was calculated by comparing the threshold cycle (Ct) at which product initially appeared above background according to: 2−(ΔCt), where ΔCt is the difference between PT and VC values.
In Situ Hybridization.
Two monkeys were anesthetized with ketamine and sodium pentobarbital and perfused with 4% paraformaldehyde in 0.1 M phosphate buffer (pH 7.2). Brains were removed, blocked, postfixed for 4 h at 4°C, cryoprotected, and frozen in dry ice. Microtome-cut sections were placed in fixative at 4°C for at least 7 days.
Riboprobes were generated from PCR fragments cloned into pCR4-TOPO vector (Invitrogen). Probes were labeled by in vitro transcription by using [α-33P]UTP or [α-35S]UTP. Sections were incubated in hybridization solution containing anti-sense cRNA probes, at 60°C, and washed in: 4× saline sodium citrate (SSC) at 60°C, twice, 30 min (1× SSC = 0.15 M NaCl/0.015 M sodium citrate, pH 7.0); ribonuclease A (0.02 mg/ml in 0.01 M Tris·HCl buffer, pH 8.0/1 mM EDTA/2.9% NaCl) at 45°C, 1 h; and 2× SSC, room temperature, twice, 30 min. Sections were then exposed to radiographic film at 4°C and counterstained with thionin. Sense-strand RNA probes were used as controls.
Immunocytochemistry.
Sections were placed directly in 0.1 M phosphate buffer, pH 7.4, at 4°C. Standard immunocytochemical techniques were used for immunolabeling and visualization with 3,3′ diaminobenzidine 4 HCl. For double-immunofluorescent labeling, sections were exposed to a mixture of primary antibodies, followed by a mixture of fluorescent-labeled secondary antibodies (Alexa-488, -568, or -647; Molecular Probes, Eugene, OR), and counterstained with Hoechst 33342 (Molecular Probes).
The following monoclonal or polyclonal antibodies were used: calbindin, parvalbumin, GABA (Sigma, St. Louis, MO), α type II calcium/calmodulin-dependent protein kinase, glial fibrillary acidic protein-(GFAP), myelin basic protein, myelin/oligodendrocyte-specific protein, chondroitin sulfate proteoglycan (NG2; Chemicon, Temecula, CA), prospero-related homeobox 1 (Prox1; Covance, Berkeley, CA), Tcf7l2 (Upstate Biotechnology, Lake Placid, NY, and Zymed, San Francisco, CA), SPP1 (Osteopontin; Developmental Studies Hybridoma Bank, Iowa City, IA), and SPARC (Osteonectin; US Biologicals, Swampscott, MA).
Thalamic nuclei were identified by cytoarchitectural features observed in Nissl-stained sections (ref. 7; www.brainmaps.org). Autoradiographic images were captured by using a Nikon D1X camera (Nikon, Melville, NY). Brightfield photomicrographs were obtained by using a Nikon Eclipse 1000 microscope equipped with a Quantix CCD camera (Photometrics, Tuscon, AZ). Fluorescent images were obtained with a Zeiss (Oberkochen, Germany) LSM 510 META laser-scanning confocal microscope. Z-stacked images were collected, collapsed, saved as TIFF files, imported into Adobe Photoshop, and labeled in Adobe Illustrator (Adobe Systems, San Jose, CA).
Supplementary Material
Acknowledgments
We thank Xiao-Hong Fan, Phong Nguyen, and Malalai Yusufzai for technical assistance. This work was supported by the National Institutes of Health (Grants NS21377 and NS39094) and the W. M. Keck Program in Neuroscience Imaging. K.D.M. is the recipient of a young investigator award from the National Alliance for Research in Schizophrenia and Depression and the Sunshine from Darkness Gala. The monoclonal antibody MPIIIB101 was developed by Michael Solursh and Ahnders Franzen under the auspices of the National Institute of Child Health and Human Development.
Abbreviations
- ECM
extracellular matrix
- CM
centre médian nucleus
- FC
frontal cortex
- GO
Gene Ontology
- PT
posterior thalamus
- Pf
parafascicular nucleus
- VC
visual cortex
- dLGN
dorsal lateral geniculate nucleus.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences reported in this paper have been deposited in the GEO database (accession no. GSE6708).
This article contains supporting information online at www.pnas.org/cgi/content/full/0610742104/DC1.
References
- 1.Jones EG. Adv Neurol. 1998;77:49–71. [PubMed] [Google Scholar]
- 2.Llinás R, Ribary U, Contreras D, Pedroarena C. Philos Trans R Soc London B. 1998;353:1841–1849. doi: 10.1098/rstb.1998.0336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Llinás R, Ribary U. Ann NY Acad Sci. 2001;929:166–175. [PubMed] [Google Scholar]
- 4.Llinás RR, Paré D. Neuroscience. 1991;44:521–535. doi: 10.1016/0306-4522(91)90075-y. [DOI] [PubMed] [Google Scholar]
- 5.Puelles L, Rubenstein JL. Trends Neurosci. 1993;16:472–479. doi: 10.1016/0166-2236(93)90080-6. [DOI] [PubMed] [Google Scholar]
- 6.Puelles L, Rubenstein JL. Trends Neurosci. 2003;26:469–476. doi: 10.1016/S0166-2236(03)00234-0. [DOI] [PubMed] [Google Scholar]
- 7.Jones EG. The Thalamus. 2nd Ed. Cambridge, UK: Cambridge Univ Press; 2006. [Google Scholar]
- 8.Jan YN, Jan LY. Neuron. 2003;40:229–242. doi: 10.1016/s0896-6273(03)00631-7. [DOI] [PubMed] [Google Scholar]
- 9.Tessier-Lavigne M. Harvey Lect. 2002;98:103–143. [PubMed] [Google Scholar]
- 10.Jones EG, Rubenstein JL. J Comp Neurol. 2004;477:55–80. doi: 10.1002/cne.20234. [DOI] [PubMed] [Google Scholar]
- 11.Nakagawa Y, O'Leary DD. J Neurosci. 2001;21:2711–2725. doi: 10.1523/JNEUROSCI.21-08-02711.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huntsman MM, Leggio MG, Jones EG. J Neurosci. 1996;16:3571–3589. doi: 10.1523/JNEUROSCI.16-11-03571.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jones EG, Tighilet B, Tran BV, Huntsman MM. J Comp Neurol. 1998;397:371–393. doi: 10.1002/(sici)1096-9861(19980803)397:3<371::aid-cne5>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- 14.Benson DL, Isackson PJ, Hendry SH, Jones EG. J Neurosci. 1991;11:1540–1564. doi: 10.1523/JNEUROSCI.11-06-01540.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Benson DL, Isackson PJ, Gall CM, Jones EG. Neuroscience. 1992;46:825–849. doi: 10.1016/0306-4522(92)90188-8. [DOI] [PubMed] [Google Scholar]
- 16.Liu XB, Jones EG. Proc Natl Acad Sci USA. 1996;93:7332–7336. doi: 10.1073/pnas.93.14.7332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lein ES, Zhao X, Gage FH. J Neurosci. 2004;24:3879–3889. doi: 10.1523/JNEUROSCI.4710-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao X, Lein ES, He A, Smith SC, Aston C, Gage FH. J Comp Neurol. 2001;441:187–196. doi: 10.1002/cne.1406. [DOI] [PubMed] [Google Scholar]
- 19.Zirlinger M, Kreiman G, Anderson DJ. Proc Natl Acad Sci USA. 2001;98:5270–5275. doi: 10.1073/pnas.091094698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lein ES, Callaway EM, Albright TD, Gage FH. J Comp Neurol. 2005;485:1–10. doi: 10.1002/cne.20426. [DOI] [PubMed] [Google Scholar]
- 21.Kawasaki H, Crowley JC, Livesey FJ, Katz LC. J Neurosci. 2004;24:9962–9970. doi: 10.1523/JNEUROSCI.2165-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hirai H, Pang Z, Bao D, Miyazaki T, Li L, Miura E, Parris J, Rong Y, Watanabe M, Yuzaki M, Morgan JL. Nat Neurosci. 2005;8:1534–1541. doi: 10.1038/nn1576. [DOI] [PubMed] [Google Scholar]
- 23.Cho EA, Dressler GR. Mech Dev. 1998;77:9–18. doi: 10.1016/s0925-4773(98)00131-2. [DOI] [PubMed] [Google Scholar]
- 24.Korinek V, Barker N, Willert K, Molenaar M, Roose J, Wagenaar G, Markman M, Lamers W, Destree O, Clevers H. Mol Cell Biol. 1998;18:1248–1256. doi: 10.1128/mcb.18.3.1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee YJ, Swencki B, Shoichet S, Shivdasani RA. J Biol Chem. 1999;274:1566–1572. doi: 10.1074/jbc.274.3.1566. [DOI] [PubMed] [Google Scholar]
- 26.Charron F, Tessier-Lavigne M. Development (Cambridge, UK) 2005;132:2251–2262. doi: 10.1242/dev.01830. [DOI] [PubMed] [Google Scholar]
- 27.Christiansen JH, Coles EG, Wilkinson DG. Curr Opin Cell Biol. 2000;12:719–724. doi: 10.1016/s0955-0674(00)00158-7. [DOI] [PubMed] [Google Scholar]
- 28.Patapoutian A, Reichardt LF. Curr Opin Neurobiol. 2000;10:392–399. doi: 10.1016/s0959-4388(00)00100-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ciani L, Salinas PC. Nat Rev Neurosci. 2005;6:351–362. doi: 10.1038/nrn1665. [DOI] [PubMed] [Google Scholar]
- 30.Pleasure SJ. Trends Neurosci. 2001;24:69–71. doi: 10.1016/s0166-2236(00)01722-7. [DOI] [PubMed] [Google Scholar]
- 31.Hall AC, Lucas FR, Salinas PC. Cell. 2000;100:525–535. doi: 10.1016/s0092-8674(00)80689-3. [DOI] [PubMed] [Google Scholar]
- 32.Krylova O, Messenger MJ, Salinas PC. J Cell Biol. 2000;151:83–94. doi: 10.1083/jcb.151.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Luo ZG, Wang Q, Zhou JZ, Wang J, Luo Z, Liu M, He X, Wynshaw-Boris A, Xiong WC, Lu B, et al. Neuron. 2002;35:489–505. doi: 10.1016/s0896-6273(02)00783-3. [DOI] [PubMed] [Google Scholar]
- 34.Zhou CJ, Pinson KI, Pleasure SJ. J Neurosci. 2004;24:7632–7639. doi: 10.1523/JNEUROSCI.2123-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jones EG, Hendry SHC, Liu XB, Hodgins S, Potkin SG, Tourtellotte WW. J Neurosci Methods. 1992;44:133–144. doi: 10.1016/0165-0270(92)90006-y. [DOI] [PubMed] [Google Scholar]
- 36.Bunney WE, Bunney BG, Vawter MP, Tomita H, Li J, Evans SJ, Choudary PV, Myers RM, Jones EG, Watson SJ, et al. Am J Psychiatry. 2003;160:657–666. doi: 10.1176/appi.ajp.160.4.657. [DOI] [PubMed] [Google Scholar]
- 37.Choudary PV, Molnar M, Evans SJ, Tomita H, Li JZ, Vawter MP, Myers RM, Bunney WE, Jr, Akil H, Watson SJ, et al. Proc Natl Acad Sci USA. 2005;102:15653–15658. doi: 10.1073/pnas.0507901102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Anderle P, Duval M, Draghici S, Kuklin A, Littlejohn TG, Medrano JF, Vilanova D, Roberts MA. BioTechniques. 2003;(Suppl:36–44) [PubMed] [Google Scholar]
- 39.Draghici S, Khatri P, Bhavsar P, Shah A, Krawetz SA, Tainsky MA. Nucleic Acids Res. 2003;31:3775–3781. doi: 10.1093/nar/gkg624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Khatri P, Draghici S, Ostermeier GC, Krawetz SA. Genomics. 2002;79:266–270. doi: 10.1006/geno.2002.6698. [DOI] [PubMed] [Google Scholar]
- 41.Murray KD, Isackson PJ, Jones EG. Neuroscience. 2003;122:407–420. doi: 10.1016/j.neuroscience.2003.07.015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.