Abstract
Stability of the mitochondrial genome is controlled by nuclear loci. In plants, nuclear genes suppress mitochondrial DNA rearrangements during development. One nuclear gene involved in this process is Msh1. Msh1 appears to be involved in the suppression of illegitimate recombination in plant mitochondria. To test the hypothesis that Msh1 disruption leads to the type of mitochondrial DNA rearrangements associated with naturally occurring cytoplasmic male sterility in plants, a transgenic approach for RNAi was used to modulate expression of Msh1 in tobacco and tomato. In both species, these experiments resulted in reproducible mitochondrial DNA rearrangements and a condition of male (pollen) sterility. The male sterility was, in each case, heritable, associated with normal female fertility, and apparently maternal in its inheritance. Segregation of the transgene did not reverse the male sterile phenotype, producing stable, nontransgenic male sterility. The reproducible transgenic induction of mitochondrial rearrangements in plants is unprecedented, providing a means to develop novel cytoplasmic male sterile lines for release as non-GMO or transgenic materials.
Keywords: MutS homolog, nonhomologous recombination, RNA interference, tobacco, tomato
Cytoplasmic male sterility (CMS) is a widespread trait among both wild and domesticated plant species, manifested as the inability of an otherwise phenotypically normal plant to shed viable pollen. The male sterile trait arises with rearrangements of the mitochondrial genome and demonstrates a maternal inheritance pattern (1). From an agricultural perspective, this form of male sterility is valuable to the hybrid seed industry as a means of generating cross-pollinated seed without the need for labor-intensive hand emasculations (2) and as a strategy for preventing pollen escape in transgenic crops. In natural populations, CMS is thought to participate in the gynodioecious mating strategy of several plant species (3, 4).
The CMS trait is associated with a variety of mitochondrial DNA rearrangements, and no two CMS mutations described to date have been identical. In general, these mutations arise from low frequency, illegitimate recombination, or nonhomologous end joining activity within the mitochondrial genome, each giving rise to an expressed ORF comprised of chimeric sequences (1, 5). Plant mitochondrial genomes are known to undergo abundant recombination activity. However, there is evidence to suggest that CMS mutations might not arise de novo in a mitochondrial population at the time male sterility occurs, but rather may already be present at low copy number before their specific amplification (6, 7).
Plant mitochondrial genomes are distinctive in their highly variable size and multipartite structures relative to the smaller, more compact circular genomes of mammalian mitochondria. This redundant, multipartite organization can presumably result in heterogeneous mitochondrial populations (heteroplasmy) within the plant (8). Although predominant DNA configurations may be retained throughout plant development at approximately equimolar levels cell to cell, certain mitochondrial DNA forms are maintained at unusually low levels, estimated in one study at one copy per every 100–200 cells (6). These substoichiometric mitochondrial DNA forms can be up- or down-regulated in relative copy number within one plant generation, a process termed substoichiometric shifting (SSS) (9). Whether this modulation involves de novo recombination is not known. CMS mutations are sometimes contained in regions of the mitochondrial genome regulated by the SSS process (2). When present in high copy number, the mutation confers pollen sterility, but when shifted to low copy number, the plant reverts to male fertility.
Several years ago, it was shown that the mitochondrial SSS process is controlled by nuclear genes (10). More recently, a nuclear gene in Arabidopsis was cloned that directly influences mitochondrial SSS (11). The gene, designated Msh1, encodes a homolog of the Escherichia coli mismatch repair component MutS at its N terminus and contains a carboxyl-terminal domain homolog of a GIY-YIG-type homing endonuclease essential to MSH1 function (12). Msh1 loci display a high degree of amino acid sequence homology across plant families (12), but no Msh1 homolog has been identified in mammalian genomes to date. A single copy of the Msh1 gene is present in the Arabidopsis genome, and its protein targets to both mitochondria and plastids (11, 13). Disruption of the Arabidopsis Msh1 locus results in green-white leaf variegation and amplification of a particular mitochondrial DNA configuration (14). Subsequent introduction of the wild-type Msh1 allele via pollination does not appear to reverse the phenotype.
The Msh1-associated SSS process observed in Arabidopsis does not result in male sterility of the plant, indicating that not all SSS events necessarily give rise to a CMS phenotype. However, evidence suggests that a wide range of plant species harbor CMS mutations or the potential for such mutations within their mitochondrial genomes. In this study, expression of Msh1 was suppressed in tobacco and tomato by using an RNAi strategy to test the feasibility of transgenic induction of CMS. These experiments were designed to test the hypotheses that substoichiometric DNA forms within the mitochondrial genome can be selectively amplified by disruption of Msh1 expression, and that this selective amplification can give rise to CMS.
Results
Suppressed Msh1 Expression in Tobacco and Tomato by RNAi.
The level of expression of Msh1 appears to be extremely low in plants, with markedly amplified expression levels in flower bud tissues (V. Shedge and S.A.M., unpublished data). This observation complicates the confirmation of reduced Msh1 transcript levels in RNAi transgenic lines. Therefore, two independent transformation experiments each were implemented in tobacco and tomato, with 28–35 independent transformants per experiment, as well as careful phenotypic analysis over several subsequent generations to confirm heritability. In the case of tobacco, the two experiments used Nicotiana tabacum, cv. Xanthi. In tomato, experiments were carried out in two different cultivars of Solanum lycopersicum, the first cv. Moneymaker and the second cv. Rutgers. The RNAi construction was designed to target a sequence that is conserved between the tomato and tobacco Msh1 sequences (ref. 12 and data not shown). For tomato, a vector containing the UidA (GUS) reporter gene minus the RNAi construction was introduced as a negative experimental control. In tobacco, an independent transgene, the Pseudomonas syringae HopU1 Type III effector transgene, was introduced to Xanthi, and the regenerated population of 10 independent transformants was monitored for the appearance of a male sterility phenotype as negative control. These controls were developed from arbitrarily selected transgenic constructions to test for the occurrence of male sterility under our plant transformation and plant regeneration conditions. Table 1 presents data from both transgenic tobacco and tomato experiments.
Table 1.
Evaluation of transgenic plant populations for male sterility and leaf variegation (in parentheses)
| Population | No. of plants | Self progeny |
No. of plants | Test cross* results |
||||
|---|---|---|---|---|---|---|---|---|
| Fertile | Semisterile | Male sterile | Fertile | Semisterile | Male sterile | |||
| Tobacco, Xanthi | ||||||||
| Exp. 1 T0† | 28 | 26 | 2 | 0 | ||||
| Exp. 2 T0 | 28 | 23 | 5 | 0 | ||||
| Exp. 1 T1, plant 23 | 50 | 33 | 16 | 1 | 48 | 38 | 8 | 2 |
| Exp. 2 T1 | ||||||||
| Plant 2 | 20 | 16 | 3 | 1 | ||||
| Plant 6 | 20 | 10 | 8 | 2 | ||||
| Plant 7 | 19 | 12 | 5 | 2 | ||||
| Plant 12 | 29 | 9 | 8 | 3 | ||||
| Exp. 1 T2 | ||||||||
| Plant 23-5 | 50 | 3 | 24 | 23 | ||||
| Plant 23-32 | 40 | 10 | 16 | 14 | ||||
| Plant 23-32 × Xanthi | 40 | 10 | 16 | 14 | ||||
| Xanthi × plant 23-32 | 54 | 45 | 7 | 2 | ||||
| Tomato | ||||||||
| Moneymaker T0 | 31 | 26 | 5 (5) | 0 | ||||
| Rutgers T0 | 35 | 32 | 3 (3) | 0 | ||||
| Rutgers T1 | ||||||||
| Plant 17 | 20 (14) | 18 (12) | 2 (2) | 0 | ||||
| Plant 20 | 15 (11) | 12 (8) | 3 (3) | 0 | ||||
| Rutgers T2 | ||||||||
| Plant 17-12 | 10 (7) | 0 | 6 (4) | 4 (3) | ||||
| Plant 20-4 | 18 (16) | 6 (4) | 12 (12)‡ | |||||
Semisterility in tobacco is defined as dramatic reduction or absence of visible pollen on the anthers of some plants, greatly reduced capsule size, and reduced seed set. Full male sterility is the absence of visible pollen on some plants and fully collapsed seed capsules with no seed set. In tomato, semisterility is defined as reduced pollen shed and poor (5–10% of normal) seed set in fruit. Full male sterility is characterized by high rates of flower drop, delayed fruit set, and seed set at 1–2% of normal.
*Test cross progeny derive from pollination with wild-type pollen.
†T0 plants are confirmed transformants.
‡This population was analyzed for leaf variegation and sterility-associated mitochondrial shifting only, with plants in the Fertile column demonstrating no mitochondrial shift and plants in the Male sterile column demonstrating evidence of mitochondrial SSS.
In both tobacco experiments, a small number of semisterile plants were obtained in the T0 generation (Table 1). The observed tobacco male sterile phenotype was characterized by (2-week) delayed flowering, anthers that often appeared devoid of pollen (although some male sterile plants produced abundant inviable pollen), occasional petaloid anthers, and fully or partially collapsed seed capsules (Fig. 1). The pollen shed on male sterile plants was determined to be inviable based on unsuccessful test crosses to wild-type Xanthi. By the T2 generation, extremely subtle leaf variegation was also evident on a few leaves in ≈10% of the plants (Fig. 1D). Individual semisterile T0 plants from experiment 1 (plant no. 23) and experiment 2 (plant nos. 2, 6, 7, and 12) were selected for test cross and/or progeny testing. Male sterility was detected in an increasing proportion of the population for each generation. This observation suggests that multiple generations are needed to complete the cytoplasmic sorting required to shift the mitochondrial DNA population to the altered configuration. The male sterility phenotype was not reversed in progeny produced with wild-type pollen (Table 1 and data not shown). Successful pollination of the male sterile progeny produced normal seed set and indicated that the selected plants were female fertile. By the T2 generation of the tobacco population 23-32, 75% of progeny showed partial or full male sterility.
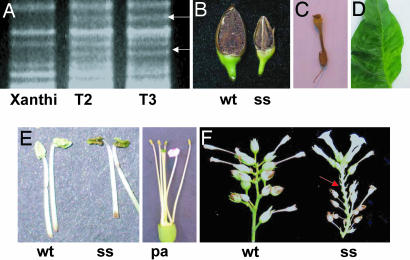
Fig. 1.
Evidence of transgenically induced CMS in tobacco. (A) Prepared mitochondrial DNA from Xanthi [wild-type (wt)], T2 male sterile plant 23-5-39, and T3 male sterile plant 23-32-4-17, digested with PstI and SstII and fractionated by 0.6% agarose gel electrophoresis. Upper polymorphic band, 6.5 kb; lower band, 5.5 kb (designated by arrows). (B) Seed capsules from Xanthi (wt) and the semisterile progeny plant no. 4 from Xanthi × P-23-32 (ss). In this case the transgene was transmitted through pollen to effect the sterility phenotype. The small size of the capsule is associated with dramatically reduced seed set, but plants classified as semisterile with small capsules do produce very small amounts of viable, selfed seed. Fully male sterile plants produce fully collapsed, detached capsules. (C) Fully collapsed, detached seed capsule characteristic of male sterile tobacco plants. (D) Evidence of subtle leaf variegation in a tobacco transformant. Variegation was not evident until the T2 generation, was infrequent, and was not restricted to fully male sterile plants. (E) Detached anthers from Xanthi (wt), the Xanthi semisterile transgenic T0 plant no. 23 (ss), and a petaloid anther (pa). Several but not all of the tobacco male sterile and semisterile plants were characterized by the production of extremely small amounts of visible pollen. (F) Detached floral branches from Xanthi (wt) and the semisterile progeny plant no. 4 from Xanthi × P-23-32 (ss), in which the transgene was transmitted through pollen. Semisterility is characterized by both collapsed capsules that immediately detach (arrow) and small capsules that produce minute amounts of seed (see B). Fully male sterile plants produce only collapsed capsules that immediately detach.
A separate transgenic tobacco population of 10 independent transformants, containing the P. syringae HopU1 Type III effector transgene (16), was developed in parallel and used as a negative control to assess the frequency of male sterility conditioned by our tissue culture, selection, and regeneration procedures. No evidence of male sterility was observed in this control population (data not shown).
Quantitative real-time (RT)-PCR analysis of several selected tobacco and tomato transformants demonstrated variation in Msh1 transcript suppression levels (Fig. 2). To further confirm that the CMS phenotype and mitochondrial DNA rearrangements observed in tobacco were the consequence of the transgene, we selected the male sterile T1 plant 23-32 from experiment 1 for reciprocal crossing to wild-type Xanthi. Although the selected plant appeared fully sterile (set little or no selfed seed), crosses with its pollen produced ≈80 seed from 10 crossed Xanthi flowers. Results shown in Table 1 indicate that progeny from Xanthi pollinated by plant 23-32 produced two fully male sterile and seven semisterile plants in a population of 54 plants. This frequency of male sterility induction is comparable to that observed in our original T1 populations to provide compelling evidence of transgenically induced CMS. The reciprocal of this cross (23-32 × Xanthi) produced 14 fully male sterile and 16 partially sterile plants of 40 plants, consistent with maternal inheritance of the male sterility trait.
Fig. 2.
Assay for reduced Msh1 transcript levels in transgenic tomato (A) and tobacco (B) lines. Shown are quantitative RT-PCR amplifications with Msh1-specific primers, giving a fragment of 629 bp, versus an 18S internal control amplification, giving a fragment of 315 bp. RNA was prepared from young leaves of tomato lines T17-12 (Rutgers transgenic), MM-12 (Moneymaker transgenic), and Rutgers (wild type) and from tobacco lines 23-5, 23-32, and Xanthi (wild type). Results shown are from two independent experiments using different numbers of PCR cycles because of the low abundance of MSH1 transcript. M designates molecular weight marker lanes.
In tomato, some of the transformants of both cultivars demonstrated striking white-green leaf variegation resembling that observed in msh1 mutants of Arabidopsis (Fig. 3B). The MSH1 protein has been shown to be dual targeted to both mitochondria and plastids in tomato (12), likely accounting for the plastid phenotype; MSH1 protein targeting behavior in tobacco has not yet been tested. Two of the Rutgers T1 male sterile plants, designated T17-12 and T20-4, have been test crossed and progeny-tested, to date, for more detailed segregation and phenotypic analysis. Male sterility in tomato was observed as delayed flowering, increased flower drop, deformed anthers, and dramatically reduced selfed fruit and seed set, although parthenocarpic (seedless) fruit set was evident (Fig. 3 and data not shown). As was the case in tobacco, the male sterility trait increased in intensity and in plant numbers each generation, with nearly 100% of the T2 generation appearing fully or partially male sterile. In no case was partial or full male sterility observed in a population of 12 independent transformants for the uidA (GUS) transgene (data not shown). All male sterile tomato and tobacco plants used for analysis were tested and found to contain a single copy of the transgene (data not shown).
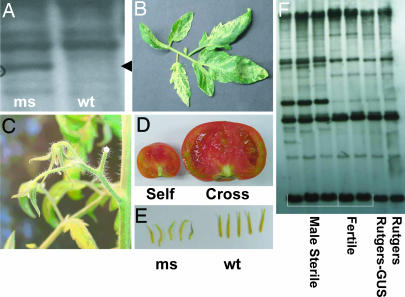
Fig. 3.
The male sterility phenotype observed in transgenic tomato lines. (A) DNA gel blot hybridization analysis of total genomic DNA from Rutgers (wt) and Rutgers male sterile (ms) T0 transgenic line 17 digested with PstI/SalI. The mitochondrial DNA probe, derived from A. thaliana BAC clone T5E7, encompasses eight mitochondrial genes and/or ORFs. Evidence of DNA polymorphism in the male sterile line is shown by an arrowhead. (B) Some leaves from a transgenic Rutgers T1 semisterile plant display a green-white variegation pattern that appears to be associated with the mitochondrial DNA rearrangement shown in A. (C) The male sterility phenotype is associated with bud drop, the premature senescence of flowers after their opening. (D) The male sterile Rutgers transgenic plants produce parthenocarpic fruit or fruit containing a small number of seeds upon self-pollination (Self). Pollination of these plants with wild-type Rutgers pollen produces normal seed set (Cross). (E) Anther morphology is modified in the male sterile transgenic plants (ms) relative to those from wild-type Rutgers (wt). (F) DNA gel blot analysis of T1 transgenic tomato plants. The male sterile selections, T17-12, T17-15, and T20-4, are partially sterile and variegated and contain a single copy of the transgene. The male fertile selections, T17-17 and T20-5, are nonvariegated and lack the transgene. Rutgers-GUS is a control transformant that contains the uidA transgene. Total genomic DNA was double-digested with PstI and SstII and hybridized with a DNA probe that encompasses mitochondrial genes atp9, nad1, nad5, and rps13.
Transgenically Induced Male Sterility Is Associated with Mitochondrial DNA Rearrangements.
In tobacco transformants, mitochondrial DNA rearrangement was evident in male sterile progeny by the T2 generation (Fig. 1A). The rearrangement was observed by restriction endonuclease analysis of purified tobacco mitochondrial DNA fractionated by gel electrophoresis. The observed tobacco mitochondrial DNA rearrangement was stably inherited to the T3 generation.
Mitochondrial DNA rearrangement was also evident in tomato, with apparently identical mitochondrial DNA changes evident in both Rutgers and Moneymaker transformants (Fig. 3 A and F). The DNA rearrangement identified in Rutgers did not show perfect cosegregation with leaf variegation, but did cosegregate with the sterility phenotype (fertile segregants lacked the mitochondrial rearrangements as shown in Fig. 3F). This lack of perfect correlation with variegation may be due, at last in part, to incomplete penetrance of the variegation phenotype.
The Male Sterility and SSS Phenotype Showed Maternal Inheritance in Subsequent Generations.
To test the inheritance of mitochondrial rearrangements and the male sterility phenotype, test crosses were performed, pollinating male sterile transgenic lines with pollen from the wild type in both tomato and tobacco. In both cases, segregation for the transgene was observed, while the vast majority of progeny remained male sterile (Table 2). These observations demonstrate the maternal inheritance pattern of the male sterility phenotype and mitochondrial rearrangements and suggest that it should be feasible to retain stable CMS, once induced, in the absence of the transgene. Additional generations of testing will be needed to confirm this assumption.
Table 2.
Segregation of the transgene and male sterility in male sterile plants after pollination by wild type
| Population | No. of plants | Fertile | Semisterile | Male sterile |
|---|---|---|---|---|
| Tobacco, Xanthi | ||||
| Exp. 1, T3 | ||||
| Plant 23-32-4 | 20 (9) | 3 (0) | 15 (8) | 2 (1) |
| Plant 23-32-26 | 20 (10) | 1 (0) | 12 (4) | 7 (6) |
| Plant 23-32-32 | 20 (13) | 2 (0) | 14 (10) | 4 (3) |
| Tomato | ||||
| Rutgers T3 | 24 (0) | 6 (0) | 12 (0) | 6 (0) |
| Plant 17-12-5 | ||||
| Plant 17-12-15 | 18 (12) | 4 (2) | 5 (2) | 9 (8) |
Numbers in parentheses represent plants containing the transgene.
Discussion
Male sterility phenotypes in both tomato and tobacco derived, in each case, from two independent transformation experiments. In both species, male sterility was heritable, increasing in phenotype intensity and in non-Mendelian proportions of the population in subsequent generations. This observation is consistent with expectations of nuclear-controlled mitochondrial SSS and subsequent cytoplasmic sorting. The observations of apparently identical mitochondrial rearrangements in Moneymaker and Rutgers RNAi lines, and what appears to be one type of rearrangement in the tobacco transformants, are consistent with SSS activity to amplify mitochondrial sterility sequences that were already present within the genome. It is important to note that the observed mitochondrial polymorphisms are not necessarily the CMS-causing rearrangements, but only evidence of mitochondrial SSS activity.
The experiments found evidence of mitochondrial DNA rearrangement and a leaf variegation phenotype similar to that observed in msh1 mutants of Arabidopsis. Not all male sterile plants showed variegation, but they all appeared to be female fertile, and the sterility and SSS phenotypes were not reversed by segregation of the transgene or pollination with wild-type pollen in experiments conducted to date. However, we did observe fully or partially male fertile progeny deriving from a male sterile plant in T1 populations. Although the proportion of the population demonstrating partial male fertility declined in T2 and T3 generations, we still observed a percentage of the population by T3 that showed some level of pollen viability. We interpret this pattern to reflect a multigeneration, cytoplasmic sorting process. The observation of markedly lower male fertility in each subsequent generation suggests that it should be feasible to reach 100% male sterility.
We observed no evidence of partial or full male sterility arising in tomato or tobacco plants transformed with unrelated, arbitrarily selected transgenes. More importantly, however, these transformation experiments were conducted within the University of Nebraska Center for Biotechnology Plant Transformation Center where tomato (Rutgers and Moneymaker) and tobacco (Xanthi) transformations are conducted on a routine basis with a wide variety of transgene constructions, and male sterility is not observed with the transformation and regeneration procedures that were used for this study (T. Clemente, personal communication).
The observations presented, taken together, provide evidence of transgenic induction of CMS as a consequence of Msh1 suppression. Although additional testing will be necessary, we predict that the transgenically induced mitochondrial rearrangement is not reversible, similar to the effect of msh1 mutation in Arabidopsis. If this is the case, segregation of the transgene once male sterility is achieved should permit retention of the stably altered cytoplasm and could allow the release of novel CMS materials generated by using this strategy as nontransgenic lines. Although transgenic traits are gaining acceptance by the agricultural community worldwide, the ability to develop novel CMS crop lines that can be released without the transgene would be of value in many important export crops.
Past strategies for the transgenic induction of male sterility in crops have met with mixed success. We suggest that four features will be key to successful deployment of a male sterility system for hybrid seed production: cytoplasmic inheritance to facilitate the recovery of 100% male sterile plants each breeding cycle, an efficient genetic strategy for fertility restoration in F1 populations, ready transferability of the system to most major crop species, and the ability to release the hybrid in a nontransgenic form. To date, no method has been reported that meets all of these goals. Recently, an interesting strategy for CMS induction was reported by using chloroplast transformation and overexpression of a gene for β-ketothiolase in tobacco (17). However, difficulties with chloroplast transformation in most major crops, the persistent requirement of a highly expressed transgene, and the unavailability of a strategy for efficient fertility restoration could complicate its implementation (18).
The extent to which the transgenic approach described in this report will be useful in other crops should be tested in soybean, millet, and maize. For those crops in which transformation is currently infeasible, selection of Msh1 mutants may provide an effective, alternative approach. The accompanying leaf variegation phenotype served as a useful early generation phenotypic marker for the purposes of our study; this variegation was reduced or disappeared completely in more advanced generations.
CMS has been reported previously in both tobacco and tomato. Sources studied in tobacco have resulted from interspecific hybridization (19) and from in vitro culture (20). In tomato, CMS was also observed as a consequence of interspecific hybridization (21). Both cell culture and alloplasmy routinely give rise to mitochondrial DNA rearrangement and SSS activity and may involve reduced Msh1 expression.
The development of hybrids using CMS in some crops requires not only a stable, nonreverting source of male sterility, but also the identification of a nuclear genotype that reversibly suppresses the male sterility trait. Nuclear fertility restorer genes have been the focus of intense investigation over the past few years, with five recently cloned restorer loci shown to contain pentatricopeptide repeats (2, 22–26). Pentatricopeptide repeat proteins, numbering over 400 in Arabidopsis, are thought to be involved in transcript processing events within the mitochondrial and plastid genomes and, in the case of fertility restoration, likely direct processing of CMS-associated transcripts (27). The means by which CMS was induced in this study, implementing illegitimate recombination that is known to occur naturally in plant mitochondria, should lend itself to the identification of natural restorers already present within the species, most likely in undomesticated materials. It will now be feasible to screen various tobacco and tomato genotypes as pollinators for their ability to effect fertility restoration to the newly derived CMS mutants. In those crops grown for vegetative products rather than seed, including carrot, onion, or sugar beet, the system we have described should be of benefit without any added requirement of a restorer.
Materials and Methods
RNAi Construction and Transformation Procedure.
A segment encoding amino acids 651–870 of the MSH1 protein was derived from a tomato EST sequence (12) by using the primer sequences TOM-CD1F (5′-CGCAGGTATCACGAGGCAAGTGCTAAGG-3′) and TOM-CD1R (5′-ATCCCCAAACAGCCAATTTCGTCCAGG-3′) and cloned in forward and reverse orientation, separated by an intron sequence. The base vector, pUCRNAi-intron, which harbors the second intron of the Arabidopsis small nuclear riboprotein (At4g02840), was provided by H. Cerutti (University of Nebraska, Lincoln, NE). The CaMV 35S promoter and transcription terminator regulate expression of the construction and the neomycin phosphotransferase II (npt II) reporter gene, and the insert is flanked by right border and left border integration sequences. Agrobacterium tumefaciens stain C58C1/pMP90 (28) was used for transformation in tobacco (29) and tomato (30).
Quantitative RT-PCR Assay of Msh1 Transcript Levels in Transformants.
Total RNA from young leaves was isolated by using TRIzol (Sigma, St. Louis, MO), as recommended by the manufacturer and then treated with DNA-free DNase (Ambion, Austin, TX) to remove any contaminating DNA. Prepared RNA (1 μg) was used to synthesize cDNA by using the Ambion RETROscript First Strand Synthesis kit for RT-PCR. Quantitative RT-PCR was done by using the Ambion QuantumRNA 18S Internal Standard kit. For these experiments, a 1:9 primer:competimer ratio was used with 59°C annealing temperature and 24 PCR cycles. Primers designed from the tomato Msh1 cDNA sequence were TomMSF3 (5′-cctacttgggtggcaactgggttgaaagtt-3′) and TomMSR3 (5′-tctttgcatctgtcttgaatgtgatagc-3′).
Detection of Mitochondrial DNA Polymorphisms.
Tobacco mitochondrial DNA preparations (31) and tomato total genomic DNA preparations (10–15 μg) were fractionated by agarose gel electrophoresis (0.6% agarose in 1× TBE buffer at 50 V/cm) and blotted to Hybond-N (GE Healthcare, Little Chalfont, U.K.) for DNA gel blot hybridization. The probe used for tomato was Arabidopsis thaliana BAC clone T5E7 and a PCR-amplified tobacco mitochondrial fragment that spans the region containing genes atp9, nad1, nad5, and rps13. Probes were 32P-labeled by using random priming (15) for autoradiographic exposure.
Acknowledgments
Plant transformations were conducted in the University of Nebraska Center for Biotechnology Plant Transformation Core Facility by Ms. Shirley Sato and Dr. Tom Clemente. The RNAi cloning vector was provided by Dr. Heriberto Cerutti. We thank Ms. Nikki Westercamp for technical support and Drs. Alan Christensen, Maria Arrieta-Montiel, Michael Fromm, and Tom Clemente for helpful discussions. This work was supported by a grant to S.A.M. from the Biotechnology Research Development Corporation.
Abbreviations
- SSS
substoichiometric shifting
- CMS
cytoplasmic male sterility.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
References
- 1.Hanson MR, Bentolila S. Plant Cell. 2004;16:154–169. doi: 10.1105/tpc.015966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mackenzie SA. In: Plant Breeding Reviews. Janick J, editor. Vol 25. New York: Wiley; 2005. pp. 115–138. [Google Scholar]
- 3.Delph LF, Touzet P, Bailey MF. Trends Ecol Evol. 2007;22:17–24. doi: 10.1016/j.tree.2006.09.013. [DOI] [PubMed] [Google Scholar]
- 4.Mackenzie SA. In: Diversity and Evolution of Plants. Henry RJ, editor. UK: CABI, Oxon; 2005. pp. 69–80. [Google Scholar]
- 5.Schnable PS, Wise RP. Trends Plant Sci. 1998;3:175–180. [Google Scholar]
- 6.Arrieta-Montiel M, Lyznik A, Woloszynska M, Janska H, Tohme J, Mackenzie S. Genetics. 2001;158:851–864. doi: 10.1093/genetics/158.2.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kanazawa A, Tsutsumi N, Hirai A. Genetics. 1994;138:865–870. doi: 10.1093/genetics/138.3.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barr CM, Neiman M, Taylor DR. New Phytol. 2005;168:39–50. doi: 10.1111/j.1469-8137.2005.01492.x. [DOI] [PubMed] [Google Scholar]
- 9.Small ID, Isaac PG, Leaver CJ. EMBO J. 1987;6:865–869. doi: 10.1002/j.1460-2075.1987.tb04832.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mackenzie S, Chase C. Plant Cell. 1990;2:905–912. doi: 10.1105/tpc.2.9.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Abdelnoor RV, Yule R, Elo A, Christensen A, Meyer-Gauen G, Mackenzie S. Proc Natl Acad Sci USA. 2003;100:5968–5973. doi: 10.1073/pnas.1037651100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abdelnoor RV, Christensen AC, Mohammed S, Munoz-Castillo B, Moriyama H, Mackenzie SA. J Mol Evol. 2006;63:165–173. doi: 10.1007/s00239-005-0226-9. [DOI] [PubMed] [Google Scholar]
- 13.Christensen AC, Lyznik A, Mohammed S, Elowsky CG, Elo A, Yule R, Mackenzie SA. Plant Cell. 2005;10:2805–2816. doi: 10.1105/tpc.105.035287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sakamoto W, Kondo H, Murata M, Motoyoshi F. Plant Cell. 1996;8:1377–1390. doi: 10.1105/tpc.8.8.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feinberg AP, Vogelstein B. Anal Biochem. 1983;132:6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- 16.Buell CR, Joardar V, Lindeberg M, Selengut J, Paulsen IT, Gwinn ML, Dodson RJ, DeBoy RT, Durkin AS, Kolonay JF, et al. Proc Natl Acad Sci USA. 2003;100:10181–10186. doi: 10.1073/pnas.1731982100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Raiz ON, Daniell H. Plant Physiol. 2005;138:1232–1246. doi: 10.1104/pp.104.057729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chase CD. Trends Plant Sci. 2006;11:7–9. doi: 10.1016/j.tplants.2005.11.003. [DOI] [PubMed] [Google Scholar]
- 19.Hakansson G, Glimelius K. Mol Gen Genet. 1991;229:380–388. doi: 10.1007/BF00267459. [DOI] [PubMed] [Google Scholar]
- 20.Chetrit P, Rios R, De Paepe R, Vitart V, Gutierres S, Vedel F. Curr Genet. 1992;21:131–137. doi: 10.1007/BF00318472. [DOI] [PubMed] [Google Scholar]
- 21.Melchers G, Mohri Y, Watanabe K, Wakabayashi S, Harada K. Proc Natl Acad Sci USA. 1992;89:6832–6836. doi: 10.1073/pnas.89.15.6832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bentolila S, Alfonso AA, Hanson MR. Proc Natl Acad Sci USA. 2002;99:10887–10892. doi: 10.1073/pnas.102301599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brown GG, Formanova N, Jin H, Wargachuk R, Dendy C, Patil P, Laforest M, Zhang J, Cheung WY, Landry BS. Plant J. 2003;35:262–272. doi: 10.1046/j.1365-313x.2003.01799.x. [DOI] [PubMed] [Google Scholar]
- 24.Koizuka N, Imai R, Fujimoto H, Hayakawa T, Kimura Y, Kohno-Murase J, Sakai T, Kawasaki S, Imamura J. Plant J. 2003;34:407–415. doi: 10.1046/j.1365-313x.2003.01735.x. [DOI] [PubMed] [Google Scholar]
- 25.Wang Z, Zou Y, Li X, Zhang Q, Chen L, Wu H, Su D, Chen Y, Guo J, Luo D, et al. Plant Cell. 2006;18:676–687. doi: 10.1105/tpc.105.038240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Klein RR, Klein PE, Mullet JE, Minx P, Rooney WL, Schertz KF. Theor Appl Genet. 2005;111:994–1012. doi: 10.1007/s00122-005-2011-y. [DOI] [PubMed] [Google Scholar]
- 27.Wise RP, Pring DR. Proc Natl Acad Sci USA. 2002;99:10240–10242. doi: 10.1073/pnas.172388899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koncz C, Schell J. Mol Gen Genet. 1986;204:383–396. [Google Scholar]
- 29.Horsch RB, Fry JE, Hoffmann NL, Eichholtz D, Rogers SG, Fraley RT. Science. 1985;227:1229–1231. [Google Scholar]
- 30.McCormick SM, Neidermeyer J, Fry J, Barnason A, Horsch R, Fraley RT. Plant Cell Rep. 1986;5:81–84. doi: 10.1007/BF00269239. [DOI] [PubMed] [Google Scholar]
- 31.Mackenzie S. In: Plant Molecular Biology Manual. Gelvin S, Shilperoort R, Verma DP, editors. Vol 3. The Netherlands: Kluwer, Dordrecht; 1994. pp. 1–12. [Google Scholar]