SUMMARY
While human plasma represents an attractive sample for disease biomarker discovery, the extreme complexity and large dynamic range in protein concentrations present significant challenges for characterization, candidate biomarker discovery, and validation. Herein, we describe a strategy that combines immunoaffinity subtraction and subsequent chemical fractionation based on cysteinyl peptide and N-glycopeptide captures with 2D-LC-MS/MS to increase the dynamic range of analysis for plasma. Application of this “divide-and-conquer” strategy to trauma patient plasma significantly improved the overall dynamic range of detection and resulted in confident identification of 22,267 unique peptides from four different peptide populations (cysteinyl peptides, non-cysteinyl peptides, N-glycopeptides, and non-glycopeptides) that covered 3654 different proteins with 1494 proteins identified by multiple peptides. Numerous low-abundance proteins were identified, exemplified by 78 “classic” cytokines and cytokine receptors and by 136 human cell differentiation molecules. Additionally, a total of 2910 different N-glycopeptides that correspond to 662 N-glycoproteins and 1553 N-glycosylation sites were identified. A panel of the proteins identified in this study is known to be involved in inflammation and immune responses. This study established an extensive reference protein database for trauma patients, which provides a foundation for future high-throughput quantitative plasma proteomic studies designed to elucidate the mechanisms that underlie systemic inflammatory responses.
Keywords: trauma, inflammation, human plasma, proteomics, immunoaffinity subtraction, cysteinyl peptide enrichment, N-glycopeptide capture, chemical fractionation, LC-MS/MS
INTRODUCTION
The search for disease specific biomarkers from various human biofluids (e.g., plasma/serum,1-3 cerebrospinal fluid,4 bronchoalveolar lavage fluid,5 synovial fluid,6 nipple aspirate fluid,7 saliva,8 and urine9) is gaining increasing attention due to significant advances in genomic and proteomic technologies and their potential for discovering novel disease biomarkers. An advantage of using human plasma for biomarker discovery is that plasma samples are readily obtained from patients. More importantly, because almost all cells in the body communicate with blood either directly or indirectly, and tissue-specific proteins are released into the blood stream upon cell damage and cell death, the onset or presence of most disease states can potentially be determined by measuring either the altered presence or the differential abundances of proteins in the blood plasma.
Although progress has been made towards quantitatively studying the plasma proteome,10-13 significant effort still remains focused on broadly characterizing the plasma protein constituents. These constituents include a handful of major proteins such as albumin that dominate the plasma protein content, as well as other tissue leakage proteins that are generally present only at extremely low levels. As a result, robust and specific detection of the broad spectrum of proteins that span such a wide dynamic range in concentration challenges current analytical technologies. Various separation approaches (e.g., prefractionation of intact plasma proteins,14-17 utilization of solution isoelectric focusing,18 and ultra-high-efficiency separations19,20 at the peptide level and multidimensional fractionation on both the protein and peptide levels21) have been applied to address this challenge. Efforts have also been made to characterize subsets of the plasma proteome (e.g., the low molecular weight subproteome22 and the N-glycoproteome23). Additionally, the removal of albumin, immunoglobulins, and a group of high-abundance proteins in plasma/serum has been demonstrated to be very effective for attaining more comprehensive proteome coverage.20, 23-26 Recently developed multicomponent immunoaffinity subtraction systems, based on mammalian27, 28 and avian29 antibodies, are now available for highly efficient, reproducible, and reversible depletion of high-abundance proteins in plasma/serum samples. The common concern of potentially losing a subset of nontargeted proteins with these methods by way of protein-protein interactions (e.g., the “sponge effects” of albumin30) can be addressed by studying the “interactomes.”29
Herein, we report on an alternative strategy for profiling the human plasma proteome that combines immunoaffinity subtraction and subsequent chemical fractionation via solid-phase capture of cysteinyl peptides and N-glycopeptides with two-dimensional (2D) LC-MS/MS to extend the dynamic range of analysis and increase proteome coverage. The advantages afforded by dividing a complex proteome into several subproteomes include 1) reduced complexity of the subproteome samples (particularly, enriched cysteinyl peptide and N-glycopeptide samples), which allows more low-abundance proteins to be identified; 2) the complementary nature of different subproteome fractions, which significantly improves overall proteome coverage; and 3) the relative simplicity, efficiency, and reproducibility afforded by these fractionation methods, which make them amenable to automation. Application of this strategy to characterize blood plasma obtained from human trauma patients resulted in broad proteome coverage that included many low-abundance proteins (e.g., cytokines) and proteins involved in both inflammation, a hallmark of many human diseases,31, 32 and the immune response. Previous inflammation studies have relied heavily on gene expression analysis of blood leukocyte populations obtained from trauma patients.33, 34 An extensive reference plasma proteome database for trauma has been established from this study, which provides the basis for subsequent comparative quantitative proteomic studies aimed at revealing the underlying mechanisms of inflammatory response in trauma patients.
EXPERIMENTAL PROCEDURES
Immunoaffinity Subtraction
The human blood plasma samples were supplied by the Department of Surgery at the University of Florida College of Medicine, which serves as the Sample Collection and Coordination Site for a multicentered clinical study (Inflammation and the Host Response to Injury). Approval for the conduct of this programmatic research was obtained from the Institutional Review Boards of the University of Florida College of Medicine and the Pacific Northwest National Laboratory in accordance with federal regulations. Plasma samples were pooled from 6 severe trauma patients and 1 healthy subject, and the protein concentration was determined by Coomassie Protein Assay (Pierce, Rockford, IL) to be 60 mg/mL. Twelve plasma proteins — albumin, IgG, α1-antitrypsin, IgA, IgM, transferrin, haptoglobin, α1-acid glycoprotein, α2-macroglobulin, apolipoprotein A-I, apolipoprotein A-II, and fibrinogen — that constitute up to 96% of the total protein mass of human plasma were removed in a single step by using the prepacked 2-mL Seppro™ MIXED12 affinity LC column (GenWay Biotech, San Diego, CA) on an Agilent 1100 series HPLC system (Agilent, Palo Alto, CA) per the manufacturer’s instruction. A total of 2625 μL (25 μL × 105 injections) plasma was subjected to MIXED12-depletion. The flow-through fractions were pooled and concentrated under reduced pressure to one tenth of the original volume and then desalted by using a prepacked PD-10 column (Amersham Biosciences, Piscataway, NJ) equilibrated with 50 mM NH4HCO3. The desalted sample was further concentrated, and the total protein amount was determined by Coomassie Protein Assay to be 8.85 mg.
Plasma Protein Digestion
One third of the proteins from the depleted plasma sample were directly subjected to tryptic digestion prior to cysteinyl peptide enrichment (CPE). The proteins were denatured and reduced in 50 mM Tris buffer (pH 8.2), 8 M urea, 10 mM dithiothreitol (DTT) for 1 h at 37 °C. The resulting protein mixture was diluted 10 fold with 20 mM Tris buffer (pH 8.2), and then sequencing grade modified porcine trypsin (Promega, Madison, WI) was added at a trypsin:protein ratio of 1:50 (w/w). The sample was incubated overnight at 37 °C. The following day, the tryptic digest sample was loaded on a 1-mL SPE C18 column (Supelco, Bellefonte, PA) and washed with 4 mL of 0.1% trifluoroacetic acid (TFA)/5% acetonitrile (ACN). Peptides were eluted from the SPE column with 1 mL of 0.1% TFA/80% ACN and lyophilized. Peptide samples were stored at -80 °C until CPE treatment.
Fractionation via Capture of Cysteinyl Peptides
Unless otherwise noted, all solutions used in this step were degassed to prevent oxidation of the thiol content. The tryptic digest was reduced with 5 mM DTT in 80 μL of 50 mM Tris buffer (pH 7.5), 1 mM EDTA (coupling buffer) for 30 min at 37 °C, after which the samples were diluted to 400 μL by adding coupling buffer and then split into four 100 μL aliquots. Thiopropyl Sepharose 6B thiol-affinity resin (4 × 100 μL; Amersham Biosciences) was prepared from dried powder per the manufacturer’s instruction. Briefly, the dried powder was rehydrated in water for 15 min and washed by 50 bed volumes of water, followed by 50 bed volumes of coupling buffer in a Handee Mini-Spin column (Pierce). The reduced peptide sample was then incubated with the resin for 1 h at room temperature with gentle mixing, and the unbound portion (non-cysteinyl peptides) was collected by spinning the column at low speed. The resin was washed in the spin column sequentially with the following solutions: 0.5 mL of 50 mM Tris buffer (pH 8.0), 1 mM EDTA (washing buffer), 2 M NaCl, 80% ACN/0.1% TFA solution, and washing buffer. To release the captured cysteinyl peptides, 100 μL of 20 mM DTT freshly prepared in washing buffer was added to the resin and incubated for 30 min at room temperature. The resin was further washed with 100 μL of 80% ACN. The pH of the combined eluates was adjusted to 8.0 and the sample was alkylated with 80 mM iodoacetamide for 30 min at room temperature and in the dark. Both the eluted cysteinyl peptide samples and the unbound non-cysteinyl peptide samples were desalted by using a SPE C18 column and then lyophilized.
Fractionation via Capture of N-Linked glycopeptides
Hydrazide resin (Bio-Rad, Hercules, CA) and a method similar to that previously reported23, 35 were employed for capturing glycoproteins. Two-thirds of the concentrated MIXED12 flow-through fraction was diluted 10 fold in coupling buffer (100 mM sodium acetate and 150 mM NaCl, pH 5.5) and oxidized in 15 mM sodium periodate at room temperature for 1 h in the dark with constant shaking. The sodium periodate was subsequently removed by using a prepacked PD-10 column equilibrated with coupling buffer. Four 1-mL portions of hydrazide resin were washed 5 times with coupling buffer; the oxidized protein sample was split into 4 equal aliquots and then each aliquot was added to a portion of resin and incubated overnight at room temperature. The following day, the non-glycoproteins were removed by washing the resin briefly 3 times with 100% methanol and then 3 times with 8 M urea in 0.4 M NH4HCO3. The glycoproteins were denatured and reduced in 8 M urea and 10 mM DTT at 37 °C for 1 h. Protein cysteinyl residues were alkylated with 20 mM iodoacetamide for 90 min at room temperature. After washing sequentially with 8 M urea and 50 mM NH4HCO3, the resin was resuspended as a 20% slurry in 50 mM NH4HCO3, after which sequencing grade trypsin (Promega, Madison, WI) was added at a 1:100 (w/w) trypsin:protein ratio (based on the initial plasma protein concentration of 65 mg/mL). The sample was then digested on-resin overnight at 37 °C. After centrifuging at 3000 × g for 5 min, the trypsin-released peptides (non-glycopeptides) were collected from the supernatants andlyophilized. The resin was further washed extensively with the following 3 different solutions: 2 M NaCl, 100% methanol, and 50 mM NH4HCO3. The resin was resuspended as a 50% slurry in 50 mM NH4HCO3, and the N-glycopeptides were released by incubating the resin with peptide N-glycosidase F (PNGase F; New England Biolabs, Beverly, MA) at a ratio of 1 μL of PNGase F per 100 μL of plasma for 4 h at 37 °C. The released deglycosylated peptides were cleaned by using an SPE C18 column before being lyophilized.
Strong Cation Exchange (SCX) Peptide Fractionation
All of the enriched cysteinyl peptides and deglycosylated peptides, and 300 μg each of non-cysteinyl peptides and non-glycopeptides were individually reconstituted with 300 μL of 10 mM ammonium formate (pH 3.0)/25% ACN and fractionated by strong cation exchange (SCX) chromatography on a Polysulfoethyl A 200 mm × 2.1 mm column (PolyLC, Columbia, MD) that was preceded by a 10 mm × 2.1 mm guard column. The separations were performed on an Agilent 1100 series HPLC system (Agilent) at a flow rate of 200 μL/min, and with mobile phases that consisted of 10 mM ammonium formate (pH 3.0)/25% ACN (A) and 500 mM ammonium formate (pH 6.8)/25% ACN (B). After loading 300 μL of sample onto the column, the gradient was maintained at 100% A for 10 min. Peptides were separated by using a gradient from 0 to 50% B over 40 min, followed by a gradient of 50-100% B over 10 min. The gradient was then held at 100% B for 10 min. A total of 30 fractions were collected for each peptide population, and each fraction was dried under vacuum. The fractions for each population were dissolved in 30 μL of 25 mM NH4HCO3, and 10 μL of each fraction was analyzed by capillary LC-MS/MS.
Reversed-Phase Capillary LC-MS/MS Analyses
A custom-built high-pressure capillary LC system36 coupled on-line to a linear ion trap mass spectrometer (LTQ; ThermoElectron) via an in-house-manufactured electrospray ionization interface was used to analyze peptide samples. The reversed-phase capillary column was prepared by slurry packing 3-μm Jupiter C18 bonded particles (Phenomenex, Torrence, CA) into a 65-cm long, 150 μm-i.d. ×360 μm-o.d. fused silica capillary (Polymicro Technologies, Phoenix, AZ) that incorporated a retaining stainless steel screen in an HPLC union (Valco Instruments Co., Houston, TX). The mobile phases, which consisted of 0.2% acetic acid and 0.05% TFA in water (A) and 0.1% TFA in 90% ACN/10% water (B), were degassed on-line by using a vacuum degasser (Jones Chromatography Inc., Lakewood, CO). After loading 10 μL of peptides onto the column, the mobile phase was held at 100% A for 20 min. An exponential gradient elution was achieved by increasing the mobile phase composition in a stainless steel mixing chamber from 0 to 70% B over 150 min. To identify the eluting peptides, the linear ion trap mass spectrometer was operated in a data-dependent MS/MS mode (m/z 400-2000) in which each full MS scan was followed by ten MS/MS scans. The 10 most intense precursor ions were dynamically selected in order of highest to lowest intensity, and then subjected to collision-induced dissociation; a normalized collision energy setting of 35% and a dynamic exclusion duration of 1 min were used. The temperature of the heated capillary and the ESI voltage were 200 °C and 2.2 kV, respectively.
Data Analysis
The SEQUEST37 algorithm (ThermoFinnigan) was used to independently search all MS/MS spectra against the human International Protein Index (IPI) database (Version 3.05 that consists of 49,161 protein entries; available online at http://www.ebi.ac.uk/IPI) and the reversed human IPI protein database. Tandem MS peaks were generated by extract_msn.exe, part of the SEQUEST software package. Dynamic carboxamidomethylation of cysteine and oxidation of methionine were used to identify cysteinyl peptides, non-cysteinyl peptides, and non-glycopeptides; another PNGase F-catalyzed conversion of asparagine to aspartic acid at the site of carbohydrate attachment was added to identify the formerly N-linked glycopeptides.
Because false positive peptide/protein identifications are a common concern in proteomics investigations, we developed a set of criteria based on the reversed database approach for filtering the raw data to limit false positive identifications to <5%. The reversed human protein database was created by reversing the order of the amino acid sequences for each protein, and the percentage of false positive peptide identifications was estimated as previously described38 by dividing the number of peptides identified from the reversed database search by the number of peptides identified from the normal database search. Only the cysteine-containing and the NXS/T-motif-containing peptides were used to estimate false positive identifications for cysteinyl peptides and formerly N-linked glycopeptides, respectively. (An in silico assessment of the number of cysteine-containing and NXS/T-motif-containing peptides among all tryptic peptides shows the number of these kinds of peptides in the normal and reversed databases to be similar; thus, the number of random false positive identifications is assumed to be similar for both databases.)
Criteria that would yield an overall confidence of over 95% at the unique peptide level were established for filtering raw peptide identifications. Table 1 summarizes the cross correlation score (Xcorr) values for the different peptide populations; a delta correlation (ΔCn) value of 0.10 was used to determine these values. Two additional ΔCn cutoff values of 0.05 and 0.16 were applied to reduce false negative identifications, while maintaining a 95% level of confidence for peptide assignments. For ΔCn ≥0.05, the minimum acceptable Xcorr value was raised to achieve a comparable percentage of false positive rate identifications, and similarly, for ΔCn ≥0.16, the minimum acceptable Xcorr value was reduced. In an attempt to remove redundant protein entries, Protein Prophet software was used as a clustering tool to group similar or related protein entries into a “protein group.” 39 All peptides that passed the filtering criteria were assigned an identical probability score of 1 and entered into the software program (solely for clustering analysis) to generate a final list of nonredundant proteins or protein groups. One protein ID was randomly selected to represent each corresponding protein group that contains member database entries.
Table 1.
SEQUEST filtering criteria used for peptide identificationa.
| Peptide | Charge state | Xcorr | Tryptic ends | Other constraint |
|---|---|---|---|---|
| Cysteinyl peptidesb | 1+ | ≥1.5 | Fully | Cys |
| 1+ | ≥2.7 | Partially | Cys | |
| 2+ | ≥2.2 | Fully | Cys | |
| 2+ | ≥3.4 | Partially | Cys | |
| 3+ | ≥2.9 | Fully | Cys | |
| 3+ | ≥4.1 | Partially | Cys | |
| N-linked glycopeptidesc | 1+ | ≥1.5 | Fully | NXS/T |
| 1+ | ≥2.1 | Partially | NXS/T | |
| 2+ | ≥1.8 | Fully | NXS/T | |
| 2+ | ≥3.3 | Partially | NXS/T | |
| 3+ | ≥2.6 | Fully | NXS/T | |
| 3+ | ≥4.2 | Partially | NXS/T | |
| Non-cysteinyl peptides | 1+ | ≥1.5 | Fully | None |
| Non-glycopeptides | 1+ | ≥3.0 | Partially | None |
| 2+ | ≥2.7 | Fully | None | |
| 2+ | ≥3.7 | Partially | None | |
| 3+ | ≥3.3 | Fully | None | |
| 3+ | ≥4.5 | Partially | None |
All Δ Cn are required to be ≥0.10.
The presence of at least one cysteine residue was required for all peptides.
The presence of at least one NXS/T motif was required for all peptides.
Gene Ontology (GO) component, function, and process terms extracted from text-based annotation files downloaded from the European Bioinformatics Institute ftp site (ftp://ftp.ebi.ac.uk/pub/databases/GO/goa/HUMAN) were used to categorize the identified proteins. Further biological interpretation in the context of gene ontology was performed with the aid of GOstat.40 Protein transmembrane helices were determined by TMHMM, a prediction model based on a hidden Markov model (http://www.cbs.dtu.dk/services/TMHMM/).
Pathway and network analysis was performed by using the Ingenuity Pathways Knowledge Base (IPKB)41. Canonical signal transduction and metabolic pathways were examined among the proteins identified. Queries were also made to obtain cellular localization information of the detected proteins from the IPKB. The significant functional networks among the extracellular proteins were computed as described elsewhere41.
Enzyme-Linked Immunosorbent Assay (ELISA)
Specific plasma protein levels in the pooled trauma patient plasma sample were determined in duplicate using Quantikine ELISA kits (R&D Systems, Minneapolis, MN) following manufacturer’s instructions. Each sample was analyzed at two different dilutions to determine the optimal dilution. A plasma sample from a healthy subject obtained from Golden West Biologicals (Temecula, CA) was also assayed for comparison.
RESULTS
High Dynamic Range Proteomic Profiling Strategy
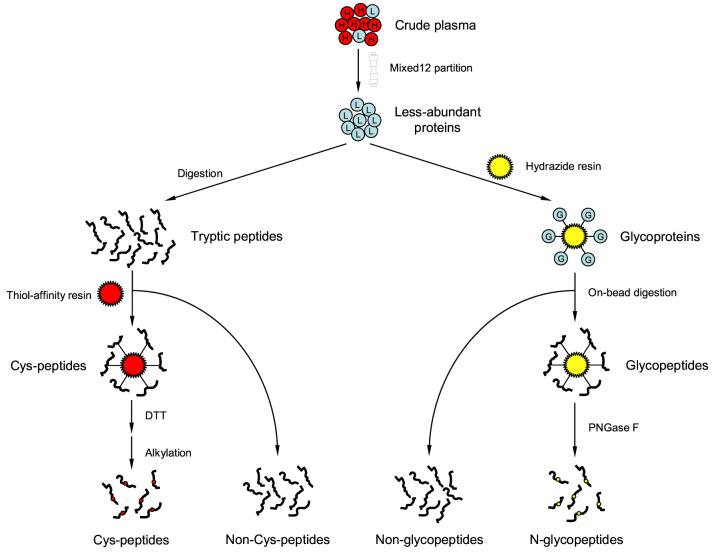
Figure 1 schematically illustrates the sample processing and fractionation strategy developed to globally characterize the trauma patient plasma proteome. High-abundance proteins were first removed by immunoaffinity subtraction. The 12 proteins targeted by the MIXED12 column in this step constitute up to 95% of the total protein mass in human plasma, and our results showed that the column removed the majority of these proteins; 5.6% of the total protein mass in the pooled trauma patient plasma remained after the depletion step. Following the depletion, the less abundant proteins in the flow-through fraction were split into two aliquots, which were subjected to solid-phase chemical fractionation schemes to enrich both cysteinyl peptides and N-glycopeptides. The cysteinyl peptides were captured by incubating the tryptic peptides with a thiol-affinity resin, Thiopropyl Sepharose, upon which mixed disulfide bonds formed between the resin and the cysteinyl peptides. Non-cysteinyl peptides were collected immediately after incubation, and the cysteinyl peptides were recovered by incubating the resin with a low molecular weight thiol, DTT, followed by alkylation. Another aliquot of proteins was first oxidized by periodate, which converted the hydroxyl groups on adjacent carbon atoms in the glycoprotein carbohydrates to aldehydes. The glycosylated proteins were then captured on the hydrazide resin via formation of covalent hydrazone bonds between the newly converted aldehyde groups and the hydrazide groups on the resin. The captured glycoproteins were denatured, reduced, alkylated, and digested in situ, after which the non-glycopeptides were collected from the supernatant using brief centrifugation. The N-glycopeptides were specifically released by incubating the resin with PNGase F.
Figure 1.
Schematic representation of the sample processing and fractionation used to characterize the trauma patient plasma proteome. High-abundance proteins were firstly removed using immunoaffinity subtraction. The resulting less-abundant proteins were split and submitted individually for solid-phase cysteinyl peptide and N-glycoprotein captures. Non-cysteinyl peptides and non-glycopeptides generated at the same time were also collected. All 4 different peptide populations were then fractionated by SCX chromatography and each fraction was analyzed by capillary LC-MS/MS.
Peptide and Protein Identifications
Thirty SCX fractions of each peptide population were analyzed by LC-MS/MS, and the resulting spectra were searched against the human IPI protein database by using SEQUEST. Only those peptide identifications that passed the filtering criteria in Table 1 were considered as confident identifications. The numbers of confident peptide and protein identifications obtained from the 4 different populations are summarized in Table 2. A total of 22,267 unique peptides (81.8% fully tryptic) were identified from the 4 different peptide populations, which corresponds to 3654 nonredundant proteins, including 1494 multi-peptide proteins (Supplemental Table 1) and 2160 single-peptide proteins (Supplemental Table 2). Note, the non-cysteinyl peptide sample contributed the largest set of peptide identifications (9951) and the N-glycopeptide fraction contributed the smallest set of peptide identifications (2910).
Table 2.
Number of peptide and protein identifications obtained from the 4 different peptide populations.
| Fraction | Unique peptides | Non-redundant proteins | Multi-peptide proteins |
|---|---|---|---|
| Cys | 7617 | 1977 | 870 (44.0%) |
| Non-Cys | 9951 | 1914 | 872 (45.6%) |
| N-glyco | 2910 | 662 | 382 (57.7%) |
| Non-glyco | 7656 | 1486 | 620 (41.7%) |
| Total | 22267 | 3654 | 1494 (40.9%) |
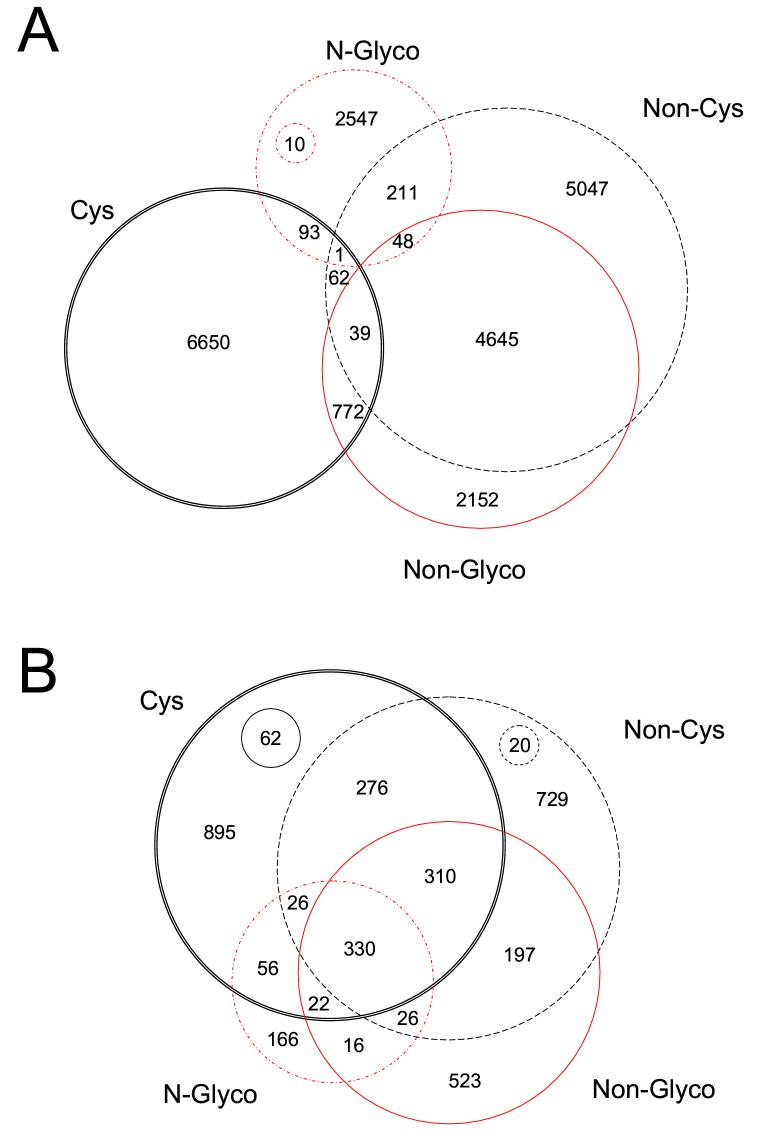
Of the 3654 proteins identified from the pooled trauma patient plasma, 1341 proteins (36.7%) were present in more than one peptide population, and 330 proteins (9.0%) were observed in all 4 peptide populations (Fig. 2B), which demonstrates the complementary nature of the 4 different types of peptide identifications for obtaining higher proteome coverage. 662 distinct N-glycoproteins were identified from the 2910 N-glycopeptides. The overlap between peptides identified in the 4 peptide populations (Fig. 2A) shows that the cysteinyl peptide and N-glycopeptide fractions share only a small percentage of identifications with the other peptide fractions. Although the non-cysteinyl peptide and non-glycopeptide fractions exhibit much greater overlap in peptide identifications, these fractions still provided additional proteome coverage (e.g., 523 proteins only from non-glycopeptides and 729 proteins only from non-cysteinyl peptides). Interestingly, the same sequences for 12.5% of the N-glycopeptides were also found in 94 cysteinyl peptides, 58 non-glycopeptides, and 260 non-cysteinyl peptides, which indicates that these N-glycosylation sites are only partially modified and that both N-glycosylated and unmodified peptides from different fractions are detected.
Figure 2.
Diagram of peptides (A) and proteins (B) identified in multiple peptide populations. All overlaps are shown (2-way, 3-way, and 4-way) for all four peptide populations. The numbers in the small circles indicate the number of proteins that are common only to the two fractions at the opposite positions in the diagram. Cys, cysteinyl peptides; Non-Cys, non-cysteinyl peptides; N-Glyco, N-glycopeptides; Non-Glyco, non-glycopeptides.
Confidence of Identifications
To illustrate the stringency of our approach for limiting peptide false discovery rate (FDR) to <5%, the filtering criteria developed in this study were compared to two sets of published criteria21, 42 previously used to filter plasma/serum MS/MS data. The three different sets of criteria were applied to filter the SEQUEST identifications for all 4 peptide populations and false positive rates resulted from each set of criteria were assessed using the reversed database approach38. The results of this evaluation are summarized in Table 3. Our filtering criteria generated the smallest number of peptide and protein identifications, consistent with the significantly lower percentage of false positive identifications (∼4%). Using the criteria recommended by the human proteome organization (HUPO)42, peptide identifications increased 1.4-fold and protein identifications increased 2.2-fold, which yielded a lower percentage of proteins with multiple peptide “hits” and increased the overall FDR at the peptide level to ∼25%. The application of the criteria recently reported by Hood et al.21 resulted in 18,958 nonredundant protein identifications; however, FDR at the peptide level was ∼66%. The common feature between our criteria and the HUPO criteria is that only fully and partially tryptic peptides are considered; however, the HUPO criteria uses much lower Xcorr cutoff values for the partially tryptic peptides. The criteria used by Hood et al. are similar to two other previously reported sets of criteria22, 25 in which chymotryptic and elastic peptides, as well as those peptides that had high Xcorr scores, but no proteolytic end were also considered. Clearly, our filtering criteria are significantly more stringent when compared with the two sets of previously published criteria. Furthermore, the cysteinyl peptide and N-glycopeptide identifications were specifically enriched through reliable solid-phase chemical tagging, which further increases the overall confidence level for our data.
Table 3.
Number of peptide and protein identifications from the present study obtained by filtering with different criteria.
| Filter | Peptides identified | Proteins identifieda | Multi-peptide proteins | Avg. peptides per protein | Estimated false discovery rateb |
|---|---|---|---|---|---|
| Reversed database | 22267 | 3654 | 1494 (40.9%) | 6.1 | ∼4% |
| HUPO | 30524 | 7928 | 2850 (35.9%) | 3.9 | ∼25% |
| Hood et al. | 66839 | 18958 | 11653 (61.5%) | 3.5 | ∼66% |
Non-redundant protein identifications.
Calculated at unique peptide level based on reversed database evaluation (Ref. 38).
The confidence of our current filtering criteria was also compared with the independent statistical model Peptide Prophet program43 by analyzing the same subset of datasets applying both approaches. Histograms of Xcorr distributions in peptides identified by reversed database approach showed that low FDR can be achieved using filtering criteria listed in Table 1 (Supplemental Figure 1). Similarly, high-confidence peptide identification can be obtained at a certain Peptide Probability (pcomp) level (Supplemental Figure 2). 6279 unique peptides were identified from the reversed database approach (≥95% confidence), while 6341 unique peptides were identified from Peptide Prophet (pcomp≥0.95), among which 5973 (∼95%) peptides were found in common. Note that the estimated FDR predicted by Peptide Prophet is 0.3% (pcomp≥0.95), further supporting the stringency of the current reversed database filtering criteria. After processing of both sets of peptides by Protein Prophet, a total of 1301 proteins and 1254 proteins were identified for the reversed database approach and Peptide Prophet, respectively. The high percentage overlap at the peptide level and the comparable numbers of protein identifications reflect the overall high quality of the currently reported data.
Dynamic Range of Detection and the Detection of Low-Abundance Proteins
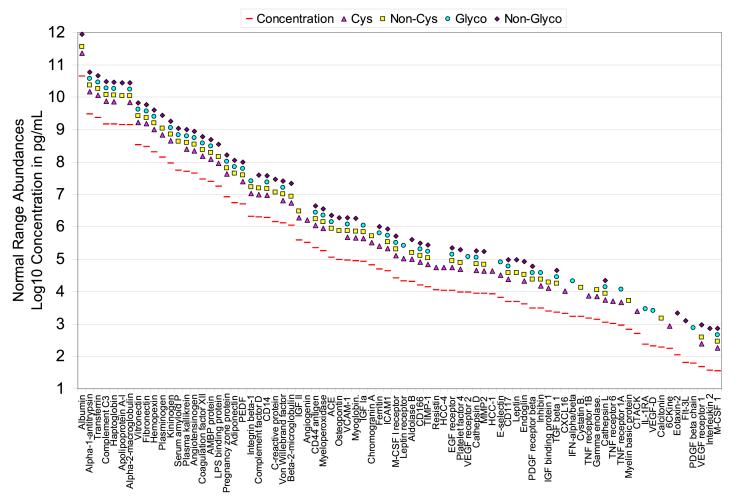
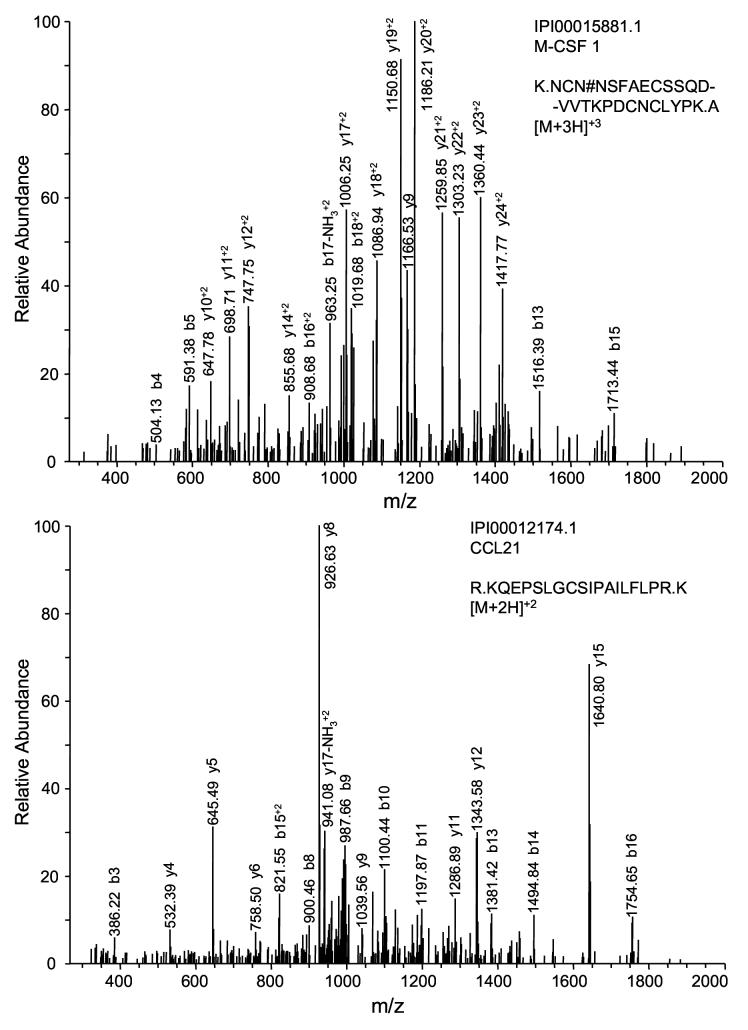
Figure 3 plots reference protein concentrations under normal conditions for 80 selected proteins identified in this study as well as their peptide distribution within the four peptide populations. It is obvious that most of the high-abundance proteins and proteins having a mid-range concentration (e.g., from μg/mL to ng/mL level) were identified from multiple peptide populations, while many of low-abundance proteins (e.g., cytokines) were identified from only one peptide population. It is speculated that some protein concentrations in trauma patients may be significantly different from those under normal conditions as a result of inflammatory responses. To test this hypothesis and evaluate the overall dynamic range of measurements that resulted from this study, six identified low-abundance proteins that are related to inflammatory response were assayed by ELISA. The results showed that the protein levels from the trauma patient plasma pool are higher for most of these cytokines or cytokine receptors than those measured from a normal individual except small inducible cytokine A21 (CCL21); however, all six proteins are still present in the 0.5∼20 ng/mL range for the trauma patient plasma samples (Table 4). An estimated overall dynamic range of detection of >107 was achieved by applying the combined approach of immunoaffinity subtraction, solid-phase chemical tagging of cysteinyl peptides and N-glycopeptides, and 2D-LC-MS/MS. This claim is justified by the confident identification of a series of ng/mL range proteins in this study, exemplified by macrophage colony stimulating factor (M-CSF) and CCL21 (Figure 4, top panel and bottom panel, respectively) that are both presenting at 1 ng/mL level in trauma patient plasma. In addition, many tissue leakage proteins in the μg/mL to ng/mL concentration range were readily detected in multiple peptide populations, which provide a solid basis for candidate disease biomarker (e.g., cancer biomarker) discovery when applying this strategy.
Figure 3.
Reference concentrations for 80 selected proteins identified from trauma patient plasma. The protein concentration is plotted on a log scale spanning 11 orders of magnitude. The reference concentrations were obtained from Refs. 1 and 11, and all data were based on healthy human subjects. It should be noted that the actual detected concentrations for the trauma sample may be significantly different from these reference concentrations. Peptide population from which those proteins were identified was labeled using the following scheme: cysteinyl peptides, triangles; non-cysteinyl peptides, squares; N-glycopeptides, circles; non-glycopeptides, diamonds. PEDF, pigment epithelium-derived factor; IGF, insulin-like growth factor; ACE, angiotensin-converting enzyme; VCAM, vascular cell adhesion molecule; ICAM, intercellular adhesion molecule; M-CSF, macrophage colony stimulating factor; TIMP, tissue inhibitor of metalloprotease; HCC, hemofiltrate CC chemokine; EGF, epidermal growth factor; VEGF, vascular endothelial growth factor; MMP, matrix metalloprotease; PDGF, platelet-derived growth factor; TGF, transforming growth factor; CXCL16; CXC-chemokine ligand 16; IFN, interferon; TNF, tumor necrosis factor; CTACK, cutaneous T cell-attracting chemokine; IL-1RA, interleukin 1 receptor antagonist; 6CKine, 6-cysteine chemokine; Flt-3L, fms-like tyrosine kinase 3 ligand.
Table 4.
Concentrations of selected proteins in the pooled trauma patient sample and a normal plasma sample measured by ELISAa.
| Protein | Concentration (pg/mL) | Standard Curve R2value |
|---|---|---|
| Trauma PDGF-B | 2807 | 0.9972 |
| Normal PDGF-B | Not detectable | |
| Trauma IL-1RA | 19666 | 0.9936 |
| Normal IL-1RA | 1179 | |
| Trauma VEGF R1 | 556 | 0.9987 |
| Normal VEGF R1 | 232 | |
| Trauma M-CSF | 1042 | 0.9975 |
| Normal M-CSF | 157 | |
| Trauma CCL21 | 1165 | 0.9873 |
| Normal CCL21 | 1115 | |
| Trauma TNF R1 | 6014 | 0.9989 |
| Normal TNF R1 | 945 |
Cytokine nomenclature and categorization are from http://www.rndsystems.com and Ref. 57. PDGF-B, platelet-derived growth factor B chain; IL-1RA, interleukin 1 receptor antagonist; VEGF R1, vascular endothelial growth factor receptor 1; M-CSF, macrophage-colony stimulating factor; CCL21, small inducible cytokine A21; TNF R1, tumor necrosis factor receptor superfamily member 1A.
Figure 4.
Representative MS/MS spectra of low-abundance proteins identified in this study. The top panel shows a deglycosylated peptide identified from M-CSF (1.0 ng/mL) and the bottom panel shows a cysteinyl peptide identified from small inducible cytokine A21 (1.1 ng/mL).
Cytokines are a diverse family of soluble immunomodulatory proteins and peptides that can be produced by every nucleated cell type in the body. Seventy-eight “classic” cytokines and cytokine receptors, e.g., tumor necrosis factor receptor (TNF R), interleukin (IL), gp130, platelet-derived growth factor (PDGF), vascular endothelial growth factor (VEGF), and transforming growth factor-β (TGF-β), and chemokines were identified in this study and are categorized in Table 5.
Table 5.
Categorization of 78 “classic” cytokines and cytokine receptors identified in this studya.
| Family | Features | Cytokines | Receptors |
|---|---|---|---|
| TNF | Contain highly conserved carboxy terminal domains; can induce receptor trimerization influencing signaling pathways | TNF RI (p55), TNF RII (p75), LT-β R, Fas (CD95), TRAIL R3, CD27, TNF R21 (DR6) | |
| γc | Receptors contain a common γchain (γc) | IL-2 | |
| IL-4 & IL-13 | Bind to shared heteromultimeric receptor complexes | IL-13 Rα1 | |
| βc | Receptors contain a common βchain (βc) | IL-5 Rα, GM-CSF Rα | |
| IL-1 | A superfamily of proteins comprised of IL-1 and IL-18 members. | IL-1RA | IL-1 RI, IL-1 RII, , IL-1R AP, IL-18 BP |
| IGF | Share sequence homology with the insulin family of proteins | IGF1a, IGF2 | IGF RII |
| HGF & MSP | Contain a 4-kringle domain and a pseudo-serine protease domain that lacks enzyme activity | Met | |
| FGF | Heparin-binding polypeptides | FGF R1, FGF R2, FGF R4 | |
| Neutrophic factors | Induce signal transduction through ligand-induced dimerization and activation of trk receptors | trk B | |
| PDGF, VEGF, PlGF | Dimeric angiogenic factors containing an 8-cysteine motif | PDGF-B, VEGF-D | PDGF Rβ, VEGF R1, VEGF R2, VEGF R3, Neuropilin-1, Neuropilin-2 |
| EGF | Contain at least one extracellular EGF structural unit (conserved 6-cysteine motif that forms 3 disulfide bonds) | EGF R | |
| gp130 | Receptors of the IL-6 superfamily are homologous to or contain the gp130 subunit as the common signaling component | OB | Ob R, OSM R, IL-6 Rα, IL-6 Rβ (gp130), CNTF Rα |
| SCF, Flt-3L, M-CSF | Contain a 4-helix bundle structure in the extracellular domain and 4 conserved cysteines; receptors are tyrosine kinases | Flt-3L, M-CSF | c-kit, Flt-3, M-CSF R |
| MK & PTN | Products of retinoic acid-responsive genes; developmentally-regulated molecules | RPTPβ | |
| TGF-β | Contain a highly conserved 7-cysteine domain that forms a characteristic cysteine knot | TGF-β1, TGF-β3, BMP-1, BMP-3, GDF-2, Inh β C, Inh β E | TGF-β RII, TGF-β RIII, BMPR-II, GFRα2, Act RII, |
| CC chemokine | Target population include: lymphoid cells (T cells and NK cells), monocytes and PMNs | HCC-1, MIP-1d , HCC-4, 6Ckine, Eot-2, CTACK, | CCR5, CCR10 |
| CXC chemokine | Target population include: predominantly PMNs, but also T and B cells | PF4, CXCL16 | |
| Unique | IL-32, MIF | GH R, IFN-α/α Rα, IFN- α/β Rβ, IL-10 Rα, IL-10 Rβ, IL-17 R, IL-23 R, IL-31 Rα, TLR4 |
Cytokine nomenclature and categorization are from http://www.rndsystems.com and Ref. 57. TNF, tumor necrosis factor; LT-β, lymphotoxin beta; TRAIL, TNF-related apoptosis-inducing ligand; DR6, death receptor 6; IL, interleukin; GM-CSF, granulocyte macrophage colony stimulating factor; IL-1RA, interleukin 1 receptor antagonist; IL-1R AP, interleukin 1 receptor accessory protein; IL-18 BP, interleukin 18 binding protein; IGF, insulin-like growth factor; HGF, hepatocyte growth factor; MSP, macrophage stimulating protein; Met, hepatocyte growth factor receptor; FGF, fibroblast growth factor; trk B, neurotrophic tyrosine kinase receptor type 2; PDGF, platelet-derived growth factor; VEGF, vascular endothelial growth factor; PlGF, placenta growth factor; EGF, epidermal growth factor; OB, leptin; OSM, oncostatin M; CNTF, ciliary neurotrophic factor; SCF, stem cell factor; Flt-3L, fms-like tyrosine kinase 3 ligand; M-CSF, macrophage-colony stimulating factor; c-kit, SCF receptor; MK, midkine; PTN, pleiotrophin; RPTPβ, receptor-type tyrosine-protein phosphatase beta; TGF, transforming growth factor; BMP, bone morphogenetic protein; GDF, growth/differentiation factor; Inh β C, inhibin beta C chain; Inh β E, inhibin beta E chain; GFR, glial-derived neurotrophic factor receptor; Act, activin; NK cells, natural killer cells; PMN, polymorphonuclear leukocytes; HCC, hemofiltrate CC (a class of chemokines); MIP, macrophage inflammatory protein; 6Ckine, 6-cysteine chemokine; Eot, eotaxin; CTACK, cutaneous T cell-attracting chemokine; CCR, CC chemokine receptor; PF4, platelet factor 4; CXCL16, CXC-chemokine ligand 16; MIF, macrophage migration inhibitory factor; GH R, growth hormone receptor; IFN, interferon; TLR, toll-like receptor.
The immune system works through leukocytes interacting with each other, other cells, tissue matrices, infectious agents, and other antigens. These interactions are mediated by cell-surface glycoproteins and glycolipids (cell differentiation molecules, or CD antigens) that are frequently cleaved from the cell surface by protease activity. A total of 136 out of the 288 (47.2%) known human protein CD antigens (available at http://www.hlda8.org/CD1toCD339.htm) were detected in this study (Supplemental Table 3). Good coverage was obtained for the CD antigens routinely detected with anti-leukocyte monoclonal antibodies and used to characterize the cell surface immunophenotypes of different leukocyte subpopulations (e.g., B-cells, helper T-cells, cytotoxic T-cells, and natural killer cells). In 90.4% of the identified CD antigens each had at least one predicted transmembrane domain, while among all other proteins, only 15.5% had predicted transmembrane domain(s). This finding is consistent with the fact that the majority of CD antigens are believed to be membrane-associated molecules.
Gene Ontology (GO) and Pathway Analysis of the Detected Proteins
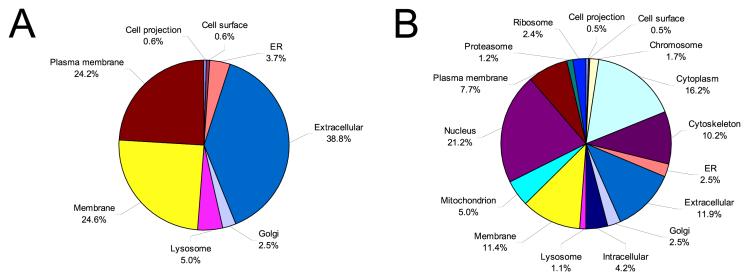
Figure 5 shows the categories of proteins identified from this study in terms of cellular location based on gene ontology analysis. Comparison of cellular components for N-glycoproteins and the other proteins identified shows major differences. The majority of N-glycoproteins (Figure 5A) are predicted to be extracellular/secreted proteins (38.8%) and membrane-associated proteins (48.8%), while all other proteins (Figure 5B) are predicted to distribute more evenly across all cellular locations. None of the N-glycoproteins identified are from the nucleus, cytoplasm, mitochondrion, ribosome, proteasome, and cytoskeleton, which is consistent with the biological functions of N-linked glycoproteins.23 The high percentage of intracellular proteins in this dataset indicates that large numbers of proteins present in plasma may result from different levels of cellular leakage.
Figure 5.
Comparison of protein categorization using GO component terms. Major component categories are shown for N-glycoprotein identifications (A) and all other protein identifications (B).
Further GOstat analyses that compared the distribution of GO terms of identified proteins with the entire human IPI database revealed over- and under-represented molecular functions and biological processes (data not shown). Over-represented molecular function categories included hematopoietin/interferon-class cytokine receptor activity, insulin-like growth factor (IGF) binding, VEGF receptor activity, metallopeptidase activity, protease inhibitor, extracellular matrix structural constituent, lipid binding and transporter activity, polysaccharide binding, receptor protein kinase activity, and oxidoreductase activity. In the GO comparison of biological processes, proteins involved in response to wound, regulation of body fluids, complement activation, and proteolysis categories appeared over-represented among the proteins identified. These findings reflect certain distinguishing features of the trauma patient plasma proteome, e.g., the presence of many inflammation and immune response-related proteins, exemplified by the proteins listed in Table 6. These proteins include acute phase reactants, cytokines and growth factors, complement proteins and coagulation factors, hormones, extracellular matrix proteins, cell adhesion molecules, and secreted proteases and protease inhibitors, in addition to other proteins and immunoglobulins.
Table 6.
Selected inflammation-related proteins identified in this study.
| Protein name | Function |
|---|---|
| Acute Phase Reactants | |
| Alpha-1-acid glycoprotein 1, 2 | Binding protein |
| Alpha-1-antitrypsin | Protease inhibitor |
| Alpha-2-antiplasmin | Coagulation cascade |
| Alpha-2-macroglobulin | Inhibitor of several serum proteases |
| C-reactive protein | Opsonin |
| Ceruloplasmin | Copper transport protein |
| Ferritin | Iron transport |
| Fibrinogen | Clotting formation |
| Fibronectin | Clotting formation |
| Haptoglobin | Scavenger |
| Hemopexin | Heme binding |
| Heparin cofactor-2 | Proteinase inhibitor |
| Kallikrein | Vascular permeability and dilatation |
| LPS binding protein | Innate immunity |
| Manganese superoxide dismutase | Antioxidant |
| Mannose-binding protein C | Innate immunity |
| Plasminogen | Clotting, fibrinolysis |
| Plasminogen activator inhibitor-1 | Protease inhibitor |
| Serum amyloid A | Scavenger |
| Serum amyloid P | Formation of IgG immune complexes |
| Cytokines and Growth Factors | |
| Interleukin 2 | Growth factor |
| Macrophage colony stimulating factor | Colony stimulation |
| Tumor necrosis factor receptor 1, 2 | Anti-inflammatory |
| Interleukin 1 receptor 2 | Anti-inflammatory |
| Interleukin 6 receptor | Anti-inflammatory |
| CD14 | Anti-inflammatory |
| Interleukin 1 receptor antagonist | Anti-inflammatory |
| Macrophage inflammation protein 1 | Chemokine |
| Eotaxin | Chemokine |
| Hemofiltrate CC chemokine 1 | Chemokine |
| Transforming growth factor β | Growth factor |
| Vascular endothelial growth factor | Growth factor |
| Insulin-like growth factor 1, 2 | Growth factor |
| Platelet-derived growth factor | Growth factor |
| Complement Proteins and Coagulation Factors | |
| C2, C3, C4, C5, C9 | Complement component |
| Coagulation Factor V, VII∼XIII | Coagulation protein |
| Protein C | Degrades coagulation proteins |
| Von Willebrand factor | Coagulation protein |
| Thrombomodulin | Binds thrombin |
| Antithrombin III | Coagulation inhibitor |
| Tissue factor pathway inhibitor | Coagulation inhibitor |
| Hormones | |
| Renin | Fluid regulation |
| Leptin | Energy balance |
| IGF binding proteins 1∼6 | Carrier proteins |
| Extracellular Matrix Proteins | |
| Laminin 1∼4 | Matrix protein |
| Collagen I, III, IV | Matrix protein |
| Nidogen | Cell-cell communication |
| Fibronectin | Matrix protein |
| Thrombospondin I, II, IV | Cell adhesion |
| Vitronectin | |
| Tenascin | |
| Cell Adhesion Molecules | |
| Intercellular adhesion molecule 1∼3 | Cell adhesion |
| Activated leukocyte adhesion molecule | Cell adhesion |
| Contactin 1∼4 | Cell adhesion |
| Mucosal addressin cell adhesion molecule | Cell recruitment |
| Neural cell adhesion molecule | Cell adhesion |
| Vascular cell adhesion molecule 1 | Cell adhesion |
| Integrin α , β | Cell adhesion |
| E-selectin | Cell adhesion |
| L-selectin | Cell adhesion |
| P-selectin | Cell adhesion |
| Secreted Proteases and Protease Inhibitors | |
| Matrix metalloprotease 2, 9, 13, 14 | Collagenases, elastases and stromelysins |
| Bone morphogenetic protein 1 | Regulatory protein |
| Metalloproteinase inhibitor 1, 2 | Endogenous inhibitors of collagenases and MMPs |
| Cathepsin B, D, F | Regulatory protease |
| Other Proteins and Immunoglobulins | |
| Albumin | Osmotic regulation |
| Transthyretin | Protein carrier |
| Transferrin | Iron binding |
| Retinol-binding protein | Protein carrier |
| Immunoglobulin gamma, alpha, mu | Host defense |
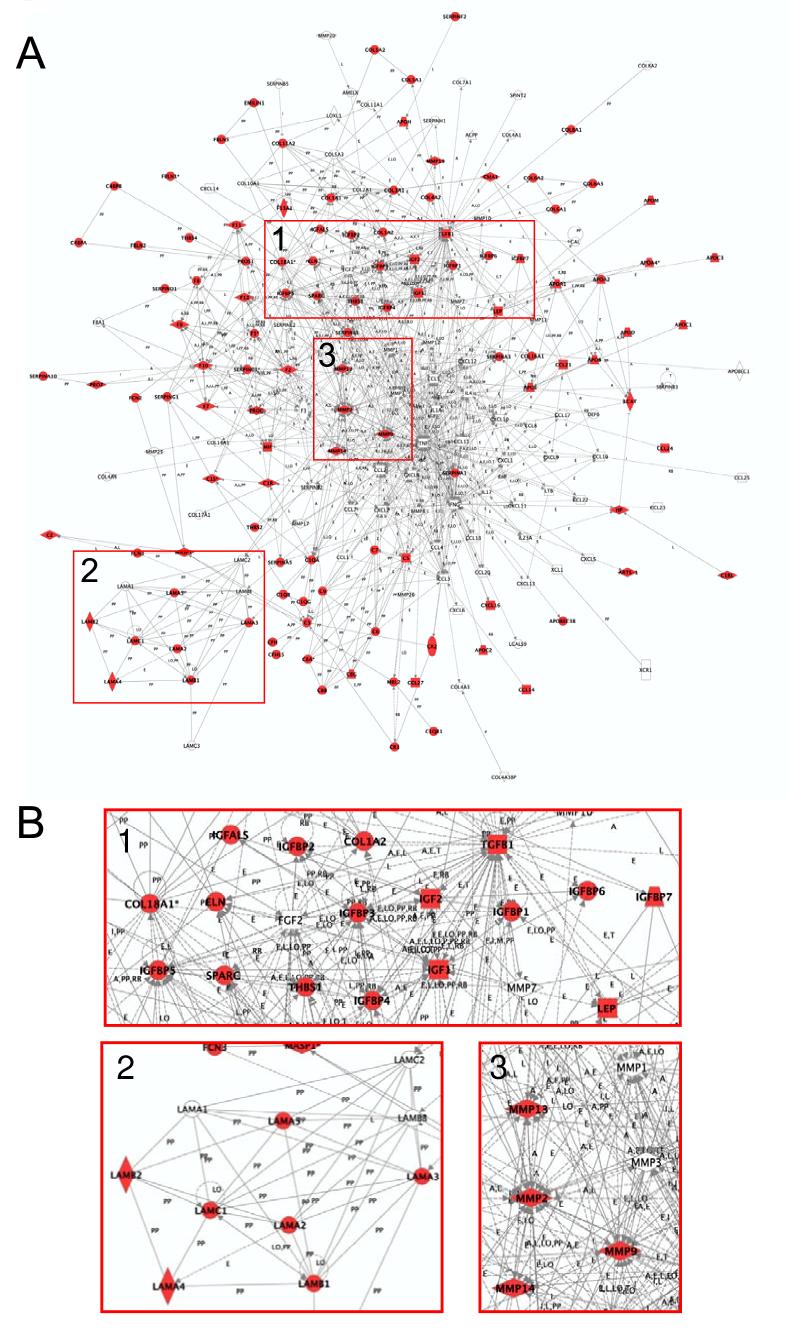
Pathway analysis revealed the significant representation of specific signaling pathways, e.g., NF-κB signaling (inflammation and immune regulation), apoptosis signaling, ERK/MAPK signaling, and Wnt/β-catenin signaling (data not shown). As an example, Figure 6A shows a global representation of the extracellular proteins that are involved in immune response, which comprises a network of 193 proteins and their interactions. In total, 113 of the 193 (58.5%) known players were identified. Figure 6B further illustrates the coverage of specific regions of this network and highlights the IGF and IGF binding proteins, laminins, and matrix metalloproteinases.
Figure 6.
Network representation of the extracellular pathways that are involved in immune response. (A) The network consists of 193 genes with 113 identified in this study. (B) Selected regions of the network (B) highlight several groups of proteins. Group 1, IGF and IGF binding proteins. Group 2, laminins. Group 3, matrix metalloproteinases.
DISCUSSION
Specific biomarkers for diagnosis/prognosis of disease and for monitoring disease progression and response to therapy have been clinically applied to screen patient tissues and blood samples, as well as used to develop therapeutics and segment the population for specific treatments.44 Proteomics is increasingly being used in this field to describe and enumerate the systematic changes in the protein constituency of a cell, to generate lists of proteins that change in expression as a cause or consequence of disease, and more importantly, to characterize the information flow through the intra- and extra-cellular molecular protein networks that interconnect organ and circulatory systems. These networks are expected to provide new targets for therapeutics and to reveal the dynamic biological changes that give rise to new candidate biomarkers.45 Because of its constant perfusion through tissues within the body, blood plasma is anticipated to contain ample information regarding these networks, and therefore, provides a basis for candidate disease biomarker discovery. However, several intrinsic features of plasma, such as an enormous dynamic range in protein concentrations of interest and extreme sample complexity and heterogeneity, hamper effective proteomic analysis.
Our strategy for analyzing blood plasma addresses these issues by combining multicomponent immunoaffinity subtraction and multiple chemical fractionations (Figure 1) with 2D-LC-MS/MS. The single-step depletion of 12 high-abundance proteins on an automated LC system significantly increases the dynamic range of detection and reduces sample heterogeneity (due to the simultaneous removal of the highly variable IgG, IgA, and IgM populations). The high efficiency CPE step further reduces sample complexity, which in turn enables detection of low-abundance proteins.46, 47 Simultaneous analysis of the non-cysteinyl peptides, generated as a “byproduct” during CPE, significantly increases proteome coverage.47, 48 The N-glycopeptide enrichment step affords yet another effective way of reducing plasma sample complexity.23, 35, 49 N-glycosylation is particularly prevalent in proteins that are secreted and located on the extra-cellular side of the plasma membrane and in proteins that are contained in various body fluids (e.g., blood plasma).50 Since the N-glycosylation sites generally fall into a consensus NXS/T sequence motif, where X represents any amino acid residue except proline,51 the availability of N-glycopeptides from each protein is limited, which provides the basis for reducing sample complexity. The performance of the immunoaffinity subtraction MIXED12 column has been previously reported to be specific, efficient, and reproducible.29 In our study, the 12 immunosubtraction-targeted proteins were specifically and effectively removed, but peptides from these proteins were still detected by MS following the depletion step (data not shown), presumably due to the initial high concentrations of these proteins. Binding of other non-target proteins to the MIXED12 column did occur, however, in a slight and fairly reproducible fashion (data not shown). Therefore, this column should be applicable to quantitative studies. The methods for cysteinyl peptide and N-glycopeptide enrichment can be integrated seamlessly into the quantitative strategies by using stable isotope labeling35, 46 (e.g., 18O labeling) and are amenable to automation. The major challenges in proteomic studies on inflammation are that, while cancer biomarkers are usually binomial, i.e., they are absent in normal and present in disease and probably do not vary extensively over short time periods, the inflammatory biomarkers are likely not a matter of presence or absence, but more of a change in concentration compared to normal and likely to change rapidly over time, which addresses the need for high-throughput temporal analysis in this field. Although the current study is qualitative, the analytical strategy proposed here is able to be directly linked to quantitative automation. Taking full advantage of the broad proteome coverage and in-depth detection dynamic range, samples from temporal studies on inflammation prepared by the current strategy, with or without stable isotope labeling, can be analyzed by LC-MS-based approaches for high-throughput biomarker discovery by using the accurate mass and time (AMT) tag approach.52
Application of this “divide and conquer” strategy significantly improved overall dynamic range of detection (estimated to be >107) for the characterization of the blood plasma proteome. In a previous report a nonredundant human plasma protein list was developed by combining proteins from 4 separate sources.53 The 46 proteins found in common among the four datasets were all relatively high-abundant “classic” plasma proteins. In the current study, some of the 330 proteins observed in all 4 of the peptide populations appear to be “classic” plasma proteins (e.g., complement components, coagulation factors, and protease inhibitors); however, there are also many proteins such as cathepsins D and L that are anticipated to be present at relatively low concentrations in plasma (<10 ng/mL at normal conditions54, 55). These results suggest the overall high dynamic range of detection applying the current strategy that allows many low-abundance proteins to be consistently detected in all 4 peptide populations. Additionally, improvements in overall proteome coverage were achieved by combining identifications from these 4 different, but complementary, peptide populations. Only very small overlap was observed between the cysteinyl peptide and non-cysteinyl peptide fractions, between the N-glycopeptide and non-glycopeptide fractions, and between the cysteinyl peptide and N-glycopeptide fractions (Figure 2A). A total of 3654 non-redundant proteins (Table 2) were confidently identified that included 662 N-glycoproteins (Fig. 2B), as well as numerous cytokines, cytokine receptors, CD antigens, and proteins involved in both inflammation and immune response processes. Approximately 63% of these non-redundant proteins were identified solely from one peptide population and included many low-abundance proteins such as IL-2 from non-glycopeptides; PDGF B chain from glycopeptides; CCL21 from cysteinyl peptides, and calcitonin from non-cysteinyl peptides.
A major challenge in the analysis of MS data is the accurate assessment of the extent of false positive identifications. Various criteria have been developed to filter raw MS/MS identifications; however, further statistical evaluation is essential to ensure high-confidence protein identifications can be derived from such analyses. For example, an initial large scale HUPO plasma proteome collaborative study assembled a list of 3020 proteins identified with two or more peptides, using data acquired on different instruments from 18 different laboratories42. The list has been recently reduced to 889 proteins (containing both multi-peptide and single-peptide protein identifications) identified with a confidence level of at least 95% using a rigorous statistical approach taking into account the length of coding regions in genes, and multiple hypothesis-testing techniques56. Our filtering criteria developed based on the reversed database searching are much more stringent compared with the early HUPO criteria (Table 3). Its stringency is also supported by the comparable results from the reversed database approach and Peptide/Protein Prophet program in terms of generating high-confidence protein identifications. Furthermore, reanalyzing the data from one of our early plasma profiling study19 using the HUPO criteria yielded 1073 proteins, the length-dependent statistical analysis yielded approximately two-fold reduction in protein identifications (433 proteins with confidence >95%)56. Similarly, for the data presented in this study the reversed database analysis also resulted in ∼2-fold fewer protein identifications compared to those identifications obtained if the HUPO criteria was applied (3654 vs. 7928 proteins using previous HUPO criteria, Table 3), suggesting an approximate comparable level of confidence for protein identifications obtained between the reversed database criteria and the recently published length-dependent statistical analysis56. These comparisons between independent statistical approaches reflect the overall high quality of the currently reported data obtained by using the novel combined approach; however, the single-peptide protein identifications will have higher FDR compared to the multi-peptide proteins; therefore, these single-peptide proteins are listed separately in Supplemental Table 2.
The relatively low-abundance cytokines present in plasma mediate not only host responses to invading organisms, tumors and trauma, but also maintain our capacity of daily survival in our germ-laden environment57. The detection of cytokines in disease states (e.g., inflammation) may provide very useful diagnostic and/or therapeutic tools; for example, IL-1 receptor antagonist has been shown to play a role in systemic host responses and improve survival in septic shock58, 59, and some cytokine receptors are also being evaluated for the anti-inflammation effects60. Our strategy enabled the detection of members of all major cytokine families (Table 5), which demonstrates the applicability of this strategy for discovering cytokine inflammation biomarkers in quantitative studies.
An area of contention surrounding biomarker discovery is whether a single protein marker or a panel of biomarkers should be used for disease diagnosis and therapeutic treatment. An increasingly common view is that the use of a single biomarker lacks the required sensitivity and specificity when applied to a heterogeneous population; however, these limitations may be overcome by utilizing panels of biomarkers.61 As in cancer, the dysfunctional or malignant cell growth may result from the changes in multiple members of the deranged protein signal transduction pathways. Therefore, an understanding of the pathways and networks that involve plasma proteins released from the cells would facilitate the development of a disease biomarker panel for clinical applications. The pathway analysis reveals that our dataset indeed provides extensive coverage for important signaling pathways (e.g., NF-κB signaling pathway) and protein networks involved in inflammatory and innate immune responses. Such coverage suggests the potential for simultaneously monitoring the temporal changes of many protein players for a specific pathway/network when the current strategy is coupled with quantitative methodologies (e.g., stable isotope 18O labeling).
Supplementary Material
Acknowledgements
We thank Jimmy Eng at the Fred Hutchinson Cancer Research Center for his assistance with Peptide and Protein Prophet analyses. Portions of this research were supported by the National Institute of General Medical Sciences (NIGMS, Large Scale Collaborative Research Grants U54 GM-62119-02) and the NIH National Center for Research Resources (RR18522). Work was performed in the Environmental Molecular Science Laboratory, a U. S. Department of Energy (DOE) national scientific user facility located on the campus of Pacific Northwest National Laboratory (PNNL) in Richland, Washington. PNNL is a multiprogram national laboratory operated by Battelle Memorial Institute for the DOE under contract DE-AC05-76RLO-1830.
Abbreviations
- PNGase F
peptide-N-glycosidase F
- 2D
two-dimensional
- SCX
strong cation exchange liquid chromatography
- NET
normalized elution time
- AMT
accurate mass and time
- CPE
cysteinyl peptide enrichment.
REFERENCES
- 1.Anderson NL, Anderson NG. The human plasma proteome: history, character, and diagnostic prospects. Mol. Cell. Proteomics. 2002;1:845–867. doi: 10.1074/mcp.r200007-mcp200. [DOI] [PubMed] [Google Scholar]
- 2.Chen R, Pan S, Brentnall TA, Aebersold R. Proteomic profiling of pancreatic cancer for biomarker discovery. Mol. Cell. Proteomics. 2005;4:525–533. doi: 10.1074/mcp.R500004-MCP200. [DOI] [PubMed] [Google Scholar]
- 3.Jacobs JM, Adkins JN, Qian WJ, Liu T, Shen Y, Camp DG, 2nd, Smith RD. Utilizing human blood plasma for proteomic biomarker discovery. J. Proteome Res. 2005;4:1073–1085. doi: 10.1021/pr0500657. [DOI] [PubMed] [Google Scholar]
- 4.Hu Y, Malone JP, Fagan AM, Townsend RR, Holtzman DM. Comparative proteomic analysis of intra- and interindividual variation in human cerebrospinal fluid. Mol. Cell. Proteomics. 2005;4:2000–2009. doi: 10.1074/mcp.M500207-MCP200. [DOI] [PubMed] [Google Scholar]
- 5.Wattiez R, Falmagne P. Proteomics of bronchoalveolar lavage fluid. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2005;815:169–178. doi: 10.1016/j.jchromb.2004.10.029. [DOI] [PubMed] [Google Scholar]
- 6.Liao H, Wu J, Kuhn E, Chin W, Chang B, Jones MD, O’Neil S, Clauser KR, Karl J, Hasler F, Roubenoff R, Zolg W, Guild BC. Use of mass spectrometry to identify protein biomarkers of disease severity in the synovial fluid and serum of patients with rheumatoid arthritis. Arthritis Rheum. 2004;50:3792–3803. doi: 10.1002/art.20720. [DOI] [PubMed] [Google Scholar]
- 7.Varnum SM, Covington CC, Woodbury RL, Petritis K, Kangas LJ, Abdullah MS, Pounds JG, Smith RD, Zangar RC. Proteomic characterization of nipple aspirate fluid: identification of potential biomarkers of breast cancer. Breast Cancer Res. Treat. 2003;80:87–97. doi: 10.1023/A:1024479106887. [DOI] [PubMed] [Google Scholar]
- 8.Fingleton B, Menon R, Carter KJ, Overstreet PD, Hachey DL, Matrisian LM, McIntyre JO. Proteinase activity in human and murine saliva as a biomarker for proteinase inhibitor efficacy. Clin. Cancer Res. 2004;10:7865–7874. doi: 10.1158/1078-0432.CCR-04-1252. [DOI] [PubMed] [Google Scholar]
- 9.Meier M, Kaiser T, Herrmann A, Knueppel S, Hillmann M, Koester P, Danne T, Haller H, Fliser D, Mischak H. Identification of urinary protein pattern in type 1 diabetic adolescents with early diabetic nephropathy by a novel combined proteome analysis. J. Diabetes Complications. 2005;19:223–232. doi: 10.1016/j.jdiacomp.2004.10.002. [DOI] [PubMed] [Google Scholar]
- 10.Bhat VB, Choi MH, Wishnok JS, Tannenbaum SR. Comparative plasma proteome analysis of lymphoma-bearing SJL mice. J. Proteome Res. 2005;4:1814–1825. doi: 10.1021/pr0501463. [DOI] [PubMed] [Google Scholar]
- 11.Qian WJ, Jacobs JM, Camp DG, 2nd, Monroe ME, Moore RJ, Gritsenko MA, Calvano SE, Lowry SF, Xiao W, Moldawer LL, Davis RW, Tompkins RG, Smith RD. Comparative proteome analyses of human plasma following in vivo lipopolysaccharide administration using multidimensional separations coupled with tandem mass spectrometry. Proteomics. 2005;5:572–584. doi: 10.1002/pmic.200400942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Qian WJ, Monroe ME, Liu T, Jacobs JM, Anderson GA, Shen Y, Moore RJ, Anderson DJ, Zhang R, Calvano SE, Lowry SF, Xiao W, Moldawer LL, Davis RW, Tompkins RG, Camp DG, 2nd, Smith RD. Quantitative proteome analysis of human plasma following in vivo lipopolysaccharide administration using 16O/18O labeling and the accurate mass and time tag approach. Mol. Cell. Proteomics. 2005;4:700–709. doi: 10.1074/mcp.M500045-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hood BL, Lucas DA, Kim G, Chan KC, Blonder J, Issaq HJ, Veenstra TD, Conrads TP, Pollet I, Karsan A. Quantitative analysis of the low molecular weight serum proteome using 18O stable isotope labeling in a lung tumor xenograft mouse model. J. Am. Soc. Mass Spectrom. 2005;16:1221–1230. doi: 10.1016/j.jasms.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 14.Tu CJ, Dai J, Li SJ, Sheng QH, Deng WJ, Xia QC, Zeng R. High-sensitivity analysis of human plasma proteome by immobilized isoelectric focusing fractionation coupled to mass spectrometry identification. J. Proteome Res. 2005;4:1265–1273. doi: 10.1021/pr0497529. [DOI] [PubMed] [Google Scholar]
- 15.Jin WH, Dai J, Li SJ, Xia QC, Zou HF, Zeng R. Human plasma proteome analysis by multidimensional chromatography prefractionation and linear ion trap mass spectrometry identification. J. Proteome Res. 2005;4:613–619. doi: 10.1021/pr049761h. [DOI] [PubMed] [Google Scholar]
- 16.Sheng S, Chen D, Van Eyk JE. Multi-dimensional liquid chromatography separation of intact proteins by chromatographic focusing and reversed phased of the human serum proteome: optimization and protein database. Mol. Cell. Proteomics. 2006;5:26–34. doi: 10.1074/mcp.T500019-MCP200. [DOI] [PubMed] [Google Scholar]
- 17.Martosella J, Zolotarjova N, Liu H, Nicol G, Boyes BE. Reversed-phase high-performance liquid chromatographic prefractionation of immunodepleted human serum proteins to enhance mass spectrometry identification of lower-abundant proteins. J. Proteome Res. 2005;4:1522–1537. doi: 10.1021/pr050088l. [DOI] [PubMed] [Google Scholar]
- 18.Xiao Z, Conrads TP, Lucas DA, Janini GM, Schaefer CF, Buetow KH, Issaq HJ, Veenstra TD. Direct ampholyte-free liquid-phase isoelectric peptide focusing: application to the human serum proteome. Electrophoresis. 2004;25:128–133. doi: 10.1002/elps.200305700. [DOI] [PubMed] [Google Scholar]
- 19.Shen Y, Jacobs JM, Camp DG, 2nd, Fang R, Moore RJ, Smith RD, Xiao W, Davis RW, Tompkins RG. Ultra-high-efficiency strong cation exchange LC/RPLC/MS/MS for high dynamic range characterization of the human plasma proteome. Anal. Chem. 2004;76:1134–1144. doi: 10.1021/ac034869m. [DOI] [PubMed] [Google Scholar]
- 20.Shen Y, Kim J, Strittmatter EF, Jacobs JM, Camp DG, 2nd, Fang R, Tolie N, Moore RJ, Smith RD. Characterization of the human blood plasma proteome. Proteomics. 2005;5:4034–4045. doi: 10.1002/pmic.200401246. [DOI] [PubMed] [Google Scholar]
- 21.Hood BL, Zhou M, Chan KC, Lucas DA, Kim GJ, Issaq HJ, Veenstra TD, Conrads TP. Investigation of the mouse serum proteome. J. Proteome Res. 2005;4:1561–1568. doi: 10.1021/pr050107r. [DOI] [PubMed] [Google Scholar]
- 22.Tirumalai RS, Chan KC, Prieto DA, Issaq HJ, Conrads TP, Veenstra TD. Characterization of the low molecular weight human serum proteome. Mol. Cell. Proteomics. 2003;2:1096–1103. doi: 10.1074/mcp.M300031-MCP200. [DOI] [PubMed] [Google Scholar]
- 23.Liu T, Qian WJ, Gritsenko MA, Camp DG, 2nd, Monroe ME, Moore RJ, Smith RD. The human plasma N-glycoproteome analysis by immunoaffinity subtraction, hydrazide chemistry, and mass spectrometry. J. Proteome Res. 2005;4:2070–2080. doi: 10.1021/pr0502065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pieper R, Gatlin CL, Makusky AJ, Russo PS, Schatz CR, Miller SS, Su Q, McGrath AM, Estock MA, Parmar PP, Zhao M, Huang ST, Zhou J, Wang F, Esquer-Blasco R, Anderson NL, Taylor J, Steiner S. The human serum proteome: display of nearly 3700 chromatographically separated protein spots on two-dimensional electrophoresis gels and identification of 325 distinct proteins. Proteomics. 2003;3:1345–1364. doi: 10.1002/pmic.200300449. [DOI] [PubMed] [Google Scholar]
- 25.Adkins JN, Varnum SM, Auberry KJ, Moore RJ, Angell NH, Smith RD, Springer DL, Pounds JG. Toward a human blood serum proteome: analysis by multidimensional separation coupled with mass spectrometry. Mol. Cell. Proteomics. 2002;1:947–955. doi: 10.1074/mcp.m200066-mcp200. [DOI] [PubMed] [Google Scholar]
- 26.Chromy BA, Gonzales AD, Perkins J, Choi MW, Corzett MH, Chang BC, Corzett CH, McCutchen-Maloney SL. Proteomic analysis of human serum by two-dimensional differential gel electrophoresis after depletion of high-abundant proteins. J. Proteome Res. 2004;3:1120–1127. doi: 10.1021/pr049921p. [DOI] [PubMed] [Google Scholar]
- 27.Pieper R, Su Q, Gatlin CL, Huang ST, Anderson NL, Steiner S. Multi-component immunoaffinity subtraction chromatography: an innovative step towards a comprehensive survey of the human plasma proteome. Proteomics. 2003;3:422–432. doi: 10.1002/pmic.200390057. [DOI] [PubMed] [Google Scholar]
- 28.Zolotarjova N, Martosella J, Nicol G, Bailey J, Boyes BE, Barrett WC. Differences among techniques for high-abundant protein depletion. Proteomics. 2005;5:3304–3313. doi: 10.1002/pmic.200402021. [DOI] [PubMed] [Google Scholar]
- 29.Huang L, Harvie G, Feitelson JS, Gramatikoff K, Herold DA, Allen DL, Amunngama R, Hagler RA, Pisano MR, Zhang WW, Fang X. Immunoaffinity separation of plasma proteins by IgY microbeads: meeting the needs of proteomic sample preparation and analysis. Proteomics. 2005;5:3314–3328. doi: 10.1002/pmic.200401277. [DOI] [PubMed] [Google Scholar]
- 30.Liotta LA, Ferrari M, Petricoin E. Clinical proteomics: written in blood. Nature. 2003;425:905. doi: 10.1038/425905a. [DOI] [PubMed] [Google Scholar]
- 31.Coussens LM, Werb Z. Inflammation and cancer. Nature. 2002;420:860–867. doi: 10.1038/nature01322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weiner HL, Selkoe DJ. Inflammation and therapeutic vaccination in CNS diseases. Nature. 2002;420:879–884. doi: 10.1038/nature01325. [DOI] [PubMed] [Google Scholar]
- 33.Biberthaler P, Bogner V, Baker HV, Lopez MC, Neth P, Kanz KG, Mutschler W, Jochum M, Moldawer LL. Genome-wide monocytic mRNA expression in polytrauma patients for identification of clinical outcome. Shock. 2005;24:11–19. doi: 10.1097/01.shk.0000163394.93467.77. [DOI] [PubMed] [Google Scholar]
- 34.Feezor RJ, Baker HV, Xiao W, Lee WA, Huber TS, Mindrinos M, Kim RA, Ruiz-Taylor L, Moldawer LL, Davis RW, Seeger JM. Genomic and proteomic determinants of outcome in patients undergoing thoracoabdominal aortic aneurysm repair. J. Immunol. 2004;172:7103–7109. doi: 10.4049/jimmunol.172.11.7103. [DOI] [PubMed] [Google Scholar]
- 35.Zhang H, Li X.-j., Martin DB, Aerbersold R. Identification and quantification of N-linked glycoproteins using hydrazide chemistry, stable isotope labeling and mass spectrometry. Nat. Biotechnol. 2003;21:660–665. doi: 10.1038/nbt827. [DOI] [PubMed] [Google Scholar]
- 36.Shen Y, Zhao R, Belov ME, Conrads TP, Anderson GA, Tang K, Pasa-Tolic L, Veenstra TD, Lipton MS, Smith RD. Packed capillary reversed-phase liquid chromatography with high-performance electrospray ionization Fourier transform ion cyclotron resonance mass spectrometry for proteomics. Anal. Chem. 2001;73:1766–1775. doi: 10.1021/ac0011336. [DOI] [PubMed] [Google Scholar]
- 37.Eng JK, McCormack AL, Yates JR. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 38.Qian WJ, Liu T, Monroe ME, Strittmatter EF, Jacobs JM, Kangas LJ, Petritis K, Camp DG, 2nd, Smith RD. Probability-based evaluation of peptide and protein identifications from tandem mass spectrometry and SEQUEST analysis: The human proteome. J. Proteome Res. 2005;4:53–62. doi: 10.1021/pr0498638. [DOI] [PubMed] [Google Scholar]
- 39.Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 2003;75:4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- 40.Beissbarth T, Speed TP. GOstat: find statistically overrepresented Gene Ontologies within a group of genes. Bioinformatics. 2004;20:1464–1465. doi: 10.1093/bioinformatics/bth088. [DOI] [PubMed] [Google Scholar]
- 41.Calvano SE, Xiao W, Richards DR, Felciano RM, Baker HV, Cho RJ, Chen RO, Brownstein BH, Cobb JP, Tschoeke SK, Miller-Graziano C, Moldawer LL, Mindrinos MN, Davis RW, Tompkins RG, Lowry SF. A network-based analysis of systemic inflammation in humans. Nature. 2005;437:1032–1037. doi: 10.1038/nature03985. [DOI] [PubMed] [Google Scholar]
- 42.Omenn GS, States DJ, Adamski M, Blackwell TW, Menon R, Hermjakob H, Apweiler R, Haab BB, Simpson RJ, Eddes JS, Kapp EA, Moritz RL, Chan DW, Rai AJ, Admon A, Aebersold R, Eng J, Hancock WS, Hefta SA, Meyer H, Paik YK, Yoo JS, Ping P, Pounds J, Adkins J, Qian X, Wang R, Wasinger V, Wu CY, Zhao X, Zeng R, Archakov A, Tsugita A, Beer I, Pandey A, Pisano M, Andrews P, Tammen H, Speicher DW, Hanash SM. Overview of the HUPO Plasma Proteome Project: results from the pilot phase with 35 collaborating laboratories and multiple analytical groups, generating a core dataset of 3020 proteins and a publicly-available database. Proteomics. 2005;5:3226–3245. doi: 10.1002/pmic.200500358. [DOI] [PubMed] [Google Scholar]
- 43.Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical Statistical Model To Estimate the Accuracy of Peptide Identifications Made by MS/MS and Database Search. Anal. Chem. 2002;74:5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- 44.Scaros O, Fisler R. Biomarker technology roundup: from discovery to clinical applications, a broad set of tools is required to translate from the lab to the clinic. Biotechniques. 2005;38:S30–32. doi: 10.2144/05384su01. [DOI] [PubMed] [Google Scholar]
- 45.Petricoin E, Wulfkuhle J, Espina V, Liotta LA. Clinical proteomics: revolutionizing disease detection and patient tailoring therapy. J. Proteome Res. 2004;3:209–217. doi: 10.1021/pr049972m. [DOI] [PubMed] [Google Scholar]
- 46.Liu T, Qian WJ, Strittmatter EF, Camp DG, 2nd, Anderson GA, Thrall BD, Smith RD. High throughput comparative proteome analysis using a quantitative cysteinyl-peptide enrichment technology. Anal. Chem. 2004;76:5345–5353. doi: 10.1021/ac049485q. [DOI] [PubMed] [Google Scholar]
- 47.Liu T, Qian WJ, Chen WU, Jacobs JM, Moore RJ, Anderson DJ, Gritsenko MA, Monroe ME, Thrall BD, Camp DG, 2nd, Smith RD. Improved proteome coverage by using high efficiency cysteinyl peptide enrichment: The human mammary epithelial cell proteome. Proteomics. 2005;5:1263–1273. doi: 10.1002/pmic.200401055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang H, Qian WJ, Chin MH, Petyuk VA, Barry RC, Liu T, Gritsenko MA, Mottaz HM, Moore RJ, Camp DG, 2nd, Khan AH, Smith DJ, Smith RD. Characterization of the mouse brain proteome using global proteomic analysis complemented with cysteinyl-peptide enrichment. J. Proteome Res. 2006;5:361–369. doi: 10.1021/pr0503681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang H, Yi EC, Li XJ, Mallick P, Kelly-Spratt KS, Masselon CD, Camp DG, 2nd, Smith RD, Kemp CJ, Aebersold R. High throughput quantitative analysis of serum proteins using glycopeptide capture and liquid chromatography mass spectrometry. Mol. Cell. Proteomics. 2005;4:144–155. doi: 10.1074/mcp.M400090-MCP200. [DOI] [PubMed] [Google Scholar]
- 50.Roth J. Protein N-glycosylation along the secretory pathway: relationship to organelle topography and function, protein quality control, and cell interactions. Chem. Rev. 2002;102:285–303. doi: 10.1021/cr000423j. [DOI] [PubMed] [Google Scholar]
- 51.Bause E. Structural requirements of N-glycosylation of proteins. Studies with proline peptides as conformational probes. Biochem. J. 1983;209:331–336. doi: 10.1042/bj2090331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Smith RD, Anderson GA, Lipton MS, Pasa-Tolic L, Shen Y, Conrads TP, Veenstra TD, Udseth HR. An accurate mass tag strategy for quantitative and high throughput proteome measurements. Proteomics. 2002;2:513–523. doi: 10.1002/1615-9861(200205)2:5<513::AID-PROT513>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 53.Anderson NL, Polanski M, Pieper R, Gatlin T, Tirumalai RS, Conrads TP, Veenstra TD, Adkins JN, Pounds JG, Fagan R, Lobley A. The human plasma proteome: A nonredundant list developed by combination of four separate sources. Mol. Cell. Proteomics. 2004;3:311–316. doi: 10.1074/mcp.M300127-MCP200. [DOI] [PubMed] [Google Scholar]
- 54.Hara T, Nishida H, Nakajima R. Highly sensitive detection of proteins separated by capillary zone electrophoresis using on-line chemiluminescence detection. Anal. Sci. 1994;10:823–825. [Google Scholar]
- 55.Lang TH, Willinger U, Holzer G. Soluble cathepsin-L: a marker of bone resorption and bone density? J. Lab Clin. Med. 2004;144:163–166. doi: 10.1016/j.lab.2004.06.001. [DOI] [PubMed] [Google Scholar]
- 56.States DJ, Omenn GS, Blackwell TW, Fermin D, Eng J, Speicher DW, Hanash SM. Challenges in deriving high-confidence protein identifications from data gathered by a HUPO plasma proteome collaborative study. Nat. Biotechnol. 2006;24:333–338. doi: 10.1038/nbt1183. [DOI] [PubMed] [Google Scholar]
- 57.Thomson AW, Lotze MT, editors. The cytokine handbook. 4th Ed. Academic Press; San Diego: 2003. [Google Scholar]
- 58.Fischer E, Van Zee KJ, Marano MA, Rock CS, Kenney JS, Poutsiaka DD, Dinarello CA, Lowry SF, Moldawer LL. Interleukin-1 receptor antagonist circulates in experimental inflammation and in human disease. Blood. 1992;79:2196–2200. [PubMed] [Google Scholar]
- 59.Fischer E, Marano MA, Van Zee KJ, Rock CS, Hawes AS, Thompson WA, DeForge L, Kenney JS, Remick DG, Bloedow DC, Thompson RC, Lowry SF, Moldawer LL. Interleukin-1 receptor blockade improves survival and hemodynamic performance in Escherichia coli septic shock, but fails to alter host responses to sublethal endotoxemia. J. Clin. Invest. 1992;89:1551–1557. doi: 10.1172/JCI115748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Van Zee KJ, Kohno T, Fischer E, Rock CS, Moldawer LL, Lowry SF. Tumor necrosis factor soluble receptors circulate during experimental and clinical inflammation and can protect against excessive tumor necrosis factor alpha in vitro and in vivo. Proc. Natl. Acad. Sci. U S A. 1992;89:4845–4849. doi: 10.1073/pnas.89.11.4845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Verma M, Maruvada P, Srivastava S. Epigenetics and cancer. Crit. Rev. Clin. Lab Sci. 2004;41:585–607. doi: 10.1080/10408360490516922. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.