Abstract
Naturally occurring mutations are useful in identifying domains that are important for protein function. We studied a mutation in the elastin gene, 800−3G>C, a common disease allele for SVAS (supravalvular aortic stenosis). We showed in primary skin fibroblasts from two different SVAS families that this mutation causes skipping of exons 16–17 and results in a stable mRNA. Tropoelastin lacking domains 16–17 (Δ16–17) was synthesized efficiently and secreted by transfected retinal pigment epithelium cells, but showed the deficient deposition into the extracellular matrix compared with normal as demonstrated by immunofluorescent staining and desmosine assays. Solid-phase binding assays indicated normal molecular interaction of Δ16–17 with fibrillin-1 and fibulin-5. However, self-association of Δ16–17 was diminished as shown by an elevated coacervation temperature. Moreover, negative staining electron microscopy confirmed that Δ16–17 was deficient in forming fibrillar polymers. Domain 16 has high homology with domain 30, which can form a β-sheet structure facilitating fibre formation. Taken together, we conclude that domains 16–17 are important for self-association of tropoelastin and elastic fibre formation. This study is the first to discover that domains of elastin play an essential role in elastic fibre formation by facilitating homotypic interactions.
Keywords: elastic fibre assembly, elastin, microfibril, supravalvular aortic stenosis (SVAS), tropoelastin
Abbreviations: CHX, cycloheximide; DMEM, Dulbecco's modified Eagle's medium; nTE, recombinant human normal tropoelastin; RT, reverse transcription; SVAS, supravalvular aortic stenosis
INTRODUCTION
Elastic fibres, which comprise an elastin core surrounded by fibrillin-rich microfibrils, are major insoluble extracellular matrix structures and impart elasticity to organs such as skin, lungs, ligaments and arteries. Elastin-knockout mice show a lethal obstructive arterial disease resulting from subendothelial cell proliferation and reorganization of smooth muscle [1]. Moreover, studies on mice hemizygous for the elastin gene have shown that elastin haploinsufficiency results in changes in the arterial wall structure, including thinner elastic lamellae and an increased number of smooth muscle cell layers similar to changes also observed in humans with SVAS (supravalvular aortic stenosis) [2]. Thus elastic fibres have a regulatory function during arterial development; they control proliferation of smooth muscle and stabilize arterial structure.
Elastin is secreted as a soluble protein of approx. 70 kDa, referred to as tropoelastin, which has alternating hydrophobic and cross-linking domains. There is substantial evidence that the hydrophobic domains are necessary for the self-aggregation of tropoelastin via coacervation, which is thought to concentrate and align tropoelastin molecules for cross-linking [3]. Hydrophobic domains of tropoelastin are rich in amino acids such as glycine, proline, valine and leucine, which are present in a variety of tandem repeat sequences. The tandem repeats, i.e. VGVAPG and GGLG(V/A), that resemble sequences found in other proteins that aggregate via β-sheet/β-turn structures [4,5], contribute to the polymerization of elastin [6,7]. It has been reported that exon 30, which encodes a hydrophobic domain containing the tandem repeat sequence GGLG(V/A), is an important sequence for the assembly process [7].
Hydrophilic domains of elastin are rich in lysine and alanine and are involved in cross-linking. The oxidative deamination of peptidyl lysine residues in tropoelastin is catalysed by lysyl oxidase to α-aminoadipic acid δ-semialdehyde (allysine), which can spontaneously condense with neighbouring amino groups or other peptidyl aldehydes to form covalent cross-links such as desmosine or isodesmosine. It has been reported that self-association of recombinant tropoelastin and/or elastin-derived peptides, through a phase-transition process called coacervation, facilitates the formation of such cross-links [8].
During elastic fibre formation, tropoelastin is deposited on pre-formed microfibrillar templates. Several studies have indicated that tropoelastin molecules interact directly with the microfibrillar proteins fibrillin-1 and fibrillin-2 [9,10]. Moreover, two structural proteins of elastic fibres, MAGP-1 (microfibril-associated glycoprotein 1) and fibulin-5, bind both tropoelastin and fibrillin-1 [10–12]. Earlier studies have shown that the C-terminus of tropoelastin plays an important role in the interaction with microfibrils. Exon 36 of the elastin gene encodes a critical domain for binding microfibrillar proteins [13–15]. More recently, a peptide-mapping study has shown that the N-terminus of fibrillin-1 binds not only to the C-terminus but also to the N-terminus of tropoelastin encompassing domains 2–18 [16]. However, the mechanisms of elastic fibre assembly are still not fully understood.
It is also known that elastin haploinsufficiency resulting from loss-of-function mutations in the elastin gene causes SVAS, which is characterized by a thickening of the arterial wall and either focal or diffuse narrowing of the aorta [17]. SVAS is caused by point mutations, translocations and deletions in the elastin gene [18]. In the present study, we found that a splice site mutation in intron 15 results in the skipping of exons 16–17 and the synthesis of stable mutant mRNA and protein. The aim of the present study was to elucidate the disease mechanism associated with this mutation by investigating the involvement of elastin domains 16–17 in elastic fibre formation in an in vitro model of elastic fibre assembly [19,20], and by biochemical analysis of recombinant tropoelastins.
MATERIALS AND METHODS
Mutation screening
Mutation 800−3C>G was previously described in patient 4478 ([21], family SVAS-1). The same molecular defect was identified in patient 1021 (family SVAS-55) during a screen of SVAS patients for ELN gene defects using denaturing HPLC and direct DNA sequencing (results not shown). Although the relatedness of patients 4478 and 1021 has not been established by genealogy, haplotype association 800−3C>G with a rare and likely neutral variant in intron 1 (83−53C>T) in these same individuals suggests identity by descent. Variant 83−53C>T was not found in 181 normal control individuals.
RNA preparation from cell culture
Primary skin fibroblast cultures were established from SVAS patients 4478 and 1021 with their consent. Two parallel flasks of cells were cultured to confluence in high-glucose DMEM (Dulbecco's modified Eagle's medium) (Invitrogen) containing L-glutamine, pyridoxine hydrochloride, sodium bicarbonate, Hepes and 10% FBS (foetal bovine serum) (Serologicals Corp.). At 2 days after reaching confluence, one flask of cells/sample was treated with 100 μg/ml CHX (cycloheximide) for 4 h before processing. Total cellular RNA was prepared from both CHX-treated and untreated cells in Tri Reagent® (Molecular Research Centre) according to the manufacturer's protocol.
RT (reverse transcription)–PCR and sequencing
First-strand DNA was synthesized with oligo-dT priming by using a Thermoscript kit (Invitrogen). RNA (3 μg) was reverse-transcribed at 50 °C for 60 min followed by RNase H digestion. A 4 μl aliquot of the RT reaction was used per 100 μl of PCR mixture to amplify the exon 14–18 region of the ELN gene transcript using the forward primer (5′-TGGTTACCCAACAGGGACAG-3′) and the reverse primer (5′-ACAACTGGAATCCCAGCTC-3′) with one of the primers end-labelled with [33P]ATP. PCRs were carried out to 18–32 cycles and PCR amplimers were separated on a 6% denaturing acrylamide gel next to a labelled pBR322 MspI digest. Dried gels were exposed to a phosphor screen and evaluated using a Typhoon (Amersham Biosciences) phosphorimager and Image Quant 5.2 software. PCR products were recovered from the dried gel, reamplified by PCR and cycle-sequenced using an ABI-3100 genetic analyser.
Antibodies
The antibodies used for immunofluorescence and Western blot analyses included mouse anti-elastin monoclonal antibody MAB2503 (Chemicon International), anti-His-G antibody and anti V5-epitope antibody (Invitrogen). In addition, an antibody against human fibrillin-1 (FBN-1) [22] was used. The secondary antibodies included Alexa Fluor® 488-conjugated goat anti-mouse IgG and Alexa Fluor® 546-conjugated goat anti-rabbit IgG (Molecular Probes) for immunofluorescence, and horseradish-peroxidase-conjugated anti-mouse IgG and anti-rabbit IgG (ICN/Cappel) for Western blot assay.
Stable transfections
Full-length human tropoelastin cDNA was inserted into the MluI/XbaI site of the pCI-neo vector (Promega) (FL). It should be noted that the human template was a natural splice variant lacking the sequence encoded by exon 22. To generate a human tropoelastin cDNA lacking exons 16 and 17 in the pCI-neo vector (FLΔ16–17), exons 14–18 were amplified by PCR using a partial human tropoelastin cDNA lacking exons 16 and 17 in pBluscript II, which was kindly provided by Dr Dean Y. Li (Program in Human Molecular Biology and Genetics, University of Utah). The following primers were used: forward primer, exon 14F (5′-GCTGGTTACCCAACAGGGACAG-3′) and reverse primer, exon 18R (5′-ACCGCAGCACCTGGGATCCC-3′). The exon 14F primer contains a BstEII site and the exon 18R primer contains a BamHI site. The PCR amplifications were digested with BstEII and BamHI and then ligated into the similarly cleaved FL vector. ARPE-19 cells (A.T.C.C., Manassas, VA, U.S.A.), derived from human retinal pigment epithelial tissue, were transfected with FL or FLΔ16–17 using FuGENE™ 6 transfection reagent (Roche). The transfected cells were maintained in DMEM containing 10% Cosmic Calf Serum (HyClone Laboratories) and 800 μg/ml G418 (Gibco-BRL) and were harvested after 10–14 days of selection. Clones ARPE-FL, ARPE-FLΔ16 were obtained by single cell sorting of the G418-resistant cells. The cloning method was performed as described in [20]. Cells were cultured for 14 days after confluence and then used in experiments.
Purification of recombinant human tropoelastin
Recombinant human normal tropoelastin (nTE) was prepared as described previously [19]. To generate the recombinant Δ16–17, FLΔ16–17 was amplified using forward primer 5′-GGAGGGGTCCCTGGGGCCATTCC-3′ and reverse primer 5′-TCATTTTCTCTTCCGGCCACAAG-3′. The product was inserted into a bacterial expression vector pTrcHis-TOPO (Invitrogen). Δ16–17 was obtained by overexpression from the plasmid and purified using the same method as for nTE. Purified recombinant proteins were resuspended in SDS/PAGE sample buffer including 100 mM DTT (dithiothreitol). Samples were separated by SDS/PAGE (7.5% gels) and subjected to Western blot analysis.
Treatment of cells with recombinant tropoelastins
ARPE-19 cells were plated on eight-well LabTek chamber slides (Nunc) or 60-mm-diameter dishes (Falcon) for immunofluorescence or the determination of cross-linked amino acids respectively. After 2 days of culture, these cells were treated with nTE for 8 days. This medium was changed every 2 days.
Immunofluorescence and determination of cross-linked amino acids into the matrix
The methods used for the immunofluorescence study and determination of desmosine contents have been described previously [19].
For immunofluorescence, cells were fixed and incubated with primary antibody diluted at 1:100 (MAB2503) and 1:1000 (FBN-1) in Block Ace (Dainippon Pharmaceutical Co.) overnight at 4 °C, followed by several washes of PBS (140 mM NaCl, 2.7 mM KCl, 8 mM Na2HPO4, and 1.5 mM KH2PO4, pH 7.4) and a second incubation with Alexa Fluor® 488- and Alexa Fluor® 546-conjugated secondary antibody diluted at 1:1000 in Block Ace.
The desmosine levels in the hydrolysates were determined by RIA [23]. The desmosine (pmol) was normalized to the total protein (mg) in the dishes.
Production of recombinant fibulin-5 and PET construct
Fibulin-5 cDNA has been described previously [24]. Previously reported PET [9] cDNA was kindly provided by Dr Robert P. Mecham (Washington University, St. Louis, MO, U.S.A.). Fibulin-5 and PET cDNA were inserted into the pcDNA3.1/V5-His-TOPO expression vector (Invitrogen). These plasmids were transfected into CHO-K1 cells (A.T.C.C., CCL-61). After 24 h of incubation, culture media were replaced with CHO-SFM-II (Invitrogen) and the cultures were incubated for 24–72 h. At the end of the incubation period, culture media were collected and concentrated using Amicon Ultra-15 (Millipore). The products were used in the experiments as conditioned media.
Solid-phase binding assay
Solid-phase binding assay was performed as described in [9]. Microtitre plates were coated at 4 °C overnight with 10 μg/ml nTE, Δ16–17 or BSA in bicarbonate buffer. The tropoelastin-bound fibulin-5 was detected with anti-V5-epitope antibody diluted 1:5000. Bound protein was quantified using a colorimetric assay with the TMB [8-(diethylamino)octyl-3,4,5-trimethoxybenzoate] substrate reagent for 30 min at room temperature (20 °C). The plates were read at a wavelength of 450 nm.
Coacervation
Coacervation was assayed by monitoring turbidity using light scattering at 400 nm with a UV spectrometer and JASCO V-500 software (Nihonbunkou). The cuvette holder was connected to a re-circulating water bath in order to control the temperature of samples. Light scattering by each solution was measured every 30 s while increasing the temperature at the rate of 1 °C/min from 15 to 50 °C. Both nTE and Δ16–17 were dissolved in PBS.
Electron microscopy
Grids (200-mesh) for electron microscopy were covered with a thin (10 nm) carbon layer. The carbon film on the grid was treated with n-pentylamine to make it hydrophilic. The nTE or Δ16–17 was laid on the grid in a temperature-controlled room at 30 °C and was stained with 2% uranyl acetate. The grid was observed in an electron microscope (EF-2000, Hitachi) operated at an accelerating voltage of 200 keV, as a zero-loss image with an energy-filtering device. The electron micrographs were taken using a CCD (charge-coupled device) camera with a pixel size of 24 μm at direct magnifications of 140000 or 6000, and sampling steps of 0.17 or 4 nm respectively.
Statistical analysis
All assays were performed in triplicate and were repeated at least twice to confirm observed results. Data were statistically analysed using ANOVA. The results were considered to be statistically significant when P<0.05. All data are shown as the mean value±S.E.M.
RESULTS
Mutation 800−3C>G results in the skipping of exons 16–17 and a stable mRNA
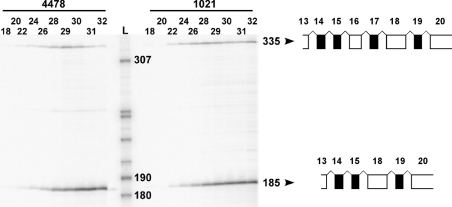
We did not see any significant difference between CHX-treated and untreated samples. RT–PCR produced a 335- and a 185-bp-long PCR amplimer of which the longer product corresponded to a normally spliced transcript of the ELN gene, containing all exons between 14 and 18 (Figure 1). Sequencing of the shorter product revealed that the 800−3C>G intronic mutation in the ELN gene caused the skipping of exons 16 and 17 in the transcripts of the mutated allele (Figure 1). Since this is an in-frame deletion, nonsense-mediated decay did not have an effect on mutant allele-specific mRNA levels. According to our RT–PCR evaluation, the mutant allele is expressed at about the same level as the normal allele in both patients. The ratio of the mutant to the wild-type product increases at higher cycle numbers, because the mutant is favoured by the PCR owing to the fact that it is approximately half the size of the wild-type amplimer. Thus comparison of the relative abundance of these amplimers is relevant at low cycle numbers. We obtained these same results in three separate experiments with either primer labelled.
Figure 1. Expression of mRNA lacking exons 16–17 in skin fibroblasts from SVAS patients.
At cycle numbers 18–24, a representative autoradiograph shows approximately equal expression of normal (335 bp) and exon-skipping (185 bp) RT–PCR products of the ELN gene in patients 4478 and 1021. The numbers on top indicate PCR cycle numbers. SVAS patient code numbers are shown above the cycle numbers. The numbers next to the DNA ladder (L) and RT–PCR products indicate DNA fragment sizes in bases. Arrowheads point to schematic representations of each RT–PCR products with exons (numbered at the top) encoding hydrophobic domains represented by open boxes and cross-link domains represented by closed boxes.
Human tropoelastin lacking domains 16–17 is secreted but shows impaired assembly with microfibrils
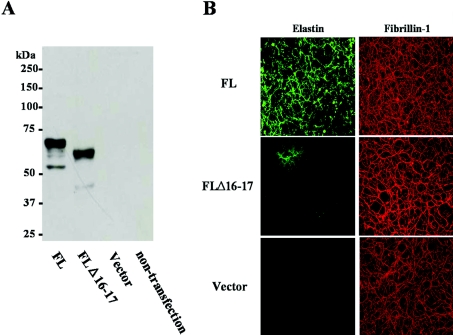
To confirm whether the tropoelastin translated from the mutant allele was stable and secreted, FLΔ16–17 was stably transfected into ARPE-19 cells (ARPE-FLΔ16–17). ARPE-19 cells were reported previously to express microfibrillar proteins but not tropoelastin [19]. This was confirmed by immunoblot analysis of conditioned media from ARPE19 transfected with an empty vector (Figure 2). The amounts of the secreted mutant (FLΔ16–17) and wild-type (FL) tropoelastins in transfected ARPE19 cells were similar (Figure 2A). The elastic fibres in ARPE-FLΔ16–17, however, were scarce compared with those in ARPE-FL (Figure 2B). The cross-linked amino acids desmosine and isodesmosine are found only in mature elastin and are routinely used as specific markers of fibre maturation and elastin quantification. When the cell cultures were assayed by RIA for desmosine, very low levels of desmosine were found in cultures of ARPE-19 cells that were either untransfected or transfected with empty vector (results not shown). Moreover, we observed that the amount of desmosine in ARPE-FLΔ16–17 was significantly lower than in ARPE-FL (1.78±0.23 pmol of desmosine/mg of total protein compared with 15.27±0.27 pmol of desmosine/mg of total protein; P=0.0003). Empty vector and the construct did not alter the expression and secretion of tropoelastin.
Figure 2. Secretion and fibre formation of the tropoelastin deletion mutant in vitro.
Wild-type (FL) or mutant elastin cDNA (FLΔ16–17) in pCi-neo expression vector (Vector) was transfected into ARPE-19 cells which did not express elastin (see Figure 3B). Immunoblot assay of the media using anti-elastin antibody showed that the mutant tropoelastin produced from the FLΔ16–17-transfectant was secreted similarly to normal tropoelastin (A). Molecular masses are given in kDa. With double immunostaining for elastin (left) and fibrillin-1 (right), although both secreted tropoelastins accumulated on to microfibrils containing fibrillin-1, the accumulation in FLΔ16–17-transfectant was diminished (B).
Quantitative fibre assembly analysis of ARPE-19 cells treated with recombinant tropoelastins
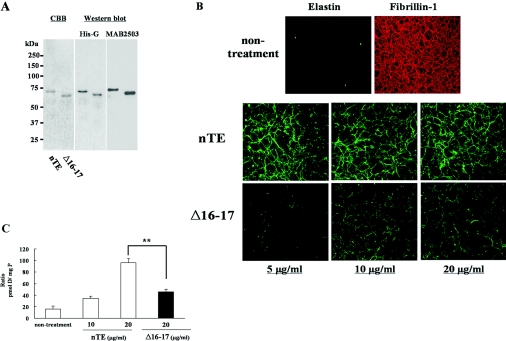
We quantitatively assessed the ability of Δ16–17 tropoelastin to participate in elastic fibre assembly with exons 16 and 17 lacking tropoelastin using our in vitro model [19]. The addition of recombinant tropoelastin into ARPE-19 cell culture medium provides a comparison of the efficiency of different tropoelastin molecules to organize into fibres. Therefore we generated recombinant human normal tropoelastin (nTE) and exons 16 and 17 lacking tropoelastin (Δ16–17). Purified recombinant tropoelastins were detected with Coomassie Blue stain and by Western blot assay using anti-His-G antibody and MAB2503 (Figure 3A).
Figure 3. Quantitative fibre assembly assay of mutant tropoelastin.
(A) Purified bacterial recombinant wild-type (nTE) and mutant (Δ16–17) human tropoelastin were detected by Coomassie Blue staining (CBB) and immunoblotting using anti-His-G antibody (His-G) and anti-elastin antibody (MAB2503). Recombinant tropoelastins were loaded at 1 μg/lane (CBB) or 500 ng/lane (Western blot assay). (B) Double immunofluorescence with MAB2503 and anti-fibrillin-1 antibody shows ARPE-19 cells form microfibril fibres containing fibrillin-1 but not elastin (non-treatment). The addition of nTE or Δ16–17 to the culture medium of ARPE-19 cells resulted in the accumulation of elastin in the extracellular matrix in a dose-dependent manner. Elastin deposition in the ARPE-19 cells treated with Δ16–17 was reduced compared with cells treated with nTE. Magnification ×200. (C) Desmosine (pmol D) was detected by radioimmunoassay and normalized to total protein (mg P). Desmosine in cultures treated with Δ16–17 were significantly reduced compared with those treated with nTE. **P<0.01.
To compare the efficiency of elastic fibre assembly in the presence of nTE and Δ16–17, the various concentrations of these tropoelastins were added to ARPE-19 culture medium. Double immunofluorescence labelling showed that the added exogenous tropoelastin assembled into elastic fibres on a microfibrillar network made by the ARPE-19 cells (Figure 3). Similar to the transfection experiments (Figure 2B), the accumulation of Δ16–17 was significantly less than that of nTE, especially at low concentrations. This result suggests that Δ16–17 has relatively poor ability to assemble into the extracellular matrix. Notably, fibrillin-1 fibres were unchanged, irrespective of the amount or kind of recombinant proteins added (results not shown). Moreover, the addition of nTE resulted in a marked increased in desmosine after 8 days in culture. Cells treated with Δ16–17 showed significantly decreased desmosine/total protein ratio compared with treatment with nTE (Figure 3C). Neither transfection of a mutant construct nor the addition of mutant tropoelastin had an effect on the expression of elastin and microfibril proteins in ARPE-19 cells (results not shown). In addition, we confirmed that there was no difference in the number of cells at the end of incubation (8 days) between cultures treated with wild-type and mutant tropoelastin (results not shown).
Molecular interaction with fibulin-5 or PET as scaffold proteins
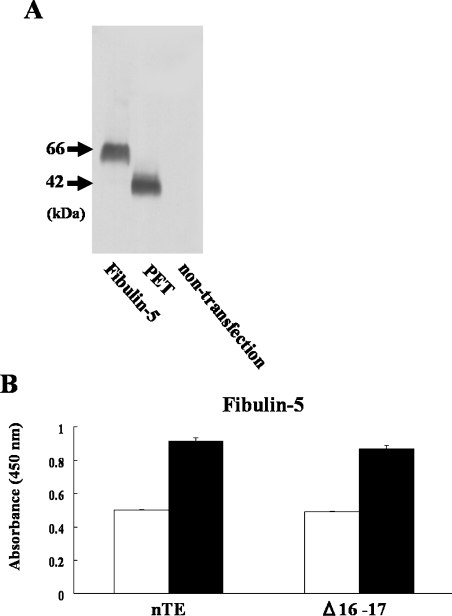
To test whether decreased deposition of Δ16–17 was caused by impaired interaction with microfibrillar proteins, we purified recombinant fibulin-5 and an N-terminal peptide of fibrillin-1 (PET), which is known to bind tropoelastin via charge–charge interactions [9]. Fibulin-5 plays an important role in elastic fibre formation [25,26]. Fibrillin-1 is also important as a scaffold protein by the binding to tropoelastin [27]. Both fibulin-5 and PET were tagged with a V5 epitope at the C-terminus, and were detected with anti-V5-epitope antibody (Figure 4A). In solid-phase binding assays, fibulin-5 bound strongly to each tropoelastin, and this binding was increased in the presence of 30 mM Ca2+ (Figure 4B). Furthermore, PET bound also to each tropoelastin efficiently (A450 values were 0.145±0.0003 for nTE and 0.118±0.002 for Δ16–17; P=0.049; background BSA A450 value was 0.071±0.0006). There was no significant difference between nTE and Δ16–17 in the molecular interaction with fibulin-5, but the interaction between Δ16–17 and PET was slightly reduced in comparison with nTE. Notably, there was no difference in the amount of nTE and Δ16–17 bound to the wells following coating as measured using an ELISA (results not shown).
Figure 4. Direct interaction of Δ16–17 with microfibril components.
(A) Using a Western blot assay using an anti-V5-epitope antibody, the molecular masses of fibulin-5 and PET from conditioned media were approx. 66 kDa and 42 kDa respectively. (B) The binding assay for each recombinant tropoelastin with fibulin-5 was performed using a solid-phase binding assay. Wells of microtitre plates were coated with equal amounts of nTE, Δ16–17 or BSA (as a negative control). Fibulin-5 bound to Δ16–17 and nTE with the same affinity: open bars, no Ca2+; closed bars, 30 mM Ca2+.
Self-assembly and polymerization of nTE and Δ16–17
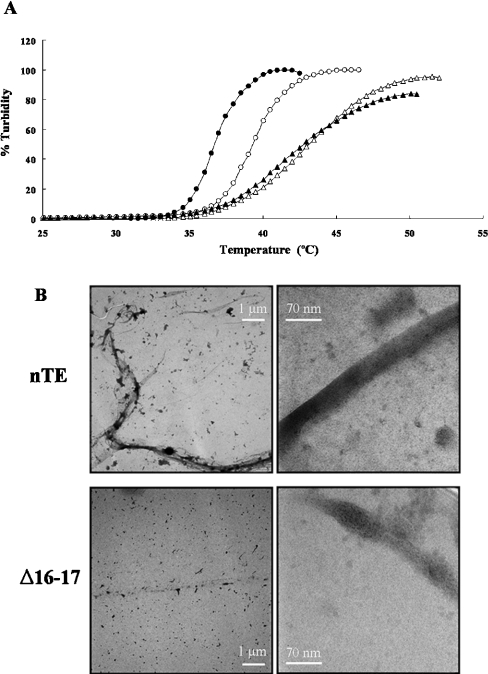
We characterized the ability of Δ16–17 to self-assemble using coacervation assays and electron microscopy. Each tropoelastin underwent coacervation, and equal concentrations of Δ16–17 coacervated at a higher temperature than did nTE. nTE showed dose-dependence for coacervation, but Δ16–17 did not: and at these concentrations, Δ16–17 showed lower incremental increase in turbidity with increase in temperature (Figure 5A). These results demonstrate impaired aggregation of Δ16–17 under physiological conditions.
Figure 5. Self-association of mutated tropoelastin.
(A) Coacervation of the nTE (circle) or Δ16–17 (triangle) measured between 25 and 50 °C using concentrations of 6.25 μM (open symbols) and 12.5 μM (closed symbols). Values for Δ16–17 and nTE were normalized to turbidity at maximum coacervation in each experiment. (B) The overviews of nTE (upper) and Δ16–17 (bottom) at the direct magnification of ×6000 (left). Elastin fibrils and aggregates stained with uranyl acetate are shown dark/black. Magnified electron micrographs of nTE and Δ16–17 at the direct magnification of ×140000 (right).
Electron micrographs (Figure 5B) showed that nTE formed long filamentous and bundled fibrils, whereas Δ16–17 formed small aggregates. The nTE fibrils were over 20 μm in total length and had submicrometer widths, indicating a substructure of thinner filamentous material. In magnified images, nTE showed filaments less than 100 nm in width, whereas Δ16–17 did not. Δ16–17 rarely showed a filamentous form. Most aggregates of Δ16–17 were short in length and were prone to produce ellipsoid/compact forms less than 100 nm. These collapsed structures may have resulted from EM fixation, rather than self association of mutant monomers. It was difficult to observe fine structure at the nanometer level in the magnified images because of tight packing of the fibrils. In the loosely packed area, some thin filamentous structures a few nanometers in width could be also observed in both nTE and Δ16–17 (results not shown). Thus Δ16–17 tropoelastin was found to form small aggregates and to reduce bundling ability.
DISCUSSION
In the present study, we have shown that a splice site mutation that commonly causes SVAS results in the skipping of exons 16–17 in the elastin mRNA, and leads to the synthesis of a stable polypeptide that is capable of forming molecular interactions with other elastic fibre proteins such as fibrillin-1 and fibulin-5 but deficient in forming homotypic interactions. These results indicate that loss of self-association properties in tropoelastin causes impaired elastin deposition.
A previous study [21] suggested that the ELN allele 800−3C>G caused the synthesis of low levels of aberrantly spliced mutant mRNA and it was speculated that the low levels of these abnormal splice products were caused by nonsense-mediated mRNA decay. In contrast, the present study shows approximately equal expression of normal mRNA and an aberrant message that lacks both exons 16 and 17. The reason for this discrepancy is that one of the oligonucleotide primers used for the RT-PCR assay in the previous study [21] was complementary to exon 17 and therefore did not detect the exon-skipping product lacking exons 16 and 17. Although extensive alternative splicing is a characteristic of elastin mRNA [28,29], exons 16 and 17 have not been observed to be subject to alternative splicing, suggesting that the corresponding domains may be important for elastin function.
Microfibrils, acting as a molecular scaffold for tropoelastin deposition, are composed of several proteins, glycoproteins and proteoglycans. Fibulin-5 is an elastin-binding protein and is essential for elastic fibre assembly because homozygous mutants show disorganization of elastic fibres, loose skin, vascular abnormalities and emphysematous lungs both in mice [25,26] and in patients with cutis laxa [30]. On the other hand, fibrillins are the principal structural components of elastic fibre-associated microfibrils. Domain-mapping studies of fibrillin-1 have demonstrated that the N-terminus of fibrillin-1, spanning exons 9–17, is a major tropoelastin-binding site [9,10]. Binding of PET and of fibulin-5 to the Δ16–17 mutant was not diminished greatly. Hence, we conclude that impaired deposition of mutant tropoelastin into the extracellular matrix was not caused by reduced binding to scaffold proteins, at least fibulin-5 and fibrillin-1.
Self-association of tropoelastin plays an important role in the alignment of tropoelastin monomers for polymeric assembly and cross-link formation [8,31,32]. Studies using tropoelastin and polypeptides based on elastin sequence showed that there was a correlation between coacervation and fibril formation by tropoelastin [33]. It has been demonstrated that the number of hydrophobic domains, their specific sequences and their context in alteration with cross-linking domains all influence self assembly of tropoelastin [6,34]. Indeed, it has been also reported that the coacervation temperature of the mutated tropoelastin derived from patients with SVAS is higher [35]. Our electron microscopic data revealed that tropoelastin lacking exons 16–17 was inefficient at forming fibrils. Recently, it has been reported that exon 30 of a hydrophobic domain contains an important sequence for the assembly process to form β-sheet structures. It was also found that a tandem repeat sequence of GGLG(V/A) encoded by exon 30 resembles sequences for the aggregation through β-sheet/β-turn structures in lamprin [4] or spidroin [5]. As shown in Table 1, the sequence encoded by exon 16 of human tropoelastin is identified by high homology with the sequence encoded by exon 30 of human tropoelastin. We propose that both exon 16 and exon 30 play an important role in the formation of β-sheet structure, which is critical for the fibre formation and self-assembly of tropoelastin [4].
Table 1. Alignment of the sequence encoded by exon 16 and 30 of human tropoelastin.
Asterisks indicate identical residues.
| Sequence | Exon |
|---|---|
| GAGAAGVLP GVGGAGVPGVPGAIP | 16 |
| GLVGAAGLGGLGVGGLGVPGVGGLG | 30 |
| **** **** ***** |
In addition to providing insights into the mechanisms of elastic fibre formation, our results are also informative for understanding the disease mechanisms responsible for SVAS. Point mutations identified in SVAS patients are frequently frameshift, nonsense and splice site mutations. Three such mutations to date have been shown to result in premature termination of the reading frame and degradation of the mutant mRNA by nonsense-mediated decay [17,36,37]. Thus these mutations led to a loss-of-function mechanism and a quantitative reduction in the deposition of insoluble elastin [17]. Our results now show that mutations in SVAS may lead to loss of function at the level of deposition and indicate that functional analysis of further SVAS mutations leading to stable elastin mRNA and protein may shed further light on to the mechanisms of elastic fibre formation.
Acknowledgments
This work was supported by the Hoshi University Science/Technology Frontier Research Base (to H.W.) and by NIH (National Institutes of Health) grant HL073703 (to Z.U.). We thank Dr Arnold Strauss and Dr Virginia V. Michels for the referral of SVAS patients and the ascertainment of fibroblast samples, and Marc Crepeau for assistance with mutational analysis. We are also grateful to Hideki Sugitani, Hayato Murata, Marie Ishida, Kayoko Shibata-Sato and Hiroyuki Oyama for their technical assistance.
References
- 1.Li D. Y., Brooke B., Davis E. C., Mecham R. P., Sorensen L. K., Boak B. B., Eichwald E., Keating M. T. Elastin is an essential determinant of arterial morphogenesis. Nature. 1998;393:276–280. doi: 10.1038/30522. [DOI] [PubMed] [Google Scholar]
- 2.Li D. Y., Faury G., Taylor D. G., Davis E. C., Boyle W. A., Mecham R. P., Stenzel P., Boak B., Keating M. T. Novel arterial pathology in mice and humans hemizygous for elastin. J. Clin. Invest. 1998;102:1783–1787. doi: 10.1172/JCI4487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vrhovski B., Jensen S., Weiss A. S. Coacervation characteristics of recombinant human tropoelastin. Eur. J. Biochem. 1997;250:92–98. doi: 10.1111/j.1432-1033.1997.00092.x. [DOI] [PubMed] [Google Scholar]
- 4.Robson P., Wright G. M., Sitarz E., Maiti A., Rawat M., Youson J. H., Keeley F. W. Characterization of lamprin, an unusual matrix protein from lamprey cartilage: implications for evolution, structure, and assembly of elastin and other fibrillar proteins. J. Biol. Chem. 1993;268:1440–1447. [PubMed] [Google Scholar]
- 5.van Beek J. D., Hess S., Vollrath F., Meier B. H. The molecular structure of spider dragline silk: folding and orientation of the protein backbone. Proc. Natl. Acad. Sci. U.S.A. 2002;99:10266–10271. doi: 10.1073/pnas.152162299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miao M., Bellingham C. M., Stahl R. J., Sitarz E. E., Lane C. J., Keeley F. W. Sequence and structure determinants for the self-aggregation of recombinant polypeptides modeled after human elastin. J. Biol. Chem. 2003;278:48553–48562. doi: 10.1074/jbc.M308465200. [DOI] [PubMed] [Google Scholar]
- 7.Kozel B. A., Wachi H., Davis E. C., Mecham R. P. Domains in tropoelastin that mediate elastin deposition in vitro and in vivo. J. Biol. Chem. 2003;278:18491–18498. doi: 10.1074/jbc.M212715200. [DOI] [PubMed] [Google Scholar]
- 8.Bellingham C. M., Lillie M. A., Gosline J. M., Wright G. M., Starcher B. C., Bailey A. J., Woodhouse K. A., Keeley F. W. Recombinant human elastin polypeptides self-assemble into biomaterials with elastin-like properties. Biopolymers. 2003;70:445–455. doi: 10.1002/bip.10512. [DOI] [PubMed] [Google Scholar]
- 9.Trask T. M., Trask B. C., Ritty T. M., Abrams W. R., Rosenbloom J., Mecham R. P. Interaction of tropoelastin with the amino-terminal domains of fibrillin-1 and fibrillin-2 suggests a role for the fibrillins in elastic fiber assembly. J. Biol. Chem. 2000;275:24400–24406. doi: 10.1074/jbc.M003665200. [DOI] [PubMed] [Google Scholar]
- 10.Rock M. J., Cain S. A., Freeman L. J., Morgan A., Mellody K., Marson A., Shuttleworth C. A., Weiss A. S., Kielty C. M. Molecular basis of elastic fiber formation: critical interactions and a tropoelastin-fibrillin-1 cross-link. J. Biol. Chem. 2004;279:23748–23758. doi: 10.1074/jbc.M400212200. [DOI] [PubMed] [Google Scholar]
- 11.Jensen S. A., Reinhardt D. P., Gibson M. A., Weiss A. S. Protein interaction studies of MAGP-1 with tropoelastin and fibrillin-1. J. Biol. Chem. 2001;276:39661–39666. doi: 10.1074/jbc.M104533200. [DOI] [PubMed] [Google Scholar]
- 12.Freeman L. J., Lomas A., Hodson N., Sherratt M. J., Mellody K. T., Weiss A. S., Shuttleworth C. A., Kielty C. M. Fibulin-5 interacts with fibrillin-1 molecules and microfibrils. Biochem. J. 2005;388:1–5. doi: 10.1042/BJ20050368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brown P. L., Mecham L., Tisdale C., Mecham R. P. The cysteine residues in the carboxy terminal domain of tropoelastin form an intrachain disulfide bond that stabilizes a loop structure and positively charged pocket. Biochem. Biophys. Res. Commun. 1992;186:549–555. doi: 10.1016/s0006-291x(05)80843-5. [DOI] [PubMed] [Google Scholar]
- 14.Hsiao H., Stone P. J., Toselli P., Rosenbloom J., Franzblau C., Schreiber B. M. The role of the carboxy terminus of tropoelastin in its assembly into the elastic fiber. Connect. Tissue Res. 1999;40:83–95. doi: 10.3109/03008209909029104. [DOI] [PubMed] [Google Scholar]
- 15.Penner A. S., Rock M. J., Kielty C. M., Shipley J. M. Microfibril-associated glycoprotein-2 interacts with fibrillin-1 and fibrillin-2 suggesting a role for MAGP-2 in elastic fiber assembly. J. Biol. Chem. 2002;277:35044–35049. doi: 10.1074/jbc.M206363200. [DOI] [PubMed] [Google Scholar]
- 16.Clarke A. W., Wise S. G., Cain S. A., Kielty C. M., Weiss A. S. Coacervation is promoted by molecular interactions between the PF2 segment of fibrillin-1 and the domain 4 region of tropoelastin. Biochemistry. 2005;44:10271–10281. doi: 10.1021/bi050530d. [DOI] [PubMed] [Google Scholar]
- 17.Urban Z., Riazi S., Seidl T. L., Katahira J., Smoot L. B., Chitayat D., Boyd C. D., Hinek A. Connection between elastin haploinsufficiency and increased cell proliferation in patients with supravalvular aortic stenosis and Williams–Beuren syndrome. Am. J. Hum. Genet. 2002;71:30–44. doi: 10.1086/341035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Milewicz D. M., Urban Z., Boyd C. Genetic disorders of the elastic fiber system. Matrix Biol. 2000;19:471–480. doi: 10.1016/s0945-053x(00)00099-8. [DOI] [PubMed] [Google Scholar]
- 19.Wachi H., Sato F., Murata H., Nakazawa J., Starcher B. C., Seyama Y. Development of a new in vitro model of elastic fiber assembly in human pigmented epithelial cells. Clin. Biochem. 2005;38:643–653. doi: 10.1016/j.clinbiochem.2005.04.006. [DOI] [PubMed] [Google Scholar]
- 20.Robb B. W., Wachi H., Schaub T., Mecham R. P., Davis E. C. Characterization of an in vitro model of elastic fiber assembly. Mol. Biol. Cell. 1999;10:3595–3605. doi: 10.1091/mbc.10.11.3595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Urban Z., Michels V. V., Thibodeau S. N., Donis-Keller H., Csiszar K., Boyd C. D. Supravalvular aortic stenosis: a splice site mutation within the elastin gene results in reduced expression of two aberrantly spliced transcripts. Hum. Genet. 1999;104:135–142. doi: 10.1007/s004390050926. [DOI] [PubMed] [Google Scholar]
- 22.Ritty T. M., Broekelmann T., Tisdale C., Milewicz D. M., Mecham R. P. Processing of the fibrillin-1 carboxyl-terminal domain. J. Biol. Chem. 1999;274:8933–8940. doi: 10.1074/jbc.274.13.8933. [DOI] [PubMed] [Google Scholar]
- 23.Starcher B. C., Mecham R. P. Desmosine radioimmunoassay as a means of studying elastogenesis in cell culture. Connect. Tissue Res. 1981;8:255–258. doi: 10.3109/03008208109152385. [DOI] [PubMed] [Google Scholar]
- 24.Sato F., Wachi H., Starcher B. C., Murata H., Amano S., Tajima S., Seyama Y. The characteristics of elastic fiber assembled with recombinant tropoelastin isoform. Clin. Biochem. 2006;39:746–753. doi: 10.1016/j.clinbiochem.2006.02.017. [DOI] [PubMed] [Google Scholar]
- 25.Yanagisawa H., Davis E. C., Starcher B. C., Ouchi T., Yanagisawa M., Richardson J. A., Olson E. N. Fibulin-5 is an elastin-binding protein essential for elastic fibre development in vivo. Nature. 2002;415:168–171. doi: 10.1038/415168a. [DOI] [PubMed] [Google Scholar]
- 26.Nakamura T., Lozano P. R., Ikeda Y., Iwanaga Y., Hinek A., Minamisawa S., Cheng C. F., Kobuke K., Dalton N., Takada Y., et al. Fibulin-5/DANCE is essential for elastogenesis in vivo. Nature. 2002;415:171–175. doi: 10.1038/415171a. [DOI] [PubMed] [Google Scholar]
- 27.Roark E. F., Keene D. R., Haudenschild C. C., Godyna S., Little C. D., Argraves W. S. The association of human fibulin-1 with elastic fibers: an immunohistological, ultrastructural, and RNA study. J. Histochem. Cytochem. 1995;43:401–411. doi: 10.1177/43.4.7534784. [DOI] [PubMed] [Google Scholar]
- 28.Fazio M. J., Olsen D. R., Kauh E. A., Baldwin C. T., Indik Z., Ornstein-Goldstein N., Yeh H., Rosenbloom J., Uitto J. Cloning of full-length elastin cDNAs from a human skin fibroblast recombinant cDNA library: further elucidation of alternative splicing utilizing exon-specific oligonucleotides. J. Invest. Dermatol. 1988;91:458–464. doi: 10.1111/1523-1747.ep12476591. [DOI] [PubMed] [Google Scholar]
- 29.Indik Z., Yeh H., Ornstein-Goldstein N., Sheppard P., Anderson N., Rosenbloom J. C., Peltonen L., Rosenbloom J. Alternative splicing of human elastin mRNA indicated by sequence analysis of cloned genomic and complementary DNA. Proc. Natl. Acad. Sci. U.S.A. 1987;84:5680–5684. doi: 10.1073/pnas.84.16.5680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Loeys B., Van Maldergem L., Mortier G., Coucke P., Gerniers S., Naeyaert J. M., De Paepe A. Homozygosity for a missense mutation in fibulin-5 (FBLN5) results in a severe form of cutis laxa. Hum. Mol. Genet. 2002;11:2113–2118. doi: 10.1093/hmg/11.18.2113. [DOI] [PubMed] [Google Scholar]
- 31.Keeley F. W., Bellingham C. M., Woodhouse K. A. Elastin as a self-organizing biomaterial: use of recombinantly expressed human elastin polypeptides as a model for investigations of structure and self-assembly of elastin. Philos. Trans. R. Soc. London Ser. B. 2002;357:185–189. doi: 10.1098/rstb.2001.1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miao M., Cirulis J. T., Lee S., Keeley F. W. Structural determinants of cross-linking and hydrophobic domains for self-assembly of elastin-like polypeptides. Biochemistry. 2005;44:14367–14375. doi: 10.1021/bi0510173. [DOI] [PubMed] [Google Scholar]
- 33.Bressan G. M., Pasquali-Ronchetti I., Fornieri C., Mattioli F., Castellani I., Volpin D. Relevance of aggregation properties of tropoelastin to the assembly and structure of elastic fibers. J. Ultrastruct. Mol. Struct. Res. 1986;94:209–216. doi: 10.1016/0889-1605(86)90068-6. [DOI] [PubMed] [Google Scholar]
- 34.Toonkool P., Jensen S. A., Maxwell A. L., Weiss A. S. Hydrophobic domains of human tropoelastin interact in a context-dependent manner. J. Biol. Chem. 2001;276:44575–44580. doi: 10.1074/jbc.M107920200. [DOI] [PubMed] [Google Scholar]
- 35.Wu W. J., Weiss A. S. Deficient coacervation of two forms of human tropoelastin associated with supravalvular aortic stenosis. Eur. J. Biochem. 1999;266:308–314. doi: 10.1046/j.1432-1327.1999.00891.x. [DOI] [PubMed] [Google Scholar]
- 36.Urban Z., Zhang J., Davis E. C., Maeda G. K., Kumar A., Stalker H., Belmont J. W., Boyd C. D., Wallace M. R. Supravalvular aortic stenosis: genetic and molecular dissection of a complex mutation in the elastin gene. Hum. Genet. 2001;109:512–520. doi: 10.1007/s00439-001-0608-z. [DOI] [PubMed] [Google Scholar]
- 37.Urban Z., Michels V. V., Thibodeau S. N., Davis E. C., Bonnefont J. P., Munnich A., Eyskens B., Gewillig M., Devriendt K., Boyd C. D. Isolated supravalvular aortic stenosis: functional haploinsufficiency of the elastin gene as a result of nonsense-mediated decay. Hum. Genet. 2000;106:577–588. doi: 10.1007/s004390000285. [DOI] [PubMed] [Google Scholar]