Abstract
MUC4 (mucin 4) is a membrane-bound mucin overexpressed in the early steps of oesophageal carcinogenesis and implicated in tumour progression. We previously showed that bile acids, main components of gastro-oesophageal reflux and tumour promoters, up-regulate MUC4 expression [Mariette, Perrais, Leteurtre, Jonckheere, Hemon, Pigny, Batra, Aubert, Triboulet and Van Seuningen (2004) Biochem. J. 377, 701–708]. HNF (hepatocyte nuclear factor) 1α and HNF4α transcription factors are known to mediate bile acid effects, and we previously identified cis-elements for these factors in MUC4 distal promoter. Our aim was to demonstrate that these two transcription factors were directly involved in MUC4 activation by bile acids. MUC4, HNF1α and HNF4α expressions were evaluated by immunohistochemistry in human oesophageal tissues. Our results indicate that MUC4, HNF1α and HNF4α were co-expressed in oesophageal metaplastic and adenocarcinomatous tissues. Studies at the mRNA, promoter and protein levels indicated that HNF1α regulates endogenous MUC4 expression by binding to two cognate cis-elements respectively located at −3332/−3327 and −3040/−3028 in the distal promoter. We also showed by siRNA (small interfering RNA) approach, co-transfection and site-directed mutagenesis that HNF1α mediates taurodeoxycholic and taurochenodeoxycholic bile acid activation of endogenous MUC4 expression and transcription in a dose-dependent manner. In conclusion, these results describe a new mechanism of regulation of MUC4 expression by bile acids, in which HNF1α is a key mediator. These results bring new insights into MUC4 up-regulation in oesophageal carcinoma associated with bile reflux.
Keywords: adenocarcinoma, Barrett's metaplasia, bile acid, hepatocyte nuclear factor 1α (HNF1α), mucin 4 (MUC4), oesophagus
Abbreviations: ChIP, chromatin immunoprecipitation; D-PBS, Dulbecco-PBS; EMSA, electrophoretic mobility-shift assay; ErbB2, erythroblastic leukaemia viral oncogene homologue 2; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; GATA-4, GATA-binding protein 4; GNa, sodium glycocholate; HNF, hepatocyte nuclear factor; MUC4, mucin 4; OA, oesophageal adenocarcinoma; RT, reverse transcriptase; siRNA, small interfering RNA; TC, taurocholic acid; TCDC, taurochenodeoxycholic acid; TDC, taurodeoxycholic acid
INTRODUCTION
The incidence of OA (oesophageal adenocarcinoma) is increasing rapidly in Western populations [1]. Despite recent advances in multimodal therapy, the prognosis for invasive OA remains poor, reflecting the natural history of this disease to disseminate early [2]. Most if not all of OA develop from Barrett's metaplasia, defined as the replacement of any portion of the normal squamous lining by a metaplastic columnar epithelium which is visible macroscopically [3]. Chronic duodeno-gastro-oesophageal reflux of acid and bile plays an important role in initiation and evolution of Barrett's metaplasia along the metaplasia–dysplasia–carcinoma sequence [4]. Although the importance of bile acids in the development of adenocarcinoma in Barrett's metaplasia is still being discussed among authors, many studies suggest that they are the main factor in initiating the process [1,5]. A better understanding of the molecular mechanisms triggered by bile acids that may contribute to development and evolution of Barrett's metaplasia into OA is thus necessary and will help identify them as key factors in oesophageal cancer pathogenesis associated with reflux.
In normal oesophagus, mucins protect the underlying mucosa against potential injuries such as reflux of gastroduodenal contents including bile acids [6]. Among the members that compose the family of mucins, membrane mucins are thought to play important roles in tumour cell biology, cell proliferation, tumour progression and metastasis [7]. MUC1 (mucin 1) and MUC4 (mucin 4) are the best-characterized membrane mucins [8,9]. They are large O-glycoproteins with an extended heavily glycosylated extracellular domain that protrudes far away from the cellular membrane and is involved in cell recognition processes. Consequently, overexpression of membrane mucins at the cancer cell surface will provide the cancer cell new biological properties by altering their interactions with immune cells, epithelial or endothelial cells or the extracellular matrix [7,9]. In the light of identifying new markers and therapeutic targets in OA, MUC4 could be of particular interest because it is known to be a mediator of tumour growth and metastasis by acting as a ligand for the receptor tyrosine kinase ErbB2 (erythroblastic leukaemia viral oncogene homologue 2) [8,10,11]. MUC4 is expressed in the oesophageal stratified squamous epithelium and during the whole OA carcinogenetic sequence [12–14]. A recent study identified MUC4 as a promising early diagnostic tumoural marker, since it was overexpressed in the high-grade dysplasia state and in adenocarcinomatous tissues [14].
In relation with (i) MUC4 overexpression in OA and (ii) the role of bile acids in induction and degeneration of Barrett's metaplasia, we recently showed that TC (taurocholic acid), TDC (taurodeoxycholic acid), TCDC (taurochenodeoxycholic acid) and glycocholic acids and GNa (sodium glycocholate) were strong activators of MUC4 expression [15]. We showed that this regulation occurred at the transcriptional level, and involved activation of phosphoinositide 3-kinase pathway. Bile acids may however mediate their effects using other pathways. Activation of HNF (hepatocyte nuclear factor) 1α and HNF4α was recently identified as a mechanism used by bile acids to activate transcription of their target genes [16,17]. The recent identification of HNF1α cis-elements in the distal promoter of MUC4 [18] was a positive argument in favour of the possible implication of that transcription factor in mediating MUC4 regulation by bile acids. In the present paper, we show for the first time that HNF1α is a key mediator of MUC4 activation by TDC and TCDC bile acids.
MATERIALS AND METHODS
Tissue sample collection and immunochemistry
Surgical specimens were collected from ten patients who underwent curative oesophagectomy for adenocarcinoma developed on Barrett's metaplasia without neoadjuvant treatment. Permission for removal of surgical samples was obtained from the Centre Hospitalier Régional et Universitaire de Lille Review Board. A consent form was obtained from each patient. The surgical specimen was quickly immersed in 10% (v/v) neutral formaldehyde solution (pH 7.4) in PBS. Samples from normal mucosa, Barrett's metaplasia and adenocarcinoma were processed for paraffin embedding. The diagnosis was assessed by two pathologists after staining the section (4 μm) with haematoxylin–eosin–saffron. Immunohistochemical studies for MUC4 expression were performed as described in [15]. Monoclonal anti-MUC4 antibody [19] was used at 1:20000 dilution. A positive control for MUC4 immunostaining was included in each set of experiments on human lung sections. Immunohistochemical studies for HNF1α and HNF4α expression were performed as described in [20]. The primary antibodies directed against human HNF1α and human HNF4α (Santa Cruz Biotechnology) were used at a 1:2000 dilution. A positive control for HNF1α and HNF4α immunostaining on human small intestine sections was included in each set of experiments.
Cell culture
The OA cell line OE33 was purchased from ECACC (European Collection of Cell Cultures) and was cultured as previously described [15]. HNF1α-expressing KATO-III cells were cultured as previously described [21]. Cells were treated with bile acids and their conjugates for 24 h as described previously [15]. Optimal concentrations for bile acids were: TC, 0.5 mM; TDC, 1 mM; TCDC, 0.5 mM; glycocholic acid, 0.5 mM; and GNa, 0.5 mM. For dose–effect experiments, a third, a half and the full concentration of each bile acid was used. To inhibit protein synthesis or transcription, cells were treated for 30 min with cycloheximide (20 μg/ml) or actinomycin D (1 μg/ml) respectively. All reagents were from Sigma unless otherwise indicated.
siRNA (small interfering RNA) assays
KATO-III cells were seeded the day before transfection at a density of 20×103 cells/well in antibiotic-free medium. Cells were transfected with 100 nM of either TCF1 (transcription factor 1; HNF1α) SMARTpool® or HNF4α SMARTpool® siRNA, using 1 μl of DharmaFECT™ 4 transfection reagent according to the manufacturer's instructions (Dharmacon). Controls included mock transfected cells, cells transfected with siCONTROL™ Non Targeting siRNA or siCONTROL™ GAPD siRNA. Bile acid treatment was performed 24 h after transfection and lasted 24 h as described above. Total RNA was isolated 48 h after transfection as described below. Each siRNA was assayed in triplicate in at least three separate experiments. PCR was performed on 5 μl of cDNA as previously described. Densitometric analysis of DNA bands was carried out using the GelAnalyst-GelSmart software (Clara Vision). Results were expressed as MUC4/GAPDH (glyceraldehyde-3-phosphate dehydrogenase) ratio.
RT (reverse transcriptase)–PCR
Total RNAs from cultured cells were prepared using the RNeasy mini-kit from Qiagen. One microgram of total RNA was used to prepare cDNA (Advantage™ RT-for-PCR kit; Clontech) as described previously [22]. PCR was performed on 5 μl of cDNA using specific pairs of primers for MUC4, HNF1α, HNF4α and 18 S or β-actin as internal controls (MWG-Biotech). Primer information is given in Supplementary Table 1 at http://www.BiochemJ.org/bj/402/bj4020081add.htm. PCRs were carried out in 50 μl final solutions as described in [23]. PCR products were analysed on 1.5% (w/v) agarose gels containing ethidium bromide run in 1×TBE (45 mM Tris/borate/1 mM EDTA) buffer. A 100 bp DNA ladder was purchased from Amersham Biosciences. RT–PCR were carried out on cDNAs from four different sets of experiments. Densitometric analysis of DNA bands was carried out as above. Results were expressed as ratio of MUC4/18 S or MUC4/β-actin.
Plasmids and expression vectors
The two pGL3-MUC4 deletion mutants used in the present study were previously described [24]. They cover the −3713/−3059 and the −3135/−2837 regions of the distal promoter. Two mutated versions of these constructs were made using the QuikChange® site-directed mutagenesis kit (Stratagene). The two HNF-binding sites identified at −3332/−3327 and −3040/−3028 were mutated in the −3713/−3059 and −3135/−2837 deletion mutants respectively. The oligonucleotides containing the mutation were designed according to the manufacturer's instructions and are shown in Supplementary Table 1. pSG5-HNF1α and pSG5-HNF4α expression vectors were a gift from Dr J. K. Divine (Washington University, St Louis, MO, U.S.A.). Plasmids used for transfection studies were prepared using the Endofree Plasmid Maxi kit (Qiagen).
Transient transfections
Transient transfection experiments were performed using Effectene® reagent (Qiagen) as previously described [21]. Luciferase activity was corrected for transfection efficiency by co-transfecting cells with pRL-TK vector (Promega, Charbonnières, France). Total cell extracts were prepared after a 48 h incubation time at 37 °C using 1×Passive Lysis Buffer (Promega) as described in the manufacturer's instruction manual. Luciferase activities were measured on a TD 20/20 luminometer (Turner Design). Each plasmid was assayed in triplicate in at least three separate experiments. In bile acid experiments, relative luciferase activity was expressed as fold activation of luciferase activity in bile acid-treated cells compared with untreated cells. In co-transfection studies, 1 μg of the pGL3-MUC4 deletion mutant was transfected with 0.25 μg of the expression vectors pSG5-HNF1α, pSG5-HNF4α or the corresponding empty vector (pSG5) as the reference. Results were expressed as fold induction of luciferase activity in cells transfected with expression vectors compared with that obtained with the empty vector. To study the effect of HNF1α and HNF4α overexpressions on endogenous MUC4 mRNA level, cells (0.5×106) were transfected with 4 μg of the expression vector of interest and cultured for 48 h before being lysed and processed for total RNA extraction as described previously [20].
Western blotting
Total cellular extracts were prepared using standard procedures and kept at −80 °C before use. Briefly, cells were scraped into the medium and centrifuged for 5 min at 1600 g. The pellet was washed twice with 1×PBS, centrifuged for 1 min at 9000 g and then resuspended in lysis buffer (50 mM Tris/HCl, pH 7.4, 1 mM EDTA, 0.25% sodium deoxycholate, 150 mM NaCl and 1% Nonidet P40) supplemented with protease inhibitors (1 μg/ml aprotinin, 1 μg/ml leupeptin and 1 mM PMSF) and phosphatase inhibitors (5 mM sodium fluoride and 5 mM sodium orthovanadate). The lysate was incubated for 30 min on ice, before being centrifuged at 16000 g for 10 min at 4 °C. The supernatant was collected for Western blotting analysis. Protein content (2 μl of total extracts) was measured in 96-well plates using the bicinchoninic acid method as described in the manufacturer's instruction manual (Pierce). For MUC4 expression analysis, proteins (20 μg) were separated on a SDS (0.1%)/agarose [2% (w/v)] gel. For β-actin expression analysis, proteins were separated using SDS/10% PAGE. Resolved proteins were transferred on to a 0.2 μm nitrocellulose membrane (Schleicher and Schuell) and subjected to the standard immunodetection procedure using specific mouse monoclonal antibodies against β-actin (Sigma A5441) and MUC4 [19]. Secondary antibodies consisted of alkaline phosphatase-conjugated IgG (Promega) for β-actin or horseradish peroxidase-conjugated anti-mouse (Pierce) for MUC4. For MUC4 detection, blots were processed with West® Pico chemiluminescent substrate (Pierce) and the signal was detected by exposing the processed blots to Hyperfilm™ ECL® (enhanced chemiluminescence; Amersham Biosciences). For β-actin detection, the blots were processed with Nitro Blue Tetrazolium and BCIP (5-bromo-4-chloroindol-3-yl phosphate) substrate (Life Technologies).
Nuclear extract preparation and EMSA (electrophoretic mobility-shift assay)
Nuclear extracts were prepared from KATO-III cells as previously described [25] and kept at −80 °C until use. Protein content was measured in 96-well plates using the bicinchoninic acid protein assay (Pierce). Identification of putative HNF-binding sites was carried out by analysing the sequence of MUC4 promoter with MatInspector V2.2 software (http://www.genomatix.de) [26]. The oligonucleotides (MWG-Biotech) were annealed before being phosphorylated at the 5′-end with [γ-32P]ATP (Amersham Biosciences) and T4 polynucleotide kinase (Roche). Wild-type and mutated oligonucleotides used as probes and competitors in EMSAs are shown in Supplementary Table 1. Nuclear protein incubation with radiolabelled probes and competitions with unlabelled probes were as described in [23]. For supershift analyses, 2 μl of anti-HNF1α (sc-6547x) or anti-HNF4α (sc-6556x) (Santa Cruz Biotechnology) antibodies were added to the proteins and left for 2 h at room temperature (22 °C) before adding the radiolabelled probe. Electrophoresis conditions and gel processing were as described previously [27].
ChIP (chromatin immunoprecipitation)
KATO-III cells (15×106 cells) were treated with 1% (v/v) formaldehyde for 10 min at room temperature and cross-links were quenched with glycine at a final concentration of 0.125 M for 5 min. Cells were rapidly rinsed with ice-cold 1× D-PBS (Dulbecco-PBS), with D-PBS containing protease inhibitors (10 μg/ml leupeptin, 10 μg/ml aprotinin, 0.2 mM EDTA and 0.5 mM PMSF), and scraped off and collected by centrifugation at 700 g for 5 min at 4 °C, before being resuspended in lysis buffer (10 mM Hepes, pH 7.9, 10 mM KCl, 1.5 mM MgCl2 and 0.1% Nonidet P40) plus protease inhibitors and incubated for 10 min on ice. Chromatin was sheared with the Bioruptor system (Diagenode). The extracts were sonicated twice for 5 pulses of 30 s each with a 30 s rest between each pulse at 200 W. After clearing by centrifugation at 10000 g for 10 min at 4 °C, the supernatant was diluted in dilution buffer (16.7 mM Tris/HCl, pH 8.0, 167 mM NaCl, 0.01% SDS, 1.1% Triton X-100 and 1.2 mM EDTA). The chromatin solution was precleared by incubation with sonicated salmon sperm DNA/BSA/Protein G–agarose gel slurry (Upstate Biotechnology) for 1 h at 4 °C with rotation followed by centrifugation at 1500 g for 1 min at 4 °C. An aliquot of the total supernatant was removed as input control and the rest was fractionated and precipitated with either 4 μg of the specific antibody or normal rabbit IgGs (Upstate Biotechnology). Immunoprecipitation was performed overnight on a rotating platform at 4 °C, Protein G–agarose gel slurry was then added (v/v) and left for another 2 h. Agarose beads were collected and washed sequentially for 4 min in Low Salt Immune Complex wash buffer, High Salt buffer and LiCl buffer (Upstate) and twice with TE buffer [10 mM Tris/HCl (pH 8.0) and 1 mM EDTA]. The complexes were eluted with 2×0.25 ml of 1% SDS/0.1 M NaHCO3 after a 15 min incubation at room temperature. Formaldehyde cross-links were reversed with 0.3 M NaCl at 65 °C overnight. Chromatin-associated proteins were digested with Qiagen protease at 50 °C for 1 h and the DNA was purified with the Wizard® DNA Clean-up System (Promega). Samples (50 ng) were then subjected to PCR analysis. Primers are indicated in Supplementary Table 1. PCR was performed in a 30 μl final volume consisting of 1×PCR buffer II, 2.5 mM MgCl2, 1.5 units of AmpliTaq Gold polymerase (Applied Biosystems) and 5 pmol of each primer. The temperature-cycling protocol consisted of 12 min of preheating at 94 °C followed by 35 cycles of 45 s of denaturation at 94 °C, 1 min of primer annealing at 50 °C, and 1 min of primer extension at 72 °C followed by a 10 min final extension at 72 °C. PCR products (15 μl) were analysed on a 2% agarose gel.
Statistics
All values in the present paper are means±S.D. When indicated, data were analysed by Mann–Whitney U test with differences P≤0.05 considered significant.
RESULTS
Expression of MUC4, HNF1α and HNF4α in human oesophageal tissues
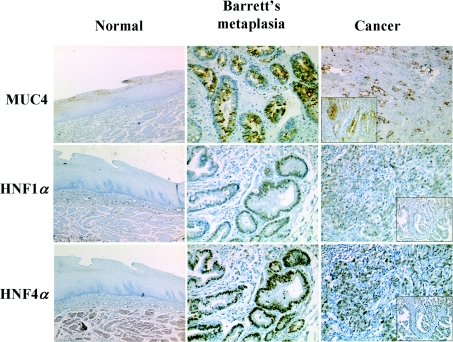
Study of MUC4 expression in human normal, metaplastic and adenocarcinomatous oesophageal tissues indicated that MUC4 was expressed in the cytoplasm of superficial epithelial cells of the normal mucosa (Figure 1). MUC4 cytoplasmic expression was found in Barrett's metaplastic tissue both in non-goblet columnar cells and goblet cells. In adenocarcinoma, MUC4 expression was diffuse and heterogeneous, independently of tumour differentiation. HNF1α and HNF4α were not expressed in normal oesophageal mucosa. They were both expressed in Barrett's metaplasia with strong nuclear staining in all cell types on the surface and glands (including goblet cells). Expression of both factors was visualized independently of tumour differentiation in well-differentiated (inset) and undifferentiated adenocarcinomatous tissues.
Figure 1. Expression of MUC4, HNF1α and HNF4α in human normal, metaplastic and adenocarcinomatous oesophageal tissue.
Immunohistochemical studies of human normal oesophageal mucosa, Barrett's metaplasia and OA, with anti-HNF1α, anti-HNF4α and anti-MUC4 antibodies. Inset: expression in well-differentiated areas of adenocarcinomatous tissues. Magnification: ×150 for normal tissues; ×250 for Barrett's neoplasia and cancer tissues.
Regulation of MUC4 expression by HNF1α and HNF4α
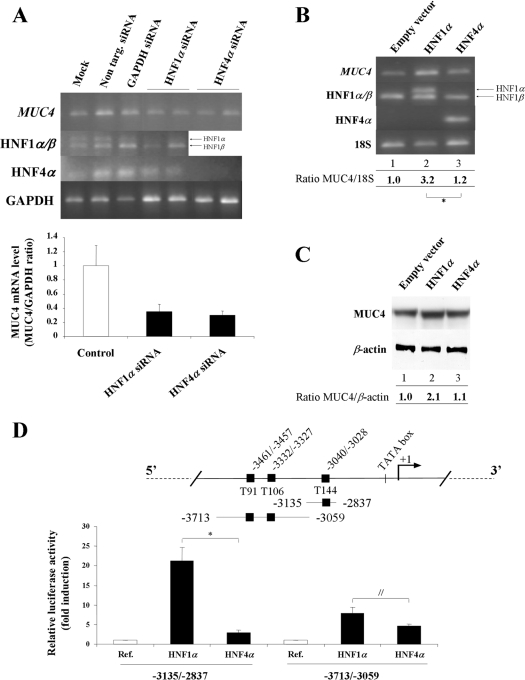
To investigate whether HNF1α and HNF4α transcription factors participate in MUC4 mRNA regulation, we combined knockdown assays with specific siRNA (Figure 2A) and overexpression experiments (Figures 2B and 2C). HNF1α and HNF4α knockdown in gastric KATO-III cells, which express these two transcription factors, led to a substantial decrease in MUC4 mRNA (65±8% and 70±6% respectively), indicating that both transcription factors regulate endogenous MUC4 expression (Figure 2A). Results from four experiments in oesophageal OE33 cells in which HNF1α and HNF4α were overexpressed showed that overexpression of HNF1α led to a 3.2-fold increase (3.2±0.4) of MUC4 mRNA (Figure 2B, lane 2), whereas overexpression of HNF4α had a significantly weaker effect (1.2±0.1-fold increase, P=0.02) (lane 3). At the protein level, we also observed an increase in MUC4 apomucin expression (2.1 times) when HNF1α was overexpressed (Figure 2C, lane 2), whereas no variation was seen in HNF4α-transfected cells (lane 3).
Figure 2. Transcriptional regulation of MUC4 by HNF1α and HNF4α in epithelial cancer cells.
(A) siRNA experiments were carried out as described in the Materials and methods section. MUC4, HNF1α, HNF4α and GAPDH mRNA levels were assessed by RT–PCR. PCR products (10 μl for MUC4 and 5 μl for HNF1α, HNF4α and GAPDH) were analysed on a 1.5% agarose gel. The lower panel shows results expressed as MUC4/GAPDH ratio. Control corresponds to the mean values from mock cells and cells transfected with non-targeting siRNA; this value was arbitrarily set to 1. (B) RT–PCR analysis of MUC4, HNF1α and HNF4α mRNA levels in OE33 cells transfected with 4 μg of pSG5 empty vector (lane 1), pSG5-HNF1α (lane 2), or pSG5-HNF4α (lane 3) expression vectors. PCR products (15 μl for MUC4 and 4 μl for HNF1α, HNF4α and 18 S) were analysed on a 1.5% agarose gel. The value corresponding to cells transfected with empty vector was arbitrarily set to 1. *P=0.02. (C) Western blotting analysis of MUC4 protein expression in total cellular extracts prepared from OE33 cells transfected under the same conditions as above. (D) Schematic representation of MUC4 distal promoter with the three HNF putative binding sites (black squares). Co-transfection experiments in OE33 cells were performed in the presence of 1 μg of MUC4 pGL3-deletion constructs −3135/−2837 or −3713/−3059 and 0.25 μg of pSG5-HNF1α or pSG5-HNF4α. ‘Ref.’ refers to the normalized luciferase activity of pGL3-MUC4 constructs transfected with the empty vector pSG5 (white bar); this value was arbitrarily set to 1. *P=0.001. //P=0.03.
Activation of MUC4 transcription at the promoter level by HNF1α was confirmed by performing co-transfection studies in OE33 cells with the −3135/−2837 and the −3713/−3059 constructs that cover the MUC4 distal promoter. The results shown in Figure 2(D) indicate that HNF1α strongly transactivated (21.2±3.4-fold activation) the −3135/−2837 region and to a lower extent the −3713/−3059 region (7.9±1.6-fold activation). The effect of HNF4α on both constructs was significantly weaker (3.0±0.6- and 4.7±0.4-fold activation, P=0.001 and P=0.03 respectively).
Identification of two HNF1α cis-elements in MUC4 distal promoter
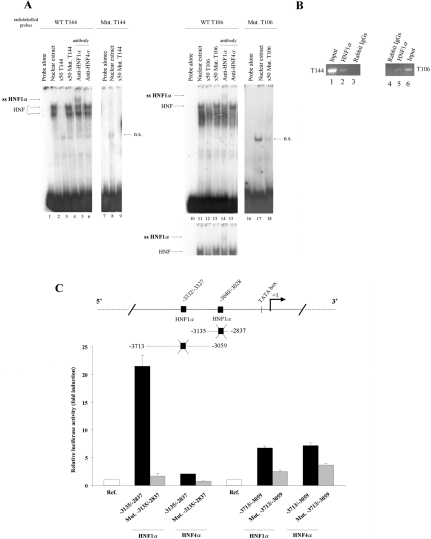
Having shown that HNF1α and HNF4α were transactivating the MUC4 distal promoter, we undertook to identify their binding sites using EMSA. Three putative HNF-binding sites are present in the MUC4 distal promoter (Figure 2D). When radiolabelled probes T144 (−3040/−3028) and T106 (−3332/−3327) were incubated with nuclear extracts, two shifted bands were visualized with T144 probe (Figure 3A, lane 2), whereas only one major retarded band was seen with T106 probe (lane 11). Specificity of the protein–DNA complexes was confirmed by the loss of the shifted bands when unlabelled competition was performed with a 50 times excess of either unlabelled T144 (lane 3) or T106 (lane 12) DNA probes. As expected, competition with a 50 times excess of unlabelled mutated T144 (lane 4) or T106 (lanes 13) probes did not modify the pattern of the shift. We further confirmed the direct involvement of HNF DNA motifs in the complex formation by using radiolabelled mutated T144 and T106 probes in which the HNF-binding sequence was mutated (see Supplementary Table 1). In this case, no binding with nuclear proteins was observed indicating that these nucleotides are directly mediating HNF binding to the MUC4 promoter (lanes 8 and 17 respectively). Finally, direct implication of HNF1α in the binding to the T144 and T106 probes was proved by the supershift obtained when a specific anti-HNF1α antibody was added to the reaction mixture (lanes 5 and 14). No supershift was observed when anti-HNF4α antibody was added (lanes 6 and 15). The third putative HNF-binding site T91 (−3461/−3457) did not bind HNF1α or HNF4α (results not shown). In vivo binding of HNF1α to the chromatin region covering the HNF1α cis-elements identified at −3040/−3028 (T144) and −3332/−3327 (T106) was confirmed by ChIP analysis (Figure 3B, lanes 2 and 5). The direct implication of these two HNF cis-elements in mediating HNF1α and HNF4α activation of the MUC4 promoter was then assessed by carrying out transfections in OE33 cells in the presence of mutated forms of the −3135/−2837 and −3713/−3059 pGL3-MUC4 promoter constructs (Figure 3C). Mutation of the T144 HNF cis-element in the −3135/−2837 construct abolished transactivating effects of HNF1α and HNF4α. Mutation of the T106 site in the −3713/−3059 region led to a 75±4% (HNF1α) and 50±5% (HNF4α) decrease in luciferase activity.
Figure 3. Identification of two HNF1α cis-elements and study of their functionality in regulating MUC4 distal promoter.
(A) Identification of HNF cis-elements by EMSA. Nuclear extracts (8 μg) from HNF1α-expressing KATO-III cells were incubated with radiolabelled T144 (lanes 1–6), mutated T144 (lanes 7–9), T106 (lanes 10–15), or mutated T106 (lanes 16–18) DNA probes. Lanes 1, 7, 10 and 16, radiolabelled probe alone. Lanes 2, 8, 11 and 17, incubation of T144, mutated T144, T106 or mutated T106 probes with nuclear proteins. Lane 3, cold competition with 50 times excess of unlabelled T144. Lanes 4 and 9, cold competition with 50-fold excess of unlabelled Mut. T144. Lane 12, cold competition with 50-fold excess of unlabelled T106. Lanes 13 and 18, cold competition with 50 times excess of unlabelled Mut. T106 probe. Lanes 5 and 14, supershift analyses with anti-HNF1α. Lanes 6 and 15, supershift analysis with anti-HNF4α. DNA–protein complexes (HNF) and supershifts (ssHNF1α) are indicated by arrows on the left side of the autoradiograms. n.s., non-specific. (B) ChIP analysis of HNF1α binding to chromatin covering the T144 (left panel) and T106 (right panel) binding sites using specific primers (Supplementary Table 1; see http://www.BiochemJ.org/bj/402/bj4020081add.htm). Input (lanes 1 and 6), anti-HNF1α (lanes 2 and 5) and rabbit IgGs (lanes 3 and 4). PCR products (15 μl) were analysed on 2% agarose gels. (C) Site-directed mutagenesis of the T144 and T106 HNF cis-elements in the −3135/−2837 or −3713/−3059 constructs and their effects on their regulation by HNF1α or HNF4α. ‘Ref.’ refers to the normalized luciferase activity of wild-type or mutated pGL3-MUC4 constructs co-transfected with the empty vector pSG5 (white bar). This value was arbitrarily set to 1.
MUC4 regulation by bile acids occurs at the transcriptional level
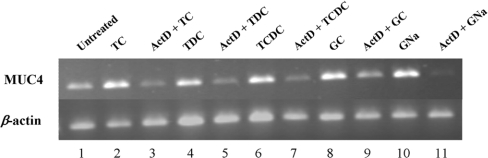
Before studying the role of HNF1α in mediating MUC4 activation by TC, TDC, TCDC, glycocholic acid and GNa bile acids, previously identified as activators of MUC4 expression [15], we checked whether that process occurred at the transcriptional level. To this aim, oesophageal cancer cells were pretreated with actinomycin D or cycloheximide before bile acid exposure. MUC4 mRNA expression was then assessed by RT–PCR. The results indicate that activation of MUC4 expression by bile acids occurred at the transcriptional level, since MUC4 mRNA level returned to basal level when cells were pretreated with actinomycin D (Figure 4). This process did not require de novo protein synthesis since pretreatment of OE33 cells with cycloheximide did not modify the level of MUC4 mRNA (results not shown).
Figure 4. Effect of actinomycin D pretreatment on MUC4 regulation by bile acids in OE33 cells.
Cells were pretreated with actinomycin D (ActD) for 30 min before a 24 h bile acid incubation. MUC4 and β-actin mRNA levels were studied by RT–PCR and compared with cells incubated with bile acids only. PCR products (15 and 4 μl respectively) were analysed on a 1.5% agarose gel. GC, glycocholic acid.
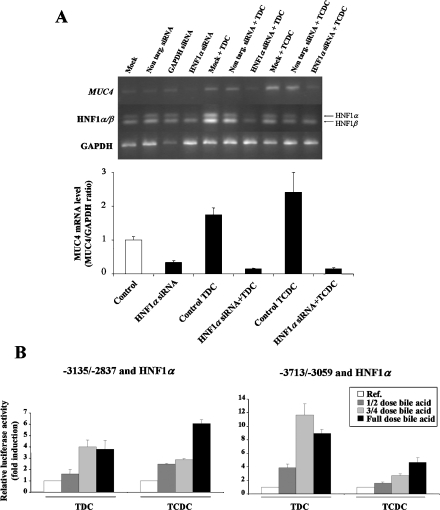
HNF1α mediates MUC4 up-regulation by TDC and TCDC bile acids
To demonstrate that HNF1α was involved in bile acid regulation of MUC4 endogenous expression and promoter activity, we combined knockdown assays (Figure 5A) and co-transfection experiments (Figure 5B). HNF1α knockdown led to a substantial decrease in MUC4 mRNA (66±5% decrease, Figure 5A). We then focused our studies on TDC and TCDC since overexpression of HNF1α did not influence MUC4 activation by TC, glycocholic acid or GNa (results not shown). Treatment of cells with TDC and TCDC enhanced the basal expression level of MUC4 mRNA (1.8±0.2 and 2.4±0.6 times respectively) (Figure 5A). Knockdown of HNF1α strongly inhibited TDC- and TCDC-mediated increase in MUC4 mRNA level (91±2% and 94±3% respectively). This indicates that HNF1α mediates endogenous MUC4 expression in response to these bile acids. At the promoter level, we found that overexpression of HNF1α followed by cell treatment with full dose of TDC and TCDC resulted in a 3.8±0.8-fold and a 6.1±0.3-fold activation of the −3135/−2837 deletion fragment (Figure 5B, left panel). Studies on the −3713/−3059 region of the MUC4 distal promoter (Figure 5B, right panel) again indicated that overexpression of HNF1α followed by full concentration of TDC (1 mM) or TCDC (0.05 mM) induced a strong activation of MUC4 promoter (9.0±0.6- and 4.7±0.5-fold respectively). Moreover, studies performed with different concentrations of bile acids indicated that HNF1α mediation of TDC and TCDC effects on the two MUC4 promoter fragments were dose-dependent (Figure 5B).
Figure 5. HNF1α-mediated up-regulation of MUC4 transcription by TDC and TCDC is dose-dependent.
(A) siRNA experiments were carried out as described in the Materials and methods section. MUC4, HNF1α and GAPDH mRNA levels were assessed by RT–PCR. PCR products (10 μl for MUC4 and 5 μl for HNF1α and GAPDH) were analysed on a 1.5% agarose gel. The lower panel shows results expressed as MUC4/GAPDH ratio. Control corresponds to the mean values from mock cells and cells transfected with non-targeting siRNA; this value was arbitrarily set to 1. (B) Co-transfection experiments in OE33 cells were performed in the presence of 1 μg of pGL3-MUC4 constructs −3135/−2837 or −3713/−3059 and 0.25 μg of pSG5-HNF1α expression vector. Cells were then incubated for 24 h with one-half, three-quarters or full concentration of TDC or TCDC (1 and 0.5 mM respectively). ‘Ref.’ refers to the normalized luciferase activity of pGL3-MUC4 constructs co-transfected with HNF1α expression vector without bile acid treatment; this value was arbitrarily set to 1.
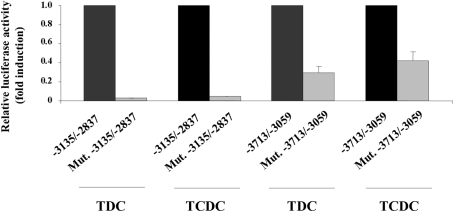
Finally, we demonstrated that the two HNF cis-elements identified in Figure 3 were necessary for HNF1α-mediated up-regulation of MUC4 promoter by TDC and TCDC. To this end, we compared MUC4 promoter activity in cells co-transfected with HNF1α expression vector and wild-type pGL3-MUC4 constructs followed by TDC or TCDC treatment with that in cells transfected under the same conditions with the mutated forms of MUC4 promoter (Figure 6). The mediation by HNF1α was completely abolished when co-transfecting the mutated form of the −3135/−2837 construct (grey bars). When using the −3713/−3059 mutated construct, promoter transactivation mediated by HNF1α decreased for more than a half (70±6% for TDC and 60±9% for TCDC).
Figure 6. HNF1α cis-elements are directly implicated in MUC4 regulation by TDC and TCDC.
Co-transfection experiments in OE33 cells were performed in the presence of 1 μg of pGL3-MUC4 constructs −3135/−2837, Mut. −3135/−2837, −3713/−3059 or Mut. −3713/−3059 and 0.25 μg of pSG5-HNF1α expression vector. Cells were then treated for 24 h with TDC or TCDC. The values obtained in cells transfected with wild-type pGL3-MUC4 constructs were arbitrarily set to 1.
DISCUSSION
In this paper, we demonstrate that activation of MUC4 expression by TDC and TCDC bile acids is dose-dependent and is mediated via binding of HNF1α to two cognate cis-elements in the MUC4 distal promoter. Bile acids are a component of duodeno-gastro-oesophageal reflux often associated with development and/or promotion of OA on Barrett's metaplasia [4]. Thus our results identify these two bile acids as potent inducers of MUC4 expression during oesophageal carcinogenesis associated with bile reflux. Since these two bile acids were previously shown to be especially toxic under in vivo conditions [28] and that TDC is a secondary bile acid found in higher concentration in patients with Barrett's oesophagus compared with asymptomatic patients [28], they may be considered as tumour promoters and potential therapeutic targets.
In a previous study, we had demonstrated that MUC4 regulation by TC, TDC, TCDC, glycocholic acid and GNa bile acids in OA cells involved phosphoinositide 3-kinase signalling pathway and to a lower extent mitogen-activated protein kinase, protein kinase A and protein kinase C pathways [15]. In the present study, we show that induction of MUC4 expression may occur via an alternative pathway involving HNF1α, this mechanism being restricted to TDC and TCDC. The fact that bile acids are able to activate MUC4 expression using different pathways indicates that it is an important mechanism which necessitates alternative pathways in order to increase the capacity of the oesophageal cancer cell to respond to bile acids. We could not show HNF1α mediation for DC, glycocholic acid and GNa, suggesting that other mechanisms are involved. Nuclear factor κB and transcription factors of the activating transcription factor-1/cAMP-response-element-binding protein family have been described to mediate phosphoinositide 3-kinase effects [29]. Further investigations will be needed to answer that question. Although HNF4α is able to regulate endogenous MUC4 expression, we could not identify an HNF4-binding element, which suggests that the regulation is indirect, most likely involving participation of cofactors as has already been shown for HNF4 [30]. Moreover, HNF4α has no functional role in mediating MUC4 regulation by TDC and TCDC. Taken together, our in vitro and ex vivo results showing co-expression of HNF1α and MUC4 in Barrett's metaplasia and adenocarcinoma strongly suggest that HNF1α plays a major role in controlling MUC4 expression during oesophageal carcinogenesis.
HNF1α is a homoeodomain-containing transcription factor expressed in the liver, kidney, intestine, stomach and pancreas [31]. Inactivation of HNF1α has been associated with the development of human liver adenomas and some hepatocarcinomas, suggesting a potential role for HNF1α as a tumour suppressor gene in these diseases [32]. In the present study, we did not find loss of HNF1α expression during the oesophageal carcinogenetic sequence. On the contrary, expression was induced in Barrett's metaplasia and adenocarcinoma. This suggests that HNF1α plays a fundamental role in the progression of oesophageal carcinogenesis. Moreover, HNF1α is known to regulate the expression of several intestine-specific genes (lactase and sucrase isomaltase) [33,34], and to control intestinal cell differentiation [35], a mucosa in which MUC4 is expressed [36]. Since intestinal differentiation of oesophageal mucosa, secondary to bile acid reflux, is a characteristic step towards development of OA derived from Barrett's metaplasia, it may explain the strong HNF1α expression in Barrett's metaplasia mucosa with concomitant activation of MUC4. A quantitative study of the potential diagnostic value of HNF1α expression during the carcinogenetic sequence leading to oesophagus adenocarcinoma in a higher number of patients would be interesting to investigate.
HNF1α binds the consensus sequence GGTTAATNATTAAC-(A/C) [30,37]. The two cis-elements identified in MUC4 promoter are very similar to that consensus sequence (CTTAATAAACATC at −3332/−3327 and GTGGAATATTAAC at −3040/−3028). Mutation of some nucleotides in the −3040/−3028 element completely abolished binding and transactivation of MUC4 by HNF1α, indicating that HNF1α alone is sufficient to convey MUC4 activation via this site. Mutation of the other cis-element at −3332/−3327 inhibited HNF1α binding but transactivation was partially affected, suggesting probable involvement of a cofactor. Among transcription factors known to synergize with HNF1α [34] and for which we found putative binding sites in the distal promoter of MUC4 [18] are Cdx-2 and GATA-4 (GATA-binding protein 4). Moreover Cdx transcription factors are also associated with OA developed on Barrett's metaplasia [38,39]. Preliminary results, however, did not suggest additive or synergistic effects between these three transcription factors in oesophageal OE33 cancer cells (G. Piessen and I. Van Seuningen, unpublished work). These synergistic activities may thus be restricted to intestinal cells [34].
The pattern of mucin expression is tissue-specific and changes during neoplastic progression suggest an important role for mucins in tumour growth and progression and have recently justified their use as reliable phenotypic markers [40,41]. During the different steps of the OA carcinogenetic sequence, MUC4, a high-molecular-mass membrane-bound mucin, is overexpressed in the high-grade dysplasia state and in adenocarcinomatous tissues and is presently evaluated by us and other groups as a promising early diagnostic tumour marker [12–14]. Moreover, membrane-bound mucins have been associated with both steric protection of epithelial surfaces and cellular signalling functions [7,41,42]. MUC4 is a mediator of tumour growth and metastatic properties of cancer cells by acting as a ligand for the receptor tyrosine kinase ErbB2 [8,10,11]. ErbB2 is an oncogene overexpressed in 11–35% of OA derived from Barrett's metaplasia [43,44], and tumours overexpressing ErbB2 have a poorer survival prognosis [43]. Recently, we showed that MUC4–ErbB2 interaction may play a role in the progression/differentiation balance of pancreatic tumour cells [11]. Obviously, this duality of function could be of interest in terms of therapeutic application with MUC4 in OA and could also explain the variations of MUC4 prognostic value in the upper aerodigestive tract (good prognosis) [41] or in pancreatic adenocarcinoma (poor prognosis) [45] published in the literature. Dual biological activity has been proposed for the MUC4 mucin, since it may be expressed either as a membrane-associated protein or as a soluble mucin [7,8,46]. The soluble form, which may be produced by either alternative splicing or proteolysis, contributes to maintain a pool of secreted MUC4 that participates in epithelial protection [46]. Interestingly, in the present study MUC4 expression was found both at the apical surface and in the cytoplasm of oesophageal cancer cells. This pattern of expression has already been described for MUC4 in physiological fluids [46] as well as in other tumour locations [47] and may thus represent a more general pattern of expression of MUC4 that translates its dual role in epithelial cell biology.
In conclusion, we have demonstrated that HNF1α is a strong activator of MUC4 expression, and that it specifically mediates MUC4 up-regulation by TDC and TCDC bile acids. This novel mechanism of regulation of MUC4 may account for its up-regulation during OA associated with bile reflux. In the future, the development of an animal model of induced oesophageal carcinogenesis will allow us to evaluate the exact role of MUC4 overexpression in the development of oesophagus adenocarcinoma on Barrett's metaplasia associated with reflux, the consequences on the biological properties of oesophageal cancer cells and to identify potential molecular targets that could have a therapeutic value.
Online data
Acknowledgments
This paper is dedicated to Dr Jean-Pierre Aubert, Director of our Laboratory, who died in September 2005. He initiated the collaboration between surgeons and scientists of the unit in order to stimulate translational research and promote potential clinical implications. We thank Dr J. K. Divine (Washington University, St. Louis, MO, U.S.A.) for the gift of pSG5-HNF1α and pSG5-HNF4α expression vectors. This work was funded by a grant from la Ligue Nationale contre le Cancer (comité du Pas-de-Calais). A. V. is the recipient of an Inserm-Région Nord-Pas de Calais Ph.D. fellowship.
References
- 1.Wild C. P., Hardie L. J. Reflux, Barrett's oesophagus and adenocarcinoma: burning questions. Nat. Rev. Cancer. 2003;3:676–684. doi: 10.1038/nrc1166. [DOI] [PubMed] [Google Scholar]
- 2.Mariette C., Balon J. M., Piessen G., Fabre S., Van Seuningen I., Triboulet J. P. Pattern of recurrence following complete resection of esophageal carcinoma and factors predictive of recurrent disease. Cancer. 2003;97:1616–1623. doi: 10.1002/cncr.11228. [DOI] [PubMed] [Google Scholar]
- 3.Wijnhoven B. P., Tilanus H. W., Dinjens W. N. Molecular biology of Barrett's adenocarcinoma. Ann. Surg. 2001;233:322–337. doi: 10.1097/00000658-200103000-00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jankowski J. A., Harrison R. F., Perry I., Balkwill F., Tselepis C. Barrett's metaplasia. Lancet. 2000;356:2079–2085. doi: 10.1016/S0140-6736(00)03411-5. [DOI] [PubMed] [Google Scholar]
- 5.Fein M., Peters J. H., Chandrasoma P., Ireland A. P., Oberg S., Ritter M. P., Bremner C. G., Hagen J. A., DeMeester T. R. Duodenoesophageal reflux induces esophageal adenocarcinoma without exogenous carcinogen. J. Gastrointest. Surg. 1998;2:260–268. doi: 10.1016/s1091-255x(98)80021-8. [DOI] [PubMed] [Google Scholar]
- 6.Corfield A. P., Myerscough N., Longman R., Sylvester P., Arul S., Pignatelli M. Mucins and mucosal protection in the gastrointestinal tract: new prospects for mucins in the pathology of gastrointestinal disease. Gut. 2000;47:589–594. doi: 10.1136/gut.47.4.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hollingsworth M. A., Swanson B. J. Mucins in cancer: protection and control of the cell surface. Nat. Rev. Cancer. 2004;4:45–60. doi: 10.1038/nrc1251. [DOI] [PubMed] [Google Scholar]
- 8.Carraway K. L., Ramsauer V. P., Haq B., Carothers Carraway C. A. Cell signaling through membrane mucins. BioEssays. 2003;25:66–71. doi: 10.1002/bies.10201. [DOI] [PubMed] [Google Scholar]
- 9.Gendler S. J. MUC1, the renaissance molecule. J. Mamm. Gland Biol. Neoplasia. 2001;6:339–353. doi: 10.1023/a:1011379725811. [DOI] [PubMed] [Google Scholar]
- 10.Singh A. P., Moniaux N., Chauhan S. C., Meza J. L., Batra S. K. Inhibition of MUC4 expression suppresses pancreatic tumor cell growth and metastasis. Cancer Res. 2004;64:622–630. doi: 10.1158/0008-5472.can-03-2636. [DOI] [PubMed] [Google Scholar]
- 11.Fauquette V., Perrais M., Cerulis S., Jonckheere N., Ducourouble M.-P., Aubert J.-P., Pigny P., Van Seuningen I. The antagonistic regulation of human MUC4 and ErbB-2 genes by the Ets protein PEA3 in pancreatic cancer cells: implications for the proliferation/differentiation balance in the cells. Biochem. J. 2005;386:35–45. doi: 10.1042/BJ20040706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guillem P., Billeret V., Buisine M.-P., Flejou J.-F., Lecomte-Houcke M., Degand P., Aubert J.-P., Triboulet J.-P., Porchet N. Mucin gene expression and cell differentiation in human normal, premalignant and malignant esophagus. Int. J. Cancer. 2000;88:856–861. doi: 10.1002/1097-0215(20001215)88:6<856::aid-ijc3>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- 13.Arul G. S., Moorghen M., Myerscough N., Alderson D. A., Spicer R. D., Corfield A. P. Mucin gene expression in Barrett's oesophagus: an in situ hybridisation and immunohistochemical study. Gut. 2000;47:753–761. doi: 10.1136/gut.47.6.753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bax D. A., Haringsma J., Einerhand A. W., Van Dekken H., Blok P., Siersema P. D., Kuipers E. J., Kusters J. G. MUC4 is increased in high grade intraepithelial neoplasia in Barrett's oesophagus and is associated with a proapoptotic Bax to Bcl-2 ratio. J. Clin. Pathol. 2004;57:1267–1272. doi: 10.1136/jcp.2004.017020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mariette C., Perrais M., Leteurtre E., Jonckheere N., Hemon B., Pigny P., Batra S., Aubert J.-P., Triboulet J.-P., Van Seuningen I. Transcriptional regulation of human mucin MUC4 by bile acids in oesophageal cancer cells is promoter-dependent and involves activation of the phosphatidylinositol 3-kinase signalling pathway. Biochem. J. 2004;377:701–708. doi: 10.1042/BJ20031132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jung D., Kullak-Ublick G. A. Hepatocyte nuclear factor 1 alpha: a key mediator of the effect of bile acids on gene expression. Hepatology. 2003;37:622–631. doi: 10.1053/jhep.2003.50100. [DOI] [PubMed] [Google Scholar]
- 17.Shimamoto Y., Ishida J., Yamagata K., Saito T., Kato H., Matsuoka T., Hirota K., Daitoku H., Nangaku M., Yamagata K., et al. Inhibitory effect of the small heterodimer partner on hepatocyte nuclear factor-4 mediates bile acid-induced repression of the human angiotensinogen gene. J. Biol. Chem. 2004;279:7770–7776. doi: 10.1074/jbc.M310577200. [DOI] [PubMed] [Google Scholar]
- 18.Jonckheere N., Pigny P., Hémon B., Ducourouble M.-P., Perrais M., Aubert J.-P., Van Seuningen I. Cell-specific transcriptional regulation of human mucin gene MUC4 by HNF-1/-3/-4, Cdx-1/-2 and GATA-4 transcription factors involved in the development of the gut endoderm. Gastroenterology. 2002;122:247–248. [Google Scholar]
- 19.Swartz M. J., Batra S. K., Varshney G. C., Hollingsworth M. A., Yeo C. J., Cameron J. L., Wilentz R. E., Hruban R. H., Argani P. MUC4 expression increases progressively in pancreatic intraepithelial neoplasia. Am. J. Clin. Pathol. 2002;117:791–796. doi: 10.1309/7Y7N-M1WM-R0YK-M2VA. [DOI] [PubMed] [Google Scholar]
- 20.Van der Sluis M., Melis M. H., Jonckheere N., Ducourouble M.-P., Buller H. A., Renes I., Einerhand A. W., Van Seuningen I. The murine Muc2 mucin gene is transcriptionally regulated by the zinc-finger GATA-4 transcription factor in intestinal cells. Biochem. Biophys. Res. Commun. 2004;325:952–960. doi: 10.1016/j.bbrc.2004.10.108. [DOI] [PubMed] [Google Scholar]
- 21.Perrais M., Pigny P., Buisine M. P., Porchet N., Aubert J.-P., Van Seuningen-Lempire I. Aberrant expression of human mucin gene MUC5B in gastric carcinoma and cancer cells. Identification and regulation of a distal promoter. J. Biol. Chem. 2001;276:5386–5396. doi: 10.1074/jbc.M010534200. [DOI] [PubMed] [Google Scholar]
- 22.Van Seuningen I., Perrais M., Pigny P., Porchet N., Aubert J.-P. Sequence of the 5′-flanking region and promoter activity of the human mucin gene MUC5B in different phenotypes of colon cancer cells. Biochem. J. 2000;348:675–686. [PMC free article] [PubMed] [Google Scholar]
- 23.Mesquita P., Jonckheere N., Almeida R., Ducourouble M.-P., Serpa J., Silva E., Pigny P., Silva F. S., Reis C., Silberg D., et al. Human MUC2 mucin gene is transcriptionally regulated by Cdx homeodomain proteins in gastrointestinal carcinoma cell lines. J. Biol. Chem. 2003;278:51549–51556. doi: 10.1074/jbc.M309019200. [DOI] [PubMed] [Google Scholar]
- 24.Perrais M., Pigny P., Ducourouble M.-P., Petitprez D., Porchet N., Aubert J.-P., Van Seuningen I. Characterization of human mucin gene MUC4 promoter: importance of growth factors and proinflammatory cytokines for its regulation in pancreatic cancer cells. J. Biol. Chem. 2001;276:30923–30933. doi: 10.1074/jbc.M104204200. [DOI] [PubMed] [Google Scholar]
- 25.Van Seuningen I., Ostrowski J., Bustelo X. R., Sleath P. R., Bomsztyk K. The K protein domain that recruits the interleukin 1-responsive K protein kinase lies adjacent to a cluster of c-Src and Vav SH3-binding sites. Implications that K protein acts as a docking platform. J. Biol. Chem. 1995;270:26976–26985. doi: 10.1074/jbc.270.45.26976. [DOI] [PubMed] [Google Scholar]
- 26.Cartharius K., Frech K., Grote K., Klocke B., Haltmeier M., Klingenhoff A., Frisch M., Bayerlein M., Werner T. MatInspector and beyond: promoter analysis based on transcription factor binding sites. Bioinformatics. 2005;21:2933–2942. doi: 10.1093/bioinformatics/bti473. [DOI] [PubMed] [Google Scholar]
- 27.Perrais M., Pigny P., Copin M. C., Aubert J. P., Van Seuningen I. Induction of MUC2 and MUC5AC mucins by factors of the epidermal growth factor (EGF) family is mediated by EGF receptor/Ras/Raf/extracellular signal-regulated kinase cascade and Sp1. J. Biol. Chem. 2002;277:32258–32267. doi: 10.1074/jbc.M204862200. [DOI] [PubMed] [Google Scholar]
- 28.Nehra D., Howell P., Williams C. P., Pye J. K., Beynon J. Toxic bile acids in gastro-oesophageal reflux disease: influence of gastric acidity. Gut. 1999;44:598–602. doi: 10.1136/gut.44.5.598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tullai J. W., Schaffer M. E., Mullenbrock S., Kasif S., Cooper G. Identification of transcription factor binding sites upstream of human genes regulated by the phosphatidylinositol 3-kinase and MEK/ERK signaling pathways. J. Biol. Chem. 2004;279:20167–20177. doi: 10.1074/jbc.M309260200. [DOI] [PubMed] [Google Scholar]
- 30.Cereghini S. Liver-enriched transcription factors and hepatocyte differentiation. FASEB J. 1996;10:27–282. [PubMed] [Google Scholar]
- 31.Pontoglio M. Hepatocyte nuclear factor 1, a transcription factor at the crossroads of glucose homeostasis. J. Am. Soc. Nephrol. 2000;11:S140–S143. [PubMed] [Google Scholar]
- 32.Bluteau O., Jeannot E., Bioulac-Sage P., Marques J. M., Blanc J. F., Bui H., Beaudoin J. C., Franco D., Balabaud C., Laurent-Puig P., et al. Bi-allelic inactivation of TCF1 in hepatic adenomas. J. Nat. Genet. 2002;32:312–315. doi: 10.1038/ng1001. [DOI] [PubMed] [Google Scholar]
- 33.Van Wering H. M., Huibregtse I. L., Van der Zwan S. M., de Bie M. S., Dowling L. N., Boudreau F., Rings E. H., Grand R. J., Krasinski S. D. Physical interaction between GATA-5 and hepatocyte nuclear factor-1alpha results in synergistic activation of the human lactase-phlorizin hydrolase promoter. J. Biol. Chem. 2002;277:27659–27667. doi: 10.1074/jbc.M203645200. [DOI] [PubMed] [Google Scholar]
- 34.Boudreau F., Rings E. H., Van Wering H. M., Kim R. K., Swain G. P., Krasinski S. D., Moffett J., Grand R. J., Suh E. R., Traber P. G. Hepatocyte nuclear factor-1 alpha, GATA-4, and caudal related homeodomain protein Cdx2 interact functionally to modulate intestinal gene transcription. Implication for the developmental regulation of the sucrase-isomaltase gene. J. Biol. Chem. 2002;277:31909–31917. doi: 10.1074/jbc.M204622200. [DOI] [PubMed] [Google Scholar]
- 35.Traber P. G., Silberg D. G. Intestine-specific gene transcription. Annu. Rev. Physiol. 1996;58:275–297. doi: 10.1146/annurev.ph.58.030196.001423. [DOI] [PubMed] [Google Scholar]
- 36.Audie J. P., Janin A., Porchet N., Copin M. C., Gosselin B., Aubert J. P. Expression of human mucin genes in respiratory, digestive, and reproductive tracts ascertained by in situ hybridization. J. Histochem. Cytochem. 1993;41:1479–1485. doi: 10.1177/41.10.8245407. [DOI] [PubMed] [Google Scholar]
- 37.Ryffel G. U. Mutations in the human genes encoding the transcription factors of the hepatocyte nuclear factor (HNF)1 and HNF4 families: functional and pathological consequences. J. Mol. Endocrinol. 2001;27:11–29. doi: 10.1677/jme.0.0270011. [DOI] [PubMed] [Google Scholar]
- 38.Wong N. A., Wilding J., Bartlett S., Liu Y., Warren B. F., Piris J., Maynard N., Marshall R., Bodmer W. F. CDX1 is an important molecular mediator of Barrett's metaplasia. Proc. Natl. Acad. Sci. U.S.A. 2005;102:7565–7570. doi: 10.1073/pnas.0502031102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kazumori H., Ishihara S., Rumi M. A., Kadowaki Y., Kinoshita Y. Bile acids directly augment caudal related homeobox gene Cdx2 expression in oesophageal keratinocytes in Barrett's epithelium. Gut. 2006;55:16–25. doi: 10.1136/gut.2005.066209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Van Seuningen I., Pigny P., Perrais M., Porchet N., Aubert J. P. Transcriptional regulation of the 11p15 mucin genes. Towards new biological tools in human therapy, in inflammatory diseases and cancer? Front. Biosci. 2001;6:D1216–D1234. doi: 10.2741/seuning. [DOI] [PubMed] [Google Scholar]
- 41.Weed D. T., Gomez-Fernandez C., Yasin M., Hamilton-Nelson K., Rodriguez M., Zhang J., Carraway K. L. MUC4 and ErbB2 expression in squamous cell carcinoma of the upper aerodigestive tract: correlation with clinical outcomes. Laryngoscope. 2004;114:1–32. doi: 10.1097/00005537-200408001-00001. [DOI] [PubMed] [Google Scholar]
- 42.Dekker J., Rossen J. W., Büller H. A., Einerhand A. W. The MUC family: an obituary. Trends Biochem. Sci. 2002;27:126–131. doi: 10.1016/s0968-0004(01)02052-7. [DOI] [PubMed] [Google Scholar]
- 43.Flejou J. F., Paraf F., Muzeau F., Fekete F., Henin D., Jothy S., Potet F. Expression of c-erbB-2 oncogene product in Barrett's adenocarcinoma: pathological and prognostic correlations. J. Clin. Pathol. 1994;47:23–26. doi: 10.1136/jcp.47.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Walch A., Specht K., Bink K., Zitzelsberger H., Braselmann H., Bauer M., Aubele M., Stein H., Siewert J. R., Hofler H., et al. Her-2/neu gene amplification, elevated mRNA expression, and protein overexpression in the metaplasia–dysplasia–adenocarcinoma sequence of Barrett's esophagus. Lab. Invest. 2001;81:791–801. doi: 10.1038/labinvest.3780289. [DOI] [PubMed] [Google Scholar]
- 45.Moniaux N., Andrianifahanana M., Brand R. E., Batra S.K. Multiple roles of mucins in pancreatic cancer, a lethal and challenging malignancy. Br. J. Cancer. 2004;9:1633–1638. doi: 10.1038/sj.bjc.6602163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Carraway K. L., Perez A., Idris N., Jepson S., Arango M., Komatsu M., Haq B., Price-Schiavi S. A., Zhang J., et al. Muc4/sialomucin complex, the intramembrane ErbB2 ligand, in cancer and epithelia: to protect and to survive. Prog. Nucleic Acid Res. Mol. Biol. 2002;71:149–185. doi: 10.1016/s0079-6603(02)71043-x. [DOI] [PubMed] [Google Scholar]
- 47.Tamada S., Shibahara H., Higashi M., Goto M., Batra S. K., Imai K., Yonezawa S. MUC4 is a novel prognostic factor of extrahepatic bile duct carcinoma. Clin. Cancer Res. 2006;15:4257–4264. doi: 10.1158/1078-0432.CCR-05-2814. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.