Abstract
The adaptive response to amino acid limitation in mammalian cells inhibits global protein synthesis and promotes the expression of proteins that protect cells from stress. The arginine/lysine transporter, cat-1, is induced during amino acid starvation by transcriptional and post-transcriptional mechanisms. It is shown in the present study that the transient induction of cat-1 transcription is regulated by the stress response pathway that involves phosphorylation of the translation initiation factor, eIF2 (eukaryotic initiation factor-2). This phosphorylation induces expression of the bZIP (basic leucine zipper protein) transcription factors C/EBP (CCAAT/enhancer-binding protein)-β and ATF (activating transcription factor) 4, which in turn induces ATF3. Transfection experiments in control and mutant cells, and chromatin immunoprecipitations showed that ATF4 activates, whereas ATF3 represses cat-1 transcription, via an AARE (amino acid response element), TGATGAAAC, in the first exon of the cat-1 gene, which functions both in the endogenous and in a heterologous promoter. ATF4 and C/EBPβ activated transcription when expressed in transfected cells and they bound as heterodimers to the AARE in vitro. The induction of transcription by ATF4 was inhibited by ATF3, which also bound to the AARE as a heterodimer with C/EBPβ. These results suggest that the transient increase in cat-1 transcription is due to transcriptional activation caused by ATF4 followed by transcriptional repression by ATF3 via a feedback mechanism.
Keywords: activating transcription factor, amino acid response element, amino acid transporter, eIF2 phosphorylation, stress response gene, transcriptional regulation
Abbreviations: AARE, amino acid response element; AP-1, activating protein-1; AS, asparagine synthetase; ATF, activating transcription factor; bZIP, basic leucine zipper protein; C/EBP, CCAAT/enhancer-binding protein; ChIP, chromatin immunoprecipitation; CHOP, C/EBP homology protein; CRE, cAMP-response-element; CREB, CRE-binding protein; DMEM, Dulbecco's modified Eagle's medium; eIF2, eukaryotic initiation factor-2; EMSA, electrophoretic mobility-shift assay; ER, endoplasmic reticulum; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; IRES, internal ribosome entry site; LUC, luciferase; MEF, mouse embryonic fibroblast; RT, reverse transcriptase; SNAT2, sodium-coupled neutral amino acid transporter 2; Sp1, stimulating protein-1; UTR, untranslated region; xCT, cystine/glutamate transporter
INTRODUCTION
Amino acids enter cells via transmembrane transporters. These transporters are specific for amino acids with similar chemical properties [1]. The expression of some amino acid transporters, including System A and System Y+, is regulated by amino acid availability [2]. There are four System Y+ transporters: cat-1, cat-2a, cat-2b and cat-3 that transport the cationic amino acids, arginine and lysine [2]. cat-1 is widely expressed in the plasma membrane of mammalian cells, where it takes up these essential amino acids, both for protein synthesis and for the synthesis of NO (nitric oxide) from arginine [3]. Because of the essential functions of cat-1, the regulation of its expression is important for cellular homoeostasis.
Our previous studies have shown that cat-1 gene expression is subject to adaptive regulation at three distinct levels during amino acid starvation: transcription, mRNA degradation and translation (reviewed in [2]). Each of these mechanisms leads to increased cat-1 protein levels during amino acid starvation and suppression in amino acid-sufficient cells. The regulation is not limited to the substrate amino acids, lysine and arginine; limitation of any essential amino acid induces cat-1 gene expression [4]. This feature of cat-1 gene regulation prompted us to demonstrate that the induction of cat-1 gene transcription during limited amino acid supply is part of the general response of cells to nutritional stress [5].
The initial cellular response to nutritional stress caused by amino acid limitation is the phosphorylation of the α subunit of translation initiation factor eIF2 (eukaryotic initiation factor-2) [6]. This phosphorylation decreases the translation of most mRNAs by inhibiting the delivery of the initiator Met-tRNA (methionyl-tRNA), to the initiation complex. However, eIF2α phosphorylation also causes increased translation of some mRNAs, including the transcription factor ATF (activating transcription factor) 4 [7,8], which directly or indirectly induces transcription of genes that promote cell survival or apoptosis [9]. The importance of this transcription factor in the response to unfolded proteins in the endoplasmic reticulum has been demonstrated in ATF4−/− cells, which are impaired in expressing genes involved in amino acid import, glutathione biosynthesis and resistance to oxidative stress [10,11].
We have previously shown that the cat-1 gene has a TATA-less promoter and that it contains an AARE (amino acid response element), TGATGAAAC, within the first exon [4]. AARE-like sequences in the promoters of other genes have been shown to be responsible for enhanced transcription during amino acid limitation (reviewed in [12]). These genes include AS (asparagine synthetase) [13,14], CHOP [C/EBP (CCAAT/enhancer-binding protein) homology protein] [13,15], SNAT2 (sodium-coupled neutral amino acid transporter 2) [16], and the cystine/glutamate transporter, xCT [17]. The AAREs are located either in the promoter (AS, CHOP, xCT), within exons (cat-1) or within introns (SNAT2) and are expected to function in a position-independent manner in transiently transfected chimeric genes [12]. The function of these elements as transcriptional regulators is similar, but not identical in the different genes. For example it has been shown that the AS gene is regulated by an AARE element and a second region that is separated by 11 nucleotides [18]. In contrast, the AARE element in the CHOP gene, which is similar to the AS element, does not require an additional unit [13].
Regulation of transcription from other promoters that contain AARE-like elements during amino acid starvation has been shown to involve ATF4, ATF3 and ATF2, which are members of the bZIP (basic leucine zipper protein) family of transcription factors. These proteins function as homo- or hetero-dimers (reviewed in [19,20]). ATF4 has been reported to function mainly as a transcriptional activator although it may also act as a repressor [19]. ATF3 is a stress-inducible gene and its expression correlates well with cellular damage, such as cardiac ischaemia and liver injury [21,22]. The functions of ATF3 vary from protection to apoptotic, depending on the extracellular signals [23]. ATF4 has been shown to be a transcriptional activator of both CHOP and AS [12] and ATF2 is an activator of CHOP [15], whereas ATF3 represses transcription in transient transfection experiments [24,25]. However, recent reports using cells with mutations in these transcription factors suggest that the mechanism of transcriptional induction of AARE-containing genes may be more complex than previously thought. In fact, studies in ATF3−/− cells showed that CHOP was not induced by amino acid starvation [26], suggesting that promoters with similar AAREs respond differently to stress caused by amino acid starvation.
We have shown in our previous studies that amino acid starvation induces translation of the cat-1 mRNA via a mechanism that requires eIF2 phosphorylation [27]. This phosphorylation indirectly mediates formation of an RNA structure in the cat-1 mRNA leader that functions as an IRES (internal ribosome entry site) under conditions of amino acid starvation [28]. Activation of the IRES requires translation of a small open reading frame within the cat-1 mRNA leader, thus causing a conformational change of the leader, leading to formation of the active IRES [29]. Interestingly, this open reading frame begins within the transcriptional control element TGATGAAAC, which raised the hypothesis that transcriptional and translational control of cat-1 gene regulation are co-ordinated processes. Because translational control requires eIF2 phosphorylation, we decided to determine whether this pathway is also required for transcriptional activation during amino acid limitation.
In the present study we examined the role of eIF2 phosphorylation and the function of the AARE in the stimulation of cat-1 gene transcription during amino acid starvation. We show that eIF2 phosphorylation and the increased ATF4 levels during amino acid starvation parallel transcriptional activation of the cat-1 gene via a mechanism that involves binding of ATF4 to the AARE as a heterodimer, possibly with C/EBPβ. We also show that ATF3, which is stimulated during amino acid starvation by ATF4, binds the cat-1 AARE and represses transcription. These results suggest that the AARE regulates cat-1 transcription during amino acid starvation via a feedback mechanism of ATF4-mediated induction followed by ATF3-mediated repression, and this co-ordinated regulation requires eIF2 phosphorylation.
MATERIALS AND METHODS
Cells and cell culture
C6 rat glioma cells were maintained in DMEM (Dulbecco's modified Eagle's medium, high glucose formulation; Invitrogen) supplemented with 10% fetal bovine serum. Mouse embryonic fibroblasts with and without homozygous deletions of the ATF4 (a gift from T. Townes, Department of Biochemistry and Molecular Biology, University of Alabana at Birmingham, Birmingham, Alabana, U.S.A.) and ATF3 genes (a gift from Tsonwin Hai, Center for Molecular Neurobiology, The Ohio State University, Columbus, Ohio, U.S.A.) and the eIF2α S51A mutation (a gift from R. Kaufman, Department of Biological Chemistry, University of Michigan, Ann Arbor, Michigan, U.S.A.) were maintained in DMEM (high glucose formulation) supplemented with 4 mM glutamine, 10 μM 2-mercaptoethanol and 15% fetal bovine serum [26]. This medium contained all essential amino acids. The culture medium was replaced 12 h prior to transfection.
Fed cells were incubated in DMEM with 10% fetal bovine serum dialysed against PBS. Starved cells were incubated in KRB (Krebs–Ringer bicarbonate) supplemented with 10% dialysed fetal bovine serum (Figures 1, 2 and 3C–3E) or for all other experiments in DMEM that did not contain cysteine, methionine and glutamine (Invitrogen). We have previously demonstrated similar regulation of the cat-1 gene in the two types of medium [4].
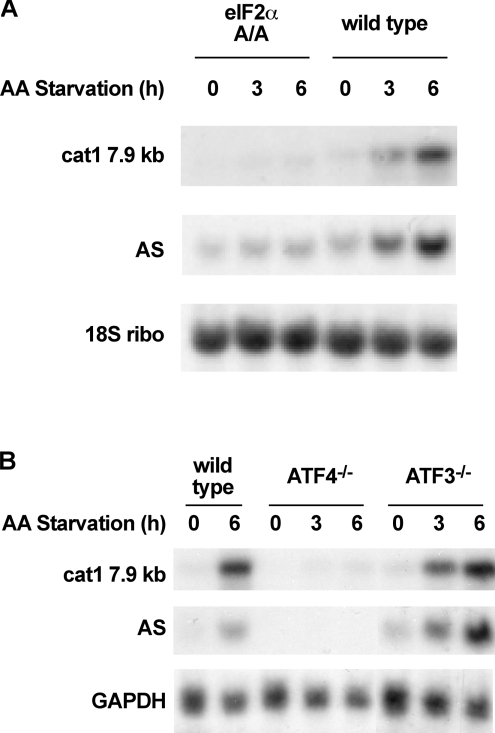
Figure 1. Induction of the cat-1 mRNA level in MEF cells by amino acid starvation is inhibited by mutations in eIF2α and ATF4 but not ATF3.
Wild-type and mutant MEF cells were starved of amino acids for the indicated times and RNA levels were determined by Northern blot analysis as described in the Materials and methods section for the following RNAs: cat-1, AS, GAPDH and 18S ribosomal RNA as a loading control. (A) Analysis of wild-type MEF and cells homozygous for the eIF2α S51A mutation (A/A). (B) Analysis of wild-type MEF and cells with homozygous disruptions in ATF3 or ATF4.
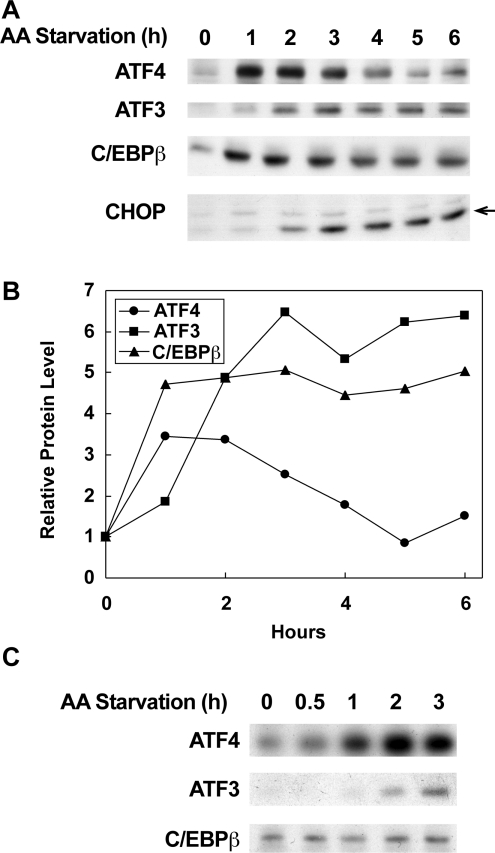
Figure 2. Time course of bZIP transcription factor accumulation during amino acid starvation in rat C6 glioma cells.
(A) C6 glioma cells were starved of amino acids for the indicated times and protein levels in nuclear extracts were determined by Western blotting as described in the Materials and methods section. The arrow indicates the location of the CHOP protein. (B) Protein levels in (A) were quantified and normalized to the level in cells prior to starvation. (C) Protein levels in whole cell extracts were analysed by Western blotting.
Plasmids and transfections
The cat-1 promoter vector, PA1.4/UTR was generated as described in [4]. Mutations in this vector were created using PCR-directed mutagenesis. The 3x-AARE constructs were prepared by ligating an oligonucleotide containing three copies of the AARE into the SalI enhancer site of the pGL3-promoter vector (Promega). The following transcription factors were expressed from transfected plasmids: ATF3 in pCG (from T. Hai), ATF4 in pcDNA3.1 (from J. Alam), and C/EBPβ in pcDNA3.1 [30]. Plasmid DNA was transfected into C6 rat glioma cells using the Fugene® 6 kit (Roche) under normal growth conditions. Transient transfections were performed with 1.5×105 cells/well in 24-well plates with 200 ng of reporter plasmid and 200 ng of transcription factor plasmids. When more than one protein was expressed, the total amount of DNA was kept constant using Escherichia coli β-galactosidase in pcDNA3.1. Stably-transfected cells were generated by co-transfecting an expression vector containing the neomycin gene and selecting in 0.1% G418.
EMSA (electrophoretic mobility-shift assay)
A double-stranded DNA oligonucleotide containing the wild-type cat-1 AARE (Figure 6A) was radiolabelled using T4 polynucleotide kinase and [γ-32P]dATP. His-tagged recombinant ATF3, ATF4 and C/EBPβ were purified from bacteria as described in [31,32]. The labelled oligonucleotide (1.5 pmol) was incubated with 100 ng of recombinant protein in 10 μl of 20 mM Hepes/KOH (pH 7.4), 100 mM NaCl, 1 mM EDTA, 2.5% glycerol and 0.01 mg/ml poly(dI-dC) for 1 h at 4 °C. Competitor oligonucleotides were added in 10-fold excess. Antibodies (5 μg; Santa Cruz Biotechnology) were added to some reaction mixtures and incubated for 1 h at 4 °C prior to addition of the labelled oligonucleotide. Products were resolved on 4% non-denaturing polyacrylamide gels containing 0.5X TBE [44.5 mM Tris/HCl (pH8), 44.5 mM boric acid and 1 mM disodium EDTA]. Gels were dried and analysed using a Phosphorimager (Molecular Dynamics).
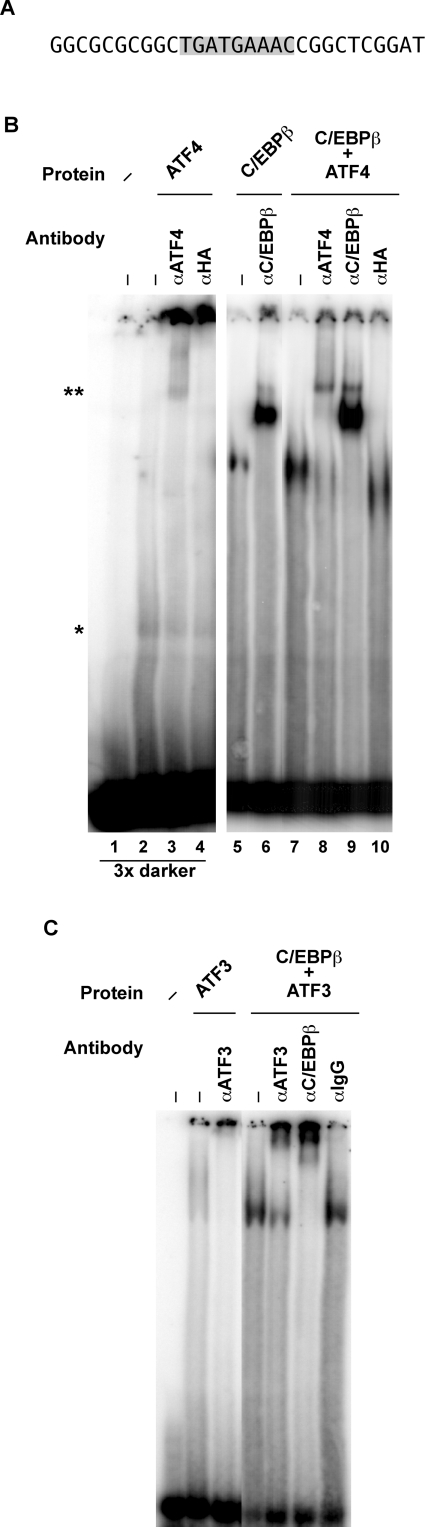
Figure 6. Recombinant ATF4 and C/EBPβ proteins bind to the cat-1 AARE in vitro.
EMSA assays were performed by incubating a 32P-labelled double-stranded oligonucleotide containing the AARE with the indicated recombinant proteins and antibodies. Samples were analysed by gel electrophoresis as described in the Materials and methods section. (A) Sequence of the oligonucleotide probe. (B and C) Samples contained the indicated recombinant proteins (100 ng each) and antibodies as described in the Materials and methods section. In (B), the image for lanes 1–4 is 3 times darker than the other lanes to show the positions of the complex with ATF4 (*) and the supershifted complex (**).
RT (reverse transcriptase)–PCR
For real-time PCR, cDNAs from MEF cells were generated using the antisense primers to cat-1 (5′-GTTCTTGACTTCTTCCCCTGTGG-3′) and β-actin (5′-GGTCATCTTTTCACGGTTGGC-3′). Real-time PCR was performed using an iCycler (BioRad) and SYBR® Green PCR Core Reagents (Applied Biosystems) according to the manufacturer's instructions using the antisense primers and the sense primers: 5′-CTTTGGATTCTCTGGTGTCCTGTC-3′ (cat-1) and 5′-CTGGCACCACACCTTCTACAATG-3′ (β-actin). Samples were analysed in triplicate and the cat-1 signal was normalized to the β-actin signal in each sample.
ChIP (chromatin immunoprecipitation) analysis
C6 glioma cells (2.5×107) were grown on 150-mm dishes and cultured under fed or starved conditions and cross-linked with 1% formaldehyde in medium for 10 min at room temperature (22 °C). The cross-linking was quenched with 0.125 M glycine for 5 min. Cells were then washed with PBS, nuclear extracts were prepared [33], sonicated for 30 s, and placed on ice. The size of the fragments (100–500 bp) was verified by heating an aliquot at 65 °C for 2–3 h, extracting the DNA, and electrophoresis was performed. The extracts were incubated for 3 h with Protein A–agarose beads, centrifuged (3000 g for 5 min at 4 °C) and the supernatants were mixed with immunoprecipitation buffer [0.02% SDS, 2% Triton X-100, 4 mM EDTA, 40 mM Tris/HCl (pH 8) and 300 mM NaCl] and 2 μg of antibodies to ATF3, ATF4, C/EBPβ, Sp1 (stimulating protein-1) or RNA polymerase II (N-20; Santa Cruz Biotechnology) was added. These samples were rotated overnight at 4 °C. Then Protein A and Protein G Dynabeads (Dynal Biotech) were added and samples were incubated for 3 h at room temperature with rotation. Beads were collected with a magnet, washed once with immunoprecipitation buffer, once with wash buffer I [0.1% SDS, 1% Triton X-100, 2 mM EDTA, 40 mM Tris/HCl (pH 8) and 50 mM NaCl], and once with wash buffer II [0.1% SDS, 1% Triton X-100, 2 mM EDTA, 40 mM Tris/HCl (pH 8) and 500 mM NaCl] and then incubated with proteinase K (100 μg/ml) for 2 h at 50 °C. Samples were then incubated at 65 °C for 5–6 h to reverse the DNA–protein cross-links. DNA was purified (QIAquick gel extraction kit, Qiagen) and used for PCR analysis. Fragments of the cat-1 promoter and 3′-UTR (untranslated region), as well as fragments of the β-actin and myogenin promoters were amplified (see the Supplementary Table at http://www.BiochemJ.org/bj/402/bj4020163add.htm for primer sequences and PCR parameters).
Analytical procedures
mRNAs were detected by Northern blotting using 32P-labelled DNA probes for the chimeric cat-1/LUC (luciferase) mRNA, 18S ribosomal RNA and endogenous mRNAs [cat-1 and GAPDH (glyceraldehyde-3-phosphate dehydrogenase)] as previously described [4]. Autoradiograms were scanned and quantified using NIH (National Institutes of Health) Image software. LUC activity in cell extracts was assayed using the Promega LUC assay system in a 96-well microplate reader. ATF3, ATF4, CHOP and C/EBPβ were detected on Western blots using polyclonal antibodies from Santa Cruz Biotechnology, a horseradish peroxidase-con-jugated secondary antibody and a chemiluminescent substrate.
RESULTS
The induction of cat-1 mRNA by amino acid starvation requires phosphorylation of eIF2α and expression of the transcription factor ATF4
We have previously shown that cat-1 gene transcription is induced by amino acid starvation and we have identified an AARE element, which is located in the first exon of the gene, as a sequence required for this induction [4]. To extend these studies, we examined the signalling pathways and transcription factors that regulate transcription from the AARE. First, we examined the role played by phosphorylation of the translation initiation factor, eIF2α, because phosphorylation of this protein by GCN2 (positive general control of transcription-2) kinase is a key signal in the response to amino acid deprivation [34]. This was accomplished by comparing the induction of cat-1 mRNA expression by amino acid starvation in wild-type mouse embryonic fibroblasts and cells with a mutation in eIF2α [35] that changes Ser51, which is the target of phosphorylation, to an alanine residue (eIF2α A/A). The 7.9 kb cat-1 mRNA, which is the sole species found in mice, was induced by starvation in wild-type but not the mutant cells (Figure 1A), indicating that phosphorylation of eIF2α is required for induction. Similar results were seen for AS, whose induction is expected to require eIF2α phosphorylation [10,36].
Next, we examined the role of the bZIP transcription factors ATF3 and ATF4 in the regulation of cat-1 mRNA expression. Induction of eIF2α phosphorylation induces the translation of ATF4 and C/EBPβ mRNAs. Increased levels of ATF4 protein regulate transcription to cause increased expression of ATF3 and other bZIP factors. The roles of ATF3 and ATF4 in the induction of cat-1 mRNA expression were examined using mouse embryo fibroblasts with disruptions in these genes. cat-1 mRNA was induced by amino acid starvation in wild-type cells. However, no induction of cat-1 mRNA was seen in cells with mutant ATF4 (Figure 1B). In contrast, disruption of the ATF3 gene had no effect on cat-1 mRNA expression during 6 h of starvation. In agreement with previous biochemical studies [36], it is shown here genetically, that the expression of AS was induced in wild-type but not in ATF4−/− cells whereas amino acid starvation stimulated AS expression in ATF3−/− cells (Figure 1B). In contrast with the regulation of AS and cat-1, the induction of SNAT2 mRNA levels by starvation was increased in both wild-type and mutant cells [37] and no induction of CHOP mRNA was observed in either mutant cell line [26], consistent with differential regulation of genes that contain AAREs. These results demonstrate that ATF4 is required for activation of cat-1 gene expression by amino acid starvation. In contrast ATF3 is not required for induction, although this protein does have an inhibitory effect on cat-1 gene transcription (see below).
As the next step in understanding the role of bZIP transcription factors in the regulation of cat-1 transcription, we examined the levels of these proteins during amino acid starvation [15,38]. Western blot analysis of nuclear extracts showed that the level of ATF4 increased by 3.5-fold after 1 h of starvation and then declined slowly after 2 h, reaching baseline levels by 5 h (Figures 2A and 2B). The level of ATF3 increased more slowly than ATF4 during starvation but the magnitude of the increase was greater (6-fold). It reached a peak at 3 h and remained elevated. Finally, the level of the 35 kDa form of C/EBPβ (also termed LAP), which is a transcriptional activator, also increased by 1 h and remained elevated. These results are consistent with previous findings [38] and with the idea that changes in the levels of these transcription factors regulate cat-1 transcription. Analysis of these proteins in whole cell extracts revealed that the levels of ATF3 and ATF4 increased during starvation with time courses similar to those seen in nuclear extracts (Figure 2C). In contrast, the level of the 35 kDa form of C/EBPβ in whole cell extracts did not change, consistent with the idea that phosphorylation of this protein [39,40] may regulate its activity during amino acid deprivation.
To obtain direct proof for the binding of these transcription factors to the cat-1 gene in cultured cells, we performed ChIP analysis. Chromatin from starved and fed cells was immunoprecipitated with antibodies to each protein and a DNA fragment containing the AARE of the endogenous cat-1 gene was amplified by PCR (Figure 3A). A weak band was seen in samples that did not contain antibody or were immunoprecipitated with pre-immune serum (results not shown), confirming the specificity of the immunoprecipitations. When samples were precipitated with an antibody to the RBP2 subunit of RNA polymerase II, there was a weak signal from fed cells and a strong band at 2 h, with the signal returning to baseline by 9 h. This shows for the first time the transient activation of cat-1 gene transcription during amino acid starvation.
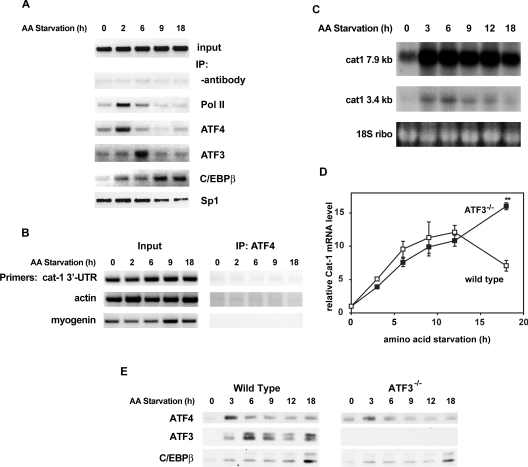
Figure 3. bZIP transcription factors associate with the cat-1 promoter during amino acid starvation.
(A and B) C6 glioma cells were starved of amino acids for the indicated times and ChIP was performed with antibodies to RNA polymerase II (Pol II), Sp1, ATF4, ATF3 and C/EBPβ as described in the Materials and methods section. In (A), PCR was used to amplify bases −37 to +156, relative to the cat-1 transcription start site. Samples that used input DNA as a template are also shown. In (B), samples precipitated with ATF4 antibody were amplified with the indicated primers. Representative gels are shown. (C) Time course of endogenous cat-1 mRNA accumulation during amino acid starvation. Cells were starved of amino acids for the indicated times and the 3.4 and 7.9 kb forms of the cat-1 mRNA were determined by Northern blot analysis. These two forms arise by the use of alternate polyadenylation sites. 18S ribosomal RNA was measured by ethidium bromide staining as a loading control. (D) ATF3 is required for a decrease in cat-1 transcription late in starvation. Wild-type and ATF3−/− MEF cells were starved for amino acids and the level of cat-1 mRNA was analysed by reverse transcription and real-time PCR as described in the Materials and methods section. The means±S.E.M. from three experiments is shown. **Significantly different from wild type (P<.001). (E) Wild-type and ATF3−/− MEF cells were starved of amino acids for the indicated times and protein levels in nuclear extracts were determined by Western blotting as described in the Materials and methods section.
We also examined the association of transcription factors with the cat-1 promoter. The transcription factor Sp1 interacted with the cat-1 promoter at all times (Figure 3A), consistent with our finding that the TATA-less promoter of the cat-1 gene supports basal transcription via Sp1 (results not shown). In contrast, the interaction between ATF4 and the promoter was induced by amino acid starvation with a pattern similar to that observed for RNA polymerase II. The time course of association was also similar to that observed for the level of ATF4 protein in the nucleus (Figures 2B and 2C). This suggests that the level of ATF4 may control its association with the promoter and strongly supports the role of ATF4 as a regulator of cat-1 transcription. Amino acid starvation also induced association of ATF3 and C/EBPβ with the cat-1 promoter. ATF3 showed strong association at 6 h, after the peak of transcription, and then declined. C/EBPβ was bound throughout the time course, with strongest binding at 9–18 h. For both of these proteins, the time course of association with the promoter was different than the time course of protein accumulation. This suggests that the amount of these proteins is not the sole factor that controls their association with the promoter.
Several control experiments were performed to demonstrate the specificity of the ChIP experiments (Figure 3B). Samples were precipitated with ATF4 antibody and amplified with primers for the 3′-UTR of the cat-1 gene (78 kb downstream of the transcription start site), the β-actin promoter or the myogenin promoter. In all cases very weak signals were seen and amino acid starvation did not cause a change in the signals. These results demonstrate the specificity of our ChIP result and support our conclusion that cat-1 transcription is regulated by specific interactions of bZIP transcription factors with the promoter.
The transient transcriptional activation of the cat-1 gene during amino acid starvation is also demonstrated by the accumulation of the 3.4 kb cat-1 mRNA in rat C6 glioma cells (Figure 3C). The rat gene contains an alternative polyadenylation site that generates two mRNAs. The longer 7.9 kb mRNA contains an element in the 3′-UTR that stabilizes this species during amino acid starvation [33]. The shorter 3.4 kb species lacks this element and has a short half-life. Therefore its levels are a more accurate reflection of the rate of transcription. It is shown in the present study that the 3.4 kb mRNA increased at 3–6 h of starvation, suggesting that transcriptional activation occurs at this time and declined thereafter (Figure 3C). Because the mouse gene does not have an alternative polyadenylation site, we only detected the longer mRNA in mouse fibroblasts (Figure 1).
ATF3 has been suggested to act as a transcriptional repressor of the AS gene and to be required for transcriptional activation of CHOP [26], both genes containing similar AAREs [41]. Consequently, we tested whether ATF3 acts as an activator or inhibitor of cat-1 transcription by comparing the time course of cat-1 mRNA accumulation during amino acid starvation in wild-type and ATF3−/− cells using real time PCR (Figure 3D). In wild-type cells, the cat-1 mRNA level increased during starvation, reached a peak at 9 h and then declined. In the mutant cells, the mRNA level showed a similar increase during the first 9 h, but then continued to increase, in agreement with our hypothesis that the late induction of ATF3 repressed cat-1 transcription. In order to demonstrate that the persistence of cat-1 mRNA levels in ATF3−/− cells was not due to overexpression of stimulatory transcription factors, we analysed nuclear extracts of ATF3−/− and wild-type cells by Western blotting (Figure 3E). Levels of ATF4 and C/EBPβ were similar in mutant and wild-type cells, with ATF4 showing a transient increase and then declining, whereas C/EBPβ increased at later times. This result supports the conclusion that the increased expression of cat-1 mRNA in mutant cells is due to the absence of repression by ATF3. In wild-type MEF cells, ATF3 appears as two bands in the Western blot. These could represent proteins synthesized from alternatively spliced mRNAs, which have been reported in cultured human cells [25]. In contrast, only one ATF3 band is seen in the blots of C6 glioma cells (Figure 2), possibly reflecting a difference between murine and rat cells.
cat-1 transcription is activated by ATF4 and C/EBPβ and is repressed by ATF3
In order to carry out a detailed analysis of the role of the AARE and the bZIP transcription factors in the regulation of the cat-1 promoter, we studied chimeric constructs containing the cat-1 promoter in transfected cells. We also examined the effects of expressing individual transcription factors from transfected plasmids. These studies used a construct, PA1.4/UTR that contains 1.4 kb of DNA upstream of the transcription start site and the 5′-UTR of the cat-1 mRNA coupled to a LUC open reading frame (Figure 4A). Overexpression of ATF4 or C/EBPβ caused a 6- to 8-fold increase in LUC activity expressed from wild-type PA1.4/UTR (Figure 4B). Transfection with larger amounts of ATF4 did not give this increase (results not shown). This is consistent with our previous report [4] and agrees with the idea that high levels of ATF4 can cause ‘squelching’ by sequestering elements of the transcription machinery. When ATF4 and the 35 kDa activating form of C/EBPβ were expressed together, there was an additional increase in reporter activity, suggesting that they work in an additive or synergistic fashion. In contrast, ATF3 suppressed transcriptional activation of the cat-1 promoter by ATF4 and C/EBPβ. The negative effect of ATF3 was dominant, because low levels of transcription were seen when this protein was expressed together with ATF4, C/EBPβ or the two proteins together. ATF3 inhibition was seen at low levels of transfected DNA (50–200 ng), suggesting that this is authentic inhibition, rather than transcriptional squelching.
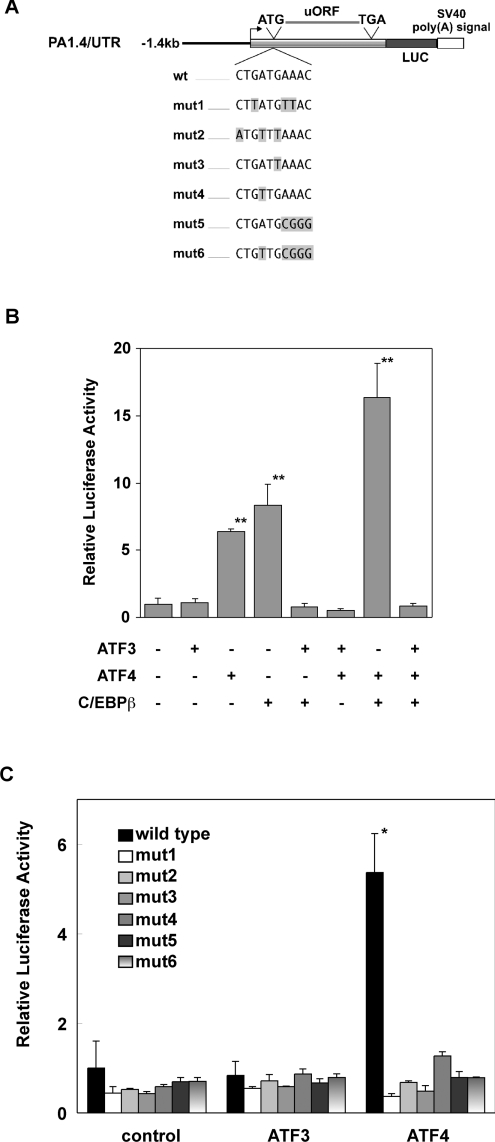
Figure 4. bZiP transcription factors regulate cat-1 gene transcription through the AARE.
(A) Constructs used in the present study. The PA1.4/UTR constructs contain 1.4 kb of cat-1 genomic DNA upstream of the transcription start site, the entire cat-1 mRNA leader and the LUC open reading frame. The transcription start site (arrow) and the open reading frame in the mRNA leader (upstream open reading frame) are shown. Point mutations are shown by shaded boxes. (B and C) C6 cells were transiently transfected with wild-type PA1.4/UTR (B) or the indicated PA1.4/UTR constructs (C) and plasmids encoding transcription factors. LUC activity was measured in extracts from fed cells. The graphs show the means±S.E.M. of LUC activity/μg protein from three independent samples normalized to the values in cells transfected with wild-type PA1.4/UTR without transcription factors. Significant differences from samples without transfected transcription factor are indicated: *P<0.01, **P<0.001.
The AARE is essential for induction by amino acid starvation
We have previously shown that the cat-1 AARE, which is located in the first exon of the gene, is required for the activation of transcription by amino acid starvation [4]. In order to determine whether the effects of these transcription factors are mediated by the cat-1 AARE, we examined whether these proteins affected transcription from constructs with mutant AAREs (Figure 4C). ATF4 activated transcription in the wild-type construct but not in mut1–mut6. This result indicates that the regulation of cat-1 transcription by ATF4 requires the entire AARE because mutations in the 5′ half (mut2–mut4) and 3′ half (mut5 and mut6) abolish the stimulation by ATF4. Mutations within the AARE do not affect translation of a LUC reporter in chimeric mRNA during amino acid sufficiency [4].
ATF4 is necessary for activation of cat-1 transcription during amino acid starvation
The results in Figure 1(B) show that ATF4 is required for the induction of cat-1 gene transcription during amino acid starvation. As an additional test of this conclusion, we examined transcription from the chimeric cat-1 constructs in ATF4−/− MEF cells. Cells were transiently transfected with PA1.4/UTR and LUC expression was measured after 9 h in fed or amino acid-starved conditions. In wild-type cells, LUC activity was increased 2.5-fold by starvation (results not shown), consistent with our previous observation that the rat cat-1 promoter is induced in mouse cells. In contrast, LUC activity in amino acid-starved ATF4−/− cells was only 54% that in fed cells, supporting the conclusion that ATF4 is required for induction of cat-1 transcription by amino acid starvation.
The cat-1 AARE confers regulation on a heterologous promoter
Our studies suggest that the AARE is essential for regulation of cat-1 transcription by amino acid starvation. To prove that the AARE is responsible for this regulation, its ability to function in a heterologous promoter was tested. Three tandem repeats of this element were placed into a vector that contains an SV40 promoter and a LUC reporter. The resulting 3x-AARE construct has the AARE element inserted downstream of the LUC open reading frame (Figure 5A). This construct was transfected into C6 glioma cells and the effect of amino acid starvation on AARE-mediated transcription was determined by performing LUC assays. LUC expression from the 3x-AARE construct was increased 3.5-fold after 9 h of amino acid starvation (results not shown). In contrast, starvation did not increase LUC expression from the vector or from the 3x-AARE constructs carrying the AARE mutations mut1–mut5. We also determined the mRNA levels by RT–PCR. mRNA from the 3x-AARE constructs increased by 3-fold during 6 h of starvation, whereas mRNA constructs containing the mutant AAREs decreased (results not shown).
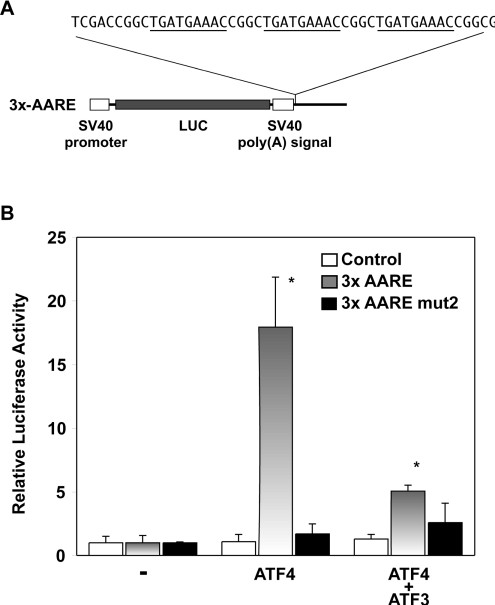
Figure 5. Transcription of a heterologous promoter containing three copies of the cat-1 AARE is induced by amino acid starvation.
(A) The 3x-AARE construct contains three repeats of the AARE (underlined) in the enhancer site of the pGL3-promoter vector. Point mutations mut1–mut6 were also introduced into the AARE repeats. (B) C6 glioma cells were transiently transfected with the indicated 3x-AARE constructs and expression plasmids. The LUC activity in fed cells was measured and normalized to the level in cells transfected with the pGL3-promoter vector without cat-1 promoter sequences. The graph shows relative LUC activity normalized to samples without transfected transcription factor cDNA. Data from three independent samples are shown. *Significantly different from cells without transcription factor (P<0.01).
As a final test of the function of the cat-1 AARE in a heterologous promoter, we examined the regulation of the 3x-AARE construct by transfected transcription factors. Transcription from the 3x-AARE construct was strongly activated by ATF4 (Figure 5B). In addition, as expected, ATF3 prevented transcriptional activation by ATF4. In agreement with our findings on the AARE in the cat-1 promoter, the mut2 AARE did not support transcriptional activation in the heterologous promoter. Unfortunately, we were unable to test the regulation of the AARE by C/EBPβ because transcription from the control vector was activated by this protein, indicating that there are binding sites on this vector (results not shown). These results demonstrate that much of the regulation of the cat-1 promoter by amino acid starvation is due to the AARE. This sequence confers responsiveness to amino acid starvation on a heterologous promoter. Moreover, it is likely that the AARE acts as an enhancer because it can function >1 kb downstream of the transcription start site. Finally, the regulation of the 3x-AARE construct by transfected ATF4 and ATF3 strongly suggests that these proteins regulate cat-1 transcription by binding to this element.
ATF3 and ATF4 bind to the cat-1 AARE in vitro as heterodimers with C/EBPβ
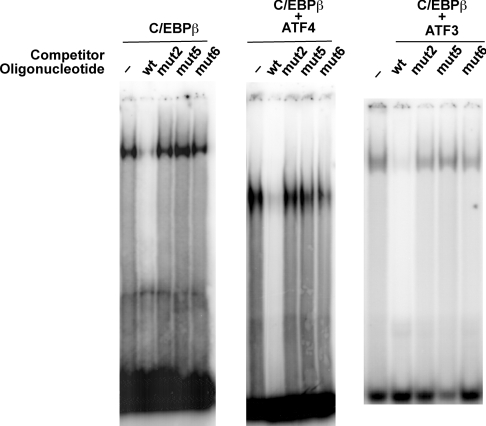
Our results suggest that transcription from the cat-1 promoter is regulated by binding of ATF4 and ATF3 transcription factors to the AARE. bZIP proteins bind to DNA as dimers. Homodimers of the CREB (cAMP-response-element-binding protein)/ATF family bind preferentially to a palindrome of two inverted half sites [19]. In contrast, heterodimers of CREB/ATF and C/EBP family members can bind CRE (cAMP-response-element) 5′-half sites with the 3′-half being a CRE-related site. The cat-1 AARE is one of these CRE/ATF composite sites, which could bind heterodimers more stably than homodimers [42]. We therefore expected cat-1 transcriptional control to occur via binding of heterodimers. To test this hypothesis, we determined the sequence-specific binding of ATF4, ATF3 and C/EBPβ to a radioactive oligonucleotide containing the AARE (Figure 6A) using an EMSA. Supershifts with antibodies were used to confirm the specificity of the complexes.
ATF4 bound very weakly to the cat-1 oligonucleotide (Figure 6B). A faint band that was supershifted specifically with an anti-ATF4 antibody was seen only with long exposure, suggesting that homodimers of ATF4 are unlikely to regulate cat-1 gene transcription via the AARE. Stronger binding was observed for C/EBPβ (Figure 6B). This complex was specific because it was supershifted by an antibody to C/EBPβ but not by antibodies to ATF3 or actin (results not shown). A strong complex was also observed when C/EBPβ and ATF4 were added together. This complex is most likely due to binding of an ATF4–C/EBPβ heterodimer because most of the complexes were supershifted by antibodies to both proteins, but not by an irrelevant antibody. We conclude that because ATF4 binds strongly to the cat-1 AARE with C/EBPβ, but shows weak binding by itself, it is likely that ATF4 stimulates cat-1 transcription in vivo by binding as a heterodimer with another bZIP transcription factor, most likely from the C/EBP family.
Similar results were observed with ATF3 and C/EBPβ (Figure 6C). ATF3 bound to the AARE weakly by itself, but formed a strong complex with C/EBPβ. The existence of a heterodimeric complex was established by supershift experiments, which resulted in large complexes with antibodies that migrated close to the top of the gel. The complexes with ATF3 and C/EBPβ were completely supershifted by an anti-C/EBPβ antibody and partially supershifted by an anti-ATF3 antibody. This suggests that the complexes are mixtures of ATF3–C/EBPβ heterodimers and C/EBPβ homodimers.
The specificity of the DNA–protein complexes was confirmed in competition experiments with wild-type oligonucleotides and with mutants (mut2, mut5 and mut6), which do not support ATF4-mediated transcription in transfected cells (Figure 4C and [4]). These experiments showed that the binding of C/EBPβ as well as ATF4–C/EBPβ and ATF3–C/EBPβ heterodimers was competed by a 10-fold excess of wild-type oligonucleotide (Figure 7). In contrast, no competition was seen with mut2 or mut5, and mut6 competed weakly for C/EBPβ and ATF4–C/EBPβ, indicating that the proteins bind specifically to the AARE. Because mut2 has changes in the left half of the AARE, whereas mut5 has changes in the right half, these results demonstrate that the entire AARE is required for sequence-specific binding of these proteins. In contrast with these results, homodimers of either ATF4 or ATF3 bound to the mutant AAREs, supporting the conclusion that the weak binding of these transcription factors is not sequence specific (results not shown).
Figure 7. Homodimers and heterodimers of ATF4 and ATF3 with C/EBPβ bind wild-type but not mutant AARE's.
EMSA assays were performed by incubating recombinant proteins with the 32P-labelled oligonucleotide containing the wild-type AARE and a 10-fold excess of the indicated wild-type and mutant unlabelled oligonucleotides. Experimental analysis is as described in the legend to Figure 6 and in the Materials and methods section.
DISCUSSION
Cells respond to the stress of amino acid starvation by activating a gene expression program that either protects them from stress or leads to apoptosis [43,44]. This adaptive response involves the phosphorylation of eIF2α, which causes translation attenuation of most messages and increases translation of some transcription factors, including ATF4 and the C/EBPs [7,8,45]. The increased ATF4 induces transcription of genes that promote either protection from stress (e.g. amino acid transporters, proteins involved in glutathione biosynthesis and in resistance to oxidative stress) or cell death [e.g. transcription factors such as CHOP and TRB3 (tribbles 3)] [46].
In the present study, we show that induction of cat-1 gene transcription during amino acid starvation requires phosphorylation of eIF2, which is a master regulator of cellular stress responses. This result, combined with our previous studies, shows that this signalling pathway controls cat-1 gene expression in two ways during amino acid starvation. eIF2 phosphorylation causes increased cat-1 gene transcription (the present study) and also increases cat-1 mRNA translation by stimulating the activity of the inducible IRES [29]. It is very interesting that cat-1 transcriptional and translational induction use the same sequence in the DNA and mRNA respectively. The AARE (TGATGAAAC), which is in the first exon of the gene, contains the upstream open reading frame initiation codon; mutation of A to T in the ATG initiation codon abolished transcriptional control (mut4) and translational control [27]. A third mode of regulation increases the lifetime of the cat-1 mRNA during amino acid starvation [33]. Together, these mechanisms stimulate cat-1 protein expression during starvation at a time when global protein synthesis is inhibited.
We also show that the induction of cat-1 transcription during amino acid starvation is driven by ATF4 and that ATF4 induces cat-1 transcription by associating with the AARE as a heterodimer, most likely with one of the C/EBPs. Although ATF4 and C/EBPβ can activate cat-1 gene transcription in vivo and these proteins can bind to the cat-1 AARE as heterodimers in vitro, we do not know whether C/EBPβ is the interaction partner that functions with ATF4 to induce transcription during amino acid starvation. ATF4 forms heterodimers with many members of the AP-1 (activating protein-1) and C/EBP family [47]. Moreover, C/EBPβ is subject to post-translational modifications in vivo [39,40], so it is also possible that formation of ATF–C/EBP heterodimers is regulated by these post-translational modifications.
In contrast with the positive effect of ATF4 on cat-1 gene transcription, ATF3 attenuates the stimulation of transcription by ATF4 during prolonged starvation, most likely by replacing ATF4 on the AARE. Transcriptional activation of the cat-1 promoter during amino acid limitation is transient. In the present study we show that ATF4 drives increased cat-1 transcription and ATF3 attenuates this increase at later times. Because ATF4 increases the expression of ATF3 [26], the attenuation of transcriptional activation by ATF3 is a remarkable negative feedback mechanism. This feedback can limit cat-1 mRNA accumulation under limited amino acid supply and could spare ATF4 for transcriptional control of target genes [10]. Our data exclude the possibility that ATF3 stimulates cat-1 transcription during the early stages of amino acid starvation, as has been reported for CHOP [25,26].
The association of ATF4 and ATF3 with the cat-1 AARE in ChIP assays paralleled their levels in the nucleus (Figure 2). Therefore the levels of these transcription factors are probably key factors in the regulation of cat-1 gene transcription during amino acid starvation. However, C/EBPβ binding was highest at 9–18 h of starvation when ATF4 and ATF3 binding and cat-1 transcription were low. C/EBPβ homodimers bind to the AARE in a sequence-specific manner (Figure 7), so it is possible that binding of C/EBPβ homodimers to the cat-1 promoter during prolonged starvation may maintain a low level of transcription. It is also possible that C/EBPβ inhibits transcription under these conditions by binding as a heterodimer with another bZIP protein.
The cat-1 AARE works as an enhancer-like element, capable of inducing transcription from a heterologous promoter when it is placed >1 kb downstream of the transcription start site. Because the AARE functions in a heterologous promoter, other cat-1 sequences are not required for the induction. However, the extent of activation in the heterologous promoter (3-fold) was less than the activation in the endogenous promoter (5- to 15-fold). Consequently, the position of the AARE or other elements of the TATA-less cat-1 promoter may play a role in the induction.
Activation of transcription by AARE elements has been reported for other genes, including AS [13,14], CHOP [13,15], SNAT2 [16], the cystine/glutamate transporter xCT [17] and ATF3 [48]. All AAREs have similar sequences. At the 5′-end there is TGAT, which is half a CRE or ATF/CRE element [49]. At the 3′-end AAREs have the consensus sequence G(C/A)AA(C/T), which is half a C/EBP binding site. It is therefore believed that AAREs are composites of ATF and C/EBP half-sites. An interesting feature of the AAREs is that their sequences are not identical, differing by one or two nucleotides. In addition, the regulatory region in the AS gene is more complex, containing an AARE and a second element (GTTACA), which are separated by 11 nucleotides [18].
The AAREs are recognized by the bZIP family of proteins [12], which contain a transactivating domain, a basic DNA-binding domain and a leucine-zipper dimerization domain. Subfamilies within this group are AP-1, CREB/ATF and C/EBP. ATF4 and ATF3 are members of the CREB/ATF family [19]. Formation of homo- or hetero-dimers by these proteins creates functional diversity [47]. Moreover, their ability to form stable dimers upon binding to the DNA target is probably a key determinant in discriminating between similar target sequences [42]. Podust et al. [42] suggested that the disordered basic region of C/EBPβ may allow binding of ATF4–C/EBPβ heterodimers to a broad range of half site CREs, whereas binding of heterodimers to a CRE site was preferred over homodimers. Our conclusion that cat-1 transcription is controlled by binding of ATF4 and ATF3 to the AARE as heterodimers with a C/EBP family member is in agreement with these reports.
Transcriptional control of the cat-1 gene by amino acid starvation is similar to the AS gene but different than the CHOP and SNAT2 genes which have AAREs similar to cat-1. In the present study we show genetically that cat-1 and AS induction requires eIF2α phosphorylation. Their induction is also dependent on ATF4 and independent of ATF3. CHOP is not induced in ATF4−/− cells but it is also not induced in ATF3−/− cells. However, its induction requires phosphorylation of ATF2 [15]. On the other hand, SNAT2 is induced in ATF3−/− and ATF4−/− cells [37] even though it has the same AARE as CHOP. It is therefore concluded that subtle differences in the AAREs or interactions with other regulatory elements may confer specificity for binding of bZIP heterodimers that regulate transcription during stresses that involve eIF2α phosphorylation.
Transcriptional control via ATF4, C/EBPβ and ATF3 has important common features. All three proteins are induced by stresses such as amino acid limitation, ER (endoplasmic reticulum) stress or oxidative stress by a mechanism that involves eIF2α phosphorylation [34]. It is therefore expected that cat-1 and other stress-response genes will be regulated by similar mechanisms. The exact nature of the regulation of each gene will depend on the sequence of the ATF composite site, which will specify the bZIP dimers that bind to the AARE. Genes that are regulated in this way, including cat-1, AS, CHOP and SNAT2 are discussed above. Other types of regulation by bZIP factors have also been observed. For example, ATF4 can act as a negative regulator of CRE-dependent transcription [19]. A second example is the regulation of Grp78 (BiP) during the ER stress response [50]. Grp78 transcription increases during ER stress via a mechanism that involves binding of ATF6 to an ER stress response element [51]. An AARE-like element (TGACGTGA) within the promoter plays a role in induction by binding ATF4 [50]. This site consists of two halves of an ATF/CREB site.
Our findings suggest that gene activation during stress is regulated by the concentrations of ATF4, ATF3 and C/EBPs. In cells with high C/EBP levels, transcription of genes that contain canonical C/EBPβ sites will be favoured. When the concentration of the ATF proteins increases, heterodimer formation will be favoured and the transcription of genes with composite ATF–C/EBP sites will be targeted. This explanation is supported by the transient induction of these genes during stress.
In conclusion, we show that cat-1 is a transcriptional target of the adaptive response to amino acid starvation. The gene is expressed in nearly all mammalian tissues except the liver [2] and it is an essential gene, as knockout animals die soon after birth [52]. Consequently, the regulation that we have characterized in cultured cells is likely to be part of the stress response of cat-1 expressed in a wide range of cells and tissues. It is expected that the details of the stress response will vary, depending on the repertoire of transcription factors expressed in each cell type. The activation of both cat-1 transcription (the present study) and translation [28] depend on the phosphorylation of eIF2α. As would be expected for cells with limited amino acids, gene activation is transient and involves a sophisticated network of transcription factors that heterodimerize and control transcription by discriminating between DNA-binding sites with small sequence variations. Studies on individual genes containing AAREs will increase our understanding on how these elements control transcription during different stress conditions that involve eIF2 phosphorylation.
Online data
Acknowledgments
We thank Dr Tsonwin Hai for providing expression plasmids, Dr Tim Townes, Dr Tsonwin Hai, and Dr Randal Kaufman for ATF4−/−, ATF3−/− and eIF2α A/A mouse embryonic fibroblasts respectively. Dr Michael Kilberg and Dr Hong Chen provided valuable advice about the ChIP assay. We also thank Haiyan Liu for preparing some of the DNA constructs. This work was supported by RO1-DK60596, RO1-DK53307 to M. H. and by RO1-CA103867 to C.-M. C. A. B. L. was supported by 5T32-DK07319.
References
- 1.Palacin M., Estevez R., Bertran J., Zorzano A. Molecular biology of mammalian plasma membrane amino acid transporters. Physiol. Rev. 1998;78:969–1054. doi: 10.1152/physrev.1998.78.4.969. [DOI] [PubMed] [Google Scholar]
- 2.Hatzoglou M., Fernandez J., Yaman I., Closs E. Regulation of cationic amino acid transport: the story of the CAT-1 transporter. Annu. Rev. Nutr. 2004;24:377–399. doi: 10.1146/annurev.nutr.23.011702.073120. [DOI] [PubMed] [Google Scholar]
- 3.Li C., Huang W., Harris M. B., Goolsby J. M., Venema R. C. Interaction of the endothelial nitric oxide synthase with the CAT-1 arginine transporter enhances NO release by a mechanism not involving arginine transport. Biochem. J. 2005;386:567–574. doi: 10.1042/BJ20041005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fernandez J., Lopez A. B., Wang C., Mishra R., Zhou L., Yaman I., Snider M. D., Hatzoglou M., Hatzolgou M. Transcriptional control of the arginine/lysine transporter, cat-1, by physiological stress. J. Biol. Chem. 2003;278:50000–50009. doi: 10.1074/jbc.M305903200. [DOI] [PubMed] [Google Scholar]
- 5.Kimball S. R., Jefferson L. S. Amino acids as regulators of gene expression. Nutr. Metab. 2004;1:3. doi: 10.1186/1743-7075-1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jefferson L. S., Kimball S. R. Amino acid regulation of gene expression. J. Nutr. 2001;131:S2460–S2466. doi: 10.1093/jn/131.9.2460S. [DOI] [PubMed] [Google Scholar]
- 7.Lu P. D., Harding H. P., Ron D. Translation reinitiation at alternative open reading frames regulates gene expression in an integrated stress response. J. Cell Biol. 2004;167:27–33. doi: 10.1083/jcb.200408003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vattem K. M., Wek R. C. Reinitiation involving upstream ORFs regulates ATF4 mRNA translation in mammalian cells. Proc. Natl. Acad. Sci. U.S.A. 2004;101:11269–11274. doi: 10.1073/pnas.0400541101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harding H. P., Novoa I., Zhang Y., Zeng H., Wek R., Schapira M., Ron D. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol. Cell. 2000;6:1099–1108. doi: 10.1016/s1097-2765(00)00108-8. [DOI] [PubMed] [Google Scholar]
- 10.Harding H. P., Zhang Y., Zeng H., Novoa I., Lu P. D., Calfon M., Sadri N., Yun C., Popko B., Paules R., et al. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol. Cell. 2003;11:619–633. doi: 10.1016/s1097-2765(03)00105-9. [DOI] [PubMed] [Google Scholar]
- 11.Roybal C. N., Hunsaker L. A., Barbash O., Vander Jagt D. L., Abcouwer S. F. The oxidative stressor arsenite activates vascular endothelial growth factor mRNA transcription by an ATF4-dependent mechanism. J. Biol. Chem. 2005;280:20331–20339. doi: 10.1074/jbc.M411275200. [DOI] [PubMed] [Google Scholar]
- 12.Kilberg M. S., Pan Y. X., Chen H., Leung-Pineda V. Nutritional control of gene expression: how mammalian cells respond to amino acid limitation. Annu. Rev. Nutr. 2004;25:59–85. doi: 10.1146/annurev.nutr.24.012003.132145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bruhat A., Averous J., Carraro V., Zhong C., Reimold A. M., Kilberg M. S., Fafournoux P. Differences in the molecular mechanisms involved in the transcriptional activation of the CHOP and asparagine synthetase genes in response to amino acid deprivation or activation of the unfolded protein response. J. Biol. Chem. 2002;277:48107–48114. doi: 10.1074/jbc.M206149200. [DOI] [PubMed] [Google Scholar]
- 14.Guerrini L., Gong S. S., Mangasarian K., Basilico C. Cis- and trans-acting elements involved in amino acid regulation of asparagine synthetase gene expression. Mol. Cell. Biol. 1993;13:3202–3212. doi: 10.1128/mcb.13.6.3202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Averous J., Bruhat A., Jousse C., Carraro V., Thiel G., Fafournoux P. Induction of CHOP expression by amino acid limitation requires both ATF4 expression and ATF2 phosphorylation. J. Biol. Chem. 2004;279:5288–5297. doi: 10.1074/jbc.M311862200. [DOI] [PubMed] [Google Scholar]
- 16.Palii S. S., Chen H., Kilberg M. S. Transcriptional control of the human sodium-coupled neutral amino acid transporter system A gene by amino acid availability is mediated by an intronic element. J. Biol. Chem. 2004;279:3463–3471. doi: 10.1074/jbc.M310483200. [DOI] [PubMed] [Google Scholar]
- 17.Sato H., Nomura S., Maebara K., Sato K., Tamba M., Bannai S. Transcriptional control of cystine/glutamate transporter gene by amino acid deprivation. Biochem. Biophys. Res. Commun. 2004;325:109–116. doi: 10.1016/j.bbrc.2004.10.009. [DOI] [PubMed] [Google Scholar]
- 18.Zhong C., Chen C., Kilberg M. S. Characterization of the nutrient-sensing response unit in the human asparagine synthetase promoter. Biochem. J. 2003;372:603–609. doi: 10.1042/BJ20030076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hai T., Hartman M. G. The molecular biology and nomenclature of the activating transcription factor/cAMP responsive element binding family of transcription factors: activating transcription factor proteins and homeostasis. Gene. 2001;273:1–11. doi: 10.1016/s0378-1119(01)00551-0. [DOI] [PubMed] [Google Scholar]
- 20.van Dam H., Castellazzi M. Distinct roles of Jun: Fos and Jun: ATF dimers in oncogenesis. Oncogene. 2001;20:2453–2464. doi: 10.1038/sj.onc.1204239. [DOI] [PubMed] [Google Scholar]
- 21.Okamoto Y., Chaves A., Chen J., Kelley R., Jones K., Weed H. G., Gardner K. L., Gangi L., Yamaguchi M., Klomkleaw W., et al. Transgenic mice with cardiac-specific expression of activating transcription factor 3, a stress-inducible gene, have conduction abnormalities and contractile dysfunction. Am. J. Pathol. 2001;159:639–650. doi: 10.1016/S0002-9440(10)61735-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Allen-Jennings A. E., Hartman M. G., Kociba G. J., Hai T. The roles of ATF3 in glucose homeostasis: a transgenic mouse model with liver dysfunction and defects in endocrine pancreas. J. Biol. Chem. 2001;276:29507–29514. doi: 10.1074/jbc.M100986200. [DOI] [PubMed] [Google Scholar]
- 23.Yan C., Jamaluddin M. S., Aggarwal B., Myers J., Boyd D. D. Gene expression profiling identifies activating transcription factor 3 as a novel contributor to the proapoptotic effect of curcumin. Mol. Cancer Ther. 2005;4:233–241. [PubMed] [Google Scholar]
- 24.Fawcett T. W., Martindale J. L., Guyton K. Z., Hai T., Holbrook N. J. Complexes containing activating transcription factor (ATF)/cAMP- responsive-element-binding protein (CREB) interact with the CCAAT/enhancer-binding protein (C/EBP)-ATF composite site to regulate Gadd153 expression during the stress response. Biochem. J. 1999;339:135–141. [PMC free article] [PubMed] [Google Scholar]
- 25.Pan Y., Chen H., Siu F., Kilberg M. S. Amino acid deprivation and endoplasmic reticulum stress induce expression of multiple activating transcription factor-3 mRNA species that, when overexpressed in HepG2 cells, modulate transcription by the human asparagine synthetase promoter. J. Biol. Chem. 2003;278:38402–38412. doi: 10.1074/jbc.M304574200. [DOI] [PubMed] [Google Scholar]
- 26.Jiang H. Y., Wek S. A., McGrath B. C., Lu D., Hai T., Harding H. P., Wang X., Ron D., Cavener D. R., Wek R. C. Activating transcription factor 3 is integral to the eukaryotic initiation factor 2 kinase stress response. Mol. Cell. Biol. 2004;24:1365–1377. doi: 10.1128/MCB.24.3.1365-1377.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fernandez J. M., Yaman I., Sarnow P., Snider M. D., Hatzoglou M. Regulation of internal ribosomal entry site-mediated translation by phosphorylation of the translation initiation factor eIF2α. J. Biol. Chem. 2002;277:19198–19205. doi: 10.1074/jbc.M201052200. [DOI] [PubMed] [Google Scholar]
- 28.Fernandez J. M., Yaman I., Merrick W. C., Koromilas A. E., Wek R. C., Sood R., Hensold J. O., Hatzoglou M. Regulation of internal ribosome entry site-mediated translation by eukaryotic initiation factor-2α phosphorylation and translation of a small upstream open reading frame. J. Biol. Chem. 2002;277:2050–2058. doi: 10.1074/jbc.M109199200. [DOI] [PubMed] [Google Scholar]
- 29.Yaman I., Fernandez J., Liu H., Caprara M., Komar A. A., Koromilas A. E., Zhou L., Snider M. D., Scheuner D., Kaufman R. J., Hatzoglou M. The zipper model of translational control: a small upstream ORF is the switch that controls structural remodeling of an mRNA leader. Cell. 2003;113:519–531. doi: 10.1016/s0092-8674(03)00345-3. [DOI] [PubMed] [Google Scholar]
- 30.Lincoln A. J., Monczak Y., Williams S. C., Johnson P. F. Inhibition of CCAAT/enhancer-binding protein α and β translation by upstream open reading frames. J. Biol. Chem. 1998;273:9552–9560. doi: 10.1074/jbc.273.16.9552. [DOI] [PubMed] [Google Scholar]
- 31.He C. H., Gong P., Hu B., Stewart D., Choi M. E., Choi A. M., Alam J. Identification of activating transcription factor 4 (ATF4) as an Nrf2-interacting protein. Implication for heme oxygenase-1 gene regulation. J. Biol. Chem. 2001;276:20858–20865. doi: 10.1074/jbc.M101198200. [DOI] [PubMed] [Google Scholar]
- 32.Parkin S. E., Baer M., Copeland T. D., Schwartz R. C., Johnson P. F. Regulation of CCAAT/enhancer-binding protein (C/EBP) activator proteins by heterodimerization with C/EBPγ (Ig/EBP) J. Biol. Chem. 2002;277:23563–23572. doi: 10.1074/jbc.M202184200. [DOI] [PubMed] [Google Scholar]
- 33.Yaman I., Fernandez J., Sarkar B., Schneider R. J., Snider M. D., Nagy L. E., Hatzoglou M. Nutritional control of mRNA stability is mediated by a conserved AU-rich element that binds the cytoplasmic shuttling protein HuR. J. Biol. Chem. 2002;277:41539–41546. doi: 10.1074/jbc.M204850200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wek R. C., Jiang H. Y., Anthony T. G. Coping with stress: eIF2 kinases and translational control. Biochem. Soc. Trans. 2006;34:7–11. doi: 10.1042/BST20060007. [DOI] [PubMed] [Google Scholar]
- 35.Scheuner D., Song B., McEwen E., Liu C., Laybutt R., Gillespie P., Saunders T., Bonner-Weir S., Kaufman R. J. Translational control is required for the unfolded protein response and in vivo glucose homeostasis. Mol. Cell. 2001;7:1165–1176. doi: 10.1016/s1097-2765(01)00265-9. [DOI] [PubMed] [Google Scholar]
- 36.Kilberg M. S., Pan Y. X., Chen H., Leung-Pineda V. Nutritional control of gene expression: how mammalian cells respond to amino acid limitation. Annu. Rev. Nutr. 2005;25:59–85. doi: 10.1146/annurev.nutr.24.012003.132145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gaccioli F., Huang C. C., Wang C., Bevilacqua E., Franchi-Gazzola R., Gazzola G. C., Bussolati O., Snider M. D., Hatzoglou M. Amino acid starvation induces the SNAT2 neutral amino acid transporter by a mechanism that involves eukaryotic initiation factor 2α phosphorylation and cap-independent translation. J. Biol. Chem. 2006;281:17929–17940. doi: 10.1074/jbc.M600341200. [DOI] [PubMed] [Google Scholar]
- 38.Chen H., Pan Y. X., Dudenhausen E. E., Kilberg M. S. Amino acid deprivation induces the transcription rate of the human asparagine synthetase gene through a timed program of expression and promoter binding of nutrient-responsive basic region/leucine zipper transcription factors as well as localized histone acetylation. J. Biol. Chem. 2004;279:50829–50839. doi: 10.1074/jbc.M409173200. [DOI] [PubMed] [Google Scholar]
- 39.Hanlon M., Sturgill T. W., Sealy L. ERK2- and p90(Rsk2)-dependent pathways regulate the CCAAT/enhancer-binding protein-β interaction with serum response factor. J. Biol. Chem. 2001;276:38449–38456. doi: 10.1074/jbc.M102165200. [DOI] [PubMed] [Google Scholar]
- 40.Ghosh A. K., Bhattacharyya S., Mori Y., Varga J. Inhibition of collagen gene expression by interferon-gamma: novel role of the CCAAT/enhancer binding protein β (C/EBPβ) J. Cell Physiol. 2006;207:251–260. doi: 10.1002/jcp.20559. [DOI] [PubMed] [Google Scholar]
- 41.Chen C., Dudenhausen E. E., Pan Y. X., Zhong C., Kilberg M. S. Human CCAAT/enhancer-binding protein β gene expression is activated by endoplasmic reticulum stress through an unfolded protein response element downstream of the protein coding sequence. J. Biol. Chem. 2004;279:27948–27956. doi: 10.1074/jbc.M313920200. [DOI] [PubMed] [Google Scholar]
- 42.Podust L. M., Krezel A. M., Kim Y. Crystal structure of the CCAAT box/enhancer-binding protein β activating transcription factor-4 basic leucine zipper heterodimer in the absence of DNA. J. Biol. Chem. 2001;276:505–513. doi: 10.1074/jbc.M005594200. [DOI] [PubMed] [Google Scholar]
- 43.Marciniak S. J., Yun C. Y., Oyadomari S., Novoa I., Zhang Y., Jungreis R., Nagata K., Harding H. P., Ron D. CHOP induces death by promoting protein synthesis and oxidation in the stressed endoplasmic reticulum. Genes Dev. 2004;18:3066–3077. doi: 10.1101/gad.1250704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jiang H. Y., Wek R. C. Phosphorylation of the α-subunit of the eukaryotic initiation factor-2 (eIF2α) reduces protein synthesis and enhances apoptosis in response to proteasome inhibition. J. Biol. Chem. 2005;280:14189–14202. doi: 10.1074/jbc.M413660200. [DOI] [PubMed] [Google Scholar]
- 45.Calkhoven C. F., Muller C., Leutz A. Translational control of C/EBPα and C/EBPβ isoform expression. Genes Dev. 2000;14:1920–1932. [PMC free article] [PubMed] [Google Scholar]
- 46.Ohoka N., Yoshii S., Hattori T., Onozaki K., Hayashi H. TRB3, a novel ER stress-inducible gene, is induced via ATF4–CHOP pathway and is involved in cell death. EMBO J. 2005;24:1243–1255. doi: 10.1038/sj.emboj.7600596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hai T., Curran T. Cross-family dimerization of transcription factors Fos/Jun and ATF/CREB alters DNA binding specificity. Proc. Natl. Acad. Sci. U.S.A. 1991;88:3720–3724. doi: 10.1073/pnas.88.9.3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wolfgang C. D., Liang G., Okamoto Y., Allen A. E., Hai T. Transcriptional autorepression of the stress-inducible gene ATF3. J. Biol. Chem. 2000;275:16865–16870. doi: 10.1074/jbc.M909637199. [DOI] [PubMed] [Google Scholar]
- 49.Hai T., Wolfgang C. D., Marsee D. K., Allen A. E., Sivaprasad U. ATF3 and stress responses. Gene Expr. 1999;7:321–335. [PMC free article] [PubMed] [Google Scholar]
- 50.Luo S., Baumeister P., Yang S., Abcouwer S. F., Lee A. S. Induction of Grp78/BiP by translational block: activation of the Grp78 promoter by ATF4 through and upstream ATF/CRE site independent of the endoplasmic reticulum stress elements. J. Biol. Chem. 2003;278:37375–37385. doi: 10.1074/jbc.M303619200. [DOI] [PubMed] [Google Scholar]
- 51.Ye J., Rawson R. B., Komuro R., Chen X., Dave U. P., Prywes R., Brown M. S., Goldstein J. L. ER stress induces cleavage of membrane-bound ATF6 by the same proteases that process SREBPs. Mol. Cell. 2000;6:1355–1364. doi: 10.1016/s1097-2765(00)00133-7. [DOI] [PubMed] [Google Scholar]
- 52.Perkins C. P., Mar V., Shutter J. R., del Castillo J., Danilenko D. M., Medlock E. S., Ponting I. L., Graham M., Stark K. L., Zuo Y., et al. Anemia and perinatal death result from loss of the murine ecotropic retrovirus receptor mCAT-1. Genes Dev. 1997;11:914–925. doi: 10.1101/gad.11.7.914. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.