Abstract
Bromodomains are present in many chromatin-associated proteins such as the SWI/SNF and RSC chromatin remodelling and the SAGA HAT (histone acetyltransferase) complexes, and can bind to acetylated lysine residues in the N-terminal tails of the histones. Lysine acetylation is a histone modification that forms a stable epigenetic mark on chromatin for bromodomain-containing proteins to dock and in turn regulate gene expression. In order to better understand how bromodomains read the ‘histone code’ and interact with acetylated histones, we have tested the interactions of several bromodomains within transcriptional co-activators with differentially acetylated histone tail peptides and HAT-acetylated histones. Using GST (glutathione S-transferase) pull-down assays, we show specificity of binding of some bromodomains to differentially acetylated H3 and H4 peptides as well as HAT-acetylated histones. Our results reveal that the Swi2/Snf2 bromodomain interacts with various acetylated H3 and H4 peptides, whereas the Gcn5 bromodomain interacts only with acetylated H3 peptides and tetra-acetylated H4 peptides. Additionally we show that the Spt7 bromodomain interacts with acetylated H3 peptides weakly, but not with acetylated H4 peptides. Some bromodomains such as the Bdf1-2 do not interact with most of the acetylated peptides tested. Results of the peptide experiments are confirmed with tests of interactions between these bromodomains and HAT-acetylated histones. Furthermore, we demonstrate that the Swi2/Snf2 bromodomain is important for the binding and the remodelling activity of the SWI/SNF complex on hyperacetylated nucleosomes. The selective recognition of the bromodomains observed in the present study accounts for the broad effects of bromodomain-containing proteins observed on binding to histones.
Keywords: acetylation, bromodomain, histone, SWI/SNF
Abbreviations: DTT, dithiothreitol; GST, glutathione S-transferase; HAT, histone acetyltransferase; TAP, tandem affinity purification
INTRODUCTION
The structure of the chromatin places an effective barrier on the ability of transcription machinery to access its binding sites. Many studies have described conserved protein complexes whose function it is to modify the chromatin structure and relieve its repressive effect (reviewed in [1–5]). They include ATP-dependent chromatin remodelling enzymes, as well as enzymes that post-translationally modify the N-terminal histone tails by acetylation, methylation, phosphorylation, sumoylation, ubiquitination and ADP-ribosylation [6–11]. Covalent modification of histone proteins has a well-known role in gene expression. Some of these histone modifications are short-lived, but others may persist through cell divisions and are thought to serve as stable epigenetic memory known as the ‘histone code’. These stable markers on histone tails act as binding sites for transcriptional co-activator proteins that may further modify the chromatin structure.
One evolutionarily conserved motif found in many transcriptional co-activators, including HATs (histone acetyltransferases; such as SAGA), chromatin remodelling complexes (such as SWI/SNF and RSC), and general transcription factors (such as TAFII250) is the bromodomain. It is a small domain that has been shown to interact with acetylated lysine residues in N-terminal tails of histones H3 and H4 in vitro [12–19]. It can also recognize and bind to acetylated non-histone proteins such as MyoD, the HIV-1 Tat and the p53 transcription factor [20–24]. Using immobilized template assays, we have previously shown that bromodomains were necessary for the anchoring of the SWI/SNF and the SAGA complexes to acetylated promoter nucleosomes [25,26]. In experiments in which SWI/SNF was recruited to nucleosomal templates, the retention of SWI/SNF required both acetylated histones and the Swi2/Snf2 bromodomain. Moreover, recognition by a few bromodomains of different acetylation patterns has been demonstrated in vitro [16,27–30]. Consistent with these in vitro experiments, mutant phenotypes were observed for an Swi2/Snf2 bromodomain deletion when combined with mutations in the SAGA HAT complex, and the Swi2/Snf2 bromodomain was required for the presence of SWI/SNF at the SUC2 promoter [26]. Furthermore, using fluorescence resonance energy transfer, it has been demonstrated that bromodomain proteins show selective recognition of acetylated histones in vivo [31]. More recently we have demonstrated the requirement of the Swi2/Snf2 bromodomain for the functional activity of the complex on SAGA-acetylated nucleosomes [32]. While the loss of the Swi2/Snf2 bromodomain has no effect on the remodelling and octamer transfer activity of unmodified nucleosomes, the bromodomain-deleted SWI/SNF has much reduced activity on SAGA-acetylated nucleosomes compared with the wild-type. Additionally, we have shown the displacement of SAGA by the SWI/SNF complex on acetylated nucleosomes and the requirement of the Swi2/Snf2 bromodomain for this activity. These data illustrate a novel and significant role of the Swi2/Snf2 bromodomain in remodelling of acetylated promoter nucleosomes and in displacing SAGA from promoters.
The evidence thus far suggests that bromodomains act as targeting modules to anchor chromatin-modifying proteins on promoters. Since bromodomains within various proteins have structural diversity, the present study investigated the interaction of some of these bromodomains with differentially acetylated histones in order to determine the preferred acetylated lysine residue as a target for their binding. We were particularly interested in determining the binding affinity of the various bromodomains within co-activator proteins to HAT-modified histone tails. Our results reveal selective recognition of acetylated histones by different bromodomains. Specifically, we show different sensitivities of binding of bromodomains to various acetylated H3 and H4 peptides as well as HAT-acetylated histones. Additionally, using hyperacetylated histones, we demonstrate that the bromodomain-deleted SWI/SNF complex cannot bind or remodel acetylated nucleosomes as efficiently as the wild-type. Together these data show that bromodomains have differing abilities to bind to and exert their modifying effects on acetylated nucleosomes.
EXPERIMENTAL
Cloning and purification of GST (glutathione S-transferase) fusion proteins
Seven DNA fragments encoding bromodomains within various yeast transcriptional co-activator proteins were PCR amplified from yeast genomic DNA and cloned into the BamHI and EcoRI sites of the GST expression vector, pGEX-2T, as described in the GST gene fusion system handbook (Amersham Biosciences). Table 1 in the supplementary data (http://www.BiochemJ.org/bj/402/bj4020125add.htm) shows the sequences of all of the primers used for cloning. The clones were screened for the insert using both PCR and restriction enzyme analysis. GST fusion proteins were then expressed in bacteria and purified using Glutathione Sepharose 4B beads as described in the manufacturer's protocol (Amersham Biosciences), except that protease inhibitors (PMSF, leupeptin, pepstatin A and aprotinin) and DTT (dithiothreitol) were added to the resuspension buffer. Protein purity and amounts were checked and normalized by SDS/PAGE and Coomassie Blue staining.
GST pull-down assays with histone peptides and proteins
GST fusion proteins bound to Glutathione Sepharose 4B beads were separately incubated with acetylated histone H3 or H4 peptides in a pull-down buffer [50 mM Hepes (pH 7.5), 1 mM EDTA, 150 mM NaCl, 10% glycerol, 0.1% Tween 20, 0.5 mM DTT, 1 mM PMSF and 2 μg/ml pepstatin A] for 2 h at 4 °C while mixing on a rotation wheel. The supernatants were collected, and the beads were washed with the pull-down buffer three times and left as a 50% slurry after the final wash. The beads were loaded on to SDS/PAGE (15% gels), and the presence of acetylated peptide was detected by immunoblotting with acetyl-specific antibodies. The GST–bromodomain pull-downs with intact histones (control or hyperacetylated) were performed similarly, except that in these pull-down experiments, histone proteins served as substrates for the GST–bromodomains. Quantification of three independent experiments ±S.D. was performed by scanning the gels and using total histone signal intensity in the beads as the percentage pull-down. The GST–bromodomain pull-downs with intact HAT- acetylated histones were performed similarly, except that instead of incubation with acetylated peptides or control and hyperacetylated histones, histones were pre-acetylated with either SAGA or NuA4 HATs for 30 min at 30 °C with either non-radioactive or 3H-labelled acetyl-CoA (Amersham Biosciences) prior to incubation with the GST fusion proteins. After removing the supernatants, the beads were washed twice and the levels of interaction with the various GST fusion proteins were determined by performing either Western blot analysis or a HAT assay as described previously [33].
Purification of wild-type and mutant SWI/SNF complexes
Wild-type and Δbromodomain SWI/SNF (a strain lacking the Swi2/Snf2 bromodomain) complexes were purified from yeast whole cell extract using the TAP (tandem affinity purification) method using Snf6-TAP strains over two affinity columns as described previously [32,34–37]. Briefly, whole cell extracts were prepared from 6 litres of yeast cells grown in YPD medium [1% (w/v) yeast extract/2% (w/v) peptone/2% (w/v) glucose] and were added to IgG resin (Amersham Biosciences). The complexes were eluted from the beads by TEV (tobacco etch virus) protease (Invitrogen) cleavage in a buffer containing 10 mM Tris/HCl (pH 8), 150 mM NaCl, 0.5 M EDTA, 0.1% Nonidet P40, 10% (v/v) glycerol, 1 mM PMSF, 2 μg/ml leupeptin, 1 μg/ml pepstatin A and 1 mM DTT. Following binding to calmodulin resin (Amersham Biosciences), the complexes were eluted using a buffer containing 10 mM Tris/HCl (pH 8), 150 mM NaCl, 1 mM MgAc, 1 mM imidazole, 2 mM EGTA, 0.1% Nonidet P40, 10% (v/v) glycerol, 1 mM PMSF, 2 μg/ml leupeptin, 1 μg/ml pepstatin A and 0.5 mM DTT. Purification was monitored by Western blot analysis using an anti-TAP antibody (Open Biosystems) as well as silver staining. The same amounts of the wild-type and the Δbromodomain SWI/SNF complexes were used in both the immobilized template binding and the restriction enzyme accessibility assays after normalization of the amounts of purified protein.
Immobilized template binding assay
pG5E4-5S containing a dinucleosome length G5E4 fragment flanked on both sides by 5 S sequences was prepared as described in [38]. The G5E4-5S fragments were produced by digesting pG5E4-5S with Asp718 and end-labelling with biotin-14-dATP using Klenow. The 2.5 kb end-labelled G5E4-5S fragments were then gel purified away from the backbone by digesting with ClaI and EaeI. This array was then reconstituted by step dilution with either control or hyperacetylated histones as described previously [26]. The nucleosomal arrays were then bound to paramagnetic beads coupled to streptavidin (Dynabeads Streptavidin, Invitrogen) as described previously [26,39]. Approx. 4 nM of either the wild-type or the mutant SWI/SNF was added to 200 ng of the above template in 20 μl of binding buffer and incubated for 1 h at 30 °C. The templates were then concentrated on a magnet, the supernatant was removed, and the beads were washed twice before performing Western blot analysis. The blots were probed with antibodies against the tag (anti-TAP antibodies). A graphical representation of the ratio of the bead to the supernatant ±S.D. is shown after quantification of three independent experiments determined by scanning the gels and using the signal intensities of the Western blots.
Restriction enzyme accessibility assay
In this assay, we used a 183 bp GUB fragment as the DNA template generated by PCR using a radiolabelled 5′ primer and the pGALUSFBEND plasmid [40–42]. This DNA fragment was reconstituted into a nucleosome using either control or hyperacetylated histones. The assay was performed as described previously [32,42]. Briefly, wild-type or Δbromodomain mutant SWI/SNF complexes were added to approximately 10 ng of the 32P-labelled GUB template in a binding buffer [10 mM Hepes (pH 7.8), 50 mM KCl, 5 mM DTT, 5 mM PMSF, 5% glycerol, 0.25 mg/ml BSA and 2 mM MgCl2] in the presence or absence of 2 mM ATP. After incubation for 1 h at 30 °C, the binding reactions were treated with 10 units of SalI for 30 min at 30 °C. An equal volume of stop buffer [20 mM Tris/HCl (pH 7.5), 50 mM EDTA, 2% SDS, 0.2 mg/ml proteinase K and 1 mg/ml glycogen] was added to the reaction mixtures, and incubated at 50 °C for 1 h. Deproteinized samples were precipitated with 200 mM NaCl and 3 volumes of ethanol, and the pellet resuspended in 5 μl of the formamide dye (95% formamide, 10 mM EDTA, 0.1% xylene cyanol and 0.1% Bromophenol Blue). After heat denaturation, the samples were resolved on a 6% acrylamide (19:1 acrylamide to bis-acrylamide)/8 M urea sequencing gel at 150 V for 3 h and visualized by autoradiography. Graphical representation of the percentage cleavage ±S.D. is shown after quantification of three independent experiments determined by scanning the gels and measuring the signal intensities.
RESULTS
Bromodomains have a higher affinity for hyperacetylated histones
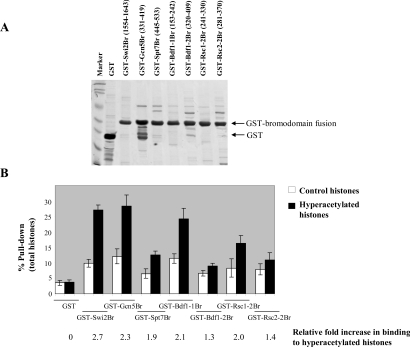
In order to determine the binding affinity of bromodomains within chromatin-modifying complexes to acetylated histones as well as acetylated lysine residues, we have cloned and purified bromodomains from several protein complexes as GST fusion proteins. Seven bromodomains from yeast co-activator proteins were cloned into a GST expression vector (pGEX-2T), including bromodomains from Swi2/Snf2 (the catalytic subunit of the SWI/SNF complex), Gcn5 and Spt7 (of the SAGA HAT), two bromodomains in the Bdf1 protein and one of the two bromodomains each in Rsc1 and Rsc2. We had difficulty in expressing and purifying to satisfaction the other bromodomains of the RSC complex. The sequences of the primers used for cloning are shown in Table 1 of the supplementary data (http://www.BiochemJ.org/bj/402/bj4020125add.htm). The GST fusion proteins were expressed in bacteria and the protein amounts and purity were monitored by SDS/PAGE (15% gels) and Coomassie Blue staining (Figure 1A). The overall purity of all of the bromodomains was good, except for the Gcn5 bromodomain, which had some degradation products. The cloned amino acid sequences of these bromodomains are shown at the top of Figure 1(A). The GST fusion bromodomain proteins bound to glutathione beads were first tested for their ability to bind to either control or hyperacetylated histones. Following incubation of the bromodomains with these histones and the separation of the supernatant from the beads, the beads were washed and the amount of total histones bound to bromodomains (the percentage pull-down) was determined by SDS/PAGE and subsequent scanning and quantification of the bands. Figure 1(B) shows the percentage pull-down of the total histones for both control and hyperacetylated histones with the different GST–bromodomain fusion proteins. Although there are differences between the binding affinities of the different bromodomains, all bromodomains had a higher affinity for hyperacetylated histones compared with control unacetylated histones. This higher affinity ranged from 1.3-fold for the Bdf1-2 bromodomain to 2.7-fold for the Swi2/Snf2 bromodomain, shown at the bottom of Figure 1(B). The Swi2/Snf2, Gcn5, Bdf1-1, Rsc1-2 and the Spt7 bromodomain had the highest increases in binding to hyperacetylated histones respectively, whereas the second bromodomains of Bdf1 and Rsc2 had a weaker binding to hyperacetylated histones (1.3-fold and 1.4-fold respectively). Although these data do not differentiate between the different histones or acetylated lysine residues, a higher affinity for total histones when they are acetylated is observed in all bromodomains tested in the present study. In general, the increased affinity of the GST–bromodomains for hyperacetylated histones confirms the importance of the bromodomain as an acetylated binding domain within co-activator proteins.
Figure 1. Purification of some of the yeast bromodomains as GST fusion proteins and their binding to hyperacetylated histones.
(A) Coomassie Blue-stained SDS/PAGE of the purified yeast bromodomains. After purification, the amounts of GST and GST–bromodomain fusion proteins were normalized and used for all of the assays. The degradation of the Gcn5 bromodomain fusion proteins in GST did not significantly affect the results since the majority of the protein was the fusion protein. The relevant regions of the seven bromodomains in transcriptional co-activators in yeast were identified based on sequence homology to the Swi2/Snf2 bromodomain as well as published literature and are shown in parentheses. (B) The yeast bromodomains preferentially bind to histones with a high degree of acetylation with different affinities. A GST pull-down assay was performed with intact control (open columns) and hyperacetylated (solid columns) HeLa histones purchased from Upstate Biotechnology. After pull-down, beads were washed and bound proteins were analysed by SDS/PAGE and quantified. The quantification of the three independent experiments ±S.D. was determined by scanning the gels and using total histone signal intensity in the beads as the percentage pull-down. The relative fold increase in binding of the GST–bromodomain fusion proteins to hyperacetylated histones compared with control histones is shown at the bottom.
Differential binding of bromodomains to acetylated H3 and H4 histone tail peptides
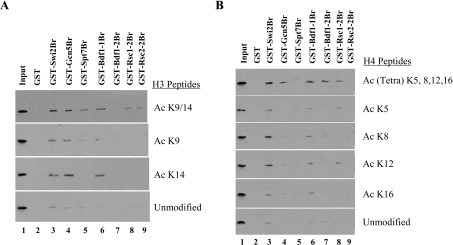
In order to study the binding abilities of the different bromodomains to acetylated H3 and H4 histone tails, the GST pull-down experiments were repeated using acetylated peptides. The GST–bromodomain proteins were individually incubated with various acetylated histone H3 or H4 peptides, followed by separation of the supernatant and the beads. After washing the beads, they were loaded on to SDS/PAGE (15% gels), and the presence of acetylated peptide was detected after immunoblotting with acetyl-specific antibodies. These experiments revealed differential binding affinities of the bromodomains with acetylated histone H3 and H4 peptides (Figure 2). For example, the Swi2/Snf2 bromodomain had a higher affinity for acetylated histone H3 Lys9 or Lys14 peptides compared with unmodified H3 peptide (Figure 2A, lane 3) and interacted with all acetylated H4 peptides as well (Figure 2B, lane 3). On the other hand, the Gcn5 bromodomain interacted specifically with acetylated H3 peptides (Figure 2A, lane 4) and only weakly with the tetra-acetylated H4 peptide (Figure 2B, lane 4). The other SAGA bromodomain (in its Spt7 subunits) only seemed to bind to H3 peptides acetylated at its Lys9 (Figures 2A and 2B, lane 5). This seems to be in contrast with the data in Figure 1(B) where we showed efficient enhancement of the binding of the Spt7 bromodomain to hyperacetylated histones compared with the control histones (a 1.9-fold increase in binding). The data in the present study are consistent with our previous observations where we have shown that the Spt7 bromodomain itself can bind to SAGA acetylated templates, but not in the context of the SAGA complex [29]. These results demonstrate that the bromodomain within the catalytic subunit of the SAGA complex is perhaps more important for recognition and binding to acetylated lysine residues in the histone tails, whereas the Spt7 bromodomain may have another function such as recognition of acetylated transcription factors or multiple lysine residues. The first bromodomain in the Bdf1 protein complex (Bdf1-1) seems to be important as an acetyl-lysine recognition module (Figures 2A and 2B, lane 6); however, its other bromodomain (Bdf1-2) only bound to tetra-acetylated H4 peptides (Figures 2A and 2B, lane 7), consistent with previously published data [20]. The two RSC bromodomains tested (Rsc1-2 and Rsc2-2) had weak binding properties to both acetylated H3 and H4 peptides. The Rsc1-2 bromodomain bound only to acetylated H3 Lys9/Lys14 peptides and some of the H4 peptides, whereas the Rsc2-2 bromodomain did not seem to interact with any acetylated H4 peptide (Figures 2A and 2B, lanes 8 and 9). Together these results argue for the selective recognition of bromodomains within co-activator proteins and may account for the differences observed in the affinities of bromodomain-containing proteins for histones.
Figure 2. Selective binding of the GST–bromodomains to acetylated histone peptides.
(A) Binding of the GST–bromodomain proteins to acetylated H3 peptides. A GST pull-down assay was performed with acetylated histone H3 peptides purchased from Upstate Biotechnology. After pull-down, beads were washed and bound proteins were analysed by SDS/PAGE and Western blotting using an acetyl-specific H3 antibody or an antibody to unmodified H3. (B) Binding of the GST–bromodomain proteins to acetylated H4 peptides. The GST pull-down assay was performed in a similar manner to (A), but in this case acetylated histone H4 peptides were used as the substrate and the immunoblotting was performed using an acetyl-specific H4 antibody. Ac K, acetylated lysine.
Differential binding of bromodomains to HAT-acetylated histones
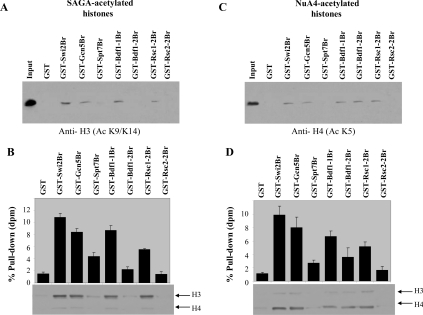
In order to understand whether these bromodomains have similar affinities for more physiologically acetylated H3 or H4 histones, we have acetylated histones with either the H3- or H4-specific HATs SAGA or NuA4 respectively, prior to the GST pull-down assay. The level of interaction with acetylated histones (the percentage pull-down) was detected both by Western blot analysis using H3 or H4 acetyl-specific antibodies (Figures 3A and 3C) and by counting the levels of radioactive 3H-acetylated lysine residues in the pull-down assay (Figures 3B and 3D). These assays revealed a selective interaction of the GST–bromodomains with SAGA- or NuA4-acetylated histones. While interactions were observed between Swi2/Snf2, Gcn5, Bdf1-1 and Rsc1-2 and to some extent Spt7 bromodomains with SAGA-acetylated histones, no such interactions were observed between the Bdf1-2 and Rsc2-2 bromodomains with these histones (Figure 3A). This was confirmed when the GST pull-down was repeated with histones that had been acetylated with SAGA in the presence of [3H]acetylCoA, followed by fluorography (bottom panel of Figure 3B) and liquid scintillation counting on filter paper (Figure 3B). All of these results point to the differential affinities of these bromodomains for SAGA-acetylated histones. When the same experiments were repeated with histones that were pre-acetylated with NuA4, the results again showed different degrees of interaction. In this case, the Bdf1-2 bromodomain bound to these NuA4-acetylated templates in pull-down assays, whereas the Spt7 and the Rsc2-2 did not (Figure 3C). This was confirmed by the radioactive 3H-acetylation pull-down assay (Figure 3D). Together, all our data point to selective recognition and specificity of interaction of the bromodomains tested with various acetylated peptides and histones.
Figure 3. Differential binding of the GST–bromodomains to HAT-acetylated histones.
(A and B) Binding of the GST–bromodomain proteins to SAGA-acetylated histones. The GST pull-down assay with HAT-acetylated histones was performed similarly, except histones were pre-acetylated with the SAGA HATs for 30 min at 30 °C with either non-radioactive or 3H-labelled acetyl-CoA (Amersham Biosciences) prior to incubation with the GST fusion proteins. After removing the supernatants, the beads were washed twice and the levels of interaction with the various GST fusion proteins were determined by performing either Western blot analysis (A) or a HAT assay (B) as described previously [33] to follow the 3H-labelled acetylated histone. The quantification of three independent pull-down experiments ±S.D. was performed by liquid-scintillation counting/filter binding of the 3H-labelled histones in the beads and is shown as the percentage pull-down with one representative fluorograph shown (bottom panel). (C and D) Binding of the GST–bromodomain proteins to NuA4-acetylated histones. The GST pull-down assay was repeated, except histones were pre-acetylated with the NuA4 HATs for 30 min at 30 °C with either non-radioactive or 3H-labelled acetyl-CoA prior to incubation with the GST fusion proteins. Using either a Western blot analysis (C) or a HAT assay (D), we analysed the level of interaction of these acetylated H4 histones with the various GST fusion proteins. The quantification of three independent pull-down experiments with these templates was performed in a similar manner to above and the percentage pull-down with one representative fluorograph is shown in the bottom panel.
The Swi2/Snf2 bromodomain is important for the binding and remodelling activities of the complex on hyperacetylated nucleosomes
Previous studies have shown that the SWI/SNF complex is recruited to promoters by yeast transcriptional activators [41–47] and is also stabilized to acetylated nucleosomes through its Swi2/Snf2 bromodomain [25,26]. Moreover, the bromodomain of Swi2/Snf2 participates in the functions of this protein complex in vivo [26]. In the present study, we show selective binding of different bromodomains to acetylated lysine residues, where the Swi2/Snf2 bromodomain is one of the most efficient in binding to various acetylated peptide tails and histones (see Figures 1–3). These results led us to investigate the role of the Swi2/Snf2 bromodomain in remodelling of acetylated nucleosomes. To test directly whether this bromodomain contributes to the binding and the functional activity of the complex on nucleosomes, we purified wild-type SWI/SNF as well as SWI/SNF from a strain lacking the Swi2/Snf2 bromodomain (termed Δbromodomain SWI/SNF), and tested them in histone binding assays as well as chromatin remodelling assays. For all of these experiments, we used wild-type and mutant SWI/SNF complexes that had been highly purified over two affinity columns using the TAP method [32,34–37]. The loss of the Swi2/Snf2 bromodomain did not affect complex integrity as detected by silver staining (results not shown). The same amounts of the wild-type and the Δbromodomain SWI/SNF complexes were used in these assays based on the normalization of the amounts of purified protein after Western blotting.
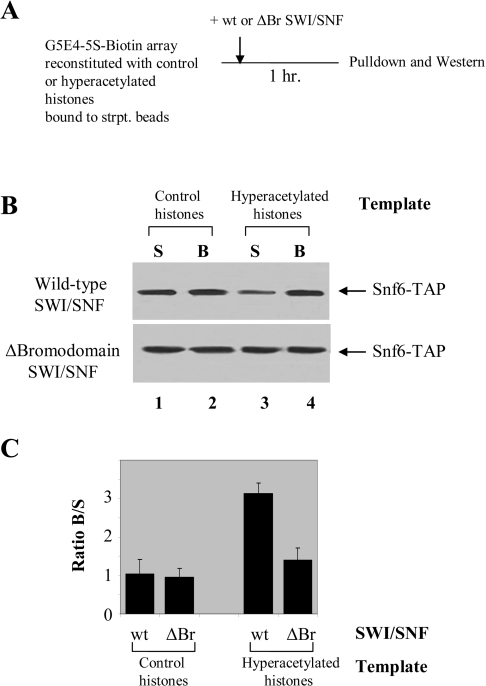
To investigate the binding abilities of the wild-type and the Δbromodomain SWI/SNF complexes to nucleosomes, we utilized an immobilized nucleosomal array template, pG5E4, which contains five Gal4 binding sites and an E4 promoter flanked on both sides by 5 S rDNA nucleosome positioning sequences [26,38]. Figure 4(A) shows the outline of the experiment. Briefly, the template was first biotinylated and reconstituted with either control or hyperacetylated histones, followed by the addition of either the wild-type or the Δbromodomain SWI/SNF complexes. After pull-down, the binding of both complexes to nucleosome arrays was detected by Western blot analysis using an anti-TAP antibody (Figure 4B), therefore testing the ability of the Swi2/Snf2 bromodomain to bind to acetylated nucleosomes. Both the wild-type and the Δbromodomain SWI/SNF complex bound equally well to the control template (Figure 4B, lanes 1 and 2). However, the wild-type SWI/SNF complex had a higher affinity for hyperacetylated templates compared with the mutant SWI/SNF complex (Figure 4B, lanes 3 and 4). Figure 4(C) shows a graphical representation of the ratio of the beads to the supernatants (B/S) for three independent immobilized binding experiments ±S.D. Again, the binding of the SWI/SNF complex to unmodified nucleosomes did not depend on the Swi2/Snf2 bromodomain of the SWI/SNF complex (Figure 4C). In contrast, when hyperacetylated nucleosome templates were used, the wild-type SWI/SNF but not the bromodomain-deleted complex could bind the templates efficiently (Figure 4C). These results show that while both an intact SWI/SNF complex and a complex that lacked the Swi2/Snf2 bromodomain can bind to unmodified promoter nucleosomes, the bromodomain-containing SWI/SNF complex binds with a greater affinity to the acetylated nucleosomes. The data clearly illustrate the importance of the Swi2/Snf2 bromodomain in binding to hyperacetylated histones and its requirement for efficient binding and anchoring of the SWI/SNF complex to these templates, leading to chromatin remodelling and subsequent gene activation.
Figure 4. The wild-type SWI/SNF complex has a higher affinity for hyperacetylated nucleosomes.
(A) Diagram of the immobilized template binding experiment. Biotinylated G5E4-5S was reconstituted with either control or hyperacetylated histones and bound to paramagnetic beads (Dynabeads) coupled to streptavidin as described in [25]. After the addition of the wild-type and Δbromodomain SWI/SNF complexes, the beads were separated from the supernatants and Western blotting was performed as described in [26] using an anti-TAP antibody. (B) Immobilized template binding assay shows that the wild-type SWI/SNF complex had a higher affinity for hyperacetylated templates compared with the Δbromodomain SWI/SNF complex (lanes 3 and 4). In contrast, both the wild-type and the Δbromodomain SWI/SNF complex bound equally well to the control template (lanes 1 and 2). (C) The quantification of three independent experiments is shown as the ratio of the bead to the supernatant (B/S) ±S.D., determined by scanning the gels and using the signal intensities of the Western blots for control and hyperacetylated histones. wt, wild-type.
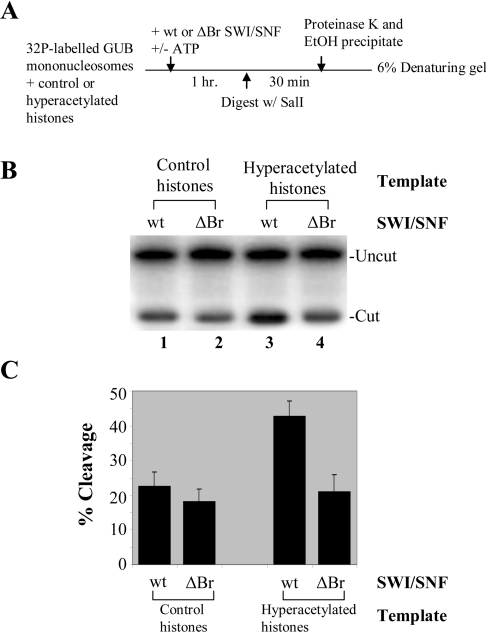
To test directly whether the increased binding of the wild-type SWI/SNF complex to hyperacetylated nucleosomes (observed in Figure 4) leads to increased remodelling activity of the complex, a restriction enzyme digestion assay was performed. The DNA used in this assay was the 183 bp GUB fragment described above. As shown in Figure 5(A), the radiolabelled GUB DNA fragment was reconstituted into a nucleosome template by octamer transfer followed by the re-modelling reaction and digestion by the restriction enzyme SalI. We analysed the ability of the SalI restriction enzyme to digest its site in approximately the middle of the 183 bp GUB nucleosomal DNA with the wild-type and the Δbromodomain SWI/SNF complexes in the presence of ATP. Figure 5(B) shows that both the wild-type and the mutant complexes increased the accessibility of the restriction enzyme to its site on the nucleosomal DNA. An equal amount of the control nucleosomal template was cleaved with either of these complexes, indicating that the deletion of the bromodomains in the Swi2/Snf2 subunit of the SWI/SNF complex did not significantly affect its remodelling activity on control (unmodified) histone templates. However, when hyperacetylated nucleosomes were used as the template, there was a 2-fold increase in remodelling activity of the wild-type SWI/SNF compared with the Δbromodomain SWI/SNF complex, indicating that the Swi2/Snf2 bromodomain is important for the efficient remodelling activity of the complex on acetylated nucleosomes. Quantification of three independent restriction enzyme accessibility experiments ±S.D. (Figure 5C) confirms these findings. In our recent publication [32], we have compared the activities of the wild-type SWI/SNF with that of the bromodomain-deleted complex on SAGA-acetylated templates and have shown that when the template was acetylated by SAGA in the presence of acetyl-CoA, reduced digestion by the SalI restriction enzyme was observed with the mutant SWI/SNF complex. In the present study, since we did not want the presence of the SAGA complex itself hindering the complete binding and remodelling of the templates by the wild-type or the mutant SWI/SNF complexes, we used hyperacetylated histones as the template instead. We have shown that when using hyperacetylated histones, there was a 2-fold increase in remodelling activity of the wild-type SWI/SNF compared with the Δbromodomain SWI/SNF complex or compared with the activity of the wild-type complex on control templates. The increase in the activity of the wild-type complex on hyperacetylated templates compared with control templates differs from when SAGA-acetylated templates were used. All these results show that the deletion of the bromodomain significantly reduced the binding as well as the functional activity of the SWI/SNF to remodel acetylated nucleosome templates.
Figure 5. The wild-type SWI/SNF complex has enhanced remodelling activity on hyperacetylated nucleosomes compared with the Δbromodomain SWI/SNF complex.
(A) Diagram of the restriction enzyme accessibility assay as described previously [36,46]. A 32P-labelled GUB fragment was reconstituted into mononucleosomes with either control or hyperacetylated histones for use in this assay. An equal amount of wild-type or Δbromodomain SWI/SNF was added to this GUB template in the presence or absence of ATP followed by SalI digestion. After stopping the digestion reaction and ethanol precipitation, the DNA was resolved on a 6% acrylamide/8 M urea sequencing gel at 150 V for 3 h. The gels were dried and visualized by autoradiography and quantified. (B) Restriction enzyme accessibility assay showing increased activity on hyperacetylated nucleosomes when the Swi2/Snf2 bromodomain is present in the complex. The uncut and cut DNA fragments are labelled. (C) The quantification of three independent experiments is shown as the percentage cleavage of the mononucleosome templates by the restriction enzyme in the presence of the wild-type or the Δbromodomain mutant chromatin-remodelling complex SWI/SNF. The amount of digestion in the absence of ATP is subtracted from that value obtained in its presence when the percentage cleavage was calculated. Quantification of the experiments shows that a similar percentage of the unmodified nucleosome template (with control histones) is cleaved with both the wild-type and the mutant complexes. However, with hyperacetylated nucleosomes as the template, there is a 2-fold increase in remodelling activity of the wild-type SWI/SNF compared with the Δbromodomain SWI/SNF complex.
DISCUSSION
In the present paper, we have studied the binding of various bromodomains within transcriptional co-activators to differentially acetylated histone tail peptides as well as HAT-acetylated histones. Moreover, we have examined the importance of the Swi2/Snf2 bromodomain in the yeast SWI/SNF complex in the binding and remodelling activity of this complex on hyperacetylated nucleosomes. We have confirmed that the bromodomain is an acetyl binding and recognition domain as all of the bromodomains tested showed increased affinity for hyperacetylated histones compared with control histones in GST pull-down assays (Figure 1B). Moreover, we have shown that different bromodomains of yeast co-activator complexes bind to acetylated H3 and H4 peptides with different affinities (Figure 2). Our results reveal that while the Swi2/Snf2 bromodomain had the strongest binding affinity to acetylated H3 and H4 peptides of all of the bromodomains tested, the other bromodomains differentially recognize the acetylated peptides. In addition, more physiological acetylation by the native yeast HATs (SAGA and NuA4) also show different degrees of binding to the bromodomains consistent with the peptide binding assay (Figure 3). Again, the Swi2/Snf2 bromodomain has the highest level of binding to both SAGA- and NuA4- acetylated histones. These data illustrate the importance of the bromodomains in binding to acetylated nucleosomes in general, and their selective binding to differentially acetylated histones, in particular. Our results reveal that the Swi2/Snf2, Gcn5, Bdf1-1, Rsc1-2 and the Spt7 bromodomain had the highest increase in binding to hyperacetylated histones, whereas the second bromodomains of Bdf1 and Rsc2 had a weaker overall binding to hyperacetylated histones. This did not seem to be the case when acetylated peptides or HAT-acetylated histones were used as the template. The two SAGA bromodomains had a higher affinity for acetylated H3 histones, whereas the Bdf1-2 bromodomain preferred binding to acetylated H4 histones consistent with previously published data [16,26]. We observed a 1.9-fold increase in binding of the Spt7 bromodomain to hyperacetylated histones compared with control, but only weak binding to acetylated H3 peptides and SAGA-acetylated histones. It is possible that the Spt7 bromodomain weakly interacts with individually acetylated lysine residues, but would have a synergistic increase in binding when multiple acetylated lysine residues are presented as the substrate, as our data shows. The differences observed between these bromodomains in binding to various acetylated histones are perhaps a result of sequence diversity that exists among them. Figure 6 compares the amino acid sequences between the different bromodomains used in the present study and shows the sequences that are identical within the bromodomains. The sequences also show the diversity among the bromodomains, which supports the idea that these bromodomains may have evolved to recognize and read distinct ‘histone codes’. The selective recognition of the bromodomains observed in the present study accounts for the broad effects of bromodomain-containing proteins observed on binding to histones.
Figure 6. Sequence homology of the different bromodomains.
The amino acid sequences of all the seven bromodomains used in the present paper are aligned and identical sequences in each of the bromodomains are shown separately in different shades of grey. The code at the bottom shows the amino acids that are identical within the bromodomains.
The fact that the Swi2/Snf2 bromodomain binds efficiently to almost all of the acetylated histone peptides suggests low selectivity of the Swi2/Snf2 bromodomain for acetylated lysine residues. This low selectivity of the Swi2/Snf2 bromodomain in recognizing the acetylated peptides could mean that the SWI/SNF complex might play an important and a more global role in interactions with acetylated promoters and in transcription regulation than previously thought. In other words, the SWI/SNF complex could potentially interact with a variety of acetylated promoters that may have different acetylation patterns. Other more selective bromodomains may be more important in regulation of specifically acetylated promoters. It is also likely that some of the other modifying proteins that have two or more bromodomains (such as SAGA, RSC and TFIID) could bind to acetylated histones with higher affinity, in a similar manner to the Swi2/Snf2 bromodomain, as a result of co-operation between two tandem bromodomains within the same complex [28,29]. It is reasonable to assume that the multiple bromodomains within one complex enables simultaneous recognition of multiple modifications of acetylated histones, resulting in enhanced overall binding to acetylated promoter nucleosomes.
We have also shown that even though SWI/SNF purified from a Swi2/Snf2 bromodomain deletion strain bound and remodelled unacetylated nucleosome templates as well as the wild-type complex, it could not bind or remodel acetylated nucleosomes efficiently (Figures 4 and 5). The results show the requirement for the Swi2/Snf2 bromodomain in anchoring the complex on acetylated templates prior to any remodelling. The difference between the ability of the wild-type and the bromodomain-deleted SWI/SNF complexes to remodel acetylated promoter nucleosomes is due to the fact that the Swi2/Snf2 bromodomain has the ability to recognize and bind to acetylated histone [26]. We have also previously shown that the Swi2/Snf2 bromodomain deletion reduces occupancy of the SWI/SNF complex at the SUC2 promoter under derepressing conditions [26]. This is consistent with our current data showing that the Swi2/Snf2 bromodomain deletion cannot bind and remodel acetylated promoter nucleosomes. It is likely that bromodomain-containing protein complexes such as SWI/SNF, RSC and TFIID bind with greater affinity to nucleosomes that are pre-acetylated by HATs such as the SAGA or NuA4 complexes [32,48–50]. Thus acetylated nucleosomes could serve as substrates for sequential and ordered binding of these complexes to the promoter, where these complexes compete with each other for binding. SAGA itself, containing two bromodomains, could also bind to the nucleosomes (through its Gcn5 bromodomain) that it has acetylated or those that are acetylated by NuA4 and compete for binding with other complexes. This competition between the bromodomain-containing proteins depends on which lysine residue is acetylated since these would serve as docking sites for the complexes. Whereas the Swi2/Snf2 bromodomain, for example, is broader in recognition and binding to acetylated lysine residues, other bromodomains are more selective in this recognition (Figure 2) and thus it is also possible that multiple complexes can bind to promoters and exert their combined effects to regulate transcription. Therefore multiple bromodomains in proteins with different specificities and multiple complexes with bromodomains, as well as other protein motifs (such as the chromodomain or the SANT domain), add further complexity to histone recognition. Multiple post-translational modifications within the histone tails and multiple DNA-/protein-interacting domains within all the different co-activators together could provide a mechanism by which protein complexes can read the ‘histone code’ and exert control over chromatin function and gene regulation.
Online data
Acknowledgments
We thank Philippe Prochasson and Jerry L. Workman for their help and advice in conducting these experiments. This work was supported by grants from the UAE University (02-03-8-11/04 and 02-03-8-11/05).
References
- 1.Workman J. L., Kingston R. E. Alteration of nucleosome structure as a mechanism of transcriptional regulation. Annu. Rev. Biochem. 1998;67:545–579. doi: 10.1146/annurev.biochem.67.1.545. [DOI] [PubMed] [Google Scholar]
- 2.Brown C. E., Lechner T., Howe L., Workman J. L. The many HATs of transcription coactivators. Trends Biochem. Sci. 2000;25:15–19. doi: 10.1016/s0968-0004(99)01516-9. [DOI] [PubMed] [Google Scholar]
- 3.Peterson C. L., Workman J. L. Promoter targeting and chromatin remodeling by the SWI/SNF complex. Curr. Opin. Genet. Dev. 2000;10:187–192. doi: 10.1016/s0959-437x(00)00068-x. [DOI] [PubMed] [Google Scholar]
- 4.Vignali M., Hassan A. H., Neely K. E., Workman J. L. ATP-dependent chromatin-remodeling complexes. Mol. Cell. Biol. 2000;20:1899–1910. doi: 10.1128/mcb.20.6.1899-1910.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Johnson C. N., Adkins N. L., Georgel P. Chromatin remodeling complexes: ATP-dependent machines in action. Biochem. Cell Biol. 2005;83:405–417. doi: 10.1139/o05-115. [DOI] [PubMed] [Google Scholar]
- 6.Strahl B. D., Allis C. D. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 7.Jenuwein T., Allis C. D. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 8.Gill G. Something about SUMO inhibits transcription. Curr. Opin. Genet. Dev. 2005;15:536–541. doi: 10.1016/j.gde.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 9.Kimura A., Matsubara K., Horikoshi M. A decade of histone acetylation: marking eukaryotic chromosomes with specific codes. J. Biochem. (Tokyo) 2005;138:647–662. doi: 10.1093/jb/mvi184. [DOI] [PubMed] [Google Scholar]
- 10.Martin C., Zhang Y. The diverse functions of histone lysine methylation. Nat. Rev. Mol. Cell Biol. 2005;6:838–849. doi: 10.1038/nrm1761. [DOI] [PubMed] [Google Scholar]
- 11.Shilatifard A. Chromatin modifications by methylation and ubiquitination: implications in the regulation of gene expression. Annu. Rev. Biochem. 2006;75:243–269. doi: 10.1146/annurev.biochem.75.103004.142422. [DOI] [PubMed] [Google Scholar]
- 12.Ornaghi P., Ballario P., Lena A. M., Gonzalez A., Filetici P. The bromodomain of Gcn5p interacts in vitro with specific residues in the N-terminus of histone H4. J. Mol. Biol. 1999;287:1–7. doi: 10.1006/jmbi.1999.2577. [DOI] [PubMed] [Google Scholar]
- 13.Jacobson R. H., Ladurner A. G., King D. S., Tjian R. Structure and function of a human TAFII250 double bromodomain module. Science. 2000;288:1422–1425. doi: 10.1126/science.288.5470.1422. [DOI] [PubMed] [Google Scholar]
- 14.Owen D. J., Ornaghi P., Yang J. C., Lowe N., Evans P. R., Ballario P., Neuhaus D., Filetici P., Travers A. A. The structural basis for the recognition of acetylated histone H4 by the bromodomain of histone acetyltransferase gcn5p. EMBO J. 2000;19:6141–6149. doi: 10.1093/emboj/19.22.6141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ladurner A. G., Inouye C., Jain R., Tjian R. Bromodomains mediate an acetyl-histone encoded antisilencing function at heterochromatin boundaries. Mol. Cell. 2003;11:365–376. doi: 10.1016/s1097-2765(03)00035-2. [DOI] [PubMed] [Google Scholar]
- 16.Matangkasombut O., Buratowski S. Different sensitivities of bromodomain factors 1 and 2 to histone H4 acetylation. Mol. Cell. 2003;11:353–363. doi: 10.1016/s1097-2765(03)00033-9. [DOI] [PubMed] [Google Scholar]
- 17.Loyola A., Almouzni G. Bromodomains in living cells participate in deciphering the histone code. Trends Cell Biol. 2004;14:279–281. doi: 10.1016/j.tcb.2004.04.005. [DOI] [PubMed] [Google Scholar]
- 18.Martinez-Campa C., Politis P., Moreau J. L., Kent N., Goodall J., Mellor J., Goding C. R. Precise nucleosome positioning and the TATA box dictate requirements for the histone H4 tail and the bromodomain factor Bdf1. Mol. Cell. 2004;15:69–81. doi: 10.1016/j.molcel.2004.05.022. [DOI] [PubMed] [Google Scholar]
- 19.Yang X. J. Lysine acetylation and the bromodomain: a new partnership for signaling. BioEssays. 2004;26:1076–1087. doi: 10.1002/bies.20104. [DOI] [PubMed] [Google Scholar]
- 20.Polesskaya A., Naguibneva I., Duquet A., Bengal E., Robin P., Harel-Bellan A. Interaction between acetylated MyoD and the bromodomain of CBP and/or p300. Mol. Cell Biol. 2001;21:5312–5320. doi: 10.1128/MCB.21.16.5312-5320.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mujtaba S., He Y., Zeng L., Farooq A., Carlson J. E., Ott M., Verdin E., Zhou M. M. Structural basis of lysine-acetylated HIV-1 Tat recognition by PCAF bromodomain. Mol. Cell. 2002;9:575–586. doi: 10.1016/s1097-2765(02)00483-5. [DOI] [PubMed] [Google Scholar]
- 22.Dorr A., Kiermer V., Pedal A., Rackwitz H. R., Henklein P., Schubert U., Zhou M. M., Verdin E., Ott M. Transcriptional synergy between Tat and PCAF is dependent on the binding of acetylated Tat to the PCAF bromodomain. EMBO J. 2002;21:2715–2723. doi: 10.1093/emboj/21.11.2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mujtaba S., He Y., Zeng L., Yan S., Plotnikova O., Sachchidanand Sanchez, R., Zeleznik-Le N. J., Ronai Z., Zhou M. M. Structural mechanism of the bromodomain of the coactivator CBP in p53 transcriptional activation. Mol. Cell. 2004;13:251–263. doi: 10.1016/s1097-2765(03)00528-8. [DOI] [PubMed] [Google Scholar]
- 24.Ma B., Pan Y., Gunasekaran K., Keskin O., Venkataraghavan R. B., Levine A. J., Nussinov R. The contribution of the Trp/Met/Phe residues to physical interactions of p53 with cellular proteins. Phys. Biol. 2005;2:56–66. doi: 10.1088/1478-3975/2/2/S06. [DOI] [PubMed] [Google Scholar]
- 25.Hassan A. H., Neely K. E., Workman J. L. Histone acetyltransferase complexes stabilize swi/snf binding to promoter nucleosomes. Cell. 2001;104:817–827. doi: 10.1016/s0092-8674(01)00279-3. [DOI] [PubMed] [Google Scholar]
- 26.Hassan A. H., Prochasson P., Neely K. E., Galasinski S. C., Chandy M., Carrozza M. J., Workman J. L. Function and selectivity of bromodomains in anchoring chromatin-modifying complexes to promoter nucleosomes. Cell. 2002;111:369–379. doi: 10.1016/s0092-8674(02)01005-x. [DOI] [PubMed] [Google Scholar]
- 27.Pamblanco M., Poveda A., Sendra R., Rodriguez-Navarro S., Perez-Ortin J. E., Tordera V. Bromodomain factor 1 (Bdf1) protein interacts with histones. FEBS Lett. 2001;496:31–35. doi: 10.1016/s0014-5793(01)02397-3. [DOI] [PubMed] [Google Scholar]
- 28.Dey A., Chitsaz F., Abbasi A., Misteli T., Ozato K. The double bromodomain protein Brd4 binds to acetylated chromatin during interphase and mitosis. Proc. Natl. Acad. Sci. U.S.A. 2003;100:8758–8763. doi: 10.1073/pnas.1433065100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kasten M., Szerlong H., Erdjument-Bromage H., Tempst P., Werner M., Cairns B. R. Tandem bromodomains in the chromatin remodeler RSC recognize acetylated histone H3 Lys14. EMBO J. 2004;23:1348–1359. doi: 10.1038/sj.emboj.7600143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou Y., Grummt I. The PHD finger/bromodomain of NoRC interacts with acetylated histone H4K16 and is sufficient for rDNA silencing. Curr. Biol. 2005;15:1434–1438. doi: 10.1016/j.cub.2005.06.057. [DOI] [PubMed] [Google Scholar]
- 31.Kanno T., Kanno Y., Siegel R. M., Jang M. K., Lenardo M. J., Ozato K. Selective recognition of acetylated histones by bromodomain proteins visualized in living cells. Mol. Cell. 2004;13:33–43. doi: 10.1016/s1097-2765(03)00482-9. [DOI] [PubMed] [Google Scholar]
- 32.Hassan A. H., Awad S., Prochasson P. The Swi2/Snf2 bromodomain is required for the displacement of saga and the octamer transfer of saga-acetylated nucleosomes. J. Biol. Chem. 2006;281:18126–18134. doi: 10.1074/jbc.M602851200. [DOI] [PubMed] [Google Scholar]
- 33.Eberharter A., John S., Grant P. A., Utley R. T., Workman J. L. Identification and analysis of yeast nucleosomal histone acetyltransferase complexes. Methods. 1998;15:315–321. doi: 10.1006/meth.1998.0635. [DOI] [PubMed] [Google Scholar]
- 34.Rigaut G., Shevchenko A., Rutz B., Wilm M., Mann M., Seraphin B. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 1999;17:1030–1032. doi: 10.1038/13732. [DOI] [PubMed] [Google Scholar]
- 35.Puig O., Caspary F., Rigaut G., Rutz B., Bouveret E., Bragado-Nilsson E., Wilm M., Seraphin B. The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods. 2001;24:218–229. doi: 10.1006/meth.2001.1183. [DOI] [PubMed] [Google Scholar]
- 36.Graumann J., Dunipace L. A., Seol J. H., McDonald W. H., Yates J. R., 3rd, Wold B. J., Deshaies R. J. Applicability of tandem affinity purification MudPIT to pathway proteomics in yeast. Mol. Cell. Proteomics. 2004;3:226–237. doi: 10.1074/mcp.M300099-MCP200. [DOI] [PubMed] [Google Scholar]
- 37.Lee K. K., Prochasson P., Florens L., Swanson S. K., Washburn M. P., Workman J. L. Proteomic analysis of chromatin-modifying complexes in Saccharomyces cerevisiae identifies novel subunits. Biochem. Soc. Trans. 2004;32:899–903. doi: 10.1042/BST0320899. [DOI] [PubMed] [Google Scholar]
- 38.Ikeda K., Steger D. J., Eberharter A., Workman J. L. Activation domain-specific and general transcription stimulation by native histone acetyltransferase complexes. Mol. Cell. Biol. 1999;19:855–863. doi: 10.1128/mcb.19.1.855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Carrozza M. J., Hassan A. H., Workman J. L. Assay of activator recruitment of chromatin-modifying complexes. Methods Enzymol. 2003;371:536–544. doi: 10.1016/S0076-6879(03)71040-4. [DOI] [PubMed] [Google Scholar]
- 40.Juan L. J., Utley R. T., Adams C. C., Vettese-Dadey M., Workman J. L. Differential repression of transcription factor binding by histone H1 is regulated by the core histone amino termini. EMBO J. 1994;13:6031–6040. doi: 10.1002/j.1460-2075.1994.tb06949.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Neely K. E., Hassan A. H., Wallberg A. E., Steger D. J., Cairns B. R., Wright A. P. H., Workman J. L. Activation domain-mediated targeting of the SWI/SNF complex to promoters stimulates transcription from nucleosome arrays. Mol. Cell. 1999;4:649–655. doi: 10.1016/s1097-2765(00)80216-6. [DOI] [PubMed] [Google Scholar]
- 42.Prochasson P., Neely K. E., Hassan A. H., Li B., Workman J. L. Targeting activity is required for SWI/SNF function in vivo and is accomplished through two partially redundant activator-interaction domains. Mol. Cell. 2003;12:983–990. doi: 10.1016/s1097-2765(03)00366-6. [DOI] [PubMed] [Google Scholar]
- 43.Utley R. T., Ikeda K., Grant P. A., Côté J., Steger D. J., Eberharter A., John S., Workman J. L. Transcriptional activators direct histone acetyltransferase complexes to nucleosomes. Nature. 1998;394:498–502. doi: 10.1038/28886. [DOI] [PubMed] [Google Scholar]
- 44.Vignali M., Steger D. J., Neely K. E., Workman J. L. Distribution of acetylated histones resulting from Gal4-VP16 recruitment of SAGA and NuA4 complexes. EMBO J. 2000;19:2629–2640. doi: 10.1093/emboj/19.11.2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wallberg A. E., Neely K. E., Hassan A. H., Gustafsson J.-Å., Workman J. L., Wright A. P. H. Recruitment of the SWI-SNF chromatin remodeling complex as a mechanism of gene activation by the glucocorticoid receptor t1 activation domain. Mol. Cell. Biol. 2000;20:2004–2013. doi: 10.1128/mcb.20.6.2004-2013.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Natarajan K., Jackson B. M., Zhou H., Winston F., Hinnebusch A. G. Transcriptional activation by Gcn4p involves independent interactions with the SWI/SNF complex and the SRB/mediator. Mol. Cell. 1999;4:657–664. doi: 10.1016/s1097-2765(00)80217-8. [DOI] [PubMed] [Google Scholar]
- 47.Yudkovsky N., Logie C., Hahn S., Peterson C. L. Recruitment of the SWI/SNF chromatin remodeling complex by transcriptional activators. Genes Dev. 1999;13:2369–2374. doi: 10.1101/gad.13.18.2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Agalioti T., Lomvardas S., Parekh B., Yie J., Maniatis T., Thanos D. Ordered recruitment of chromatin modifying and general transcription factors to the IFN-β promoter. Cell. 2000;103:667–678. doi: 10.1016/s0092-8674(00)00169-0. [DOI] [PubMed] [Google Scholar]
- 49.Agalioti T., Chen G., Thanos D. Deciphering the transcriptional histone acetylation code for a human gene. Cell. 2002;111:381–392. doi: 10.1016/s0092-8674(02)01077-2. [DOI] [PubMed] [Google Scholar]
- 50.Qiu H., Hu C., Yoon S., Natarajan K., Swanson M. J., Hinnebusch A. G. An array of coactivators is required for optimal recruitment of TATA binding protein and RNA polymerase II by promoter-bound Gcn4p. Mol. Cell. Biol. 2004;24:4104–4117. doi: 10.1128/MCB.24.10.4104-4117.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.