Abstract
The study of regulatory networks in human pathogens such as Cryptococcus neoformans provides insights into host-pathogen interactions that may allow for correlation of gene expression patterns with clinical outcomes. In the present study, deletion of the cryptococcal copper-dependent transcription factor 1 (Cuf1) led to defects in growth and virulence factor expression in low copper conditions. In mouse models, cuf1Δ strains exhibited reduced dissemination to the brain, but no change in lung growth, suggesting copper is limiting in neurologic infections. To examine this further, a biologic probe of available copper was constructed using the cryptococcal CUF1-dependent copper transporter, CTR4. Fungal cells demonstrated high CTR4 expression levels after phagocytosis by macrophage-like J774.16 cells and during infection of mouse brains, but not lungs, consistent with limited copper availability during neurologic infection. This was extended to human brain infections by demonstrating CTR4 expression during C. neoformans infection of an AIDS patient. Moreover, high CTR4 expression by cryptococcal strains from 24 solid organ transplant patients was associated with dissemination to the CNS. Our results suggest that copper acquisition plays a central role in fungal pathogenesis during neurologic infection and that measurement of stable traits such as CTR4 expression may be useful for risk stratification of individuals with cryptococcosis.
Introduction
Cryptococcus neoformans has emerged as a major causative agent of infection in immunocompromised hosts as a result of infections such as AIDS as well as transplant conditioning regimens and cancer chemotherapy (1). The most serious clinical manifestation of cryptococcal infection is meningoencephalitis, which is occasionally associated with cryptococcemia and is a predictor of poor outcome (2, 3). Cryptococcal pneumonia is also serious, but it is associated with a better prognosis and is more prevalent in organ transplant and hematologic malignancy patients (4–6). More recently, an immune reconstitution syndrome has been described both in AIDS patients with the availability of antiretroviral therapy (7) and in organ transplant/hematologic malignancy patients (8). Overall, AIDS-related rates of cryptococcal infection have diminished in developed countries with the advent of highly active antiretroviral therapy (HAART), but the problem has remained stubbornly persistent in transplant/hematologic patients. Clearly, a need remains for novel methods for the treatment and control of this disease.
The transition metal copper is both required for growth of fungi and highly sequestered by biologic buffers and proteins within the mammalian host (9). This suggests a scenario whereby the pathogen competes with the mammalian host for available copper stores. Unfortunately, some patients at risk for cryptococcosis such as AIDS patients may be at a particular disadvantage in the cellular battle with pathogens for copper, because these patients have been shown to have elevated levels of copper in serum (10, 11). The importance of intact copper homeostasis in the fungal pathogen C. neoformans is suggested by the copper dependency of at least 2 virulence factors, the Cu/Zn superoxide dismutase (12) and the copper-containing laccase enzyme (13, 14). In addition, copper may prove to be particularly important during fungal pathogenesis, because copper homeostasis also affects iron through a requirement for copper by the high-affinity iron transporter Fet3 (15).
While there is little doubt that copper is required for pathogens such as C. neoformans, the question remains as to whether copper is biologically limiting to the pathogen within the mammalian host. To help address this question, we studied the role for a cryptococcal homolog of copper-dependent transcription factor 1 (Cuf1) and its relationship to the Cuf1-dependent copper transporter 4 (Ctr4) during mammalian pathogenesis. These studies showed that copper is indeed limiting to the fungus during infection of brain, but not lungs of mice, the former, requiring the induction of the copper transporter during infection in both mice and humans. Furthermore, strain variation in expression levels of the CTR4 copper transporter was evident within a cohort of primary cryptococcal isolates from organ transplant patients, and that higher expression levels of this stable genetic trait were associated with brain dissemination in these patients.
Results
Identification of a copper-dependent transcription factor in C. neoformans.
Complementation of one of a series of previously described laccase-deficient mutants (JEC111; ref. 16) identified a cryptococcal homolog of the copper regulator CUF1. The cryptococcal Cuf1 protein exhibits homology within the N terminus to DNA-binding regions described for the copper transcription factors, Ace1 from Saccharomyces cerevisiae and Cuf1 from Schizosaccharomyces pombe, both of which contain a KGRP consensus sequence (ref. 17 and Figure 1A). In addition, the cryptococcal Cuf1 contains a C-terminal region and contains a cysteine-rich sequence containing the consensus putative copper-binding motif 349C–X–C–X3–C–X–C–X2–C–X2–H common to the copper-sensing transcription factor Mac1 from S. cerevisiae and Cuf1 from S. pombe (18). Identification of a cryptococcal Cuf1 homolog provides a unique tool to study the role of copper homeostasis during pathogenesis, because Cuf1 is believed to be exclusively a copper-dependent transcription factor (18, 19).
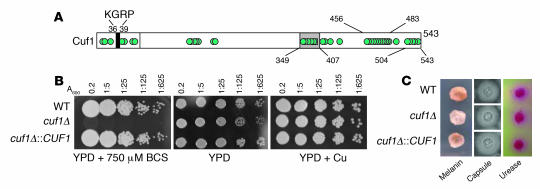
Figure 1. Deletion of CUF1 from C. neoformans yields a copper-sensitive phenotype.
(A) Schematic of the Cuf1 protein from C. neoformans shows a KGRP consensus sequence and multiple cysteine-rich domains (cysteines represented by circles). (B) Phenotypes of CUF1 mutants under various copper conditions. Cells were diluted to an A600 of 0.2, serially diluted 1:5, and applied to YPD alone or YPD supplemented with either 750 μM BCS or 10 μM CuCl2 at 37°C for 3 days. (C) Cells were incubated for melanin production as described in Methods (left panels) or on malt extract for 5 days at 30°C and examined by India ink (middle panels) or incubated on Christensen’s agar for 2 hours (right panels).
As predicted by the JEC111 mutant, a targeted cuf1Δ mutant showed decreased laccase activity and a severe growth defect at low copper concentrations, achieved through the addition of the copper chelator bathocuproinedisulfonic acid (BCS); this growth defect was restored after addition of 10 μM CuCl2 (Figure 1B), as previously described for mac1- and cuf1-null alleles of S. cerevisiae and S. pombe, respectively (17, 20). Lack of sufficient intracellular stores of copper would be expected to result in ineffective copper loading of the mitochondrial Cox1 enzyme, which was confirmed by the severe growth defect exhibited in Figure 1B (21). Growth in yeast extract/peptone/dextrose medium (YPD) without copper was mildly attenuated (Figure 1B), suggesting a mild deficiency of copper in this media; however, addition of 1 mM hydrogen peroxide or 2 μg/ml menadione caused no additional growth reduction (data not shown), suggesting no increased sensitivity to oxidant stress of the mutant. However, the cryptococcal cuf1Δ mutant showed intact expression of capsule and urease (Figure 1C), suggesting sufficient metal stores for these activities under the conditions tested.
Reduced virulence of a cryptococcal cuf1Δ mutant strain.
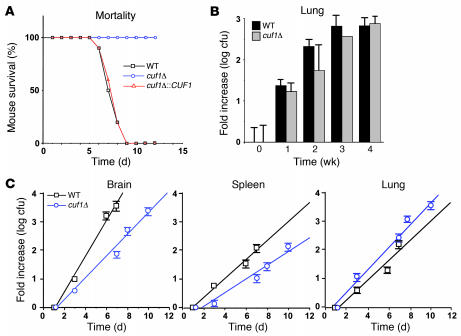
We next used the cuf1Δ strain in order to assess the virulence of a cryptococcal strain dependent on copper repletion for effective growth and virulence factor expression. As shown in Figure 2A, deletion of CUF1 resulted in attenuated virulence in an i.v. mouse model of cryptococcosis despite a very large inoculum of 106 cells; application of the same number of WT cells caused rapid development of meningoencephalitis and death (P < 0.01, cuf1Δ versus WT and cuf1Δ::CUF1). Parallel experiments comparing virulence of a lac1Δ mutant to that of the WT in the same genetic background using the same i.v. model found a smaller reduction in virulence in the lac1Δ mutant (median survival, WT, 7 days; lac1Δ, 11 days; P < 0.05 [data not shown]) as reported previously for this fungal genetic background in a different mouse model (22), which suggests that the attenuated virulence of the cuf1Δ mutant was not solely caused by its defect in laccase activity. These data show the importance of CUF1 in the expression of laccase and virulence and provide genetic evidence in support of the importance of intact copper homeostasis in cryptococcosis. To further understand the nature of the requirement for copper of the cuf1Δ mutant during pathogenesis, we assessed lung fungal burden in a pulmonary model using CBA/J mice (23). As shown in Figure 2B, WT and cuf1Δ mutant cells showed equivalent pulmonary fungal burden over 4 weeks, suggesting that efficient copper acquisition and processing is less essential to C. neoformans during pulmonary infection. Brain dissemination in the intranasal CBA/J model for all fungal strains was not sufficient to allow statistically analyzable comparisons between strains. To corroborate the similarity of growth in the lung using the i.v. model, a time course of fungal burdens was conducted after i.v. injection of 106 WT and cuf1Δ fungal cells. As shown in Figure 2C, cuf1Δ and WT cells showed equivalent growth in the lung in this second model (P = 0.49), whereas the rate of cuf1Δ growth in the brain was reduced compared with that of WT cells (P = 0.006), consistent with the attenuated virulence of the mutant shown in Figure 2A. Cells in the spleen showed a trend toward reduced growth of the mutant, but this was not statistically significant (P = 0.10).
Figure 2. Effect of CUF1 deletion on survival in mice and pulmonary fungal burden.
(A) Swiss albino mice were injected by tail vein with 106 of the indicated cells, and their progress was followed until they were moribund. (B) CBA/J mice were intranasally inoculated with 105 of the indicated cells and at the indicated time points (0 weeks corresponds to 24 hours), mice were sacrificed, and their lungs were removed and cultured for growth of fungal cells on YPD. Data are mean ± SEM cfu per organ. n = 3 per time point. Only 1 of the mice infected with cuf1Δ cells and assigned for sacrifice at 3 weeks survived. (C) Swiss albino mice were injected as in A and sacrificed at the indicated time points, after which the indicated organs were removed and cultured for growth of fungal cells on YPD.
CTR4 expression as a copper-dependent biologic probe in murine macrophages and during infection of mice.
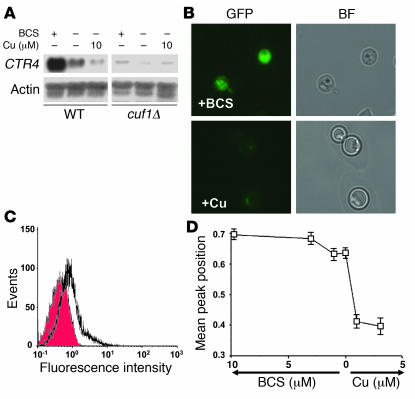
Copper-associated defects in the cuf1Δ mutant suggested that a copper transporter, such as a CTR4 homolog recently identified for use as an inducible promoter in C. neoformans (24), may show CUF1 dependence. As shown in Figure 3A, CTR4 showed a high level of CUF1-dependent activation under copper-limiting conditions as assessed by Northern blot, as described previously for S. pombe (18). Furthermore, expression of a CTR4-GFP fusion protein in WT C. neoformans under its native promoter allowed for visualization of the gene’s expression levels (Figure 3B) and quantitation by flow cytometry (Figure 3C), both of which demonstrated expression of CTR4 under copper-limiting conditions and suppression after addition of as little as 1 μM of copper salts (Figure 3D). In addition, CTR4 expression was not induced in the presence of copper after glucose starvation or incubation with oxidants such as 1 mM hydrogen peroxide or 2 μg/ml menadione (data not shown). In contrast, cuf1Δ cells expressing an equivalent CTR4-GFP construct did not exhibit copper-deficient CTR4 expression, as expected (data not shown).
Figure 3. CUF1 activates CTR4 under low copper conditions.
(A) Northern blot analysis was performed using RNA from the indicated cells grown on YPD and then incubated for 3 hours in asparagine salts with BCS or copper as indicated. (B) Epifluorescence of C. neoformans expressing a CTR4-GFP fusion protein under the native CTR4 promoter on asparagine salts as in A and supplemented with either 10 μM BCS or 3 μM CuCl2. BF, bright field. Original magnification, ×1,000. (C) Equivalent cells incubated as in B were analyzed by cell flow cytometry as described in Methods. The shaded histogram corresponds to signal from cells supplemented with Cu, and the open histogram corresponds to signal from cells treated with BCS. (D) Plot of mean peak position of epifluorescence by flow cytometry of CTR4-GFP–expressing cells incubated as in A and supplemented with the indicated concentrations of ether BCS or copper.
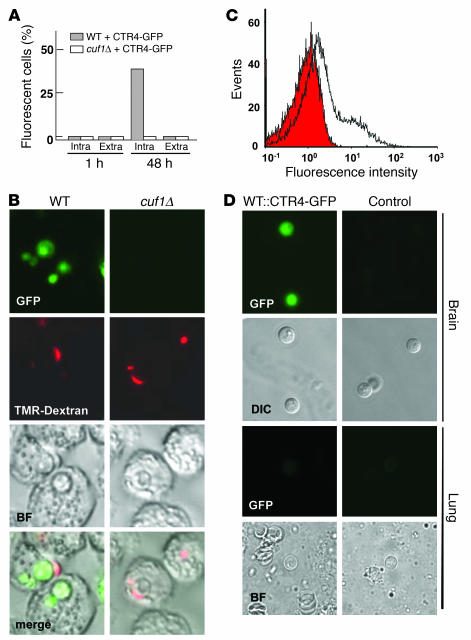
Next, CTR4-GFP expression was used to provide a biologic probe of copper availability to the fungus within the macrophage-like cell line J774.16 (25). After phagocytosis of cryptococcal cells, CTR4 was initially repressed at 1 hour as a result of exogenous copper added to the fungal growth media prior to inoculation (Figure 4A). However, after 48 hours, intracellular cryptococcal cells, demonstrated by colocalization with tetramethylrhodamine-labeled dextran, showed fluorescence indicative of CTR4 expression (P < 0.001, intracellular fungi versus all other groups; Figure 4, A and B). In contrast, phagocytosis of cryptococcal cuf1Δ mutant cells transformed with the equivalent CTR4-GFP construct in equivalent copy number did not show expression of CTR4-GFP, consistent with the notion of CUF1-dependent activation of CTR4. These data suggest that cryptococcal CTR4 activation is CUF1 dependent within phagolysosomes of the J774.16 macrophage cell line, likely a result of its copper-limiting environment. Furthermore, fungal cells recovered from mouse brains showed a high level of CTR4-GFP expression (Figure 4, C and D) compared with equivalent cells transformed with a plasmid not expressing CTR4-GFP (as a control for autofluorescence), consistent with limited copper availability during brain infection. In contrast, C. neoformans WT and cuf1Δ cells recovered from mouse lungs in a pulmonary model did not express CTR4-GFP, which suggests a greater availability of copper in this host tissue or a reduced requirement for copper by the fungus (Figure 4D). However, insufficient fungal cells were obtained from the brains of CBA/J mice to evaluate CTR4 expression.
Figure 4. Expression of CTR4 in macrophages and mouse brain demonstrates low copper availability during infection.
(A) WT or cuf1Δ cells expressing equivalent CTR4-GFP constructs under the CTR4 promoter were incubated with the macrophage-like cell line J774.16 for the indicated times; 200 intracellular (Intra) and extracellular (Extra) were scored for the presence of epifluorescence at the indicated time points. (B) Representative intracellular fungal cells from A were visualized by epifluorescence and intracellular colocalization by Tetramethylrhodamine-labeled dextran after phagocytosis as described in Methods. Original magnification, ×1,000. (C) C. neoformans cells expressing either CTR4-GFP (open histogram) or empty plasmid (shaded histogram) were injected (106 cells) by tail vein into Swiss albino mice and after sacrifice, fungal cells were harvested from their brains, and epifluorescence was measured by flow cytometry. (D) Representative cryptococcal cells from C were visualized by microscopy after recovery from the brains and lungs of intranasally injected CBA/J mice. DIC, differential interference contrast. Original magnification, ×1,000.
Expression of CTR4 in human cryptococcosis.
To determine whether the above findings in mice could be applied to human cryptococcosis, we assessed CTR4 expression in human cryptococcosis. The patient was a 42-year-old male with fatal cryptococcosis that was reported previously (26). In situ hybridization was performed with an antisense RNA strand or a sense strand (negative control) of a 237-bp fragment encoding a nonconserved region of the C terminus of C. neoformans CTR4. A positive hybridization signal was detected only in sections probed with the antisense probe (Figure 5A), whereas sections probed with the corresponding sense probe showed no hybridization to yeast forms (Figure 5B). Sections were also stained with mucicarmine specific for C. neoformans capsule (Figure 5C) and H&E (Figure 5D). These data demonstrate cryptococcal CTR4 expression during human infection, extending a requirement for efficient fungal copper acquisition to human disease.
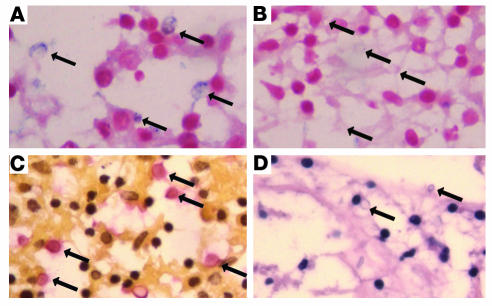
Figure 5. CTR4 expression during human cryptococcosis suggests low available copper stores during fungal infection.
(A) In situ hybridization of C. neoformans–infected brain tissue hybridized with an antisense CTR4 digoxigenin-labeled RNA probe. Arrows indicate alkaline phosphatase–positive yeast cells. (B) In situ hybridization of C. neoformans–infected brain tissue hybridized with a sense CTR4 digoxigenin-labeled RNA probe. Arrows indicate alkaline phosphatase–negative yeast cells. (C) Mucicarmine stain of C. neoformans–infected brain tissue. Arrows indicate mucicarmine-positive yeast cells. (D) H&E stain of C. neoformans–infected brain tissue. Arrows indicate unstained yeast cells. Original magnification, ×400.
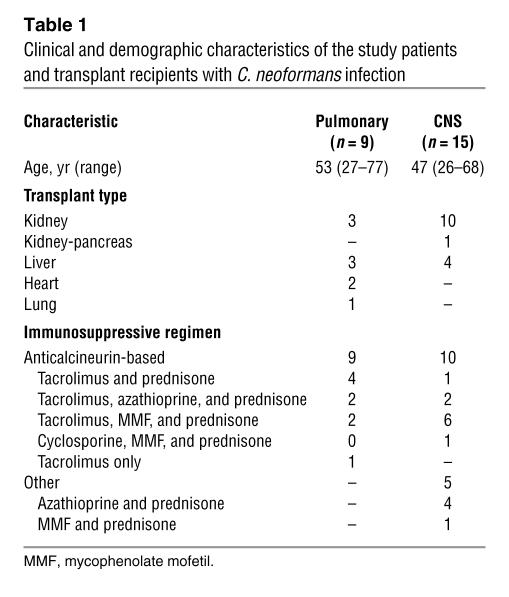
In order to further examine the role of C. neoformans CTR4 expression in human infection, we assessed CTR4 expression in 24 consecutive primary clinical isolates from patients receiving solid organ transplants; isolates were collected as part of an ongoing study of C. neoformans infection in organ transplant patients (27). All strains were established to be serotype A strains (28). The patients included in the present study were restricted to those receiving solid organ transplant with a positive serum cryptococcal antigen and positive fungal cultures from either bronchoalveolar lavage (BAL), lung biopsy, pleural fluid, or cerebrospinal fluid (CSF) (see Table 1). Those diagnosed with CNS infection had a positive CSF culture at the time of diagnosis, while those with pneumonia alone had at least 1 lumbar puncture at the time of diagnosis that yielded a negative cryptococcal antigen and negative fungal cultures. One patient that had a positive BAL culture but did not have a cryptococcal antigen performed was excluded prior to the study because of a concern that the BAL culture may represent colonization. As shown in Table 1, the immunosuppressive regimen was predominately anticalcineurin based, with 9 in the pulmonary group and 10 in the CNS group receiving such agents. Induction of fungal CTR4 was performed on primary isolates in 2 independent experiments to assess the genetic stability of the phenotype in each isolate. For our in vitro induction media, we chose asparagine salts without glucose (pH 6.5) because this provided a convenient in vitro test that could be adapted for high throughput use in later clinical trials and because this media was found to induce both CTR4 and laccase in the laboratory reference strain, H99, and laccase has also been demonstrated to be expressed during brain infection in mice (13). Only a single induction condition was chosen because of a concern that multiple comparisons between groups may reduce the statistical power of the comparison.
Table 1 .
Clinical and demographic characteristics of the study patients and transplant recipients with C. neoformans infection
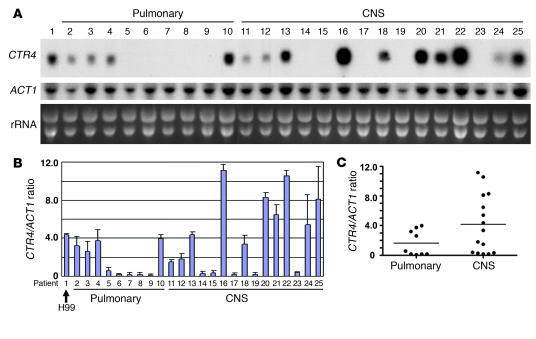
Interestingly, as shown in Figure 6, CTR4 expression varied widely between the clinical isolates (Figure 6A) but showed reproducible CTR4/ACT1 ratio values for each strain in duplicate experiments (Figure 6B). In contrast, the essential gene, ACT1, was expressed uniformly among isolates, varying less than 5%, as reflected by the intensity of ribosomal bands visualized by ethidium bromide staining of the gel. In addition, dot blots of genomic DNA from the clinical isolates showed uniform CTR4/ACT1 ratios, varying no more than 5% between isolates, which demonstrated that the wide variation in hybridization signals in the CTR4 Northern blot was not due to intrastrain variation in CTR4 sequence (data not shown). Interestingly, numerous strains causing CNS infection expressed high levels of CTR4 compared with isolates causing only pulmonary disease. In addition, the mean CTR4/ACT1 ratio was significantly elevated in these isolates compared with those causing pulmonary disease only (P = 0.043). In a secondary analysis, isolates with CTR4/ACT1 ratios greater than 4.15, the mean value of the CNS isolates, were significantly more likely to disseminate to the brain (P = 0.022, Fisher’s exact test), with an odds ratio of 16.8. In addition, because of a concern that 5 patients in the CNS infection group received different immunosuppressive regimens than did members of the pulmonary infection group (Table 1), additional analysis was performed comparing only the patients in each group receiving anticalcineurin-based regimens (i.e., excluding patients 11, 12, 14, 17, and 18 from Figure 6). According to this secondary analysis, the association of mean levels of CTR4/ACT1 with CNS infection increased in significance, as CTR4/ACT1 levels more highly correlated with dissemination to the CNS (P = 0.019). Interestingly, when isolates from representative high expressors (patients 16 and 22) and low CTR4 expressors (patients 8 and 9) were each inoculated i.v. into 10 Swiss albino mice at a 104 inoculum, mouse survival was lower and brain cfu counts at day 8 were higher in the high-expressor group than in the low-expressor group (mean ± SEM survival, patient 16, 9.1 ± 0.2 days; patient 22, 9.5 ± 0.2 days; patient 8, 20 ± 0 days; patient 9, 13 ± 1.4 days; brain cfu ± SEM, patient 16, 1.4 ± 0.5 × 107; patient 22, 1.1 ± 0.2 × 107; patient 8, 4.9 ± 0.9 × 105; patient 9, 7.8 ± 1.0 × 106; P < 0.01, survival and cfu counts between all patients with the exception of patients 16 and 22). However, because mouse models for cryptococcosis have not been validated regarding the ability to predict strain-related virulence in humans, conclusions about the contribution of CTR4 expression and virulence in these isolates must be interpreted with caution. Nevertheless, while such a result in a small sample size requires validation in larger trials for clinical applicability, it appears that CTR4 expression was predictive of dissemination toward CNS infection in this cohort of 24 clinical isolates. Because of a low number of deaths among the groups (6 deaths), we were not able to analyze for outcome by this parameter.
Figure 6. CTR4 expression varies among cryptococcal isolates, and high levels of expression confer greater ability to disseminate to the brain in human patients.
(A) Twenty-four C. neoformans primary isolates were obtained from consecutive patients (or an H99 laboratory strain) as described in the Methods. Cells were induced by inoculation in asparagine salts, and RNA was obtained and subjected to Northern blot using a fragment of the indicated gene. Ribosomal RNA (rRNA) was visualized by ethidium bromide staining of gels. (B) Northern blots were conducted in duplicate, and CTR4/ACT1 ratios were calculated for the indicated strains by densitometry. (C) Plot of CTR4/ACT1 ratios according to infected organ. Horizontal lines indicate the mean value for each group. P = 0.04.
Discussion
In the present study, isolation and deletion of a copper-dependent transcription factor, Cuf1, in the fungal pathogen C. neoformans allowed for analysis of the role of copper homeostasis during fungal infection. As expected, the cryptococcal cuf1Δ deletion strain exhibited copper-suppressible growth and virulence factor defects. More interestingly, virulence of the cuf1Δ deletion strain was attenuated compared with WT or CUF1-complemented cells using an i.v. dissemination model dependent on production of fatal meningoencephalitis (23). However, lung fungal burden of the cuf1Δ mutant after intranasal or i.v. challenge was equivalent to that of the WT, suggesting that the copper-suppressible phenotype is more associated with the development of neurologic rather than pulmonary disease. While copper has not to our knowledge been directly implicated in fungal virulence prior to these studies, the importance of effective copper acquisition and processing by pathogens has been suggested by an association in Candida albicans of the copper transporter Ctr1 and iron uptake (29), the role of C. albicans MAC1 in filamentation and invasive growth of S. cerevisiae (30), and differential expression of copper-related genes during phenotypic switching (31), traits associated with virulence in C. albicans. Indeed, competition for copper within the host may represent a general principle of microbial host defense, as increases in septic death have been observed following liver transplant in patients with Wilson disease, which is associated with copper excess (32). This finding suggests that, in addition to possible immune dysregulation effects (33), excess copper stores could tilt the balance of infection toward pathogens such as C. neoformans by providing a more ready source of the required metal.
Because transcription of CUF1 is constitutive, activating genes by a posttranslational mechanism (34), we used transcriptional levels of a copper/CUF1-dependent copper transporter, CTR4, fused to a GFP reporter gene in order to biologically probe both CUF1 activity and copper levels as sensed by the fungal pathogen. Historically, copper levels have been notoriously difficult to evaluate because the metal’s biologic availability is heavily dependent on the degree of sequestering by numerous proteins and buffers and may also be dependent on its oxidation state, making bulk measurements of tissue quantities uninformative (35). Interestingly, cryptococcal CTR4 expression was found to be highly induced during macrophage residence in a J774.16 cell line, suggesting that the availability of copper to the intracellular pathogen is insufficient for its metabolic requirements. Macrophage residence is an important environmental niche during pathogenesis of C. neoformans (36). Reducing macrophage copper content could thus be a specific defense mechanism, limiting infection by depriving the pathogen of an essential nutrient. Poor biologic availability may also be caused by an unfavorable oxidation state of the copper ion, which can be altered by macrophage-secreted molecules such as nitric oxide (37). Alternatively, minimization of copper stores in macrophages could represent a regulatory phenomenon, as copper has been shown to induce release of iron stores through a regulation of macrophage FPN1 expression (38). High expression levels of CTR4 were also exhibited by cryptococcal cells recovered from mouse brains, consistent with low levels of available copper in this organ. This could be a result of low levels of copper within macrophages containing intracellular fungi or of specific mechanisms within the brain that limit the availability of copper. It is worth noting that excesses of copper have been associated with pathologic neuronal apoptosis, which leads to Alzheimer disease, Parkinson disease, and amyotrophic lateral sclerosis (39). In addition, endocytosis by neuroendothelial cells is thought to represent a mechanism for neurodissemination of C. neoformans (40) and could represent a low-copper intracellular environment similar to that of macrophages that would require efficient fungal copper acquisition for successful brain invasion. In contrast, CTR4-GFP expression in cryptococcal cells recovered from the lung was suppressed, suggesting that copper stores may be less limiting during pulmonary infections, consistent with the pulmonary fungal burden data showing growth of cuf1Δ cells similar to that of WT cells. These effects may be caused by a requirement of higher levels of copper in lung tissue to prevent acute lung injury, which is associated with copper suppression of excessive MMP-2 and MMP-9 activity (41). Alternatively, metabolic requirements for copper by the fungus may be lower in lung tissue than in the brain, resulting in a less copper-limiting environment and reduced CTR4 expression during pulmonary infection. In addition, it is possible that heterogeneity in other phenotypes, such as cytokine response to antigen (42), could also contribute to differential CTR4 expression patterns.
Recently, there has been interest in approaches that may allow stratification of clinical outcomes based on unique genetic attributes (43). This prompted an examination whose results showed that cryptococcal CTR4 expression, under equivalent in vitro conditions, varied among patient isolates from organ transplant recipients; these expression levels correlated with a propensity for dissemination to the CNS, which is associated with poor outcome (2). This is not to say that CTR4 expression is the sole determinant of CNS dissemination in cryptococcosis. In infectious diseases such as cryptococcosis, the outcome of an infection is determined by a complex array of properties from both the host and the pathogen.
Regarding the pathogen, classical virulence studies have emphasized qualitative differences between isolates (i.e., having a capsule or not having a capsule); however, in a real-world environment, strain-to-strain variability is more a reflection of quantitative differences. Thus, to understand differences in virulence between unrelated WT strains, it may be important to characterize regulatory pathways, such as CUF1/CTR4, responsible for the natural phenotypic variations that can contribute to the ability of individual pathogens to successfully thrive in the mammalian host. Attributes that are essential for growth under all conditions, such as actin expression by the ACT1 gene (44), may be so essential that they are under strong evolutionary pressure to maintain uniform expression among strains, as demonstrated in the present study, much in the way that amino acids within the active sites of enzymes are more invariate among strains than are those in the more dispensable folding regions outside of the active site (45). Nonessential genes that are nevertheless important for survival in the host have recently been described as conveying properties of pathogenic fitness (46). Lack of an absolute requirement for survival in the environment may thus allow greater transcriptional variation in genes of pathogenic fitness, such as we observed for CTR4 among the cohort of patient isolates in the present study. It is notable that CTR4 expression levels in vitro did not predict CNS dissemination in all isolates tested. Thus, CTR4 gene expression may be one of several factors that mediate the biologically complex propensity of this organism to disseminate to the CNS. Nevertheless, high expression of genes such as CTR4 may be associated with more successful pathogen strains that are more efficient at survival in the copper-deficient conditions of the host, allowing coordinate expression of virulence factors (47) such as laccase used to identify the cvf1Δ mutant in these studies. These results were suggested by our mouse studies, in which the copper-sensitive cuf1Δ mutant cells displayed attenuated virulence despite a high fungal inoculum. However, the effect of copper acquisition appeared to be even greater during human infection, perhaps due to differences in host tissue and smaller fungal burdens expected early in infection, and points to the importance of translating animal experiments to the human condition. In conclusion, while these findings require validation in larger clinical studies for generalizability, they suggest that robust methods might be developed that allow for risk stratification of patients’ isolates based on genetic variability in the regulation of pathogenic fitness genes such as CTR4.
Methods
Fungal strains, plasmids, and media.
The C. neoformans ATCC 208821 (H99) strain was a generous gift of J. Perfect, Duke University, Durham, North Carolina, USA. Strain H99FOA19, a uracil auxotrophic mutant derived from H99 (48), was employed as a recipient strain for creating targeted gene deletion. The strains were grown in YPD (2% glucose, 1% yeast extract, 2% Bacto-peptone) or YPD agar followed by incubation in asparagine salts without glucose (1 g/l asparagines; 10 mM sodium phosphate, pH 6.5; and 0.25 g/l MgSO4) for CTR4 expression. Asparagine minimum selective media for transformant selection and detection of laccase production were previously described (49). Plasmid pCIP, containing the URA5 gene, was a kind gift of K.J. Kwon-Chung, NIH, Bethesda, Maryland, USA. Plasmids containing the hygromycin resistance gene were a gift of G. Cox, Duke University, Durham, North Carolina, USA (50).
Cell lines and culture media.
J774.16 (ATCC) is a murine macrophage-like cell line derived from a reticulum sarcoma. Cells were maintained at 37°C in 10% CO2 in DMEM (Invitrogen) that was supplemented with 10% heat-killed FCS (Harlan), Cellgrow 1× nonessential amino acids (Mediatech), Cellgrow 100 mg/ml penicillin/streptomycin, and 10% NCTC-109 medium (Invitrogen). Cell lines were used between 5 and 15 passages.
Disruption and complementation of CUF1.
Chemical methods were used to isolate a laccase-deficient mutant, JEC111 (a kind gift of J. Edman, UCSF, San Francisco, California, USA; ref. 16). The mutant was complemented by a cryptococcal complementation library (kindly provided by J. Heitman, Duke University, Durham, North Carolina, USA) as described previously (51). One plasmid was found to complement the mutant and contained a 7.4-kb fragment of a cryptococcal gene containing a single annotated sequence (annotation 177.m03070; http://www.tigr.org/tigr-scripts/euk_manatee/shared/ORF_infopage.cgi?db=cna1&orf=177.t00214).
To make the CUF1 deletion construct, a 2.7-kb genomic fragment encompassing the major part of the CUF1 open reading frame was PCR amplified and subsequently cloned into pBluescript SK (Stratagene), and a 1.3-kb PCR-amplified fragment of the C. neoformans transformation marker URA5 (52) was inserted within 2 PstI sites, resulting in a 1.3-kb disruption within CUF1. C. neoformans strain H99FOA19 was then transformed by biolistic transformation, transformants were screened by PCR (50), and the deletion was confirmed by Southern blot analysis. Digestion of WT C. neoformans DNA with SacI yielded a 7.7-kb band hybridized with a 6-kb fragment of the CUF1 gene, whereas mutant DNA digested and hybridized in the same way resulted in 2 bands (5.8 kb and 1.7 kb) because of a SacI restriction site within the URA5 transformation marker. The cuf1Δ mutant strain was then complemented using a 6-kb genomic fragment and a hygromycin resistance cassette (50) as previously described (48). Genomic insertion of the complemented gene was confirmed by Southern blot analysis of uncut DNA.
Construction of a CTR4-GFP cryptococcal expression plasmid.
The cryptococcal shuttle vector pORA-KUTAP was used to express a fusion protein between the CTR4 promoter region and a recently described GFP possessing C. neoformans codon usage (53). First, pORA-KUTAP containing the synthetic GFP upstream of the EF-1α terminator was digested with BglII and EcoRI to remove the ACT1 promoter; next, a PCR-amplified fragment of genomic DNA from H99 (using primers GCCGCCGGATCCCAGATTAAGTGAAGCAAG [CTR4-857S-Bam] and CTTGGAATTCCCCATGTTCATGTGGCTGTG [CTR4-1934A-RI]) was digested with compatible enzymes and ligated to produce pCTR4-GFP. The plasmid was recovered, verified by sequencing, linearized with SceI, and transformed into either H99FOA19 or cuf1ΔFOA cells by standard methods of electroporation (48). Transformants were selected by equivalent copy number demonstrated on uncut Southern analysis as described previously (54). For in vitro expression, cells were incubated in asparagine salts containing 2% glucose with the indicated amounts of copper (data not shown) or copper chelators, 1 mM hydrogen peroxide, or 2 μg/ml menadione as described previously (53) overnight and then examined either by deconvolution microsocopy using an IX-70 Olympus microscope or by flow cytometry. GFP fluorescence was examined in each cell using an EPICS Elite ESP flow cytometer gated to exclude aggregates using time of flight. GFP signal was collected using a 550-nm pass dichoic and a 525-nm bandpass filter.
Macrophage phagocytosis and CTR4 expression.
The J774.16 macrophage-like cell line (ATCC) was used to evaluate the ability of the C. neoformans cells to grow inside macrophages and express CTR4-GFP using a previously described method for cryptococcal phagocytosis (55). Briefly, cells were allowed to grow for 5–7 days in DMEM (Cellgro) supplemented with 10% FCS and harvested from monolayers. Macrophage concentration was adjusted to 105 cells/ml, and 100 μl of the macrophage suspension was added to each well of a 96-well plate. The cells were primed with murine IFN-γ (Sigma-Aldrich) at a concentration of 50 U/ml and were incubated at 37°C, 5% CO2 overnight. Yeast cell suspensions (107 per ml) of WT H99, WT::CTR4-GFP, and cuf1Δ::CTR4-GFP were prepared from fresh cultures in 2% glucose, asparagine salts agar containing 10 μM CuCl2 opsonized with 100% mouse serum and incubated at 37°C for 1 hour. To each well in the 96-well plate, 104 serum-opsonized cryptococcal cells were added plus 50 U IFN-γ (Sigma-Aldrich) and 1 μg LPS and a suspension of TMR-labeled dextran (1 × 106 MW; Invitrogen) to allow colocalization of fungal cells and the phagolysosome as described previously (56) and incubated at 37°C 5% CO2. After 1 hour, extracellular yeast cells and dextran were removed by washing with PBS 3 times. Cells were then examined at 1, 24, and 48 hours by epifluorescent microscopy using an IX-70 deconvolution microscope and Slidebook version 4 software (Intelligent Imaging Innovations Inc.). All experiments were performed in triplicate, and the results of 1 representative experiment are shown.
In situ hybridization.
The method of Panepinto et al. (26) was used. Briefly, formaldehyde-fixed tissue was sectioned (4-μm sections), dewaxed with xylene, and rehydrated. A 237-bp PCR-amplified fragment of a CTR4 exon from H99 genomic DNA (using the primers GCCGCCTCTAGACCCTGCGCGCCATCCTTC [CTR4+434A-Xba] and GCCGCCCTCGAGATGTTGCTCAACTTCAACACCG [CTR4+197S-Xho]) was subcloned into pBluescript SK (Stratagene), verified by sequencing, transcribed using T7 polymerase, and labeled with U-digoxigenin according to the manufacturer’s instructions (Roche Diagnostics). Tissue sections were subjected to RNA denaturation with sequential washes with 0.2 N HCl, washed twice with SSC at 70°C, blocked with buffer containing iodoacetamide and n-ethylmaleimide, and dehydrated. Serial sections were hybridized with either sense or antisense probes at 50°C overnight, then washed at the same temperature in RNAase-free buffer. Slides were incubated with a second alkaline phosphatase–conjugated secondary antibody overnight at 4°C, washed, and visualized with 5-bromo,4-chloro,3-indolylphosphate/nitroblue tetrazolium substrate and counterstained with Fast Red (Sigma-Aldrich).
Participants and sample collection.
Patients were recruited as part of an ongoing study of C. neoformans infection in organ transplant patients (57). Briefly, patients undergoing solid organ transplant who had a cryptococcal isolate from sterile fluid and a confirmatory positive cryptococcal antigen were recruited from the University of Pittsburgh Medical Center. All study protocols were approved by the respective institutional review boards at the University of Pittsburgh and the University of Illinois at Chicago. Frozen stocks of primary patient isolates were divided into 2 independent cultures and grown on YPD agar for 5 days, after which a fresh mid-log phase culture was induced for CTR4 by incubation in asparagine salts for 3 hours at 30°C as described previously (26). RNA and DNA was harvested, and Northern blot or dot blots were performed as described previously (48) with a PCR-amplified fragment of CTR4 from strain H99 using the primers CAGATTAAGTGAAGCAAG (CTR4-1) and CCCATGTTCATGTGGCTGTG (CTR4-2) or ACT1 from the same strain using the primers GCTGGTGACGACGCTCCC (ACT1-MA-F) and GCAAGGATAGAACCACCGATCC (ACT1-MA-R).
Virulence factor expression and virulence studies.
Capsule was measured by India ink microscopy (48), urease production was assessed by incubation on Christensen’s agar (58), and the method of Liu et al. (59) was used to measure laccase activity. Virulence studies were conducted according to a previously described i.v. mouse meningoencephalitis model (14) using Swiss albino mice or an intranasal model using CBA/J mice (12). Animal studies were approved by the University of Illinois at Chicago Animal Care Committee. Studies involving human tissue were approved by the University of Illinois at Chicago Office for the Protection of Research Subjects and the Institutional Review Board (protocol no. 2004-0600).
Statistics.
Statistical significance of mouse survival times was assessed by Kruskall-Willis analysis (ANOVA on Ranks, GraphPad Prism software, version 4.03) as described previously (60). Differences between expression levels in the human C. neoformans isolates were assessed by a nonparametric 2-tailed unpaired Student’s t test with Welch’s correction. An F test was performed, which showed the variances were significantly different (P = 0.022). Fisher’s exact test was performed as described previously (61). Survival in mice of the human isolates was compared using a 1-way analysis of variance with a post-test Bonferroni correction for multiple comparisons as described previously (61). In mouse mortality studies, experiments were terminated for humanitarian reasons once significance between groups was established; for statistical comparisons, time of death of the survivors was recorded as the day of experimental termination.
Acknowledgments
We would like to acknowledge J. Bennett, W. Walden, and J. Kaplan for helpful discussions. This work was supported in part by US Public Health Service grants AI45995 and AI49371 from the NIH. We also acknowledge the use of the C. neoformans Genome Project, Stanford Genome Technology Center, funded by the National Institute of Allergy and Infectious Diseases, NIH, under cooperative agreement AI47087.
Footnotes
Nonstandard abbreviations used: BAL, bronchoalveolar lavage; BCS, bathocuproinedisulfonic acid; CSF, cerebrospinal fluid; Ctr4, copper transporter 4; Cuf1, copper-dependent transcription factor 1; YPD, yeast extract/peptone/dextrose medium.
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J. Clin. Invest. 117:794–802 (2007). doi:10.1172/JCI30006
References
- 1. Casadevall, A., and Perfect, J. 1998. Cryptococcus neoformans. ASM Press. Washington, DC, USA. 541 pp. [Google Scholar]
- 2.Saag M.S., et al. Practice guidelines for the management of cryptococcal disease. Infectious Diseases Society of America. Clin. Infect. Dis. 2000;30:710–718. doi: 10.1086/313757. [DOI] [PubMed] [Google Scholar]
- 3.Jean S.S., et al. Cryptococcaemia: clinical features and prognostic factors. QJM. 2002;95:511–518. doi: 10.1093/qjmed/95.8.511. [DOI] [PubMed] [Google Scholar]
- 4.Vilchez R.A., et al. The clinical epidemiology of pulmonary cryptococcosis in non-AIDS patients at a tertiary care medical center. Medicine (Baltimore). 2001;80:308–312. doi: 10.1097/00005792-200109000-00004. [DOI] [PubMed] [Google Scholar]
- 5.Pagano L., Caira M., Fianchi L. Pulmonary fungal infection with yeasts and pneumocystis in patients with hematological malignancy. Ann. Med. 2005;37:259–269. doi: 10.1080/07853890510037374. [DOI] [PubMed] [Google Scholar]
- 6.Pagano L., Caira M., Falcucci P., Fianchi L. Fungal CNS infections in patients with hematologic malignancy. Expert Rev. Anti. Infect. Ther. 2005;3:775–785. doi: 10.1586/14787210.3.5.775. [DOI] [PubMed] [Google Scholar]
- 7.Jenny-Avital E., Abadi M. Immune reconstitution cryptococcosis after initiation of successful highly active antiretroviral therapy. Clin. Infect. Dis. 2002;35:e128–e133. doi: 10.1086/344467. [DOI] [PubMed] [Google Scholar]
- 8.Singh N., et al. An immune reconstitution syndrome-like illness associated with Cryptococcus neoformans infection in organ transplant recipients. . Clin. Infect. Dis. 2005;40:1756–1761. doi: 10.1086/430606. [DOI] [PubMed] [Google Scholar]
- 9.Rees E., Thiele D. From aging to virulence: forging connections through the study of copper homeostasis in eukaryotic microorganisms. Curr. Opin. Microbiol. 2004;7:175–184. doi: 10.1016/j.mib.2004.02.004. [DOI] [PubMed] [Google Scholar]
- 10.Jimenez-Exposito M., et al. Micronutrients in HIV-infection and the relationship with the inflammatory response. Med. Clin. (Barc.). 2002;119:765–769. [PubMed] [Google Scholar]
- 11.Moreno T., Artacho R., Navarro M., Perez A., Ruiz-Lopez M. Serum copper concentration in HIV-infection patients and relationships with other biochemical indices. Sci. Total Environ. 1998;217:21–26. doi: 10.1016/s0048-9697(98)00158-2. [DOI] [PubMed] [Google Scholar]
- 12.Cox G., et al. Superoxide dismutase influences the virulence of Cryptococcus neoformans by affecting growth within macrophages. . Infect. Immun. 2003;71:173–180. doi: 10.1128/IAI.71.1.173-180.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Williamson P.R. Biochemical and molecular characterization of the diphenol oxidase of Cryptococcus neoformans: identification as a laccase. . J. Bacteriol. 1994;176:656–664. doi: 10.1128/jb.176.3.656-664.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Salas S.D., Bennett J.E., Kwon-Chung K.J., Perfect J.R., Williamson P.R. Effect of the laccase gene CNLAC1, on virulence of Cryptococcus neoformans. . J. Exp. Med. 1996;184:377–386. doi: 10.1084/jem.184.2.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Davis-Kaplan S., Askwith C., Bengtzen A., Radisky D., Kaplan J. Chloride is an allosteric effector of copper assembly for the yeast multicopper oxidase Fet3p: an unexpected role for intracellular chloride channels. Proc. Natl. Acad. Sci. U. S. A. 1998;95:13641–13645. doi: 10.1073/pnas.95.23.13641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Torres-Guererro H., Edman J.C. Melanin-deficient mutants of Cryptococcus neoformans. . J. Med. Vet. Mycol. 1994;32:303–313. [PubMed] [Google Scholar]
- 17.Labbe S., Pena M., Fernandes A., Thiele D. A copper-sensing transcription factor regulates iron uptake genes in Schizosaccharomyces pombe. . J. Biol. Chem. 1999;274:36252–36260. doi: 10.1074/jbc.274.51.36252. [DOI] [PubMed] [Google Scholar]
- 18.Beaudoin J., Mercier A., Langlois R., Labbe S. The Schizosaccharomyces pombe Cuf1 is composed of functional modules from two distinct classes of copper metalloregulatory transcription factors. J. Biol. Chem. 2003;278:14565–14577. doi: 10.1074/jbc.M300861200. [DOI] [PubMed] [Google Scholar]
- 19.Beaudoin J., Labbe S. The fission yeast copper-sensing transcription factor Cuf1 regulates the copper transporter gene expression through an Ace1/Amt1–like recognition sequence. J. Biol. Chem. 2001;276:15472–15480. doi: 10.1074/jbc.M011256200. [DOI] [PubMed] [Google Scholar]
- 20.Jungmann J., et al. MAC1, a nuclear regulatory protein related to Cu-dependent transcription factors is involved in Cu/Fe utilization and stress resistance in yeast. EMBO J. 1993;12:5051–5056. doi: 10.1002/j.1460-2075.1993.tb06198.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Toffaletti D.L., Del Poeta M., Rude T.H., Dietrich F., Perfect J.R. Regulation of cytochrome c oxidase subunit 1 (COX1) expression in Cryptococcus neoformans by temperature and host environment. . Microbiology. 2003;149:1041–1049. doi: 10.1099/mic.0.26021-0. [DOI] [PubMed] [Google Scholar]
- 22.Pukkila-Worley R., et al. Transcriptional network of multiple capsule and melanin genes governed by the Cryptococcus neoformans cyclic AMP cascade. . Eukaryot. Cell. 2005;4:190–201. doi: 10.1128/EC.4.1.190-201.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Noverr M.C., Williamson P.R., Fajardo R.S., Huffnagle G.B. CNLAC1 is required for extrapulmonary dissemination of Cryptococcus neoformans but not pulmonary persistence. . Infect. Immun. 2004;72:1693–1699. doi: 10.1128/IAI.72.3.1693-1699.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ory J.J., Griffith C.L., Doering T.L. An efficiently regulated promoter system for Cryptococcus neoformans utilizing the CTR4 promoter. . Yeast. 2004;21:919–926. doi: 10.1002/yea.1139. [DOI] [PubMed] [Google Scholar]
- 25.Malliaris S.D., Steenbergen J.N., Casadevall A. Cryptococcus neoformans var. gattii can exploit Acanthamoeba castellanii for growth. . Med. Mycol. 2004;42:149–158. doi: 10.1080/13693786310001616500. [DOI] [PubMed] [Google Scholar]
- 26.Panepinto J., et al. The DEAD-box RNA helicase Vad1 regulates multiple virulence-associated genes in Cryptococcus neoformans. . J. Clin. Invest. 2005;115:632–641. doi: 10.1172/JCI200523048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Singh N., et al. Antifungal management practices and evolution of infection in organ transplant recipients with Cryptococcus neoformans infection. . Transplantation. 2005;80:1033–1039. doi: 10.1097/01.tp.0000173774.74388.49. [DOI] [PubMed] [Google Scholar]
- 28.Blankenship J.R., Singh N., Alexander B.D., Heitman J. Cryptococcus neoformans isolates from transplant recipients are not selected for resistance to calcineurin inhibitors by current immunosuppressive regimens. . J. Clin. Microbiol. 2005;43:464–467. doi: 10.1128/JCM.43.1.464-467.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marvin M., Mason R., Cashmore A. The CaCTR1 gene is required for high-affinity iron uptake and is transcriptionally controlled by a copper-sensing transactivator encoded by CaMAC1. . Microbiology. 2004;150:2197–2208. doi: 10.1099/mic.0.27004-0. [DOI] [PubMed] [Google Scholar]
- 30.Huang G., Nie X., Chen J. CaMac1, a Candida albicans copper ion-sensing transcription factor, promotes filamentous and invasive growth in Saccharomyces cerevisiae. . Acta. Biochim. Biophys. Sin. (Shanghai). 2006;38:213–217. doi: 10.1111/j.1745-7270.2006.00146.x. [DOI] [PubMed] [Google Scholar]
- 31.Srikantha T., Zhao R., Daniels K., Radke J., Soll D. Phenotypic switching in Candida glabrata accompanied by changes in expression of genes with deduced functions in copper detoxification and stress. . Eukaryot. Cell. 2005;4:1434–1445. doi: 10.1128/EC.4.8.1434-1445.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fagiuali S., et al. Liver transplantation: the Italian experience. Dig. Liver. Dis. 2002;34:640–648. doi: 10.1016/s1590-8658(02)80207-9. [DOI] [PubMed] [Google Scholar]
- 33.Fiszer U., et al. Gamma delta + T cells in Wilson’s disease. Int. J. Clin. Lab. Res. 1996;26:51–54. doi: 10.1007/BF02644775. [DOI] [PubMed] [Google Scholar]
- 34.Beaudoin J., Labbe S. Copper induces cytoplasmic retention of fission yeast transcription factor cuf1. Eukaryot. Cell. 2006;5:277–292. doi: 10.1128/EC.5.2.277-292.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Van Ho A., Ward D., Kaplan J. Transition metal transport in yeast. Annu. Rev. Microbiol. 2002;56:237–261. doi: 10.1146/annurev.micro.56.012302.160847. [DOI] [PubMed] [Google Scholar]
- 36.Feldmesser M., Kress Y., Novikoff P., Casadevall A. Cryptococcus neoformans is a facultative intracellular pathogen in murine pulmonary infection. . Infect. Immun. 2000;68:4225–4237. doi: 10.1128/iai.68.7.4225-4237.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lim M., Xu D., Lippard S. Visualization of nitric oxide in living cells by a copper-based fluorescent probe. Nat. Chem. Biol. 2006;2:375–380. doi: 10.1038/nchembio794. [DOI] [PubMed] [Google Scholar]
- 38.Chung J., Haile D., Wessling-Resnick M. Copper-induced ferroportin-1 expression in J774 macrophages is associated with increased iron efflux. Proc. Natl. Acad. Sci. U. S. A. 2004;101:2700–2705. doi: 10.1073/pnas.0306622101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Levenson C. Trace metal regulation of neuronal apoptosis: from genes to behavior. Physiol. Behav. 2005;86:399–406. doi: 10.1016/j.physbeh.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 40.Chang Y.C., et al. Cryptococcal yeast cells invade the central nervous system via transcellular penetration of the blood-brain barrier. Infect. Immun. 2004;72:4985–4995. doi: 10.1128/IAI.72.9.4985-4995.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lentsch A., Kato A., Saari J., Schuschke D. Augmented metalloproteinase activity and acute lung injury in copper-deficient rats. Am. J. Physiol. Lung Cell. Mol. Physiol. 2001;281:L387–L393. doi: 10.1152/ajplung.2001.281.2.L387. [DOI] [PubMed] [Google Scholar]
- 42.Kaslow D.C., Shiloach J. Production, purification and immunogenicity of a malaria transmission-blocking vaccine candidate: TBV25H expressed in yeast and purified using nickel-NTA agarose. Biotechnology (N.Y.). . 1994;12:494–499. doi: 10.1038/nbt0594-494. [DOI] [PubMed] [Google Scholar]
- 43.Zerhouni E. Translational and clinical science — time for a new vision. N. Engl. J. Med. 2005;353:1621–1623. doi: 10.1056/NEJMsb053723. [DOI] [PubMed] [Google Scholar]
- 44.Toffaletti D.L., Perfect J.R. Study of Cryptococcus neoformans actin gene regulation with a beta-galactosidase-actin fusion. . J. Med. Vet. Mycol. 1997;35:313–320. [PubMed] [Google Scholar]
- 45.Andreeva A., Murzin A. Evolution of protein fold in the presence of functional constraints. Curr. Opin. Struct. Biol. 2006;16:399–408. doi: 10.1016/j.sbi.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 46.Panepinto J., Williamson P. Intersection of fungal fitness and virulence in Cryptococcus neoformans. . FEMS Yeast Res. 2006;6:489–498. doi: 10.1111/j.1567-1364.2006.00078.x. [DOI] [PubMed] [Google Scholar]
- 47.Casadevall A., Pirofski L. The damage-response framework of microbial pathogenesis. Nat. Rev. Microbiol. 2003;1:17–24. doi: 10.1038/nrmicro732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Erickson T., et al. Multiple virulence factors of Cryptococcus neoformans are dependent on VPH1. . Mol. Microbiol. 2001;42:1121–1131. doi: 10.1046/j.1365-2958.2001.02712.x. [DOI] [PubMed] [Google Scholar]
- 49.Zhu X., Williamson P.R. A CLC-type chloride channel gene is required for laccase activity and virulence in Cryptococcus neoformans. . Mol. Microbiol. 2003;50:1271–1281. doi: 10.1046/j.1365-2958.2003.03752.x. [DOI] [PubMed] [Google Scholar]
- 50.Cox G.M., Toffaletti D.L., Perfect J.R. Dominant selection system for use in Cryptococcus neoformans. . J. Med. Vet. Mycol. 1996;34:385–391. [PubMed] [Google Scholar]
- 51.Cruz M.C., Fox D.S., Heitman J. Calcineurin is required for hyphal elongation during mating and haploid fruiting in Cryptococcus neoformans. . EMBO J. 2001;20:1020–1032. doi: 10.1093/emboj/20.5.1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Varma A., Edman J.C., Kwon-Chung K.J. Molecular and genetic analysis of URA5 transformants of Cryptococcus neoformans. . Infect. Immun. 1992;60:1101–1108. doi: 10.1128/iai.60.3.1101-1108.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu X., Hu G., Panepinto J., Williamson P. Role of a VPS41 homolog in starvation response and virulence of Cryptococcus neoformans. . Mol. Microbiol. 2006;61:1132–1146. doi: 10.1111/j.1365-2958.2006.05299.x. [DOI] [PubMed] [Google Scholar]
- 54.Zhang S., Varma A., Williamson P.R. The yeast Cryptococcus neoformans uses ‘mammalian’ enhancer sites in the regulation of the virulence gene, CNLAC1. . Gene. 1999;227:231–240. doi: 10.1016/s0378-1119(98)00590-3. [DOI] [PubMed] [Google Scholar]
- 55.Tucker S.C., Casadevall A. Replication of Cryptococcus neoformans in macrophages is accompanied by phagosomal permeabilization and accumulation of vesicles containing polysaccharide in the cytoplasm. . Proc. Natl. Acad. Sci. U. S. A. 2002;99:3165–3170. doi: 10.1073/pnas.052702799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hmama Z., et al. Quantitative analysis of phagolysosome fusion in intact cells: inhibition by mycobacterial lipoarabinomannan and rescue by an 1alpha, 25-dihydroxyvitamin D3-phosphoinositide 3-kinase pathway. J. Cell Sci. 2004;117:2131–2140. doi: 10.1242/jcs.01072. [DOI] [PubMed] [Google Scholar]
- 57.Singh N., et al. Allograft loss in renal transplant recipients with Cryptococcus neoformans associated immune reconstitution syndrome. . Transplantation. 2005;80:1131–1133. doi: 10.1097/01.tp.0000180530.17683.02. [DOI] [PubMed] [Google Scholar]
- 58.Cox G., Mukherjee J., Cole G., Casadeval A., Perfect J. Urease as a virulence factor in experimental cryptococcosis. Infect. Immun. 2000;68:443–448. doi: 10.1128/iai.68.2.443-448.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu L., Wakamatsu K., Ito S., Williamson P.R. Catecholamine oxidative products, but not melanin, are produced by Cryptococcus neoformans during neuropathogenesis in mice. . Infect. Immun. 1999;67:108–112. doi: 10.1128/iai.67.1.108-112.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Hosmer, D., and Lemeshow, S. 1999. Applied survival analysis: regression modeling of time to event data. John Wiley and Sons. New York, New York, USA. 408 pp. [Google Scholar]
- 61. Mendenhall, W. 1975. Introduction to probability and statistics. Duxbury Press. Belmont, California, USA. 460 pp. [Google Scholar]