Abstract
We describe here a novel method for generation of yeast-secreted, in vivo biotinylated recombinant antibodies, or biobodies. Biobodies are secreted by diploid yeast resulting from the fusion of two haploid yeast of opposite mating type. One yeast carries a cDNA encoding an antibody recognition sequence fused to an IgA1 hinge and a biotin acceptor site (BCCP) at the C-terminus; the other carries a cDNA encoding an E. Coli biotin ligase (BirA) fused to KEX2 golgi-localization sequences, so that BirA can catalyze the biotin transfer to the recognition sequence-fused BCCP within the yeast secretory compartment. We illustrate this technology with biobodies against HE4, a biomarker for ovarian carcinoma. Anti-HE4 biobodies were derived from clones or pools of anti-HE4-specific yeast-display scFv, constituting respectively monoclonal (mBb) or polyclonal (pBb) biobodies. Anti-HE4 biobodies were secreted directly biotinylated thus bound to labeled-streptavidin and streptavidin-coated surfaces without Ni-purification. Anti-HE4 biobodies demonstrated specificity and sensitivity by ELISA assays, flow cytometry analysis and western blots prior to any maturation; dissociation equilibrium constants as measured by surface plasmon resonance sensor were of Kd= 4.8×10-9 M and Kd =5.1×10-9 M before and after Ni-purification respectively. Thus, yeast mating permits cost-effective generation of biotinylated recombinant antibodies of high affinity.
Keywords: Antibody engineering, In vivo biotinylation, Biobody, Yeast-display scFv, Yeast-secreted proteins, Surface Plasmon Resonance Sensor
1. INTRODUCTION
Large numbers of candidate biomarkers for malignant (Etzioni et al., 2003), autoimmune (Prince, 2005), infectious (Zhang et al., 2002), neurological (Carrette et al., 2003), metabolic (Roelofsen et al., 2004; Schwegler et al., 2005) and cardiovascular (Kittleson and Hare, 2005) diseases have been generated using various high-throughput platforms (Omenn et al., 2005; Ping et al., 2005), but the production of high affinity antibodies necessary for biomarker validation remains slow and costly. New methods such as high-throughput screening by flow sorting of recombinant antibodies (scFv) displayed by yeast (Feldhaus et al., 2003) can accelerate the identification of antigen-specific scFv. However, yeast display scFv need to be converted into soluble and labeled recombinant antibodies for further use in immunoassays.
Biotinylation is a robust labeling method that modifies the lysine residues of target proteins and is routinely achieved in vitro with chemical agents (Bayer and Wilchek, 1990). The extraordinarily high affinity binding of biotin to streptavidin (dissociation equilibrium constant or Kd= 10-15M) (Green and Melamed, 1966) and extremely slow dissociation rate have been used widely in biochemistry and molecular biology. However, chemical biotinylation is not specific and thus can inactivate lysine residues critical for protein function and conformation (Smith et al., 1998). In contrast, in vivo biotinylation is a rare and highly specific posttranslational modification (Cronan, 1990) mediated by endogenous biotin ligases. E.coli contains only one such target for biotinylation: the biotin carboxyl carrier protein (BCCP), a subunit of acetyl-CoA carboxylase (Samols et al., 1988). The gene product of BirA (Barker and Campbell, 1981a), the biotin holoenzyme synthetase (BirA), catalyzes biotin activation by covalently joining biotin with ATP to form biotin-5′-adenylate, with subsequent transfer to the epsilon amino group of a specific BCCP lysine residue (Barker and Campbell, 1981b). BirA typically recognizes a large protein domain, but Schatz and colleagues have identified short peptides (Schatz, 1993; Beckett et al., 1999) that efficiently mimic BCCP biotin acceptor function.
Specific biotinylation of proteins and recombinant antibodies has been obtained in E.coli in vivo (Kipriyanov et al., 1996; Sibler et al., 1999; Santala and Lamminmaki, 2004; Warren et al., 2005) and in vitro with purified biotin ligase (Saviranta et al., 1998; Cloutier et al., 2000). Several groups have demonstrated independently that biotinylated recombinant antibodies tetramerize in presence of streptavidin and that tetramers show both higher affinity constants and lower non-specific binding than monomeric single chains (Muhlrad et al., 1992; Saviranta et al., 1998; Cloutier et al., 2000). However, current in vivo biotinylation protocols can only label one protein at a time through lengthy molecular engineering that is not compatible with high-throughput approaches. In addition, in vivo biotinylation of yeast-secreted proteins is impeded by the necessity for BirA to access the secretory compartment where secreted proteins transit. We targeted BirA anchorage to the yeast secretory compartment using localization signals from the yeast KEX2 protease. KEX2 protease modifies the yeast mating pheromone alpha-factor (MF alpha L) as it is secreted (Wilcox et al., 1992). Since the yeast-secreted proteins are under the control of the leader region of MF alpha L precursor (“alpha prepro leader”) we reasoned that KEX2 domains might target BirA to the region of the secretory compartment where it could act on BCCP-fused proteins during their secretion.
We describe here a cost- and time-effective method to generate yeast-secreted, in vivo biotinylated proteins. We illustrate this technology by producing in vivo biotinylated recombinant antibodies, or biobodies, directed against the ovarian cancer marker HE4 (Schummer et al., 1999). Anti-HE4 biobodies were validated by ELISA assays, flow cytometry analysis and western blot. We also determined by Surface Plasmon Resonance (SPR) sensor that the dissociation equilibrium constant of the anti-HE4 biobodies was 5.1×10-9 M prior to any maturation and/or purification.
2. MATERIAL AND METHODS
2.1. Constructs
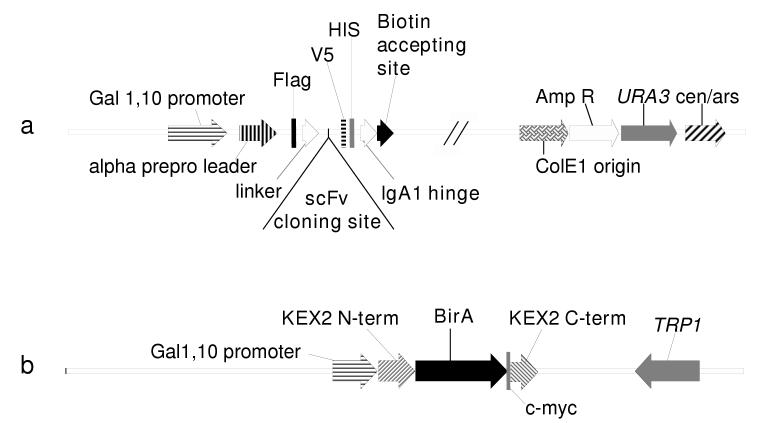
pTOR2 vector (Fig. 1a) was derived from pNL9 (the kind gift from Pacific Northwest National Laboratory). The following modifications were introduced: the HA tag (779-805bp) was replaced by a Flag tag DYKDDDK (Chubet and Brizzard, 1996) encoded by gattataaagatgacgataaa. The prolinker for human IgA1 hinge PSTPPTPSPSTPPTPSPS (Sumiyama et al., 2002) encoded by ccatcaacaccaccaactccaagtccttctactcctcctacaccttcaccatca and the biotin accepting site (BCCP) GLNDIFEAQKEWHE encoded by ggtttgaatgatatttttgaagctcaaaaaattgaatggcatgaawas were cloned in frame distal to the HIS tag (920-937 bp).
Figure 1.
Schematic representations of pTOR2 and pTOR-BIR constructs
(a) pTOR2 is derived from pNL9 by replacement of the HA tag with a Flag tag, and addition of a human IgA1 linker and of a biotin accepting site. pTOR2 confers uracil-prototrophy (URA3), autonomous replication (cen/ars), and promotes yeast secretion of V5-, flag- and HIS-tagged proteins and, when mated to strain overexpressing Golgi-localized biotin ligase, secretion of in vivo biotinylated proteins.
(b) pTOR-BIR encodes E. coli biotin ligase (BirA) fused to yeast KEX2 N- and C-terminal Golgi localization domains and confers tryptophan-prototrophy (TRP1). Both pTOR2 and pTOR-BIR vectors are under the control of galactose promoter (GAL1,10).
The E.coli biotin ligase sequence (BirA ORF II) (Howard et al., 1985) was PCR-amplified with 5′BirAHindIII (5′-ctagaagcttgccgccatgaaggataacaccgtgcc) and 3′BirAEcoR1myc (3′-gatgagtttttgttcgaattctttttctgcactacgcaggg) primers from Top 10′ E.coli DNA (Invitrogen Corporation, Carlsbad, CA) and ligated into HindIII/EcoR1 sites of pcDNA3 polylinker (Invitrogen). After sequence verification, BirA was PCR-amplified with 5′BamH1BirA (5′-ctagggatccatgaaggataacaccgtgcc) and 3′BirAcmycXho1 (5′-atatctcgagttattacagatcctcttctgatgagatg) primers and ligated into BamH1/XhoI sites of the p414-GAL1 polylinker (the kind gift of Martin Funk) between GAL1,10 promoter and CYC1 terminator (Mumberg et al., 1994) to create p414gal1-BirA-cmyc construct. To redirect BirA into the yeast secretory pathway, KEX2 leader and anchoring domain (Fuller RS, 1989; Wilcox et al., 1992) were added to p414gal1-BirA-cmyc at the 5′ and 3′ends respectively of BirA coding sequence. KEX2 leader was PCR-amplified from BY4742 yeast genomic DNA (ATCC, Manassas, VA) with KEXNfor (5′-catgactagtatgaaagtgaggaaatatattactttatgcttt) and KEXNrev (5′-attactcgagttatcacgatcgtccggaagatggaggaacatcagg) primers and was ligated into BAMH1/SpeI sites of p414gal1-BirA-cmyc to create N-KEX2-BIRA c-myc. The KEX2 C-terminal sequence was amplified from BY4742 DNA with KEXC+cmyc (5′-Ctgagaattcgaacaaaaactcatctcagaagaggatctgtctgagtacgattctactttggacaatg) and KEXCrev (5′-attactcgagttatcacgatcgtccggaagatggaggaacatcagg) primers and was ligated into EcoR1/Xho1 sites of N-KEX2--BIRA c-myc to create pTOR-BIR construct (Fig. 1b).
HE4 was amplified from an ovarian tumor cDNA with HE4 forward (5′-ctagagatctatggccttgccaacggctcga-3′) and HE4 reverse (5′-catgccgccggaggatggtccgttcaggctg-3′) primers to create a sequence flanked by BglII and SacII sites. HE4 PCR products were ligated into BglII/SacII sites of pdisplay vector (Invitrogen) in frame with IgK leader, c-myc and HA tags, and the C-terminal transmembrane anchoring domain of platelet-derived growth factor receptor (PDGFR). As negative controls for anti-HE4 biobody specificity, MPF and Mesothelin were PCR-amplified from the clone MGC:10273 IMAGE:3957372 (ATCC) and transfected into HEK293F cell lines (ATCC) to be expressed at the cell surface or secreted as a fusion protein (Meso-Ig) (Scholler et al., in press). All constructs were confirmed by sequencing.
2.2. Generation of anti-HE4 yeast-display scFv subtractive library
A yeast-display scFv library (Feldhaus et al., 2003) was first enriched for scFv that bound to 100 nM of HE4-Ig. Five hundred micrograms of HE4-Ig (the kind gift of Dr. Martha-Hayden Ledbetter (Hellstrom et al., 2003) were biotinylated with EZ-link sulfo-NHS LC biotin kit (Fisher Biotech, Fair Lawn, NJ) according to manufacturer instructions. Two successive rounds of scFv enrichment with HE4-Ig by magnetic sorting followed by three rounds of flow sorting with BD FACSAria™ cell sorter (BD Biosciences Immunocytometry Systems, San Jose, CA) were performed as described in PNNL user′s manual. All incubations were carried in 1 ml of PBE [Phosphate Buffered Saline (PBS) supplemented with 0.5% bovine serum albumin (BSA) (Sigma-Aldrich) and 4 mM EDTA (Promega Corporation, Madison WI)] for 30 min at 4°C. Yeast-display (YD) scFv were grown in synthetic selective medium containing 0.5% Casamino Acids (Fisher) (SD-CAA), 1% penicillin/streptomycin (PS) (Gibco/Invitrogen Corporation, Carlsbad, CA). Because of the presence of Candida parapsilosis in the PNNL library (http://www.sysbio.org/dataresources/candidascFvLibrary060207.pdf) 0.250 microg/ml of ketoconazole (Sigma-Aldrich, St. Louis, MO) was added after four rounds of selection. YD scFv expression was induced in selective medium supplemented with 2% galactose, 2% rafinose and 0.1% dextrose (SGR-CAA) and 1% PS.
To remove the anti-HE4-Ig YD scFv that bound to the Ig domain, YD scFv were incubated with 100 nM of biotinylated Meso-Ig and magnetically depleted. YD scFv were incubated with 50 microL of streptavidin magnetic beads (Miltenyi, Auburn, CA) for 10 min at 4°C and pelleted. After one wash with PBE, YD scFv were loaded on LD depletion column (Miltenyi). The effluent and one 7 ml-rinse containing the non-binding yeast were regrown and induced as described above. After a second round of depletion with Meso-Ig, one last round of magnetic enrichment with 50 nM of biotinylated HE4-Ig was performed to generate the anti-HE4 YD scFv subtractive library shown in Fig 2e-h. The entire procedure, including enrichment and depletion, required less than 200 micrograms of both Ig-fusion proteins.
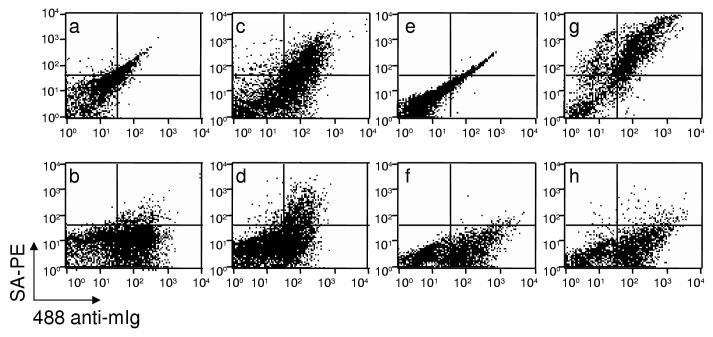
Figure 2.
Flow cytometry analysis of yeast-display scFv during and after differential sorting.
Yeast-display scFv were labeled after five enrichments with 100 nM of HE4-Ig (a-d) or after 5 enrichments followed by two rounds of depletion with 100 nM of Meso-Ig and one enrichment with 50 nM of HE4-Ig (e-h). To assess scFv expression, samples b-d and f-h were incubated with anti-cmyc mAb. To assess antigen specificity, samples were incubated with biotinylated HE4-Ig (c, g) or biotinylated Meso-Ig (d, h). All samples were labeled with SA-PE and 488 anti-mIg. Results are representative of two independent experiments.
2.3. Conversion of yeast-display scFv into yeast-secreted scFv by homologous recombination
One hundred microliters of anti-HE4 YD scFv subtractive library grown to saturation were patched in a 1 cm2 on SD-CAA plate and grown 1 day at 30°C. Yeast DNA was isolated (Hoffman and Winston, 1987) and used as a template for PCR amplification with the primers pTOR2 forward (5′-gattataaagatgacgataaaggtggtggtggttctgcta-3′) and pNL9 reverse (3′-gggttagggataggcttaccctgttgttctagaattccg-3′). The resulting 800 bp PCR products were gel-purified on a Qiaquick gel extraction column according to the manufacturer instructions (Qiagen, Valencia, CA). Ten microliters of gel-purified PCR products were cotransformed with 1 microgram of Not1/Sfi1 cut pTOR2 vector into the yeast strain BJ5464 (ATCC), also known as YVH10 (PNNL). Transformants arising by homologous recombination were selected on SD-CAA plates supplemented with 0.008% tryptophan (Sigma-Aldrich) (SD-CAA+TRP). Two hundred to two thousand colonies were recovered per transformation.
2.4. Conversion of yeast-secreted scFv into in vivo biotinylated recombinant antibodies by yeast mating.
pTOR-BIR was transformed into BJ5475 MATa ura3-52 trp1 his3 delta 200 pep4::HIS3 prb1 delta 1.6R can1 GAL (the kind gift of Elizabeth Jones). Transformants were selected on SD+CAA+0.002% uracil (Sigma-Aldrich) (SD-CAA+URA). Cells containing pTOR-BIR were spread at high density on SD-CAA+URA and grown at 30°C overnight (ON). pTOR2-scFv transformants (MATalpha trp-) and the lawn of pTOR-BIR containing cells (MATa ura-) were replica plated to a YEPD plate (rich medium), allowed to mate for 8 to 16 hours, then replica plated to a SD+CAA plate lacking uracil and tryptophan to select diploids. Selected diploid patches were replicated two more times to SD+CAA to eliminate unmated haploids. Diploid pool and diploid clones secreted in vivo biotinylated recombinant antibodies, or biobodies (Fig. 3).
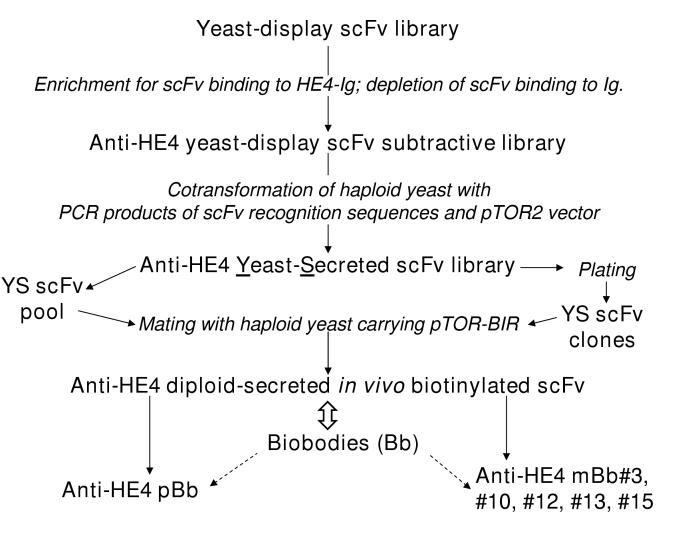
Figure 3.
Biobody method overview
A HE4-specific yeast-display scFv library was generated by enrichments for HE4-Ig-binding scFv and depletion for Ig-binding scFv. The resulting yeast-display scFv subtractive library was converted into an anti-HE4 yeast-secreted scFv library by homologous recombination in yeast between PCR fragments of scFv recognition sequences and pTOR2 vector. Uracil-prototrophic transformed yeast that secreted anti-HE4 yeast-secreted scFv were then mated with tryptophan-prototrophic yeast that carried pTOR-BIR. Resulting uracil/tryptophan-prototrophic diploid cells secreted in vivo biotinylated scFv (biobodies) of monoclonal (mBb) or polyclonal (pBb) origins.
2.4. Production and Ni-purification of biobodies.
Diploid cells were grown in 100 ml of SD-CAA medium to saturation and induced 3 days in 100 ml of YEPGR. The cultures were centrifuged 5 min at 3000 rpm to pellet the cells. Supernatants were desalted and concentrated with Vivaspin concentrators (VivaScience AG, Hannover, Germany). After adjusting to 0.3 M sodium chloride (Sigma-Aldrich) and to pH 7-8 with sodium phosphate (Na2HPO4), supernatants were incubated with HIS-Select-Nickel Affinity Gel (Sigma-Aldrich) according to manufacturer instructions. Biobodies were eluted by competition with imidazole (Fisher) into a volume of less than 1 ml and dialyzed against 1000 volume of PBS at 4°C ON in a Tube-O-Dialyzer (Upstate Cell Signaling Solutions, Charlottesville, VA). Protein concentrations were evaluated by Nanoorange Protein Quantization Kit (Invitrogen) and were routinely of 50 to 200 micrograms/ml, corresponding to a yield of 0.5 to 2 mg/L.
2.6. Flow cytometry analysis
YD scFv were incubated with 2 micrograms/ml of anti-cmyc mAb (Santa Cruz Biotechnology) and 100 nM of biotinylated HE4-Ig or Meso-Ig followed by Alexa Fluor® 488 F(ab′)2 fragment of goat anti-mouse IgG (H+L) (488 anti-mIg) (Invitrogen) diluted 1/200 and phytoerythrin-labeled streptavidin (SA-PE) (BD Pharmingen, San Diego, CA) diluted 1/100. All incubations were carried in PBE for 30 min at 4°C. Fluorescent signals were detected with a BD FACScan Cytometer.
Wild type and transfected-HEK 293 cells were incubated with 5 micrograms/ml of anti-HA mAb (Roche Applied Science, Indianapolis, IN), or 5 micrograms/ml of anti-HE4 mAb 3D8 (Hellstrom et al, 2003) followed by 488 anti-mIg, or 5 micrograms/ml of biobodies premixed with 1/500 SA-PE for 20 min at 4°C. All incubations were performed with 2-5 × 105 cells per ml, in 50 microliters of PBE, for 30 min at 4°C. Cells were washed twice with PBE between incubations.
2.7. Western blots
Samples were separated by electrophoresis on a 4-12% NuPAGE Bis Tris gel (Invitrogen) using SDS running buffer (Invitrogen). Gels were blotted to a PVDF membrane (Invitrogen) in transfer buffer (Invitrogen) with an X-cell II blot module (Invitrogen). Membranes were blocked 1 hour at room temperature in PBS supplemented with 0.05% Tween (PBST) and 3% BSA for one hour at room temperature (RT) with agitation; incubations were performed in diluent buffer (PBST supplemented with 0.5% BSA) for 30 min at RT with agitation; washes were carried out in PBST 3 times for 5 min at RT with agitation. Reduced samples were diluted 1:1 with 10 microliters of SDS loading buffer (Invitrogen) supplemented with 5% mercaptoethanol (Sigma-Aldrich) and boiled 5 min; native samples were diluted 1:1 in native running buffer (Invitrogen) and loaded without boiling. Signals were detected with Super Signal West Pico Chemiluminescent Substrate (Pierce) according to standard procedure and exposed to Kodak X-OMAT AR films (Fisher/Kodak). In some experiments signals were stripped with Western Restore™ Western Blot Stripping Buffer (Pierce).
ScFv were detected with HRP-conjugated anti-V5 mAb (Serotec, Raleigh NC). Biobodies were detected with HPR-conjugated streptavidin (BD PharMingen, San Diego, CA). When used to probe 10 cm2 membranes, 5 micrograms of biobodies were premixed with 1 microliter of SA-HRP for 20 min in 250 microliters of PBS on ice and then added to 10 ml of diluent buffer. HE4-Ig and Meso-Ig were detected with biobodies or with HRP-conjugated F(ab′)2 Fragment Goat Anti-Human IgG (H+L) polyclonal antibody (HRP-anti-hIg) (Jackson Immunoresearch Laboratory, Inc, West Grove, PA). All secondary reagents were diluted ten thousand fold in diluent buffer.
2.8. ELISA assays
ELISA assays were performed in Nunc Amino™ or Streptavidin Immobilizer™ plates (Nunc, Rochester, NY) with gentle agitation at RT. Amino plates were coated with fusion proteins diluted in bicarbonate buffer (carbonate-bicarbonate buffer capsules, Sigma-Aldrich) for one hour. PBST-prewashed streptavidin plates were coated with diploid supernatants or Ni-purified biobodies diluted in PBST for one hour. Other incubations and washes were carried in PBST for 30 min with rotation at RT. In accordance with the manufacturer′s recommendations no blocking steps were necessary with these plates since all incubations are carried in PBST. Samples were run in duplicates. Colorimetric signals were reveled with TMB One Solution (Promega), stopped with sulfuric acid (Acros, New Jersey) and read at 450 nm on a Spectra Max 250 (Molecular Devices, Sunnyvale, CA). Ig-fusion proteins were detected with 5 micrograms/ml of HRP anti hIg, or with 5 micrograms/ml of anti-HE4 mAb (3D8 or 2H5) (Hellstrom et al., 2003) followed by 1/1000 HRP rabbit anti-mouse IgG (HRP anti mIg, Serotec) or with 5 micrograms/ml of biobodies followed by 1/1000 anti-V5 or SA-HRP, or with 5 micrograms/ml or less (as indicated) of biobodies premixed for 20 min with 1/1000 of anti-V5 or SA-HRP in PBS at 4°C.
2.9. Surface plasmon resonance sensor
The SPR system used is a custom-built four-channel instrument based on the Kretschmann configuration of the attenuated total reflection method (Homola et al., 2002). The glass side of the gold-coated substrate is index matched to the prism coupler while the functionalized surface is mechanically pressed against an acrylic flow cell with a Mylar gasket. A polychromatic light beam directed through the prism and the glass substrate excites surface plasma waves at the metal-dielectric interface. The reflected light is analyzed with a spectrograph. The reflectivity spectrum is dependent upon the refractive index of analytes in proximity to the surface, allowing the detection of binding events to the surface.
SPR sensor chips, BK7 glass chips 32 mm × 15 mm × 1.5 mm (Schott), were coated with 2 nm of Cr and 55 nm of Au by electron beam evaporation. These chips were rinsed with copious amounts of absolute ethanol and water. Chips were dried by blowing with dry N2, then cleaned by UV-ozone for 20 min, and finally exhaustively rinsed with water and absolute ethanol before surface functionalization. A mixture of oligo ethylene glycol (OEG) and biotinylated alkanethiols (BAT) was dissolved in absolute ethanol. The BAT, a C15 alkanethiol chain linked to a biotin headgroup by three ethylene glycol groups, was used as the specific binding element of the SAM targeting streptavidin, while the OEG, a C10 alkanethiol chain linked to a hydroxyl headgroup by four ethylene glycol groups, created a nonfouling background (Ladd et al., 2004). The mixture had a total thiol concentration of 100 nM and consisted of a 1:9 ratio of BAT:OEG. The cleaned chips were immersed in the OEG/BAT solution overnight.
Sensor chips were then removed from the OEG/BAT solution, rinsed with copious amounts of absolute ethanol, dried with dry N2, and then mounted in the SPR instrument. All experiments were carried out at 25°C and began with flowing PBS (pH 7.4) at 50 microliters/min for 10 min. Following the establishment of a baseline, 10 micrograms/ml streptavidin was flowed over all four channels for 10 min at 50 microliters/min to prepare the surface for the binding of biobodies. After 10 min of PBS wash, either yeast supernatant or a 10 micrograms/ml purified biobody solution was directly flowed over the surface at 50 microliters/min for 5 min to immobilize the biobodies. Following 10 min of PBS wash, HE4-Ig was flowed for 10 min at 10 microliters/min, with one channel acting as a reference. PBS was flowed once again for up to 20 min to allow for the dissociation of HE4-Ig. HE4-Ig concentrations were varied from 10 ng/ml (0.2 nM) to 10 micrograms/ml (200 nM).
3. RESULTS
3.1. Isolation of anti-HE4 yeast-display scFv
A yeast-display scFv library was first enriched for scFv binding to 100 nM of HE4-Ig. After five enrichments, half of YD scFv bound to HE-4Ig (Fig. 2c) but a third of these also bound to the control protein Meso-Ig (Fig. 2d). This was expected since HE4-Ig and Meso-Ig share a 696 bp-immunoglobin sequence. To eliminate the Ig-binding scFv from the anti-HE4-Ig sublibrary, two rounds of magnetic depletion with Meso-Ig and one last enrichment with HE4-Ig were conducted as described in the Methods section. The resulting anti-HE4 yeast-display scFv subtractive library contained more than 95% scFv that bound only to HE4-Ig (Fig. 2g,h).
3.2. Diploid yeast resulting from the mating of pTOR2-scFv transformant with pTOR-BIR transformant secreted in vivo biotinylated recombinant antibodies.
To generate anti-HE4 secreted scFv, scFv recognition sequences of anti-HE4 YD scFv subtractive library were PCR-amplified and cotransformed with Not1/Sfi1-cut pTOR2 vector into the yeast strain BJ5464 to obtain a scFv library capable of secreting anti-HE4 scFv (Fig. 3). To evaluate the ability of KEX2 domains to target BirA expression to the yeast-secretory compartment, pTOR2-scFv yeast transformants were mated with yeast carrying pTOR-BIR. As controls the pTOR2-scFv yeast transformants were also mated with yeast carrying 414-GAL1 vector only or p414gal1-BirA-cmyc ligase that encodes biotin ligase without KEX2 domains. Resulting diploid supernatants were characterized by Western blot (Fig. 4a) and ELISA (Fig. 4b). All supernatants contained a V5-tagged protein of ∼42 kDA, consistent with the scFv size (Fig. 4a, upper panel). However, streptavidin-HRP could only detect the 42 kDa protein in pTOR2-scFv/pTOR-BIR diploid supernatant (Fig. 4a, lower panel, lane 4) which suggests that Golgi-targeting KEX2 domains are necessary and sufficient for proper localization and function of the E. coli biotin ligase in the yeast secretory pathway.
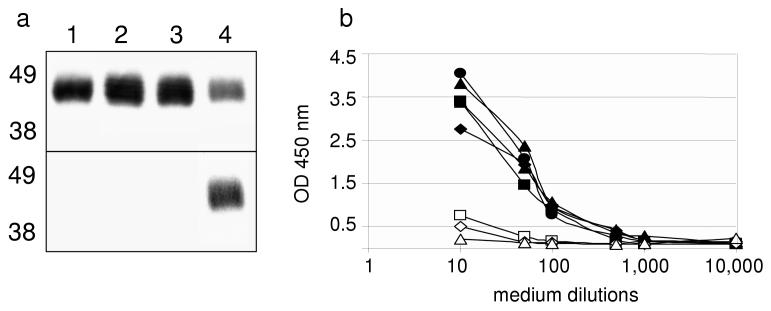
Figure 4.
Diploid supernatants contain in vivo biotinylated scFv.
(a) Western blot of supernatants from pTOR2-scFv haploid (lane 1), pTOR2-scFv/p414-GAL1 diploid (lane 2), pTOR2-scFv/p414gal1-BirA-cmyc diploid (lane 3) and pTOR-scFv/pTOR-BIR diploid (lane 4) was probed with HRP anti-V5 mAb (upper panel), stripped and reprobed with SA-HPR (lower panel). (b) Supernatants from pTOR2-scFv haploid (white squares), pTOR2-scFv/p414-GAL1 diploid (white triangles) and 5 independent pTOR2-scFv/pTOR-BIR diploids (black symbols) were incubated in streptavidin-coated wells and detected with HRP anti-V5 mAb. Yeast medium was used as back-ground control (white diamonds).
Supernatants from several independent diploids were then immobilized on streptavidin-coated wells and detected with HRP anti-V5 mAb. Only the pTOR2-scFv/pTOR-BIR diploid supernatants bound to streptavidin plates (Fig. 4b, black symbols). These results confirm that the V5-tagged protein was identical to the biotin-labeled protein and is indeed a scFv, and demonstrate that mating of haploid yeast carrying scFv fused to BCCP and yeast carrying BirA fused to Golgi-targeting KEX2 domains is necessary and sufficient to generate diploids that secrete in vivo biotinylated scFv, or biobodies.
3.3. Functional validation of biobodies
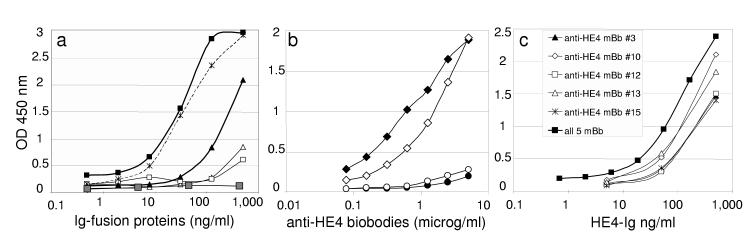
An anti-HE4 biobody corresponding to a pool of in vivo biotinylated scFv (anti-HE4 pBb, Fig. 3) were compared to anti-HE4 mAb 3D8 for its ability to detect plastic-immobilized HE4-Ig or Meso-Ig by ELISA assay (Fig. 5a). Monomeric anti-HE4 pBb incubated in HE4-Ig-coated wells and detected with HRP-conjugated anti-V5 mAb (HRP-V5 mAb, white triangles) or streptavidin (SA-HRP, white squares) generated poor colorimetric signals. However, colorimetric signals were significantly enhanced when anti-HE4 pBb was premixed with HRP anti-V5 mAb (black triangles) and, remarkably, when premixed with SA-HRP (black squares) the detection range of anti-HE4 pBb for HE4-Ig was similar to 3D8 mAb (stars on dotted line). Anti-HE4 pBb premixed with SA-HRP did not detect plastic immobilized Meso-Ig (gray squares) demonstrating the specificity of anti-HE4 biobody.
Figure 5.
Anti-HE4 biobody validation by ELISA assays.
(a) Serial dilutions of HE4-Ig were immobilized on amino wells and incubated with anti-HE4 pBb followed by HRP anti-V5 mAb (white triangles) or SA-HRP (white squares). Alternatively, immobilized HE4-Ig was incubated with anti-HE4 pBb premixed with HRP anti-V5 mAb (black triangles) or SA-HRP (black squares). As positive control, immobilized HE4-Ig was detected with anti-HE4 mAb (3D8) followed by HRP anti mIg (stars on dotted line). As negative control, serial dilutions of Meso-Ig were immobilized and incubated with anti-HE4 pBb premixed with SA-HRP (gray squares). (b) Serial dilutions of five anti-HE4 mBb were preincubated with SA-HRP together (black symbols) or separately (white symbols) before detection of 250 ng of plastic-immobilized HE4-Ig (diamonds) or Meso-Ig (circles). (c) Five mBb were coated separately or together (as indicated) on a streptavidin plate, and then incubated with serial dilutions of HE4-Ig. Captured HE4-Ig was detected with anti-HE4 mAb (2H5) followed by HRP anti mIg. All results are representative of two independent experiments; standard deviations were less than 10%.
In figure 5b, the detection ranges of five anti-HE4 mBb (mBb#3; 10; 12; 13 and 15) for 250 ng of immobilized HE4-Ig (diamonds) or Meso-Ig (circles) were compared after being premixed with SA-HRP individually (white symbols) or together (black symbols). Sensitivity increased when mBb were premixed together with SA-HRP (black diamonds) to compare with mBb premixed separately (white diamonds) when biobodies were used in limiting quantities (less than 2.5 μg/ml). This suggests that the high affinity of anti-HE4 pBb/streptavidin complexes could be due in part to a cooperative effect between the diverse biotinylated scFv species included in pBb.
Anti-HE4 mBb#3; 10; 12; 13 and 15 were also used separately or together as capture antibody in sandwich ELISA assay on streptavidin-coated plates in combination with an anti-HE4 mAb (2H5). Figure 5c shows that anti-HE4 mBb used separately or together can complement anti-HE4 mAb to detect nanograms of HE4-Ig in a sandwich ELISA assay setting.
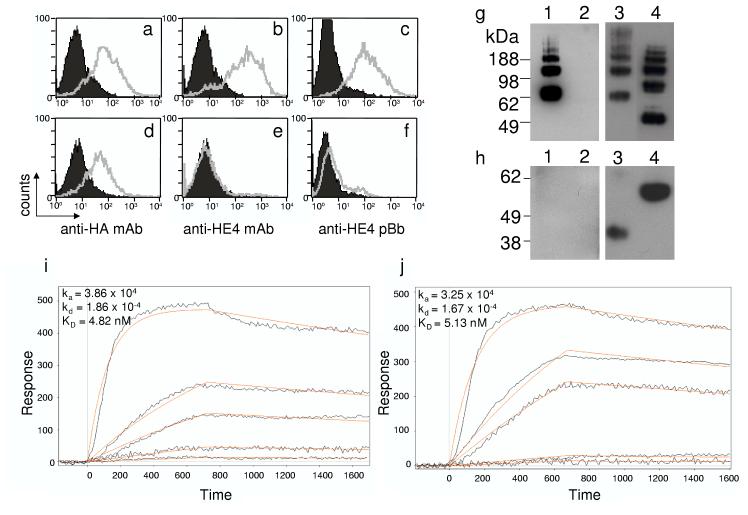
Biobodies premixed with phytoerythrin-conjugated streptavidin (SA-PE) were used for detection in flow cytometry analysis. The HEK-293F cell line was stably transfected with HE4-HA tagged (Fig. 6a-c) or, as a negative control, MPF-HA tagged cell-surface protein (Fig. 6d-f). Both transfected cell lines expressed equal amounts of HA-tags as detected with anti-HA mAb (Fig. 5a, d) but anti-HE4 mAb 3D8 and anti-HE4 pBb specifically bound to HE4-transfected cells (Fig. 5b-c vs. 5e-f).
Figure 6.
Anti-HE4 biobody validation by flow cytometry analysis, Western blots and SPR.
(a-j) Flow cytometry analysis of HE4-(a-c) and MPF-transfected (d-f) HEK 293F cells. HEK 293F cells were labeled with anti-HA mAb (a, d), anti-HE4 mAb 3D8 (b, e) and anti-HE4 pBb (c, f). As negative controls, HEK 293F cells were incubated with 488 anti-mIg only (shaded areas a-b and d-e) or SA-PE only (shared areas c and f). Results are representative of two independent experiments. (g-h) Western blots of 500 ng of native (g) or reduced (h) HE4Ig (lanes 1, 3) and Meso-Ig (lanes 2, 4) were first probed with anti-HE4 pBb premixed with SA-HRP (lanes 1-2), stripped and reprobed with HRP anti-hIg (lanes 3-4). Results are representative of three independent experiments. (i,j) SPR response curves for the detection of HE4-Ig in a range of concentrations from 0.2 nM to 200 nM. The sensorgrams were fitted to Langmuir binding isotherms with simple 1:1 interactions. Biobodies were immobilized either directly from yeast supernatant (i) or from nickel column purified anti-HE4 pBb (j). Results representative of two independent experiments.
Biobodies were used as probes for Western blots of proteins loaded in native (Fig. 5g) or reduced conditions (Fig. 5h). In native conditions several bands were detected by pBb (Fig. 5g lane 1) or HRP anti-hIg (Fig. 5g, lanes 3-4), corresponding to multi-protein complexes. Anti-HE4 pBb premixed with SA-HRP detected HE4-Ig loaded in native condition only (compare lanes 1 of fig. 5g, h). This result confirms the specificity of anti-HE4 pBb and also suggests that anti-HE4 pBb epitopes may be dependent on the tertiary structure of the protein thus altered by reducing conditions.
Finally, anti-HE4 pBb was immobilized on a SPR sensor surface after purification by nickel columns or directly from medium in which yeast had been induced to secrete Bb. SPR response curves for the binding of the anti-HE4 pBb to HE4-Ig were fit to a pseudo-first-order equation (i.e., 1:1 Langmuir isotherm) (Myszka and Morton, 1998) (Fig. 6i-j). The dissociation equilibrium constants for anti-HE4 pBb directly from unpurified and purified biobodies were Kd = 4.82×10-9 M and Kd = 5.13×10-9 M, with the association rate constants (ka) of 3.86×104 M-1 s-1 and 3.25×104 M-1s-1, and the dissociation rate constants (kd) of 1.86×10-4 s-1 and 1.67×10-4 s-1 , respectively. These results are very similar, indicating that it is not necessary to purify the biobodies prior to use with SPR sensors.
4. DISCUSSION
We demonstrated that mating of yeast carrying a cDNA encoding a recombinant antibody fused to a biotin acceptor site with yeast carrying a cDNA encoding an E. Coli biotin ligase fused to yeast Golgi-localization sequences from KEX2, results in diploid yeast able to secrete in vivo biotinylated recombinant antibodies of high affinity that we named biobodies.
Because in vivo biotinylation is highly specific for the BCCP lysine, it can be achieved without modification of critical lysine residues belonging to antibody recognition sequences and thus without functional loss of the recognition domains. The human IgA1 hinge between the recognition and the biotin accepting site sequences may also contribute to reduce scFv conformational changes due to biotinylation or to binding mediated by biotin/streptavidin interactions. Indeed, the overall affinity of the anti-HE4 scFv subtractive library displayed by yeast did not decrease after conversion into soluble in vivo biotinylated recombinant antibodies; the affinity of the anti-HE4 polyclonal biobody was found in the nanomolar range.
KEX2p is a yeast protease that modifies the mating pheromone of MATalpha yeast as it transits the secretory apparatus; KEX2p is targeted to the golgi apparatus by N-terminal and C-terminal domains. Since scFv secretion is controlled by the alpha pheromone leader sequence of pTOR2, we used the KEX2 N- and C-terminal localization signals to target BirA for the secretory compartment. While endogenous yeast biotin ligase and transfected E.coli biotin ligase without KEX2 localization domains failed to biotinylate secreted recombinant antibodies, BirA fused to KEX N- and C-domains was able to act on its target site as the biobodies were secreted.
S. cerevisiae has a number of features that are attractive for high throughput methods. First, yeast cells are able to repair gapped DNA sequences in vivo by homologous recombination. Thus, by co-transforming the yeast with pTOR2 linearized vector containing a selectable marker and an autonomously replicating sequence along with a PCR product that spans the gap in the vector, in vivo homologous recombination between the vector and the cDNA insert results in a circular plasmid containing the cDNA fused to a biotin accepting sequence that can efficiently propagate, without the necessity of in vitro ligation. In addition, gap repair makes it possible to efficiently insert multiple PCR products into pTOR2, thus to convert in a time- and cost-effective manner cDNA pools of recognition sequences into yeast-secreted recombinant antibodies, creating a reagent similar to a polyclonal antibody but from which non-specific binders can be removed as described in section 2.2. Second, the fact that yeast mates by cell fusion made it easy to combine yeast of one mating type encoding Golgi-localized BirA with yeast of the opposite mating type expressing scFv to form diploids able to secrete biobodies.
By ELISA assays, anti-HE4 biobody affinity appeared to be increased after preincubation with anti-V5, consistent with a previous report by Wang and colleagues (Wang et al., 2004), but was dramatically improved after preincubation with streptavidin, consistent with previous reports of tetramerization of biotinylated recombinant antibodies in presence of streptavidin (Muhlrad et al., 1992; Saviranta et al., 1998; Cloutier et al., 2000). In addition, the affinity of five anti-HE4 mBb increased when they were preincubated with streptavidin together instead of separately, which suggests a cooperative effect of the biobodies. Finally, five anti-HE4 mBb were successfully used as capture antibodies to complement an anti-HE4 mAb in a sandwich ELISA assay, which suggests that this method can be used to rapidly develop assays necessary for validation of serum biomarkers.
By Western blotting, anti-HE4 pBb was able to specifically detect native but not reduced HE4 protein. This implies that anti-HE4 pBb may preferentially detect conformational epitopes. It may be due to the possible limited representation of recognition sequences of this particular reagent suggested by the sequencing of 15 anti-HE4 mBb showing scFv belonging to the same vH and vL subgroups (data not shown). Alternatively, it might be an intrinsic characteristic of the yeast-display scFv library to generate scFv with preferential affinity for conformational epitopes (Weaver-Feldhaus et al., 2005). In either case, the ability of an affinity reagent to distinguish subtle conformational changes is a desirable characteristic for the accurate detection of biomarkers that often result from structural modifications of normal proteins through cleavage, aberrant transcriptions or post-translational modifications (Mahlknecht and Hoelzer, 2000; Brinkman, 2004; Venables, 2004; Kalnina et al., 2005; Santos-Rosa and Caldas, 2005).
By SPR, anti-HE4 pBb demonstrated a Kd in the nanomolar range when immobilized on a streptavidin-coated surface either from purified biobodies or directly from yeast supernatant. The same order of magnitude of dissociation equilibrium constants of purified and non-purified biobodies indicates that purification is not required for high affinity. Thus yeast supernatant can be used to directly immobilize yeast-secreted in vivo biotinylated recombinant antibodies on any streptavidin-coated surfaces, including a sensor surface. In vivo biotinylated proteins can also be produced with the same yeast-expression system. This will greatly simplify and reduce the cost of development of arrayed sensor platforms for large-scale biomarker discovery and detection.
It is conceivable that this method may also facilitate the conversion of mouse monoclonal antibodies into functional recombinant antibodies. Finally, this method may also be used to generate reagents for emerging technologies such as antibody-based proximity ligation (Fredriksson et al., 2002), antibody-based competition assays using tadpoles (Burbulis et al., 2005), bead-based assays (Vignali, 2000; Gorelik et al., 2005; Scholler et al., 2006) and protein or antibody arrays (Chen et al., 2005; Gao et al., 2005; Utz, 2005; Boozer et al., 2006). In conclusion, the yeast expression system described here allows efficient generation of directly biotinylated high-affinity reagents that are needed for a large range of applications, including evaluation of the products of large-scale proteomics discovery projects.
ACKNOWLEDGEMENTS
We thank Paul Lampe, Martin McIntosh, Beatrice Knudsen and Richard Klausner for thoughtful discussions, Kristi Schurman for manuscript editing, and Coralie S. Harmache and Colin J. Scholler for suggesting the term “biobody”. This work was supported by the Pacific Ovarian Cancer Research Consortium (POCRC)/SPORE in Ovarian Cancer (P50 CA83636, N.U.), its Developmental Research Program (N.S.), the National Science Foundation (CTS-0528605, S.J.) and the Canary Foundation.
Abbreviations:
- ScFv
single chain Fragment variable
- YD
yeast-display
- mAb
mouse monoclonal antibodies
- Bb
biobodies
- mBb
monoclonal biobodies
- pBb
polyclonal biobodies
- BCCP
biotin acceptor site
- BirA
E.Coli biotin ligase
- Ig
immunoglobulin
- ON
overnight
- RT
room temperature
- BSA
bovine serum albumin
- PBS
phosphate buffered saline
- PBST
PBS supplemented with 0.05% Tween
- PBE
PBS supplemented with BSA 0.5% and EDTA 10 mM
- PS
penicillin/streptomycin
- SD-CAA
synthetic selective medium containing 0.5% Casamino Acids
- SGR-CAA
selective medium supplemented with 2% galactose, 2% rafinose and 0.1% dextrose
- URA
uracil
- TRP
tryptophan
- PCR
polymerase chain reaction
- SPR
surface plasmon resonance
- OEG
oligo ethylene glycol
- BAT
biotinylated alkanethiols
- ml
milliliters
- L
liter
- mg
milligrams
- min
minutes
- M
molar
- nM
nanomolar
- nm
nanometers
- mm
millimeters
- °C
degree Celsius
- %
percentage
- bp
base pairs
- vH
variable Heavy chain
- vL
variable Light chain
- HRP
horse-radish peroxidase
- HRP-anti-hIg
HRP-conjugated F(ab′)2 Fragment Goat Anti-Human IgG (H+L) polyclonal antibody
- 488 anti-mIg
Alexa Fluor® 488 F(ab′)2 fragment of goat anti-mouse IgG (H+L)
- SA-PE
phytoerythrin-labeled streptavidin
- SA-HRP
HRP-labeled streptavidin.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errorsmaybe discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Barker DF, Campbell AM. The birA gene of Escherichia coli encodes a biotin holoenzyme synthetase. J Mol Biol. 1981a;146:451–67. doi: 10.1016/0022-2836(81)90042-5. [DOI] [PubMed] [Google Scholar]
- Barker DF, Campbell AM. Genetic and biochemical characterization of the birA gene and its product: evidence for a direct role of biotin holoenzyme synthetase in repression of the biotin operon in Escherichia coli. J Mol Biol. 1981b;146:469–92. doi: 10.1016/0022-2836(81)90043-7. [DOI] [PubMed] [Google Scholar]
- Bayer EA, Wilchek M. Protein biotinylation. Methods Enzymol. 1990;184:138–60. doi: 10.1016/0076-6879(90)84268-l. [DOI] [PubMed] [Google Scholar]
- Beckett D, Kovaleva E, Schatz PJ. A minimal peptide substrate in biotin holoenzyme synthetase-catalyzed biotinylation. Protein Sci. 1999;8:921–9. doi: 10.1110/ps.8.4.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boozer C, Ladd J, Chen S, Jiang S. DNA-directed protein immobilization for simultaneous detection of multiple analytes by surface plasmon resonance biosensor. Anal Chem. 2006;78:1515–9. doi: 10.1021/ac051923l. [DOI] [PubMed] [Google Scholar]
- Brinkman BM. Splice variants as cancer biomarkers. Clin Biochem. 2004;37:584–94. doi: 10.1016/j.clinbiochem.2004.05.015. [DOI] [PubMed] [Google Scholar]
- Burbulis I, Yamaguchi K, Gordon A, Carlson R, Brent R. Using protein-DNA chimeras to detect and count small numbers of molecules. Nat Methods. 2005;2:31–7. doi: 10.1038/nmeth729. [DOI] [PubMed] [Google Scholar]
- Carrette O, Demalte I, Scherl A, Yalkinoglu O, Corthals G, Burkhard P, Hochstrasser DF, Sanchez JC. A panel of cerebrospinal fluid potential biomarkers for the diagnosis of Alzheimer’s disease. Proteomics. 2003;3:1486–94. doi: 10.1002/pmic.200300470. [DOI] [PubMed] [Google Scholar]
- Chen DS, Soen Y, Stuge TB, Lee PP, Weber JS, Brown PO, Davis MM. Marked differences in human melanoma antigen-specific T cell responsiveness after vaccination using a functional microarray. PLoS Med. 2005;2:e265. doi: 10.1371/journal.pmed.0020265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chubet RG, Brizzard BL. Vectors for expression and secretion of FLAG epitope-tagged proteins in mammalian cells. Biotechniques. 1996;20:136–41. doi: 10.2144/96201pf01. [DOI] [PubMed] [Google Scholar]
- Cloutier SM, Couty S, Terskikh A, Marguerat L, Crivelli V, Pugnieres M, Mani JC, Leisinger HJ, Mach JP, Deperthes D. Streptabody, a high avidity molecule made by tetramerization of in vivo biotinylated, phage display-selected scFv fragments on streptavidin. Mol Immunol. 2000;37:1067–77. doi: 10.1016/s0161-5890(01)00023-2. [DOI] [PubMed] [Google Scholar]
- Cronan JE., Jr. Biotination of proteins in vivo. A post-translational modification to label, purify, and study proteins. J Biol Chem. 1990;265:10327–33. [PubMed] [Google Scholar]
- Etzioni R, Urban N, Ramsey S, McIntosh M, Schwartz S, Reid B, Radich J, Anderson G, Hartwell L. The case for early detection. Nat Rev Cancer. 2003;3:243–52. doi: 10.1038/nrc1041. [DOI] [PubMed] [Google Scholar]
- Feldhaus MJ, Siegel RW, Opresko LK, Coleman JR, Feldhaus JM, Yeung YA, Cochran JR, Heinzelman P, Colby D, Swers J, Graff C, Wiley HS, Wittrup KD. Flow-cytometric isolation of human antibodies from a nonimmune Saccharomyces cerevisiae surface display library. Nat Biotechnol. 2003;21:163–70. doi: 10.1038/nbt785. [DOI] [PubMed] [Google Scholar]
- Fredriksson S, Gullberg M, Jarvius J, Olsson C, Pietras K, Gustafsdottir SM, Ostman A, Landegren U. Protein detection using proximity-dependent DNA ligation assays. Nat Biotechnol. 2002;20:473–7. doi: 10.1038/nbt0502-473. [DOI] [PubMed] [Google Scholar]
- Fuller RS,BA, Thorner J. Intracellular targeting and structural conservation of a prohormone-processing endoprotease. Science. 1989;246:482–6. doi: 10.1126/science.2683070. [DOI] [PubMed] [Google Scholar]
- Gao WM, Kuick R, Orchekowski RP, Misek DE, Qiu J, Greenberg AK, Rom WN, Brenner DE, Omenn GS, Haab BB, Hanash SM. Distinctive serum protein profiles involving abundant proteins in lung cancer patients based upon antibody microarray analysis. BMC Cancer. 2005;5:110. doi: 10.1186/1471-2407-5-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorelik E, Landsittel DP, Marrangoni AM, Modugno F, Velikokhatnaya L, Winans MT, Bigbee WL, Herberman RB, Lokshin AE. Multiplexed immunobead-based cytokine profiling for early detection of ovarian cancer. Cancer Epidemiol Biomarkers Prev. 2005;14:981–7. doi: 10.1158/1055-9965.EPI-04-0404. [DOI] [PubMed] [Google Scholar]
- Green NM, Melamed MD. Optical rotatory dispersion, circular dichroism and far-ultraviolet spectra of avidin and streptavidin. Biochem J. 1966;100:614–21. doi: 10.1042/bj1000614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellstrom I, Raycraft J, Hayden-Ledbetter M, Ledbetter JA, Schummer M, McIntosh M, Drescher C, Urban N, Hellstrom KE. The HE4 (WFDC2) protein is a biomarker for ovarian carcinoma. Cancer Res. 2003;63:3695–700. [PubMed] [Google Scholar]
- Hoffman CS, Winston F. A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene. 1987;57:267–72. doi: 10.1016/0378-1119(87)90131-4. [DOI] [PubMed] [Google Scholar]
- Homola J, Dostalek J, Chen S, Rasooly A, Jiang S, Yee SS. Spectral surface plasmon resonance biosensor for detection of staphylococcal enterotoxin B in milk. Int J Food Microbiol. 2002;75:61–9. doi: 10.1016/s0168-1605(02)00010-7. [DOI] [PubMed] [Google Scholar]
- Howard PK, Shaw J, Otsuka AJ. Nucleotide sequence of the birA gene encoding the biotin operon repressor and biotin holoenzyme synthetase functions of Escherichia coli. Gene. 1985;35:321–31. doi: 10.1016/0378-1119(85)90011-3. [DOI] [PubMed] [Google Scholar]
- Kalnina Z, Zayakin P, Silina K, Line A. Alterations of pre-mRNA splicing in cancer. Genes Chromosomes Cancer. 2005;42:342–57. doi: 10.1002/gcc.20156. [DOI] [PubMed] [Google Scholar]
- Kipriyanov SM, Little M, Kropshofer H, Breitling F, Gotter S, Dubel S. Affinity enhancement of a recombinant antibody: formation of complexes with multiple valency by a single-chain Fv fragment-core streptavidin fusion. Protein Eng. 1996;9:203–11. doi: 10.1093/protein/9.2.203. [DOI] [PubMed] [Google Scholar]
- Kittleson MM, Hare JM. Molecular signature analysis: using the myocardial transcriptome as a biomarker in cardiovascular disease. Trends Cardiovasc Med. 2005;15:130–8. doi: 10.1016/j.tcm.2005.05.007. [DOI] [PubMed] [Google Scholar]
- Ladd J, Boozer C, Yu Q, Chen S, Homola J, Jiang S. DNA-directed protein immobilization on mixed self-assembled monolayers via a streptavidin bridge. Langmuir. 2004;20:8090–5. doi: 10.1021/la049867r. [DOI] [PubMed] [Google Scholar]
- Mahlknecht U, Hoelzer D. Histone acetylation modifiers in the pathogenesis of malignant disease. Mol Med. 2000;6:623–44. [PMC free article] [PubMed] [Google Scholar]
- Muhlrad D, Hunter R, Parker R. A rapid method for localized mutagenesis of yeast genes. Yeast. 1992;8:79–82. doi: 10.1002/yea.320080202. [DOI] [PubMed] [Google Scholar]
- Mumberg D, Muller R, Funk M. Regulatable promoters of Saccharomyces cerevisiae: comparison of transcriptional activity and their use for heterologous expression. Nucleic Acids Res. 1994;22:5767–8. doi: 10.1093/nar/22.25.5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myszka DG, Morton TA. CLAMP: a biosensor kinetic data analysis program. Trends Biochem Sci. 1998;23:149–50. doi: 10.1016/s0968-0004(98)01183-9. [DOI] [PubMed] [Google Scholar]
- Omenn GS, States DJ, Adamski M, Blackwell TW, Menon R, Hermjakob H, Apweiler R, Haab BB, Simpson RJ, Eddes JS, Kapp EA, Moritz RL, Chan DW, Rai AJ, Admon A, Aebersold R, Eng J, Hancock WS, Hefta SA, Meyer H, Paik YK, Yoo JS, Ping P, Pounds J, Adkins J, Qian X, Wang R, Wasinger V, Wu CY, Zhao X, Zeng R, Archakov A, Tsugita A, Beer I, Pandey A, Pisano M, Andrews P, Tammen H, Speicher DW, Hanash SM. Overview of the HUPO Plasma Proteome Project: results from the pilot phase with 35 collaborating laboratories and multiple analytical groups, generating a core dataset of 3020 proteins and a publicly-available database. Proteomics. 2005;5:3226–45. doi: 10.1002/pmic.200500358. [DOI] [PubMed] [Google Scholar]
- Ping P, Vondriska TM, Creighton CJ, Gandhi TK, Yang Z, Menon R, Kwon MS, Cho SY, Drwal G, Kellmann M, Peri S, Suresh S, Gronborg M, Molina H, Chaerkady R, Rekha B, Shet AS, Gerszten RE, Wu H, Raftery M, Wasinger V, Schulz-Knappe P, Hanash SM, Paik YK, Hancock WS, States DJ, Omenn GS, Pandey A. A functional annotation of subproteomes in human plasma. Proteomics. 2005;5:3506–19. doi: 10.1002/pmic.200500140. [DOI] [PubMed] [Google Scholar]
- Prince HE. Biomarkers for diagnosing and monitoring autoimmune diseases. Biomarkers. 2005;10(Suppl 1):44–9. doi: 10.1080/13547500500214194. [DOI] [PubMed] [Google Scholar]
- Roelofsen H, Balgobind R, Vonk RJ. Proteomic analyzes of copper metabolism in an in vitro model of Wilson disease using surface enhanced laser desorption/ionization-time of flight-mass spectrometry. J Cell Biochem. 2004;93:732–40. doi: 10.1002/jcb.20226. [DOI] [PubMed] [Google Scholar]
- Samols D, Thornton CG, Murtif VL, Kumar GK, Haase FC, Wood HG. Evolutionary conservation among biotin enzymes. J Biol Chem. 1988;263:6461–4. [PubMed] [Google Scholar]
- Santala V, Lamminmaki U. Production of a biotinylated single-chain antibody fragment in the cytoplasm of Escherichia coli. J Immunol Methods. 2004;284:165–75. doi: 10.1016/j.jim.2003.10.008. [DOI] [PubMed] [Google Scholar]
- Santos-Rosa H, Caldas C. Chromatin modifier enzymes, the histone code and cancer. Eur J Cancer. 2005;41:2381–402. doi: 10.1016/j.ejca.2005.08.010. [DOI] [PubMed] [Google Scholar]
- Saviranta P, Haavisto T, Rappu P, Karp M, Lovgren T. In vitro enzymatic biotinylation of recombinant fab fragments through a peptide acceptor tail. Bioconjug Chem. 1998;9:725–35. doi: 10.1021/bc9800217. [DOI] [PubMed] [Google Scholar]
- Schatz PJ. Use of peptide libraries to map the substrate specificity of a peptide-modifying enzyme: a 13 residue consensus peptide specifies biotinylation in Escherichia coli. Biotechnology (N Y) 1993;11:1138–43. doi: 10.1038/nbt1093-1138. [DOI] [PubMed] [Google Scholar]
- Scholler N, Crawford M, Sato A, Drescher C, O’Briant K, Kiviat N, Anderson G, Urban N. Bead-based ELISA assays for validation of ovarian cancer early detection markers. Clin Cancer Res. 2006;12:2117–2124. doi: 10.1158/1078-0432.CCR-05-2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholler N, Garvik B, Hayden-Ledbetter M, Kline T, Urban N. Development of a CA125-Mesothelin Cell Adhesion Assay as a Screening Tool for Biologics Discovery. Cancer Letters. doi: 10.1016/j.canlet.2006.03.029. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schummer M, Ng WV, Bumgarner RE, Nelson PS, Schummer B, Bednarski DW, Hassell L, Baldwin RL, Karlan BY, Hood L. Comparative hybridization of an array of 21,500 ovarian cDNAs for the discovery of genes overexpressed in ovarian carcinomas. Gene. 1999;238:375–85. doi: 10.1016/s0378-1119(99)00342-x. [DOI] [PubMed] [Google Scholar]
- Schwegler EE, Cazares L, Steel LF, Adam BL, Johnson DA, Semmes OJ, Block TM, Marrero JA, Drake RR. SELDI-TOF MS profiling of serum for detection of the progression of chronic hepatitis C to hepatocellular carcinoma. Hepatology. 2005;41:634–42. doi: 10.1002/hep.20577. [DOI] [PubMed] [Google Scholar]
- Sibler AP, Kempf E, Glacet A, Orfanoudakis G, Bourel D, Weiss E. In vivo biotinylated recombinant antibodies: high efficiency of labelling and application to the cloning of active anti-human IgG1 Fab fragments. J Immunol Methods. 1999;224:129–40. doi: 10.1016/s0022-1759(99)00016-2. [DOI] [PubMed] [Google Scholar]
- Smith PA, Tripp BC, DiBlasio-Smith EA, Lu Z, LaVallie ER, McCoy JM. A plasmid expression system for quantitative in vivo biotinylation of thioredoxin fusion proteins in Escherichia coli. Nucleic Acids Res. 1998;26:1414–20. doi: 10.1093/nar/26.6.1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumiyama K, Saitou N, Ueda S. Adaptive evolution of the IgA hinge region in primates. Mol Biol Evol. 2002;19:1093–9. doi: 10.1093/oxfordjournals.molbev.a004167. [DOI] [PubMed] [Google Scholar]
- Utz PJ. Protein arrays for studying blood cells and their secreted products. Immunol Rev. 2005;204:264–82. doi: 10.1111/j.0105-2896.2005.00251.x. [DOI] [PubMed] [Google Scholar]
- Venables JP. Aberrant and alternative splicing in cancer. Cancer Res. 2004;64:7647–54. doi: 10.1158/0008-5472.CAN-04-1910. [DOI] [PubMed] [Google Scholar]
- Vignali DA. Multiplexed particle-based flow cytometric assays. J Immunol Methods. 2000;243:243–55. doi: 10.1016/s0022-1759(00)00238-6. [DOI] [PubMed] [Google Scholar]
- Wang X, Campoli M, Ko E, Luo W, Ferrone S. Enhancement of scFv fragment reactivity with target antigens in binding assays following mixing with anti-tag monoclonal antibodies. J Immunol Methods. 2004;294:23–35. doi: 10.1016/j.jim.2004.08.005. [DOI] [PubMed] [Google Scholar]
- Warren DJ, Bjerner J, Paus E, Bormer OP, Nustad K. Use of an in vivo biotinylated single-chain antibody as capture reagent in an immunometric assay to decrease the incidence of interference from heterophilic antibodies. Clin Chem. 2005;51:830–8. doi: 10.1373/clinchem.2004.046979. [DOI] [PubMed] [Google Scholar]
- Weaver-Feldhaus JM, Miller KD, Feldhaus MJ, Siegel RW. Directed evolution for the development of conformation-specific affinity reagents using yeast display. Protein Eng Des Sel. 2005;18:527–36. doi: 10.1093/protein/gzi060. [DOI] [PubMed] [Google Scholar]
- Wilcox CA, Redding K, Wright R, Fuller RS. Mutation of a tyrosine localization signal in the cytosolic tail of yeast Kex2 protease disrupts Golgi retention and results in default transport to the vacuole. Mol Biol Cell. 1992;3:1353–71. doi: 10.1091/mbc.3.12.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Yu W, He T, Yu J, Caffrey RE, Dalmasso EA, Fu S, Pham T, Mei J, Ho JJ, Zhang W, Lopez P, Ho DD. Contribution of human alpha-defensin 1, 2, and 3 to the anti-HIV-1 activity of CD8 antiviral factor. Science. 2002;298:995–1000. doi: 10.1126/science.1076185. [DOI] [PubMed] [Google Scholar]