Abstract
The chromophore-binding properties of the higher plant light-harvesting protein CP29 have been studied by using site-directed mutagenesis of pigment-binding residues. Overexpression of the apoproteins in bacteria was followed by reconstitution in vitro with purified pigments, thus obtaining a family of mutant CP29 proteins lacking individual chromophore-binding sites. Biochemical characterization allowed identification of the eight porphyrins and two xanthophyll-binding sites. It is shown that the four porphyrin-binding sites (A1, A2, A4, and A5) situated in the central, twofold-symmetrical domain of the protein are selective for Chl-a, whereas the four peripheral sites (A3, B3, B5, and B6) have mixed Chl-a–Chl-b specificity. Within a site, porphyrin coordination by glutamine increases affinity for Chl-b as compared with glutamate. Xanthophyll site L1 is occupied by lutein, whereas site L2 can bind violaxanthin or neoxanthin. The protein is relatively stable when site L2 site is empty, suggesting that xanthophylls can be exchanged during operation of xanthophyll cycle-dependent photoprotection mechanism. Differential absorption spectroscopy allowed determination of transition energy levels for individual chromophores, thus opening the way to calculation of energy-transfer rates between Chl in higher plant antenna proteins.
Keywords: photosynthesis, chlorophyll, protein folding, CP29
Light energy for photosynthesis of green plants is collected by an antenna system composed of many homologous proteins belonging to the Lhc (light-harvesting complex) multigene family (1). These pigment–protein complexes are organized around photosynthetic reaction centers to form supramolecular complexes embedded in the thylakoid membranes and account for ≈70% of the pigments involved in photosynthesis. Understanding of energy-transfer processes in the antenna and reaction centers requires knowledge of the topological organization of subunits (2–4), of the distances between chromophores, and of their mutual transition-dipole orientation and the absorption/fluorescence energy levels. Although the resolution of the photosystem II LHC structure (LHCII) at 3.4-Å resolution (5) has allowed localization of chlorophyll (Chl)-binding sites and their relative distances, identification of transition-dipole orientation and energy levels are precluded by insufficient resolution of the structure thus far obtained or are not accessible by using structural analysis. We have used an alternative approach for the identification of the Chl-a, Chl-b, and xanthophyll molecules among the different binding sites and for determination of their individual absorption spectra: a series of mutant apoproteins was constructed by overexpression in bacteria of the Lhcb4 gene, in which individual chlorophyll-binding residues (5) were substituted for by residues unable to coordinate porphyrins. On in vitro refolding with purified pigments, proteins missing individual chromophores were obtained for seven of eight chlorophyll-binding sites and for one of the two xanthophyll-binding sites present in this antenna protein. Biochemical analysis and differential absorption spectroscopy allowed determination of the chemical nature and of the absorption properties of individual chromophores. These results represent an important step toward the understanding of energy-transfer processes within the higher plant antenna systems.
In selecting a model system for the study of Lhc proteins, LHCII, the major antenna complex of photosystem II, could be considered because its structure has been experimentally determined and therefore can be used as a guideline for mutation analysis. However, LHCII is a heterogeneous complex containing the products of many highly homologous genes (6). Also, LHCII is a heterotrimer, and influences of protein–protein interactions on the biochemical and spectroscopic characteristics of the system complicates identification of the primary effects of point mutations. Finally, four chlorophyll-binding ligands and a xanthophyll-binding site have not been identified by structural studies (5), making it difficult to design mutagenesis strategies. Among the members of the protein family, the chlorophyll–protein Lhcb4 (CP29), a minor photosystem II subunit, was chosen for this study because it is homogeneous, monomeric, and has only two xanthophylls and eight chlorophyll-binding sites, whose probable ligands are identified by homology with LHCII. Sequence homology between LHCII and CP29 in the transmembrane helices A–C and the amphipathic helix D, where chlorophyll-binding residues have been located, is very high (40–50% identity, 80% conservation), strongly suggesting a common structure.
MATERIALS AND METHODS
DNA Constructions.
Plasmids were constructed by using standard molecular-cloning procedures (7). Bacterial hosts were Escherichia coli TG1 strain (8) and SG13009 strain (9). Mutations were obtained according to ref. 10. The sequence was determined by the dideoxy method (11) by using an automated apparatus (Applied Biosystems Model 377).
Isolation of Overexpressed CP29 Apoprotein from Bacteria.
CP29 apoprotein was isolated from the SG13009 strain transformed with the CP29 constructs following a protocol previously described (12, 13).
Reconstitution of CP29 and LHCII–Pigment Complexes.
Complex-reconstitution procedures were performed as described in ref. 14. LHCII was reconstituted in the same conditions, but the Chlorophyll a/b ratio in the reconstitution mixture was set to 2.3 vs. 8.0 in CP29.
Purification of Reconstituted CP29 and LHCII.
CP29 and LHCII purification was performed by using ion-exchange chromatography (14). For determination of pigment-to-protein stoichiometry, it was necessary to obtain a fully purified protein that did not contain any residual contamination by bacterial proteins. The reconstituted CP29 was thus purified by preparative isoelectrofocusing (15) followed by ultracentrifugation in glycerol gradients (15–40% including 0.06% dodecylmaltoside and 10 mM Hepes, pH 7.6) for 12 hr at 60,000 rpm in a SW60 Beckman rotor to eliminate ampholytes.
Protein and Pigment Concentration.
The concentration of the CP29 apoprotein purified from E. coli inclusion bodies was determined by using the bicinchoninic acid assay (16). For stoichiometric (pigments/protein ratio) determination, the protein concentration was determined by using the ninhydrin method (17). Chlorophyll concentration was determined by the method described in ref. 18. HPLC analysis was performed as described in ref. 19.
Spectroscopy.
Absorption spectra were obtained by using a SLM-Aminco DW-2000 spectrophotometer at room temperature. Fluorescence excitation and emission spectra were obtained by using a Jasco-600 spectrofluorimeter. CD spectra were obtained at 8°C with a Jasco 600. Samples were in 10 mM Hepes, pH 7.6/0.06% dodecylmaltoside/20% glycerol. Chlorophyll concentrations were ≈10 μg/ml for CD and absorption measurements and 0.01 μg/ml for fluorescence measurements.
RESULTS
In vitro binding of pigments to Lhc proteins has been reported since 1987 (20, 13). Subsequent work with recombinant Lhc apoproteins has yielded pigment–protein complexes indistinguishable from the native proteins extracted from leaves (14, 21). In particular, the recombinant and native CP29 proteins were shown to bind six Chl-a, two Chl-b, and two xanthophyll molecules per polypeptide, whereas absorption, fluorescence, and CD spectra showed that pigment–pigment and pigment–protein interactions were closely reproduced (22). In this study we integrated site directed mutagenesis and in vitro reconstitution of pigment–protein complexes to identify and characterize the individual chromophores bound to the eight chlorophyll- and two xanthophyll-binding sites.
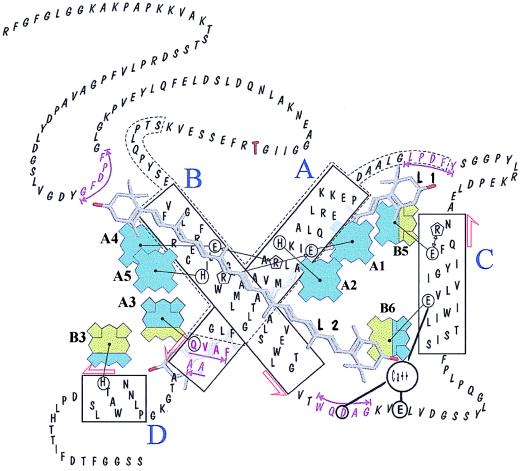
Chl-binding residues in LHCII have been localized in the α-helical domains (5), which are highly conserved in the Lhc family. Fig. 1 shows sequence comparison between LHCII and CP29 within the α-helical, pigment-binding domains, and Table 1 shows the correspondence between porphyrin-binding residues in LHCII and CP29. These are five single residues (three His, one Gln, one Glu) and three glutamates whose charge is compensated by arginines either in the same helix (Glu-174/Arg-177) or in another. The ion pairs have been proposed to have the dual function of binding pigments and of stabilizing the protein structure. Porphyrin-binding residues are fully conserved between LHCII and CP29 except for substitutions of the glutamine residue coordinating the porphyrin in site B6 of LHCII by glutamate in CP29 and the asparagine residue of site A2 by histidine.
Figure 1.
Alignment of Lhcb1 (LHCII) and Lhcb4 (CP29) protein sequences within the α-helical domains containing the Chl-binding residues. Identity (homology) is 43(80)%, 40(63)%, 41(72)%, and 40(80) % for helices A, C, B, and D, respectively.
Table 1.
Pigment-binding residues in the LHCII protein and in the maize CP29 protein
| Site | Helix | LHCII | CP29 | Mutated to |
|---|---|---|---|---|
| A1 | A/B | Glu-180/Arg-70 | Glu-213/Arg-116 | Val/Leu |
| A2 | A | Asn-183 | His-216 | Phe |
| A3 | A | Gln-197 | Gln-230 | Leu |
| A4 | B/A | Glu-65/Arg-185 | Glu-111/Arg-218 | Val/Leu |
| A5 | B | His-68 | His-114 | Phe |
| B3 | D | His-212 | His-245 | Leu |
| B5 | C/C | Glu-139/Arg-142 | Glu-174/Arg-177 | Val/Leu |
| B6 | C | Gln-131 | Glu-166 | Val, Gln |
The maize Lhcb4 cDNA (23) was used to generate a collection of mutants coding for CP29 proteins in which each Chl-binding residue was substituted by a nonbinding residue. In the case of ion-pairing ligands, individually, the interacting residues or both were mutated, as listed in Table 1. Lhcb4 clones with wild-type or mutant sequences were overexpressed in E. coli, and the recombinant apoprotein was refolded in vitro as previously described (14) to yield pigment–protein complexes whose biochemical properties are summarized in Table 2.
Table 2.
Biochemical properties of recombinant wild-type and mutant LHCII and CP29
| Recombinant protein | Binding site affected | Reconstitution efficiency, % | Chl a+b/ polypeptide | Xanthophylls | Chl/carotenoid ratio | Chl-a/b ratio | Chl-a polypeptide, no. | Chl-b polypeptide, no. | Stability at 60°C t1/2, min |
|---|---|---|---|---|---|---|---|---|---|
| CP29 | |||||||||
| Wild type | — | 100 | 8 | 2 | 4.00 | 3.00 | 6 | 2 | 4.2 |
| H245L | B3 | 94 | 7 | 2 | 3.50 | 4.35 | 5.7 | 1.3 | 2.0 |
| H216F | A2 | 75 | 7 | 2 | 3.10 | 2.53 | 5 | 2 | 4.0 |
| H114F | A5 | 53 | 7 | 2 | 3.47 | 2.42 | 5 | 2 | 0.5 |
| Q230L | A3 | 90 | 7 | 2 | 3.45 | 3.11 | 5.3 | 1.7 | 1.7 |
| R218L | A4 | 0 | |||||||
| E111V | A4 | 0 | |||||||
| E111V/R218L | A4 | 45 | 7 | 2 | 3.85 | 2.50 | 5 | 2 | 3.3 |
| E166V | B6 | 45 | 6 | 1 | 6.12 | 5.02 | 5 | 1 | 0.4 |
| E166Q | B6 | 90 | 8 | 2 | 4.00 | 2.34 | 5.6 | 2.4 | 5.2 |
| R177L | B5 | 0 | |||||||
| E174V | B5 | 55 | 7 | 1.5 | 4.75 | 3.91 | 5.6 | 1.4 | 0.5 |
| R177L/E174V | B5 | 0 | |||||||
| E213V | A1 | 0 | |||||||
| R116L | A1 | 0 | |||||||
| R116L/E213V | A1 | 0 | |||||||
| LHCII | |||||||||
| Wild type | — | 100 | 12 | 3 | 4.02 | 1.4 | 7 | 5 | ND |
| Q131E | B6 | 94 | 12 | 3 | 4.05 | 1.65 | 7.4 | 4.6 | ND |
Standard deviations: Chl-a/b ratio, 0.04 (8–12 independent measurements; Chl/xanthophyll ratio, 0.2 (six independent measurements). Reconstitution efficiency represents the yield in CP with respect to wild type. In the case of LHCII, 25% of the wild-type apoprotein used in reconstitution was recovered as CP. ND, not determined.
Stability of Mutant Proteins.
Of the 15 mutant CP29 proteins produced in this work, 7 (R218L, E111V, R177L, R177L/E174V, E213V, R116L, R116L/E213V) were either unable to refold in vitro with pigments or were dissociated during the chromatographic step of purification (Table 1). In particular, mutation of either one of the residues forming the ion pair Arg-116/Glu-213 or both to nonionic residues prevented protein folding, suggesting this ion pair is essential to the stabilization of helix A/helix B interaction. Mutation of residues E111 or R218 forming the other, symmetrical, interhelix ionic pair also (Fig. 2) prevented refolding. However, when both residues were changed to noncharged ones, a stable complex was obtained. This result indicates that ion pairing of E111 and R218 is not essential for helix–helix interactions, although the presence of noncompensated charges within the hydrophobic protein core disrupts folding. Mutation of both residues of the third ionic pair R177/E174 to L177/V174 or to L177/E174 prevented protein folding, whereas the R177/V174 mutant was stable, even though one Chl molecule is lost (Table 2). The eight mutant proteins that resisted purification (H245L, H216F, H114F, Q230L, E111V/R218L, E166V, E166Q, E174V) were stable for at least 2 days at 4°C and for several hours at room temperature as judged by absorption and fluorescence spectroscopy. The wild-type and mutant proteins were efficient in energy transfer from Chl-b to Chl-a. Heat denaturation of Chl–proteins was monitored by the half-time of decay of the red-most (679 nm) negative CD signal amplitude at 60°C. Stability was highest for wild-type CP29, whereas mutants showed decreased stability the most unstable mutants were those with decreased xanthophyll content (E174V and E166V) or lacking Chl A5 (Table 2).
Figure 2.
Molecular model of the CP29 protein obtained by homology with LHCII (5) and on the basis of mutation analysis. Tetrapyrroles are shown in dark gray for Chl-a and in light gray for Chl-b. Sites that can be occupied by either Chl-a or Chl-b have mixed filling. The portions of helices A and B showing inner homology are contoured by a broken line. Putative xanthophyll-binding sequences are underlined, and the phosphorylation site is in bold. Black lines connect chemical groups that are thought to closely interact in the folded protein.
Chl-Binding Sites. The wild-type CP29 protein bound six Chl-a, two Chl-b, and two xanthophyll molecules per polypeptide. Mutation of a Chl-a-binding site is therefore expected to change the Chl-a/b ratio from 3 to 2.5, whereas removal of a Chl-b molecule would yield a Chl-a/b ratio of 6.0. The Chl/xanthophyll ratio in both cases should change from 4.0 to 3.5 and the Chl/protein ratio from 8 to 7. Mutations at sites A2, A4, and A5 yielded proteins lacking a Chl-a chromophore because they all have Chl-a/b ratios of 2.5, Chl/xanthophyll ratios of 3.5, and Chl/protein ratios of 7. In contrast, none of the mutants exhibited characteristics expected for mutants with an empty Chl-b site (i.e., Chl-a/b = 6). Nevertheless, all mutations designed to affect pigment binding displayed decreased Chl content, confirming the interpretation of structural analysis (5).
These results suggest that sites A3, B3, B6, and B5 can bind both either Chl-a or Chl-b. Support for this conclusion is provided by the Q230L (site A3) and H245L (site B3) mutant proteins, which both bind seven Chl-and two xanthophyll molecules per polypeptide with Chl-a/b ratios of 3.10 and 4.35, respectively. This result is consistent with an average of 0.7 Chl-b and 0.3 Chl-a molecules bound to site B3 and, conversely, 0.7 Chl-a and 0.3 Chl-b in site A3 (Fig. 2).
E174V (site B5) and the E166V (site B6) mutant proteins also showed properties consistent with mixed Chl-a/Chl-b occupancy of sites B5 and B6. E174V, in fact, binds seven Chl molecules per polypeptide with a Chl-a/b ratio of 3.91, corresponding to 1.4 Chl-b and 5.6 Chl-a. The E166V mutant protein binds six Chl, suggesting that both chromophores in sites B6 and B5 are lost in this construct (see also below), whereas the Chl-a/b ratio of 5.0 shows that one Chl-a and one Chl-b are missing with respect to wild-type CP29. This observation leads to the conclusion that site B6 binds, on average, 0.6 Chl-a and 0.4 Chl-b. None of the three mutations aimed to prevent pigment binding to site A1 yielded a stable protein, thus preventing direct determination of composition at site A1. The assessments for the other sites described above, however, accounting for five Chl-a and two Chl-b molecules per polypeptide, are consistent with only Chl-a occupancy at site A1.
Site Affinity for Chl-a vs. Chl-b Can Be Modulated by the Liganding Residue.
Among Lhcb proteins, CP29 and CP26 are characterized by lower Chl-b content and have glutamate as a Chl ligand in site B6, whereas LHCII and CP24, richer in Chl-b, have glutamine in the corresponding position (2). Mixed Chl-a/Chl-b occupancy for binding sites in a Lhc protein was unexpected. To verify the binding of either Chl and to gain information on the factors modulating site affinity for Chl-a vs. Chl-b, Glu-166 was substituted by a Gln in CP29, whereas the corresponding Gln-131 residue in LHCII was mutated to glutamate. CP29 E166Q showed the same total number (eight) of Chl molecules bound per polypeptide but a higher Chl-b content with respect to the wild-type protein (Table 2). The values obtained (Table 2) were consistent with an increase of 0.4 Chl-b molecules per polypeptide and a corresponding decrease of Chl-a. LHCII proteins bind 12 Chl molecules, but the Q131E protein had an increased Chl-a content corresponding to 0.4–0.5 molecules per polypeptide (Table 2). These data confirm that site B6 can be occupied by either Chl-a or Chl-b in CP29, because an increased occupancy of this site by Chl-b can be induced by the E166Q mutation, whereas the Q131E mutation has the opposite effect in LHCII (Table 2).
Xanthophyll-Binding Sites.
Besides Chl-a and -b, Lhc proteins bind two xanthophyll molecules per polypeptide at two sites, cross-bracing helices A and B (5). Xanthophylls are essential for protein folding in both CP29 and LHCII (13, 14). The CP29 mutant protein E166V showed reduced xanthophyll content, corresponding to one xanthophyll molecule per polypeptide (Table 2). Inspection of the LHCII three-dimensional structure (5) shows that the porphyrin bound to E166 residue in site B6 is in close contact with the xanthophyll in site L2, thus suggesting that the loss of this chromophore also destabilizes the xanthophyll in site L2, leading to a protein with the L1 site occupied, while L2 remains empty. HPLC analyses of pigments extracted from the wild-type and mutant proteins show that although lutein content was not affected by the mutation, neoxanthin was almost absent, and violaxanthin decreased by 80% (Table 3). This result indicates that site L1 is occupied by lutein, whereas site L2 can be occupied by either neoxanthin or violaxanthin (Fig. 2). The properties of the E174V mutant protein, which also has reduced xanthophyll content because of impaired L1 assembly in the absence of a B5 site, support this interpretation.
Table 3.
Pigment composition of the wild-type CP29 and of two mutant proteins with decreased xanthophyll content
| CP29 | Xanthhophyll/polypeptide | Chl/xanthophyll | Lutein | Neoxanthin | Violaxanthin | Chl/polypeptide |
|---|---|---|---|---|---|---|
| Wild type | 2 | 4.04 | 0.89 | 0.47 | 0.64 | 8 |
| E166V | 1 | 6.06 | 0.84 | 0.04 | 0.12 | 6 |
| E174V | 1.46 | 4.77 | 0.83 | 0.17 | 0.46 | 7 |
Data are expressed as no. of moles per mole of polypeptide.
Spectral Characteristics of Individual Chls Within CP29 Protein.
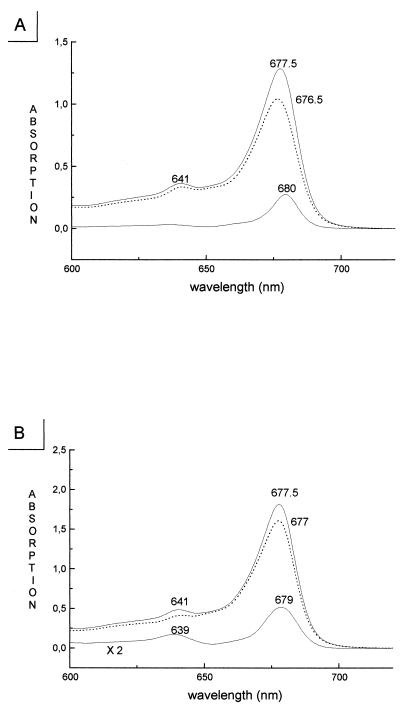
Chromophore–protein interactions modulate spectroscopic characteristics of individual Chls within the pigment–protein complex, producing multiple Chl-a and Chl-b absorption forms in the 630–680 nm range whose distribution and relative orientation of transition-moment vectors determines excitation energy flow within the protein (24). The absorption wavelengths of individual Chl molecules are difficult to determine by using spectroscopic methods because of the superimposition of their electronic transitions. The collection of point mutants described above provides a means to determine the absorption contributions of individual chl molecules by simply measuring the wild-type CP29 minus mutant difference absorption spectra. The major problem with this approach is the difficulty of normalizing the absorption spectra before subtraction. From determination of the stoichiometry of Chl-a and Chl-b to protein in wild-type and mutant proteins (Table 2), normalization of spectra could be performed for the total area over the Qy transition (630–700 nm), where there is no interference by carotenoid absorption, assuming a constant Chl-b/Chl-a extinction ratio of 0.7 (25, 22). An example of the results obtained by using this procedure is shown in Fig. 3A for the H216F mutant that we consider to lack the chromophore in site A2. The difference absorption spectrum shows a single peak at 680 nm with full-width half-maximum of 12 nm consistently with the characteristics expected for absorption by a single Chl-a molecule (26, 22). Similar results were obtained for mutants at sites A4 and A5, consistent with the loss of a Chl-a chromophore as deduced from the biochemical analyses. In the case of mutations at sites A3, B3, B5, and B6, more complex difference spectra were obtained, showing peaks in both the 630- to 660-nm range and in the 660- to 680-nm range because of Chl-b and Chl-a absorptions, respectively (22, 27). This is consistent with these sites being occupied by both Chl-a and Chl-b. As an example, the wild-type minus H245L difference spectrum is shown in Fig. 3B, which exhibits two positive peaks at 639 and 679 nm in agreement with Chl-b or Chl-a binding to site B3. Table 4 summarizes the absorption forms associated with each site within the protein. In the case of site A1, the determination is indirect because the mutant complex was unstable. Therefore, the absorption transition level of the Chl in this site was obtained by difference between wild-type spectrum and the sum of the spectra for the seven sites whose absorption was determined experimentally (data not shown). This procedure yielded a single 669-nm peak, which we attribute tentatively to Chl-a in site A1.
Figure 3.
Determination of the spectral characteristics of individual chromophores within CP29 by difference spectroscopy. (A) Absorption spectra of wild-type CP29 (Top) and of the H216F mutant (broken curve, Middle) lacking a single Chl-a molecule. The Bottom curve represents the wild-type minus mutant difference spectrum that is thought to represent the absorption spectrum of the individual Chl molecule bound to the A2 site. A single peak in the difference spectra was also obtained for mutations affecting sites A1, A2, and A5. (B) Absorption spectra of wild-type CP29 (Top) and of the H245L mutant (Middle) lacking the Chl-binding site B3 having mixed Chl-a/Chl-b occupancy. Difference spectra (Bottom curve) with peaks in both the Chl-b (630–660 nm) and the Chl-a (660–684 nm) spectral ranges were obtained with mutations affecting sites A3, B3, B5, and B6.
Table 4.
Room temperature absorption of individual chromophores in CP29
| Site | Chl species | Chl-a, % | Chl-a peak, nm | Chl-b, % | Chl-b peak, nm |
|---|---|---|---|---|---|
| A1 | a | 100 | 669 | 0 | — |
| A2 | a | 100 | 680 | 0 | — |
| A3 | a/b | 70 | 668 | 30 | 638 |
| A4 | a | 100 | 676 | 0 | — |
| A5 | a | 100 | 675 | 0 | — |
| B3 | a/b | 30 | 679 | 70 | 639 |
| B5 | a/b | 60 | 678 | 40 | 650 |
| B6 | a/b | 40 | 678 | 60 | 652 |
DISCUSSION
Chl Ligands.
The Chls of antenna complexes are noncovalently attached to the polypeptides by coordination of the central magnesium atom to polar side chains, eight of which have been identified in LHCII (5). Mutation at these sites leads to loss of Chl, confirming the assignments based on structural data. These eight residues are well conserved in CP29 (see Table 1 and Fig. 2). Exceptions are His-216 (asparagine in LHCII) and Glu-166 (glutamine in LHCII). Three glutamates, acting in both LHCII and CP29 as Chl ligands, are charge-compensated by nearby arginine residues. The CP29 sequence, however, shows no positively charged residues around Glu-166. Mutant proteins R116L, R177L, and R218L, which mimic noncompensated ions for the three other glutamate ligands in CP29, are unable to fold. We suggest that charge compensation of Glu-166 actually exists and is provided by the recently detected tightly bound Ca2+ that can be displaced by dicyclohexylcarbodiimide binding to Glu-166 (C. Jaegershold, M.C., A.M., and R.B., unpublished data) (Fig. 2). Ion pairs, besides acting in Chl binding, seem to have a crucial role in structural stabilization. Glu-111/Arg-218 and Arg-116/Glu-213 pairs lock helices A and B, but only the second pair is essential for folding, because abolition of the Glu-111/Arg-218 ionic interaction yields only a slightly increased susceptibility of the complex to heat denaturation. The Arg-177/Glu-174 ionic interaction takes places between residues located close to the stromal surface of the membrane, probably acting in the end-capping of helix C, as suggested by the inability of the double mutant 177V/174L to fold properly. In the absence of the Glu-174, Arg-177 alone can partially stabilize helix C, probably by anchoring to the hydrophilic surface (28).
Site Specificity for Chl-a and -b.
The structural differences between Chl-a and -b are too small to be detected by electron crystallographic analysis. The LHCII tetrapyrroles were tentatively attributed to Chl-a and Chl-b from the assumption that Chl-a molecules should be closer to the carotenoid molecules because they have a high probability of forming harmful triplet states that need to be quenched (5). The present analyses only partially confirm the above hypothesis. Sites A1, A2, A4, and A5, symmetrically arranged around the central two-fold axis in close contact to the xanthophylls (Fig. 2), are found to be occupied by Chl-a. However, sites A3 and B6, which lie at similar distances from the xanthophylls, can be occupied by Chl-b, similar to sites B3 and B5. This finding confirms a previous femtosecond transient-absorption study showing direct energy transfer to two Chl-b molecules on xanthophyll excitation in LHCII (29), suggesting that mixed occupancy of peripheral sites might be a property not only of CP29 but also of LHCII complex. It should be noticed that the four central Chl-a sites together with part of helices A and B represent the central core of the complex, which is conserved not only in Chl-a/b but also in Chl-a/c proteins. Helix C and the luminal parts of helices A and B are more variable, according to their ability to bind different amounts of Chl-b or even Chl-c (2). Lack of specificity in ligand binding in the peripheral sites seems to be a feature not only within different members of the protein family but also of the individual sites such that CP29 preparations, for example, appear to be a mixed population in which four sites can be occupied by either Chl-a or Chl-b with roughly equal probability. The degree of specificity of an individual site depends, at least in part, on the nature of the Chl-liganding residue, as shown by the results of mutating the residue for Chl coordination in site B6 from Glu to Gln in CP29 and, conversely, from Gln to Glu in LHCII, with Gln determining higher affinity for Chl-b than Glu. However, the protein environment must also be important in specificity as shown by the fact that sites A2, A5, and B3 have histidine ligands, and yet only site B3 can bind Chl-b. Our results are consistent with previous work with recombinant CP29 and CP24 showing that Chl-b can substitute for Chl-a to some extent and yet obtaining fully functional antenna proteins (22). However, in the absence of sufficient Chl-a, a few sites remain empty, indicating that certain sites display higher affinities (27). CP29 was found to reconstitute in vitro with the same pigment composition and spectra within a range of Chl-a/b ratios in the reconstitution mixture (21). This is somehow contrasting with the finding of four sites with mixed Chl-a/b occupancy and can be explained by hypothesizing a conformational change between sites A3/B3 and B5/B6 inducing increased affinity for Chl-b in one site on binding of Chl-a to the neighboring site. Long-range conformational change of CP29 was previously described on reversible phosphorylation of Thr-83 residue (30).
Xanthophylls: Localization and Consequences for Regulation.
Besides its role in light harvesting and energy transfer, CP29 is deeply involved in nonphotochemical quenching. This is a mechanism of regulation leading to heat dissipation of excitation energy when irradiance is in excess with respect to the capacity for electron transport (31, 32), thus protecting from photoinhibition. Genetic dissection of the nonphotochemical quenching mechanism has shown that both the rapid and slow kinetic phases of nonphotochemical quenching rise are dependent on xanthophylls (33). Moreover, it was shown that the rapid phase depends on the protonation of a group with a pK of 4.5 and the release of a divalent cation, which leads to the appearance of a binding site for zeaxanthin (34). The cation release is needed for the establishment of slow or sustained quenching (33). Mutation E166V yields a pigment–protein complex with a single xanthophyll molecule per polypeptide, and yet this complex is relatively stable, suggesting that violaxanthin-to-zeaxanthin conversion may proceed via diffusion of violaxanthin out of site L2 and replacement by zeaxanthin de-epoxidized in the thylakoid lipid phase (ref. 31; see Fig. 2). The recent finding that CP29 binds one Ca2+ to multiple acidic residues, including E166, suggests that luminal pH may control the conformation of the helix A-to-helix C loop. Thus, the diffusion of the xanthophyll molecule in site L2 would be facilitated by changes in coordination of Ca2+ to glutamate residues 138 and 143, which are part of the putative xanthophyll-binding site (Fig. 2; ref. 35). The quenching mechanism itself is likely to derive from the protonation of Glu-166, a dicyclohexylcarbodiimide-binding site (36), leading to disconnection of the Chl in site B6 and establishment of strong xanthophyll–Chl interactions (37). The presence in site L2 of xanthophylls with different conjugation lengths (i.e., with different energy levels for the lowest excited state) (38) is likely to modulate the amplitude of the quenching.
Spectral Properties of Individual Chls.
Detailed understanding of energy transfer between chromophores within a chlorophyll–protein complex requires knowledge of their mutual distances, the spectral characteristics (absorption and fluorescence energy levels) of the individual chromophores, and the mutual orientation of transition-moment vectors. Although the first parameter has been essentially elucidated by the structural studies (5), the spectral properties of each chromophore are not accessible by structural methods and are difficult to obtain by spectroscopic methods in the case of higher plant antenna because of spectral overlap. The above-described identification of Chl-a and Chl-b within the different sites also is not sufficient, because pigment–protein interactions can efficiently affect Chl-a and Chl-b electronic transitions that can be shifted within the 666- to 684-nm and 638- to 660-nm ranges, respectively, significantly influencing spectral overlap and therefore efficiency of energy transfer (39). The mutants obtained in this work allowed determination of absorption peaks for individual Chl molecules within the CP29 structure (Table 4) by simply determining wild-type minus mutant difference spectra. This simple approach is justified from previous work (22) showing that, in CP29, pigment–protein interactions rather than pigment–pigment interactions dominate in generating spectral heterogeneity. Consistently, each mutant protein was found to be depleted in a specific absorption form whose amplitude and width is consistent with the expected characteristics of a single chromophore as deduced by hole-burning experiments (26). The present results support the view that in CP29, each pigment conserves its individual characteristics on protein interaction with protein, and strong coupling with other chromophores is unlikely (22). It is not clear whether this is a general characteristic of Lhc proteins, because CP29 is a member of the family with fewer Chl molecules per polypeptide. Chromophore density in LHCII can be significantly higher, thus allowing stronger pigment–pigment interactions. For the first time, the absorption spectra of individual pigments within a Lhc protein has been resolved. The data of Table 4 are essential for the analytical dissection of the energy-transfer pathway in higher plant antenna proteins once the mutual orientation of transition-dipole vectors is determined. Orientation could be obtained by improved resolution of the complex structures beyond that of the two-dimensional crystals thus far resolved (5). An alternative approach is study the mutant collection described here by using linear and CD spectroscopy.
Acknowledgments
We thank Massimo Crimi and Roberto Simonetto (University of Verona) for discussion and help in the construction of a molecular model of CP29 and for initial work on protein denaturation. Francis-André Wollman (Paris) and Arnold Hoff (Leiden) are thanked for critically reading the manuscript. Claudio Varotto is thanked for preliminary work on recombinant LHCII. This work was supported by Progetto Nazionale Biotecnologie of the Italian Ministry of Agriculture and by “Target Program on Biotechnology” Consiglio Nazionale delle Ricerche.
ABBREVIATIONS
- Chl
chlorophyll
- CP
Chl–protein
- LHCII
light-harvesting complex of photosystem II
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1.Green B R, Pichersky E, Kloppstech K. Trends Biochem Sci. 1991;16:181–186. doi: 10.1016/0968-0004(91)90072-4. [DOI] [PubMed] [Google Scholar]
- 2.Bassi R, Sandonà D, Croce R. Physiol Plant. 1997;100:769–779. [Google Scholar]
- 3.Boekema E J, Hankamer B, Bald D, Kruip J, Nield J, Boonstra A F, Barber J, Rögner M. Proc Natl Acad Sci USA. 1995;92:175–179. doi: 10.1073/pnas.92.1.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Harrer R, Bassi R, Testi M G, Schäfer C. Eur J Biochem. 1998;255:196–205. doi: 10.1046/j.1432-1327.1998.2550196.x. [DOI] [PubMed] [Google Scholar]
- 5.Kühlbrandt W, Wang D N, Fujiyoshi Y. Nature (London) 1994;367:614–621. doi: 10.1038/367614a0. [DOI] [PubMed] [Google Scholar]
- 6.Dunsmuir P. Nucleic Acids Res. 1985;13:2503–2518. doi: 10.1093/nar/13.7.2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 8.Gibson T J. Ph.D. thesis. Cambridge, U.K.: Cambridge University; 1984. [Google Scholar]
- 9.Gottesman S, Halpern E, Trisler P. J Bacteriol. 1981;148:265–273. doi: 10.1128/jb.148.1.265-273.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yukenberg P D, Withey F, Geisselsoder J, McClary J. In: Directed Mutagenesis: A Practical Approach. McPherson M J, editor. Oxford: IRL; 1991. pp. 27–48. [Google Scholar]
- 11.Sanger F, Nicklen S, Carlson A R. Proc Natl Acad Sci USA. 1977;74:5463–5468. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nagai K, Thogersen H C. Methods Enzymol. 1987;153:461–481. doi: 10.1016/0076-6879(87)53072-5. [DOI] [PubMed] [Google Scholar]
- 13.Paulsen H, Rümler U, Rüdiger W. Planta. 1990;181:204–211. doi: 10.1007/BF02411539. [DOI] [PubMed] [Google Scholar]
- 14.Giuffra E, Cugini D, Croce R, Bassi R. Eur J Biochem. 1996;238:112–120. doi: 10.1111/j.1432-1033.1996.0112q.x. [DOI] [PubMed] [Google Scholar]
- 15.Dainese P, Hoyer-Hansen G, Bassi R. Photochem Photobiol. 1990;51:693–703. doi: 10.1111/php.1990.51.6.693. [DOI] [PubMed] [Google Scholar]
- 16.Smith P K, Krohn R I, Hermanson G T, Mallia A K, Gartner F H, Provenzano M D, Fujimoto E K, Goeke N M, Olson B J, Klenk D C. Anal Biochem. 1985;150:76–85. doi: 10.1016/0003-2697(85)90442-7. [DOI] [PubMed] [Google Scholar]
- 17.Hirs C H W. Methods Enzymol. 1967;11:325–329. [Google Scholar]
- 18.Porra R J, Thompson W A, Kriedemann P E. Biochim Biophys Acta. 1989;975:384–394. [Google Scholar]
- 19.Gilmore A M, Yamamoto H Y. J Chromatogr. 1991;543:137–145. [Google Scholar]
- 20.Plumley F G, Schmidt G W. Proc Natl Acad Sci USA. 1987;84:146–150. doi: 10.1073/pnas.84.1.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sandonà D, Croce R, Pagano A, Crimi M, Bassi R. Biochim Biophys Acta. 1998;1365:207–214. doi: 10.1016/s0005-2728(98)00068-1. [DOI] [PubMed] [Google Scholar]
- 22.Giuffra E, Zucchelli G, Sandonà D, Croce R, Cugini D, Garlaschi F M, Bassi R, Jennings R. Biochemistry. 1997;36:12984–12993. doi: 10.1021/bi9711339. [DOI] [PubMed] [Google Scholar]
- 23.Bergantino E, Sandonà D, Cugini D, Bassi R. Plant Mol Biol. 1998;36:11–22. doi: 10.1023/a:1005904527408. [DOI] [PubMed] [Google Scholar]
- 24.Hemelrijk P W, Kwa S L S, van Grondelle R, Dekker J P. Biochim. Biophys Acta. 1992;1098:159–166. [Google Scholar]
- 25.Sauer K, Lindsay-Smith J R, Schultz A J. J Am Chem Soc. 1966;88:2681–2688. [Google Scholar]
- 26.Reddy N R S, van Amerongen H, Kwa S L S, van Grondelle R, Small G J. J Phys Chem. 1994;98:4729–4737. [Google Scholar]
- 27.Pagano A, Cinque G, Bassi R. J Biol Chem. 1998;273:17154–17165. doi: 10.1074/jbc.273.27.17154. [DOI] [PubMed] [Google Scholar]
- 28.Aurora R, Rose G D. Protein Sci. 1998;7:21–38. doi: 10.1002/pro.5560070103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Connelly J P, Muller M, Bassi R, Croce R, Holzwarth A R. Biochemistry. 1997;36:281–287. doi: 10.1021/bi962467l. [DOI] [PubMed] [Google Scholar]
- 30.Croce R, Breton J, Bassi R. Biochemistry. 1996;35:11142–11148. doi: 10.1021/bi960652t. [DOI] [PubMed] [Google Scholar]
- 31.Yamamoto H, Bassi R. In: Oxygenic Photosynthesis: The Light Reactions. Ort D R, Yocum C F, editors. Dordrecht, The Netherlands: Kluwer; 1996. pp. 539–563. [Google Scholar]
- 32.Bassi R, Pineau B, Dainese P, Marquardt J. Eur J Biochem. 1993;212:297–303. doi: 10.1111/j.1432-1033.1993.tb17662.x. [DOI] [PubMed] [Google Scholar]
- 33.Niyogi K K, Grossman A R, Björkman O. Plant Cell. 1998;10:1121–1134. doi: 10.1105/tpc.10.7.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gilmore A M, Shinkarev V P, Hazlett T L, Govindjee Biochemistry. 1998;37:13582–13593. doi: 10.1021/bi981384x. [DOI] [PubMed] [Google Scholar]
- 35.Pichersky E, Jansson S. In: Oxygenic Photosyntesis: The Light Reactions. Ort D R, Yokum C F, editors. Dordrecht, The Netherlands: Kluwer; 1996. pp. 507–521. [Google Scholar]
- 36.Pesaresi P, Sandona D, Giuffra E, Bassi R. FEBS Lett. 1997;402:151–156. doi: 10.1016/s0014-5793(96)01518-9. [DOI] [PubMed] [Google Scholar]
- 37.Crofts A R, Yerkes C T. FEBS Lett. 1994;352:265–270. doi: 10.1016/0014-5793(94)00976-7. [DOI] [PubMed] [Google Scholar]
- 38.Frank H A, Cua A, Chynwat V, Young A, Gosztola D, Wasiliewsky M R. Photosynth Res. 1994;41:389–395. doi: 10.1007/BF02183041. [DOI] [PubMed] [Google Scholar]
- 39.Shipman L L, Housman D. J Photochem Photobiol. 1979;29:1163–1167. [Google Scholar]