Abstract
The total chemical synthesis of proteins has great potential for increasing our understanding of the molecular basis of protein function. The introduction of native chemical ligation techniques to join unprotected peptides next to a cysteine residue has greatly facilitated the synthesis of proteins of moderate size. Here, we describe a straightforward methodology that has enabled us to rapidly analyze the compatibility of the native chemical ligation strategy for X–Cys ligation sites, where X is any of the 20 naturally occurring amino acids. The simplified methodology avoids the necessity of specific amino acid thioester linkers or alkylation of C-terminal thioacid peptides. Experiments using matrix-assisted laser-desorption ionization MS analysis of combinatorial ligations of LYRAX-C-terminal thioester peptides to the peptide CRANK show that all 20 amino acids are suitable for ligation, with Val, Ile, and Pro representing less favorable choices because of slow ligation rates. To illustrate the method’s utility, two 124-aa proteins were manually synthesized by using a three-step, four-piece ligation to yield a fully active human secretory phospholipase A2 and a catalytically inactive analog. The combination of flexibility in design with general access because of simplified methodology broadens the applicability and versatility of chemical protein synthesis.
The introduction of unnatural chemical groups into the covalent structure of a protein molecule can provide valuable insights into questions about protein function and enzyme activity. Consequently, unnatural amino acids have been incorporated into proteins through the complementary approaches of total chemical synthesis by solid-phase peptide synthesis (SPPS) (1, 2) and in vitro translation with mis-acylated tRNA (3). Despite the success of these methods in several protein systems, technical limitations have limited their general application.
Native chemical ligation has been developed to facilitate the synthesis of proteins of moderate size (≈150 aa) (4–8). This ligation chemistry involves a chemoselective reaction between a C-terminal thioester and an N-terminal cysteine residue to yield a native peptide bond at the site of ligation (9). One limitation to this approach has been the efficient synthesis of C-terminal thioester peptides. Traditionally, these peptides have been obtained in two steps through selective alkylation of thioacid peptides (4) generated from optimized SPPS (10) by using an HF-labile thioacid linker (11, 12). However, the need for an individual-solution synthesis of a tert-butoxycarbonyl (Boc)–amino–thioacid linker for each C-terminal amino acid desired has limited the general application of this approach (12). Furthermore, deprotection and cleavage of the polypeptide chain from the resin by anhydrous HF yields the corresponding C-terminal thioacid (COSH) that requires alkylation and purification to obtain the activated thioester (COSR). As a result, only a limited set of ligation pairs has been examined by using Ala (4), Leu (13), or Gly (4, 7) as the C-terminal thioester residue from individual syntheses of amino acid thioacid linkers (12). Other approaches can be used to generate C-terminal thioester peptides directly (5, 14, 15), for example Asn, but also suffer from the requirement for synthesizing an amino acid–thioester linker of each individual amino acid.
To facilitate the synthesis of C-terminal thioester peptides, a general and straightforward procedure has been developed that generates a resin from which any desired thioester peptide can be readily synthesized. This thioester resin is a generalized and significantly simplified variant of that described by Hojo et al. (14). After chain assembly and deprotection, the resulting C-terminal activated thioester peptides can be used in native-chemical ligation without further modification. The efficacy of all 20 naturally occurring amino acids in native chemical ligation reactions has been demonstrated by synthesis and analysis of defined mixtures of peptides. The utility of this approach was demonstrated by the total chemical syntheses of 124-aa human secretory phospholipase A2 (hsPLA2) and an active-site chemical mutant in which His-47 was replaced by the isosteric β-thienyl alanine (Bta).
EXPERIMENTAL PROCEDURES
Materials.
2-(1H-Benzotriazol-1-yl)-1,1,3,3-tetramethyluronium hexafluorophosphate (HBTU) and Boc–amino acids were obtained from Nova Biochem, Boc–Arg (p-toluenesulfonyl)-OH Boc–Asn(xanthyl)-OH, and Boc–β-thienylalanine were obtained from Bachem. S-trityl-mercaptopropionic acid was obtained from Peptides International. Boc–Cys-OCH2Pam-resin and N,N-diisopropylethylamine (DIEA) were obtained from Applied Biosystems. p-Methylbenzhydrylamine (MBHA) resin was obtained from Peninsula Laboratories. 2-(Methylsulfonyl)ethyl 4-nitrophenyl carbonate was obtained from Fluka. N,N-Dimethylformamide (DMF) and HPLC-grade acetonitrile were purchased from Fischer. Trifluoroacetic acid (TFA) was obtained from Halocarbon Products (Hackensack, NJ). HF was purchased from Matheson. 1,2-Bis(heptanoylthio)-phosphatidylcholine and 5,5′-dithiobis(2-nitrobenzoic) acid (DTNB) were purchased from Cayman Chemicals (Ann Arbor, MI).
Peptide Synthesis. Peptides were prepared by manual SPPS typically on a 0.25-mmol scale by using the in situ neutralization/HBTU activation procedure for Boc chemistry as described (10). Each synthetic cycle consisted of Nα-Boc removal by a 1- to 2-min treatment with neat TFA, a 1-min DMF flow wash, a 10- to 20-min coupling time with 1.0 mmol of preactivated Boc–amino acid in the presence of excess DIEA, and a second DMF flow wash. Nα-Boc–amino acids (1.1 mmol) were preactivated for 3 min with 1.0 mmol of HBTU (0.5 M in DMF) in the presence of excess DIEA (3 mmol). After each coupling step, yields were determined by measuring residual free amine with the quantitative ninhydrin assay (16). After coupling of Gln residues, a DCM flow wash was used before and after deprotection by using TFA, to prevent possible high-temperature (TFA/DMF)-catalyzed pyrrolidone formation (10).
After chain assembly was completed, the peptides were deprotected and cleaved from the resin by treatment with anhydrous HF for 1 hr at 0°C with 4% p-cresol as a scavenger. In all cases, the imidazole side-chain 2,4-dinitrophenyl (Dnp) protecting groups remained on His residues because the Dnp-removal procedure is incompatible with C-terminal thioester groups. However, Dnp is gradually removed by thiols during the ligation reaction, yielding unprotected His. After cleavage, both peptides were precipitated with ice-cold diethylether, dissolved in aqueous acetonitrile, and lyophilized.
Trityl-Associated Mercaptopropionic Acid–Leucine (TAMPAL) Resin. Nα-Boc–Leu (4 mmol) was activated with 3.6 mmol of HBTU in the presence of 6 mmol of DIEA and coupled for 16 min to 2 mmol of MBHA resin (99.9% coupling yield). (Boc–Leu Pam resin has also been used as the initial solid support.) Next, 3 mmol of S-trityl mercaptopropionic acid was activated with 2.7 mmol of HBTU in the presence of 6 mmol of DIEA and coupled for 16 min to Leu-MBHA resin (99.9% coupling). The resulting TAMPAL resin can be used as a starting resin for polypeptide-chain assembly after removal of the trityl protecting group with two 1-min treatments with 2.5% triisopropylsilane and 2.5% H2O in TFA. The thioester bond can be formed with any desired amino acid by using standard in situ-neutralization peptide-coupling protocols for 1 hr. Treatment of the final peptide with anhydrous HF yields the C-terminal activated mercaptopropionic acid–leucine (MPAL) thioester peptides that are ready for participation in native chemical ligation.
Disulfide formation on the resin after deprotection of the trityl-protected thiol could decrease the synthetic yield. This side reaction has not been a problem, although care should be taken to avoid pulling air through the resin after trityl deprotection. Synthetic yields of hsPLA2 (all fragments synthesized on TAMPAL resin) were between 73% and 85% of the theoretical final peptide-resin weight, whereas the synthesis yield of fragment 88–124 (on Cys-phenylacetamidomethyl resin) was 72%. Moreover, when the resin was treated with 2-mercaptoethanol (5%) in DMF for 5 min after trityl removal, no increase in synthetic yield was observed.
LYRAX and CRANK Syntheses.
After trityl deprotection, 1.05 mmol of TAMPAL resin was divided into 20 glass tubes (0.05 mmol), and each was incubated for 1 hr with each of the 20 preactivated amino acids (1.1 mmol of X preactivated with 1 mmol of HBTU in the presence of 3 mmol of DIEA). After coupling, the resins were combined in four pools (pool 1, W, E, D, T, S; pool 2, R, Q, N, P, G; pool 3, F, M, I, V, A; and pool 4, Y, K, L, C, H), and LYRAX syntheses were continued in a parallel fashion (0.25 mmol of resin each, by using 1.1 mmol of amino acid preactivated with 1 mmol of HBTU in the presence of 3 mmol of DIEA). After synthesis, the LYRAX pools were Nα-acetylated, cleaved from the resin, and lyophilized. Nα-Amino CRANK peptide was synthesized on a 0.7 mmol scale on MBHA resin by using 2 mmol of preactivated amino acid per coupling (2.2 mmol of amino acid activated with 2 mmol of HBTU in the presence of 6 mmol of DIEA). After HF treatment of the peptide-resin, CRANK was lyophilized and stored at −20°C.
hsPLA2 Polypeptide Fragment Syntheses.
hsPLA2 1 (1–27), 2 (28–58), and 3 (59–87) (Fig 2A) only were synthesized on TAMPAL resin (0.25-mmol scale) to yield C-terminal MPAL-activated thioesters; peptide 4 (88–124) was synthesized on Cys-phenylacetamidomethyl resin to yield a C-terminal carboxylic acid group after deprotection and cleavage of the peptide resin (Fig. 2A). Peptides 2 and 3 were N-terminally protected with a 2-(methylsulfonyl)ethyl carbonate (Msc) group by a 2-hr incubation with a 10-fold excess of activated Msc–nitrophenyl ester in a minimal volume of DMF/5% DIEA. In a parallel synthesis, Bta-47 replaced His-47 in peptide 28–58. After deprotection and cleavage from the resin, the polypeptides were HPLC-purified, lyophilized, and stored at −20°C until use.
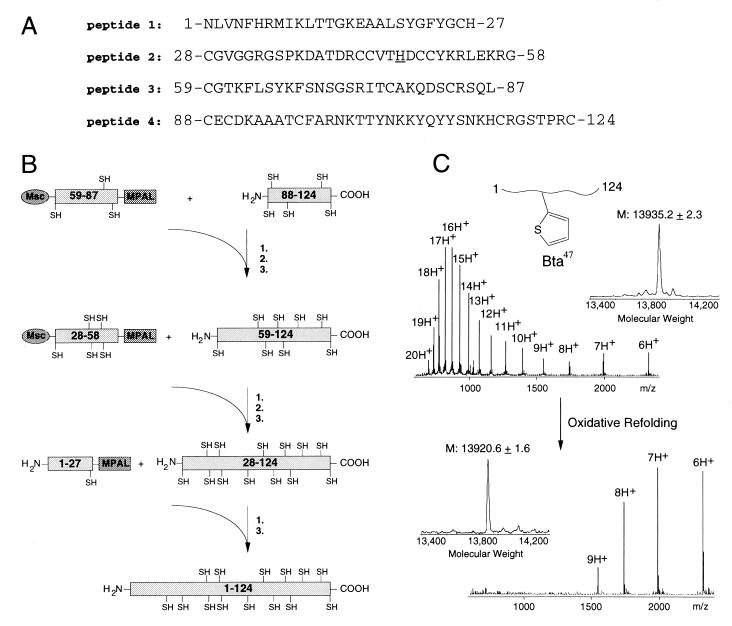
Figure 2.
Protein synthesis by multistep ligation: hsPLAA2. (A) Sequence of the four peptides comprising the 124-aa polypeptide chain of hsPLA2. Ligation sites include His(Dnp)–Cys, Gly–Cys, and Leu–Cys. In a parallel synthesis, the underlined His-47 active-site residue was replaced by the isosteric Bta to obtain a chemical hsPLA2 variant. (B) Synthetic scheme leading to the 124-aa hsPLA2 polypeptide chain. Typically, unprotected purified peptides were dissolved at 10 mg/ml in 0.1 M phosphate buffer containing 6 M guanidine and 4% benzylmercaptan and thiophenol reaching pH ≈ 7 (1). To avoid polymerization reactions, N-terminal cysteine residues of the activated thioester peptides were protected with Msc groups. After ligation, a 5-min treatment at pH 13 removed N-terminal Msc (2). HPLC yielded the purified ligation product. (3). (C) Electrospray ionization-MS of the reduced polypeptide chain of hsPLA2–Bta-47 (Upper) and the oxidized refolded enzyme variant (Lower). Mass reconstructions from the m/z ratios show a mass decrease from 13,935 Da to 13,921 Da representing the loss of 14 protons in the formation of seven internal disulfide bonds.
HPLC.
Analytical reversed-phase HPLC was performed on a Hewlett Packard HPLC 1050 system by using Vydac C-18 columns (5 μm, 0.46 × 15 cm). Semipreparative reversed-phase HPLC was performed on a Rainin HPLC system by using a Vydac C-18 column (10 μm, 1.0 × 25 cm). Linear gradients of acetonitrile in water/0.1% TFA were used to elute bound peptides. The flow rates used were 1 ml/min (analytical) and 5 ml/min (semipreparative).
MS.
Electrospray ionization MS was performed on an API-III triple quadrupole mass spectrometer (Sciex, Thornhill, ON, Canada). Peptide masses were calculated from the experimental m/z from all of the observed protonation states of a peptide by using macspec software (Sciex). Matrix-assisted laser-desorption ionization (MALDI)-MS was performed on a Dynamo DY-100 mass spectrometer (Thermo BioAnalysis, Santa Fe, NM) by using α-cyano-4-hydroxycinnamic acid (Aldrich) as a matrix. Theoretical masses of peptides and proteins were calculated by using macpromass software (Beckman Research Institute, Duarte, CA).
Native Chemical Ligation. The ligation of unprotected synthetic peptide segments was performed as follows: 0.1 M phosphate buffer (pH 8.5) containing 6 M guanidine, 4% (vol/vol) benzylmercaptan, and 4% (vol/vol) thiophenol was added to dry peptides, resulting in a final peptide concentration of 1–3 mM at a pH ≈7 (lowered because of addition of thiols and TFA from the lyophilized peptide). The ligation reaction was performed in a heating block at 37°C and was vortexed periodically to equilibrate the thiol additives. The reaction was monitored by using MALDI-MS and/or HPLC and electrospray ionization-MS until completion. LYRAX–CRANK ligations were performed at 3 mM individual LYRAX concentrations (15 mM total) and 30 mM CRANK concentrations for each of the four pools. For the hsPLA2 ligations, the following polypeptide concentrations were used: 3.4 and 2.7 mM for 59–87 and 88–124, respectively; 2.2 and 1.3 mM for 28–58 and 59–124, respectively; and 1.4 and 0.9 mM for 1–27 and 28–124, respectively.
The yields for the described hsPLA2 ligation reactions were 80–90% by HPLC, although an additional loss of 50% as a result of HPLC purification per step is typical for relatively small-scale ligations (1–2 μmol). For the described hsPLA2 ligation, we started with 6 mg of 88–124 (1.3 μmol) and obtained 1.1 mg of purified 1–124 (0.08 μmol) after three successive ligations and three HPLC purifications.
Enzymatic sPLA2-Activity Assay.
The hydrolysis of 1,2-bis(heptanoylthio)-phosphatidylcholine substrate (17) by sPLA2 was analyzed on a Cary 1-Bio double beam spectrophotometer (Varian) at 37°C as described (18). Km and kcat values were determined with enzfitter software (Elsevier-Biosoft, Cambridge, U.K.).
RESULTS AND DISCUSSION
To facilitate the synthesis of thioester peptides with a wide variety of C-terminal amino acids, a general thioester peptide-producing resin linker for Boc chemistry–SPPS was designed. Starting with the HF-labile MBHA resin, leucine was coupled followed by S-trityl mercaptopropionic acid by using standard SPPS conditions (10). The resulting TAMPAL resin can be used as a starting resin for polypeptide-chain assembly after removal of the trityl protecting group. The desired thioester bond can be generated directly on the resin (15) by using any amino acid and standard coupling conditions. This peptide–linker had been previously shown to be compatible with Boc–SPPS with glycine at the C terminus and with norleucine as a spacer to provide greater stability during synthesis (14). After HF cleavage and purification, the C-terminal MPAL–thioester peptide can be used directly in native chemical ligation after in situ thioester exchange (Fig. 1A).
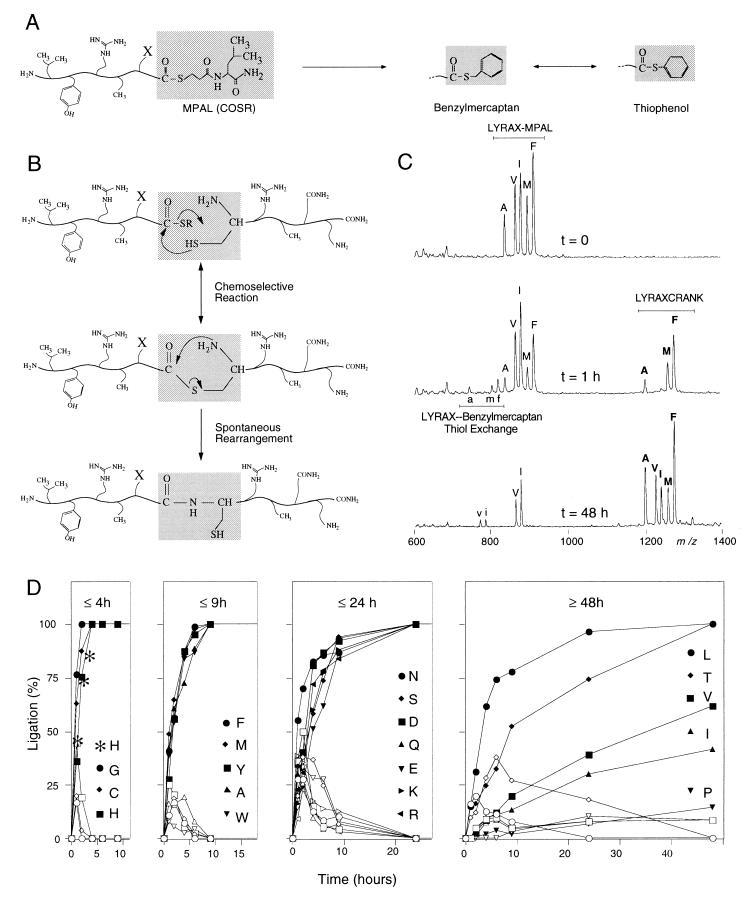
Figure 1.
Parallel native chemical ligation model study of LYRAX-to-CRANK peptide ligations, with X representing all 20 natural amino acids. (A) Under experimental conditions during native chemical ligation, LYRAX–MPAL-activated thioester peptides undergo thioester exchange reactions with benzylmercaptan and thiophenol. (B) Native chemical ligation. COSR of LYRAX–MPAL undergoes nucleophilic attack by the side chain of the N-terminal cysteine residue of CRANK, after which a rapid intramolecular rearrangement produces a native peptide bond at the site of ligation. (C) Simultaneous MALDI-MS readout of combinatorial ligations of crude LYRAX–MPAL-to-CRANK ligations featuring A, V, I, M, and F as C-terminal amino acid-activated thioester residues. (D) Determination of ligation product (filled symbols) and benzylmercaptan–thioester exchange intermediates (open symbols) as a function of time for all 20 LYRAX–MPAL C-terminal activated thioesters. C-terminal amino acids are divided into four groups, in which ligations were completed within 4, 9, or 24 hr or in 48 hr or more, respectively. Left, (t ≤ 4 hr), ∗ indicates the observed LYRAH–CRANK model-peptide ligation rate when monitored by using HPLC analysis.
The compatibility and efficiency in native chemical ligation of all 20 amino acid-COSR peptides were analyzed. Four pools of five LYRAX peptides (where X = any amino acid) were selected so that each pool would contain unique masses for every LYRAX–COSR (LYRAX–MPAL) peptide (Fig. 1A). After HF treatment, four pools of peptides, each containing five different activated LYRAX–MPAL peptides, were obtained (Fig. 1A). The purity and mass of each peptide were confirmed by using HPLC and electrospray ionization MS. Ligation reactions with the peptide, CRANK, were performed for each of the four crude peptide–thioester mixtures by using standard ligation conditions (Fig. 1B). Direct analysis of the ligation reactions was performed at various time points by using MALDI-MS without desalting. As an example, a simultaneous readout of the state of ligation to CRANK for each of the LYRAX peptides featuring C-terminal A, V, I, M, and F is shown as a function of time (Fig. 1C).
To interpret the MALDI spectra, the peak areas for each starting MPAL–thioester peptide, the intermediate benzylmercaptan–thioester exchange peptide, and the LYRAX/CRANK ligation product were calculated for every amino acid over a series of time points. These values were added to define 100% for each time point. The fractions of the starting, intermediate, and product compounds are shown as a function of time (Fig. 1D).
Although MALDI is not strictly quantitative, we have found it to be a useful tool for monitoring relative changes in similar peptides (19). To validate the MALDI results, a separate LYRAH(Dnp)–MPAL-to-CRANK ligation was performed and monitored by using HPLC (C18: 9–45% acetonitrile 1.8%/min, 0.1% TFA). The percentage of ligation was determined by using HPLC (50% ligation product after 1 hr; >80% ligation product after 4 hr) and was found to be similar to that obtained by MALDI analysis (Fig. 1D, Left).
The individual ligation reactions were divided into four groups based on time to attain 100% product formation (i.e., no starting or intermediate compounds) within 4, 9, 24, or 48 hr and longer (Fig. 1D). Interestingly, several amino acids undergo ligation at a similar rate to glycine and faster than alanine, indicating that the side chain can facilitate the ligation reaction. In particular, cysteine and histidine react at the same rate as glycine, which is the least hindered amino acid. It is possible that the thiol–cysteine side chain and the imidazole–histidine side chain participate in catalysis of the rate-limiting transthioesterification step of the ligation reaction. From these data, it can be observed that all 20 amino acids can be applied to this X–Cys ligation strategy, with Val, Ile, and Pro representing less favorable choices because of slow ligation rates.
The MALDI analysis provided several insights into the synthesis and ligation of thioester peptides. Peptides containing C-terminal proline residues are known to undergo diketopiperazine formation during chain assembly on traditional Pam resins (20). The C-terminal thioester linker would be expected to be highly susceptible to this side reaction during synthesis of the peptide LYRAP–MPAL. As expected, ≈20% of the peptide contained a 2-aa C-terminal deletion to produce LYR–MPAL. Significantly, none of the other pools of peptides contained this deletion, indicating that only C-terminal proline peptides undergo this cyclization.
The ability of thiol additives to accelerate native ligation reactions has been reported (6). The MALDI data provides a detailed look into the thioester species present during the ligation reaction. Interestingly, despite the addition of both thiophenol and benzylmercaptan, thiophenol–thioester peptides are not observed. In addition, ligation products are only observed after formation of benzyl–thioester peptides. For example, thiol exchange for LYRAV and LYRAI (Fig. 1C Bottom, v and i) as well as the corresponding ligated materials are absent after 1 hr of ligation reaction (Fig. 1C). These observations indicate that the initial MPAL–thioester peptide is not the major reactive species in the formation of the thioester intermediate of the ligation under the experimental conditions.
Activated peptides can be prone to racemization via an oxazolone rearrangement (21, 22). To investigate possible racemization of the histidine residue at the His–Cys ligation site, an individual model LYRAH–MPAL-to-CRANK ligation under the same reaction conditions was performed. The ligation product was compared with individually synthesized LYRA–lH–CRANK and LYRA–dH–CRANK by using HPLC analysis. Under our ligation conditions, the major ligation product eluted at the LYRA–lH–CRANK retention time and <2% of LYRA–dH–CRANK ligation product was observed. This observation is consistent with the absence (<1%) of racemization at Leu during a comparatively slow Leu–Cys ligation under comparable reaction conditions (13). The ligation conditions used for these model studies used 3 mM peptide concentrations, which are similar to those used for larger peptide ligations (1–5 mM).
To demonstrate this straightforward approach for protein synthesis, hsPLA2 was chosen as a model. To dissect the lipolytic activity (for a review, see ref. 23) from the anticoagulant properties of hsPLA2 (7, 24, 25, 26), a chemical analog in which the active-site residue His-47 was replaced with the isosteric, but hydrogen bonding-deficient amino acid Bta was designed.
The 124-aa polypeptide chain of hsPLA2 was assembled from four ≈30-aa peptides that could easily be assembled by manual SPPS, (Fig. 2A). The ligation sites, His-27–Cys-28, Gly-58–Cys-59, and Leu-87–Cys-88 took advantage of the knowledge gained from the model studies. Although the ligation site of Gly-25–Cys-26 could have been used, the His-27–Cys-28 site provided a stringent test for the compatibility of functional side chains with the native chemical ligation approach for protein synthesis. Peptides 1, 2, and 3 were synthesized on TAMPAL resin to yield C-terminal MPAL–thioesters, and peptide 4 was synthesized on Cys-phenylacetamidomethyl resin to yield a C-terminal carboxylic acid group after deprotection and cleavage of the peptide resin (Fig. 2A). Peptides 2 and 3 were N-terminally protected with a Msc group to prevent polymerization or cyclization of the respective peptides under ligation conditions. After deprotection and cleavage from the resin, the polypeptides were HPLC-purified, lyophilized, and stored at −20°C until use.
The individual ligation reactions for the assembly of hsPLA2 provided a test for our model studies. The ligation reaction of Msc—59–87—MPAL to 88–124 (Leu–Cys ligation site) needed 24 hr at 37°C to produce 90% ligation. This rate was as expected from the Leu–Cys ligation rate with the model peptide studies (Fig. 1). After removal of the N-terminal Msc group, the purified polypeptide 59–124 was separately ligated to two different Msc—28–58—MPAL polypeptides, one with His-47 and one with Bta-47. These ligation reactions involved a Gly–Cys ligation site, one of the most favorable according to the model peptide study (Fig. 1). Indeed, both ligation reactions were complete within 6 hr, after which removal of N-terminal Msc and HPLC yielded purified polypeptides 28–124 and 28–124—Bta-47, ready to be used in the final ligation reaction. The 1–27–MPAL to 28–124 ligations involved His(Dnp)–Cys ligation sites and were completed in 9 hr, again as expected from the model peptide study.
After HPLC purification, polypeptides 1–124 and 1–124—Bta-47 were refolded as described (26) to give hsPLA2 and the active-site variant hsPLA2–Bta-47 (Fig. 2C). The masses of the reduced and refolded phospholipases agreed well with their calculated average isotopic mass and are summarized in Table 1. For hsPLA2–His-47, the calculated molecular masses based on average isotope composition are 13,919.0 Da (reduced) and 13,905.0 Da (oxidized). For hsPLA2–Bta-47, the calculated molecular masses based on average isotope composition are 13,935.0 Da (reduced) and 13,921.0 Da (oxidized). The recombinant hsPLA2 contained Leu-8 instead of Met-8, resulting in a calculated mass decrease of 18 Da, but was shown to retain full enzymatic activity (27).
Table 1.
Molecular masses and enzyme kinetics of hsPLA2
| hsPLA2 type | Ligation | M
|
Km, mM | kcat, sec−1 | |
|---|---|---|---|---|---|
| Oxidized | Reduced | ||||
| Chemical | Two-piece | 13919.5 ± 1.6* | 13904.7 ± 0.8* | 1.0 ± 0.2* | 3.5 ± 0.3* |
| Recombinant | (Met-8 → Leu-8) | 13885.2 ± 1.0 | 0.9 ± 0.2* | 3.1 ± 0.2* | |
| Chemical | Two-piece | 0.9 ± 0.1 | 2.4 ± 0.1 | ||
| Chemical | Four-piece | 13919.5 ± 2.2 | 13904.8 ± 0.6 | 1.0 ± 0.1 | 2.5 ± 0.1 |
| Chemical (His-47 → Bta-47) | Four-piece | 13935.2 ± 2.3 | 13920.6 ± 1.6 | ND | ND |
Molecular masses were determined with electrospray ionization-ms. 1,2-Bis(heptanoylthio)-phosphatidylcholine hydrolysis was used to study the kinetic parameters of sPLA2. The kinetic data represent the average ±SD of three experiments. ND, not detectable (<0.1% of native hsPLA2).
Data from ref. 7.
When compared with recombinant hsPLA2 and synthetic hsPLA2 obtained from a two-segment ligation (7), the current four-segment hsPLA2 ligation product and refolded enzyme had the correct mass and activity and was enzymatically indistinguishable from the recombinant enzyme (Table 1). The striking shift of maximal intensity from the 16th and 17th charged state in the reduced polypeptide chain to the 7th charged state in the refolded hsPLA2–Bta-47 (Fig. 2C) is characteristic behavior of folded globular proteins compared with their denatured counterparts (28, 29).
One of the major advantages of chemical protein synthesis is the ability to introduce unnatural amino acids into the polypeptide chain. Bta is conformationally similar to histidine, but the τ-nitrogen is replaced with a methylene and the π-nitrogen with sulfur (Fig. 2). Interestingly, when this thienyl side chain was substituted for the His-47 imidazole side chain in hsPLA2, an inactive variant was obtained (Table 1). The enzymatic activity of hsPLA2–Bta-47 was below the detection limit of the assay used, i.e., at least 1,000-fold lower than hsPLA2–His-47.
These studies demonstrate that the thioester peptide producing TAMPAL resin is a useful tool for obtaining activated thioester peptides with all natural C-terminal amino acids. The results from the parallel approach for characterizing the compatibility of native ligation with all 20 amino acids in X–Cys native chemical ligation were consistent with the individual ligations in the synthesis of hsPLA2 and an enzymatically inactive hsPLA2 variant. These experiments suggest that peptides with at least 17 of the 20 C-terminal amino acids are compatible with protein synthesis by native ligation.
Although in principle, proteins of any length are attainable by using this methodology, 200 aa appears to be the practical size limit. Analysis of the ORFs encoded in the yeast genome (Saccharomyces cerevisiae) (35) indicates that 22% of these sequences are between 40 and 200 aa (≈1200 proteins). Of these proteins, 50% (600 proteins) have a cysteine residue spaced <60 amino acids apart throughout the sequence (S. B. H. Kent, personal communication). Because in this study, 85% (17/20) of X–Cys ligation sites have been shown to be compatible with native ligation, ≈40% of yeast proteins between 40 and 200 aa can be synthesized without modification. It is anticipated that nearly all of these proteins could be synthesized with one or two conservative changes to the amino acid sequence. More importantly, most protein domains are well within this size limit.
These results are of particular relevance to recently described approaches for protein semisynthesis. It has been demonstrated that biologically derived peptides can be generated with an N-terminal cysteine residue by using a unique factor X cleavage site (30). More elegantly, “expressed-protein ligation” enables C-terminal thioester peptides to be generated from biological sources by using modified inteine fusion proteins (31, 32). Our ligation studies imply that a variety of amino acids may be compatible with the protein-splicing mechanism (33). In addition to these approaches, we have recently introduced a conformationally assisted protein-ligation strategy that allows some proteins to be assembled without an N-terminal cysteine residue (34). Complementary use of our TAMPAL-resin strategy with these semisynthetic techniques will enable synthetic access to most proteins of interest.
The use of native chemical ligation has already enabled the synthesis of a variety of proteins of moderate size. The introduction of a straightforward and expanded methodology to generate the necessary α-thioester peptides should allow this approach to be useful to a wider spectrum of laboratories. The synthesis of proteins by chemical ligation should greatly enhance the application of organic chemistry to the world of proteins.
Acknowledgments
This work was supported in part by National Institutes of Health Grants HL 21544 and HL 31950 (J.H.G.) and the Skaggs Institute for Chemical Biology (P.E.D.).
ABBREVIATIONS
- Boc
tert-butoxycarbonyl
- COSR
C-terminal thioester, DIEA, N,N,-diisopropylethylamine
- DMF
N,N,-dimethylformamide
- Dnp
2,4-dinitrophenyl
- HBTU
2-(1H-benzotriazol-1-yl)-1,1,3,3-tetramethyluronium hexafluorophosphate
- hsPLA2
human secretory phospholipase A2
- MALDI
matrix-assisted laser-desorption ionization
- TAMPAL
trityl-associated mercaptopropionic acid leucine
- TFA
trifluoroacetic acid
- Bta
β-thienylalanine
- MBHA
p-methylbenzhydrylamine
- MPAL
mercaptoproprionic acid-leucine
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1.Merrifield R B. J Am Chem Soc. 1963;85:2149–2154. [Google Scholar]
- 2.Kent S B H. Annu Rev Biochem. 1988;57:957–989. doi: 10.1146/annurev.bi.57.070188.004521. [DOI] [PubMed] [Google Scholar]
- 3.Noren C J, Anthony-Cahill S J, Griffith M C, Schultz P G. Science. 1989;244:182–188. doi: 10.1126/science.2649980. [DOI] [PubMed] [Google Scholar]
- 4.Dawson P E, Muir T W, Clark-Lewis I, Kent S B H. Science. 1994;266:776–779. doi: 10.1126/science.7973629. [DOI] [PubMed] [Google Scholar]
- 5.Tam P, Lu Y-A, Liu C-F, Shao J. Proc Natl Acad Sci USA. 1995;92:12485–12489. doi: 10.1073/pnas.92.26.12485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dawson P E, Churchill M J, Ghadiri M R, Kent S B H. J Am Chem Soc. 1997;119:4325–4329. [Google Scholar]
- 7.Hackeng T M, Mounier C M, Bon C, Dawson P E, Griffin J H, Kent S B H. Proc Natl Acad Sci USA. 1997;94:7845–7850. doi: 10.1073/pnas.94.15.7845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Muir T W, Dawson P E, Kent S B H. Methods Enzymol. 1997;289:266–298. doi: 10.1016/s0076-6879(97)89052-0. [DOI] [PubMed] [Google Scholar]
- 9.Wieland T, Bokelmann E, Bauer L, Lang H U, Lau H. Liebigs Ann Chem. 1953;583:129–149. [Google Scholar]
- 10.Schnölzer M, Alewood P, Jones A, Alewood D, Kent S B H. Int J Peptide Protein Res. 1992;40:180–193. doi: 10.1111/j.1399-3011.1992.tb00291.x. [DOI] [PubMed] [Google Scholar]
- 11.Blake J. Int J Peptide Protein Res. 1981;17:273. doi: 10.1111/j.1399-3011.1981.tb01992.x. [DOI] [PubMed] [Google Scholar]
- 12.Canne L E, Walker S M, Kent S B H. Tetrahedron Lett. 1995;36:1217–1220. [Google Scholar]
- 13.Lu W, Quasim M A, Kent S B H. J Am Chem Soc. 1996;118:8518–8523. [Google Scholar]
- 14.Hojo H, Kwon Y, Kakuta Y, Tsuda S, Tanaka I, Hikichi K, Aimoto S. Bull Chem Soc Jpn. 1993;66:2700–2706. [Google Scholar]
- 15.Camarero J A, Cotton G J, Adeva A, Muir T W. J Peptide Res. 1998;51:303–316. doi: 10.1111/j.1399-3011.1998.tb00428.x. [DOI] [PubMed] [Google Scholar]
- 16.Sarin V K, Kent S B H, Tam J P, Merrifield R B. Anal Biochem. 1981;117:147–157. doi: 10.1016/0003-2697(81)90704-1. [DOI] [PubMed] [Google Scholar]
- 17.Hendrickson H S, Hendrickson E K, Dybvig R H. J Lipid Res. 1982;24:1532–1537. [PubMed] [Google Scholar]
- 18.Reynolds L R, Hughes L L, Dennis E A. Anal Biochem. 1992;204:190–197. doi: 10.1016/0003-2697(92)90160-9. [DOI] [PubMed] [Google Scholar]
- 19.Dawson P E, Fitzgerald M C, Muir T W, Kent S B H. J Am Chem Soc. 1997;119:7917–7927. [Google Scholar]
- 20.Goodman M, Steuben K C. J Am Chem Soc. 1962;84:1279. [Google Scholar]
- 21.Bergmann M, Zervass L. Biochem Z. 1928;203:280. [Google Scholar]
- 22.Jones J. The Chemical Synthesis of Peptides. Oxford, U.K.: Clarendon; 1991. pp. 62–64. [Google Scholar]
- 23.Dennis E A. J Biol Chem. 1994;269:13057–13060. [PubMed] [Google Scholar]
- 24.Kini R M, Evans H J. J Biol Chem. 1987;262:14402–14407. [PubMed] [Google Scholar]
- 25.Inada M, Crowl R M, Bekkers A C A P A, Verheij H M, Weiss J. J Biol Chem. 1994;269:26338–26343. [PubMed] [Google Scholar]
- 26.Mounier C M, Franken P A, Verheij H M, Bon C. Eur J Biochem. 1996;237:778–785. doi: 10.1111/j.1432-1033.1996.0778p.x. [DOI] [PubMed] [Google Scholar]
- 27.Franken P A, van den Berg L, Huang J, Gunyuzlu P, Lugtigheid R B, Verheij H M, De Haas G H. Eur J Biochem. 1992;203:89–98. doi: 10.1111/j.1432-1033.1992.tb19832.x. [DOI] [PubMed] [Google Scholar]
- 28.Chowdhury S K, Katta V, Chait B T. J Am Chem Soc. 1990;112:9012–9013. [Google Scholar]
- 29.Loo J A, Edmonds C G, Udseth H R, Smith R D. Anal Chem. 1990;62:693–698. doi: 10.1021/ac00206a009. [DOI] [PubMed] [Google Scholar]
- 30.Erlandson D A, Chytil M, Verdine G L. Chem Biol. 1996;3:981–991. doi: 10.1016/s1074-5521(96)90165-9. [DOI] [PubMed] [Google Scholar]
- 31.Muir T W, Sondhi D, Cole P A. Proc Natl Acad Sci USA. 1998;95:6705–6710. doi: 10.1073/pnas.95.12.6705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Evans T C, Benner J, Xu M-Q. Protein Sci. 1998;7:2256–2264. doi: 10.1002/pro.5560071103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xu M-Z, Perler F B. EMBO J. 1996;15:5146–5153. [PMC free article] [PubMed] [Google Scholar]
- 34.Beligere G S, Dawson P E. J Am Chem Soc. 1999;121:6332–6333. [Google Scholar]
- 35.Goffeau A, Barrell B G, Bussey H, Davis R W, Dujon B, Feldmann H, Galiber F, Hoheisel J D, Jacq C, Johnston J, et al. Science. 1996;274:546–567. doi: 10.1126/science.274.5287.546. [DOI] [PubMed] [Google Scholar]