Abstract
We previously have shown that adenovirus type 5 E4orf4 protein associates with protein phosphatase 2A (PP2A) and induces apoptosis in transformed cells in a p53-independent manner. Here we show that the interaction between E4orf4 and PP2A is required for induction of apoptosis by the viral protein. This conclusion is supported by a mutation analysis of E4orf4 protein, showing a correlation between the ability to bind PP2A and to induce apoptosis, and by the observation that transfection of an antisense construct of the PP2A-B55 subunit reduces expression of the PP2A-B55 subunit and inhibits induction of apoptosis by E4orf4, but not by p53. The mutant analysis also indicates that even a low level of interaction with PP2A is sufficient to initiate the E4orf4 apoptotic pathway. In addition, E4orf4 inhibits cellular transformation by various oncogenes, and this function is coupled to its ability to induce apoptosis. Furthermore, expression of oncogenes in primary cell cultures sensitizes these cells to induction of apoptosis by E4orf4. Our results suggest that E4orf4 is a potentially useful tool for cancer gene therapy.
The adenovirus type 5 E4orf4 protein has been shown to affect several cellular processes, including down-regulation of virally induced signal transduction, regulation of gene expression, and induction of apoptosis.
Previous work has shown that the adenovirus E1A proteins and cAMP cooperate to induce the accumulation of activator protein 1 (AP-1) transcription factor by activating transcription of the cellular c-fos and junB genes that encode the AP-1 components. The induced AP-1 activates transcription of early adenovirus genes through AP-1 and activating transcription factor sites in adenoviral promoters (1). The induction of AP-1 by E1A plus cAMP is counterbalanced by the 14-kDa adenovirus E4orf4 protein whose levels rise upon stimulation by E1A and cAMP (2). E4orf4 reduces AP-1 levels at both the transcription and the translation levels (2). E4orf4 activities also result in hypophosphorylation of E1A and c-fos proteins (2), and as a consequence of the cumulative E4orf4 effects, its own promoter is down-regulated as well (3). In addition to its effect on transcription and translation, E4orf4 has been reported to affect differential splicing of adenovirus late mRNA (4).
Recently, we and others have shown that E4orf4 protein induces p53-independent apoptosis in several transformed cell lines (5–7). As a result, the ability of transformed cells to form colonies was inhibited in the presence of E4orf4 (5). It has been reported that induction of apoptosis in Chinese hamster ovary cells by E4orf4 does not involve activation of zVAD-fmk-sensitive caspases (6).
When investigating the mechanisms underlying E4orf4 action, we have found that the E4orf4 protein binds several cellular proteins, one of which is protein phosphatase 2A (PP2A) (8). We and others have further shown that the E4orf4-PP2A interaction plays a role in down-regulation of stimulated transcription (3, 8) and of alternative splicing (4). Furthermore, a mutant E4orf4 protein that lost the ability to induce apoptosis also has lost its ability to bind PP2A (5). Thus, the interaction between E4orf4 and PP2A may be important for the induction of apoptosis by E4orf4.
PP2A is a serine/threonine phosphatase that plays a role in several cellular processes, including cell division, signal transduction, gene expression, and development (9). PP2A consists of three subunits. Two of them, the 36-kDa catalytic C subunit and the 63-kDa regulatory A subunit, form the core enzyme, and the B subunit binds the core enzyme to form the holoenzyme. The A and C subunits both exist as two isoforms (α and β), which are closely related, whereas the B subunit is variable, and its multiple isoforms belong to three gene families, B/B55/PR55, B′/B56/PR61, and B′′/PR72/PR130 (10–12). The B subunit also is replaceable by viral proteins, such as the simian virus 40 small t antigen and the polyomavirus small and middle T antigens (13). Adenovirus E4orf4, however, is found in a complex with the complete holoenzyme and binds directly to the B55 subunit (8).
Reversible phosphorylation plays a role in control of apoptosis, as it does in most other cellular events. Various kinases, which participate in signal transduction pathways, were shown to affect apoptosis (14), and numerous reports demonstrated the involvement of inhibitors of protein phosphatases in induction of cell death (15, 16) or in inhibition of apoptosis induced by multiple signals (17, 18). Two recent reports link PP2A more directly to the process of apoptosis. The PP2A-A subunit was reported to interact with caspase-3, one of the caspase family involved in the execution of programmed cell death. PP2A-A was cleaved by caspase-3, concurrent with increased PP2A activity toward a peptide substrate (19). PP2A also was shown to interact with Bcl-2 and affect its phosphorylation in myeloid cells after addition of survival factors (20).
We set out to determine whether the interaction of adenovirus E4orf4 protein with PP2A is required for induction of apoptosis by E4orf4. Several mutations were introduced into the E4orf4 gene, and the ability of the mutants to bind PP2A and induce apoptosis was assayed. The ability of an antisense PP2A-B construct to prevent induction of apoptosis also was tested. Because transformed cells are susceptible to E4orf4-induced apoptosis, cellular transformation may be prevented in the presence of E4orf4. Hence, we assayed the ability of wt and mutant E4orf4 proteins to prevent cellular transformation by different combinations of oncogenes. We report here that an interaction with PP2A is required, although may not be sufficient, for induction of apoptosis and inhibition of oncogenic transformation by E4orf4. Furthermore, oncogene expression sensitizes cells to killing by the E4orf4 protein.
MATERIALS AND METHODS
Plasmids and Cells. The following plasmids have been described: pCMV-E4orf4 or pCMV-A3 (5), the pCMV/neo vector and pCMV-p53 (21), pBabe-puro (22), the plasmid used for in vitro translation of PP2A-B subunit (7–1, ref. 23), BSxcThy1.2 (24), pAd5 XhoI-C (pXC15) containing the left end (1–15.5 map units) of the adenovirus type 5 genome and expressing the E1A and E1B proteins (25), pCMV-E1A (26), and pEJ6.6 encoding mutant Ha-Ras (27). The plasmid pGEM-7Zf(+) is from Promega. The plasmid expressing hemagglutinin (HA)-tagged PP2A-B was constructed by fusing 3× HA tags to the carboxyl terminus of PP2A-B55 in pBabe-puro. The PP2A-B55 subunit was subcloned from plasmid 7–1 into the pCMV/neo vector in both orientations to create pCMV-PP2A-B55 and pCMV-PP2A-B55-AS.
Human 293T cells were derived by introducing the simian virus 40 T antigen into 293 cells. 293 cells are human embryonic kidney cells that express Ad5 E1A and E1B proteins (28). 293T cells, the human non-small-cell lung carcinoma H1299 cells (29), and primary cells were cultured in DMEM supplemented with 10% FCS.
Mutagenesis.
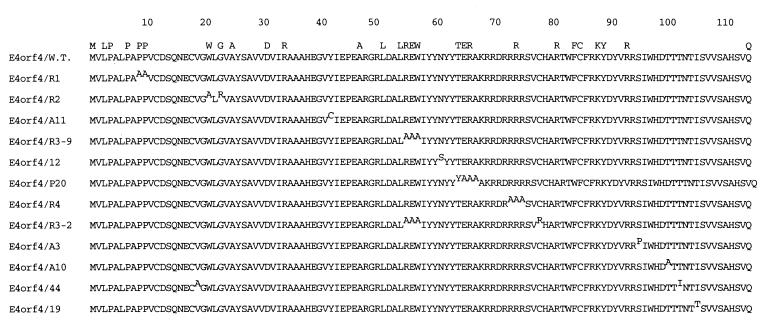
The E4orf4 mutant gene constructs were obtained by two approaches. Mutations 12, 44, 19, A3, A10, and A11 were generated by random PCR mutagenesis (30, 31), a method based on the ability of MN2+ to reduce the fidelity of Taq DNA polymerase. The template DNA plasmid, pGEM7Zf(+)-E4orf4 and the 5′ and 3′ oligonucleotide primers have been described (8). The PCR products were cloned into pGEM-7Zf(+) (Promega) for sequencing. The mutant E4orf4 genes subsequently were subcloned into the expression plasmid pCMV-Neo-Bam (21), in which they are expressed from the immediate early cytomegalovirus (CMV) promoter. Mutants R1, R2, R3–2, R3–9, R4, and P20 were obtained by site-directed PCR mutagenesis. Several amino acids, dispersed throughout the protein, are conserved in the various serotypes of adenovirus. Some of these residues were chosen as sites for targeted mutagenesis, to obtain mutations spanning the entire protein. Most of the amino acids targeted for mutagenesis were substituted to alanines. The resulting PCR products were cloned as described above. In some of the PCR products additional mutations were introduced by the PCR process. A summary of the mutations is shown in Fig. 1.
Figure 1.
Amino acid sequences of E4orf4 mutant proteins. The mutations, in superscript, are arranged from amino to carboxyl terminus. Amino acids that are conserved in several adenovirus serotypes are noted at the top.
Transfections, Immunoprecipitations, and Western Blot Analysis.
Cells were plated in 60-mm culture dishes, and transfections were carried out either by the standard method of calcium phosphate precipitation of DNA (32) for 293T cells or with Lipofectamine plus reagent (GIBCO/BRL) for all other cells. Cell extracts were prepared in lysis buffer containing 50 mM Tris⋅HCl (pH 7.4), 250 mM NaCl, 5 mM EDTA, 0.1% Triton X-100, 0.5% NP-40, 2 μg/ml leupeptin, 2 μg/ml aprotinin, and 0.5 mM PMSF. The levels of E4orf4 mutant proteins were analyzed by Western blot analysis, using a rabbit polyclonal antibody raised against E4orf4 (5). For immunoprecipitations, the antibody was covalently bound to the protein A Sepharose (Amersham Pharmacia) by dimethyl pymelimidate, as described (33). After immunoprecipitation with the anti-E4orf4 beads, the immune complexes were separated on SDS/10% or 15% polyacrylamide gels, and Western blots were stained with antibodies to the PP2A C or B55 subunits (10% gels) (5) or to E4orf4 (15% gels). Immune complexes were detected by chemiluminescence.
Phosphatase Assays.
Wild type (wt) and mutant E4orf4 proteins were immunoprecipitated from extracts of transfected cells. The immune complexes were washed twice with lysis buffer and once with PP2A buffer (50 mM imidazole, pH 7.2/0.2 mM EGTA/0.02% 2-mercaptoethanol/0.1% BSA). The immune complexes were incubated with 50 nmol peptide substrate RRA(pT)VA (34) in PP2A buffer for 60 min at 30°C. The amount of free phosphate released was measured by a color reaction generated in the presence of molybdate and malachite green (35). Okadaic acid was added to some reactions at 4 nM concentration.
In Vitro Translation and Binding to Glutathione S-Transferase Fusion Proteins.
Flow Cytometry.
Cells were cotransfected with E4orf4 mutant constructs and a plasmid expressing the Thy1.2 surface marker (24) into H1299 cells. At the indicated times, cells were harvested and stained with a fluorescein-conjugated anti-Thy1.2 antibody (PharMingen) for 30 min on ice. The cells then were washed once with PBS containing 1% calf serum and fixed with methanol for at least 30 min at −20°C. The cells subsequently were washed in PBS, resuspended in PBS containing 50 μg/ml RNase A, and incubated 30 min at 37°C. To stain the DNA, propidium iodide (Sigma) was added to a final concentration of 25 μg/ml. The DNA contents of the whole-cell population and the transfected cells were analyzed in a cell sorter (FACSCalibur, Becton Dickinson).
Fluorescence Microscopy.
Cells were subjected to fluorescence microscopy as described (5). In each experiment, 100 transfected nuclei were counted.
Cellular Transformation Assays.
Baby rat kidney cells were prepared and transfected as described (36), and the number of foci was scored 3 weeks later.
RESULTS
Generation of a Collection of E4orf4 Mutants. E4orf4 induces p53-independent apoptosis in several transformed cell lines (5–7). In Chinese hamster ovary cells, the E4orf4 apoptotic pathway was reported to be insensitive to the broad-range caspase inhibitor zVAD-fmk (6). Thus, it is of great interest to uncover the mechanisms underlying E4orf4-induced apoptosis. Because immunoprecipitation of E4orf4 from adenovirus-infected cells revealed that PP2A is a major component of the protein complex containing E4orf4 (8), we asked whether the interaction with PP2A is required for induction of apoptosis by the viral protein. Several mutant E4orf4 genes were constructed, by using various approaches (see Materials and Methods). One group of constructs contained missense mutations (Fig. 1), whereas the other group contained N- or C-terminal deletions (data not shown). The missense mutations included substitutions of 21 aa, distributed throughout the E4orf4 polypeptide. Of the 28 conserved aa in E4orf4, 11 aa were altered.
Before performing any functional assays with the mutants, we tested their ability to be expressed in cells. 293T cells were transfected with plasmids expressing the E4orf4 proteins from the immediate early CMV promoter, and E4orf4 protein expression was detected by immunoblot analysis. We used a polyclonal antibody, generated against the entire E4orf4 polypeptide (5), to facilitate detection of all mutants. Mutants containing the 12 missense mutations shown in Fig. 1 all were expressed to comparable levels (data not shown and Fig. 2A). However, none of the deletion mutants were expressed to detectable levels (data not shown), and they were not investigated further.
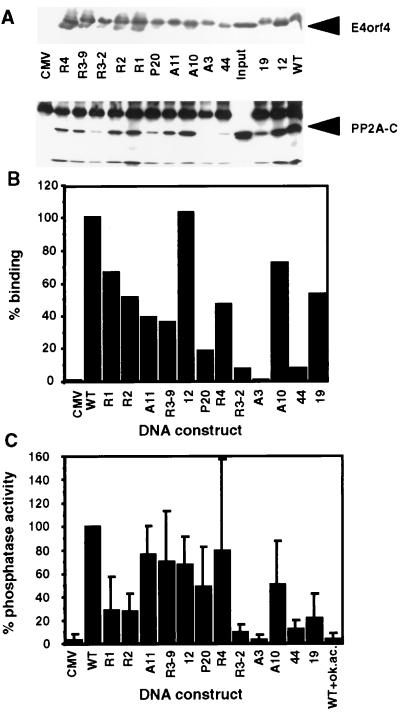
Figure 2.
Binding of PP2A to E4orf4 mutants. (A) Mutant E4orf4 proteins were immunoprecipitated from 293T cells transfected with the mutant DNA constructs or the empty CMV vector (CMV). The immune complexes were detected on a Western blot by antibodies specific for E4orf4 and the PP2A-C subunit. (B) The intensity of PP2A binding was determined by densitometry of the Western blot shown in A. The binding of PP2A to the wt E4orf4 protein was defined as 100%. Mutants are arranged according to the location of the mutations, from the amino to the carboxyl terminus. (C) E4orf4 mutant proteins were immunoprecipitated, and phosphatase activities associated with the immune complexes were determined. Phosphatase activity associated with the wt E4orf4 protein was defined as 100%. The average of four experiments is shown. Background levels were determined either in the absence of E4orf4 (CMV) or in a phosphatase reaction carried out in the presence of 4 nM okadaic acid (WT+ok. ac.).
Analysis of the Ability of the Mutant E4orf4 Proteins to Interact with PP2A. The mutated E4orf4 cDNAs were transfected into 293T cells, and the mutant E4orf4 proteins were immunoprecipitated from equal amounts of transfected cell extracts, by using the polyclonal antibody that recognizes all mutant proteins. The presence of E4orf4 mutants and PP2A in the immune complexes was analyzed by Western blots. The results shown in Fig. 2A and B indicate that although similar amounts of the E4orf4 mutant proteins were immunoprecipitated, different amounts of PP2A were present in the immune complexes. Interestingly, most mutations impaired PP2A binding to some extent. However, some mutations had a more profound effect than others. Mutant R3–2, with substitutions at amino acids 55–57 and 78, bound PP2A at 7% wt efficiency, whereas PP2A binding to mutant R3–9, with identical substitutions at amino acids 55–57, was reduced to 36% wt binding levels. Thus position 78 seems to be important to PP2A binding. The mutation at amino acid 95 in mutant A3 abolished PP2A binding completely. Mutant 44 with substitutions at amino acids 19 and 102, bound PP2A at 8% wt levels, whereas other mutations at the amino terminus of E4orf4 (R1: at amino acids 9–10; R2: at amino acids 21–23) reduced binding to 51–67% wt levels. Mutant P20 (amino acids 64–66) reduced PP2A binding to 18% wt levels. All other mutants retained intermediate (36%: mutant R3–9) to high (103%: mutant 12) levels of interaction with PP2A. Based on these results, it appears that an important determinant of the PP2A binding site is contained between amino acids 78 and 102, and amino acid 95 is crucial to the E4orf4–PP2A interaction.
We previously have reported that upon immunoprecipitation of E4orf4, the E4orf4–PP2A complex exhibits phosphatase activity (8). To further test the ability of the mutants to retain the activities of the wt protein, we assayed the levels of phosphatase activity associated with the mutant E4orf4 immune complexes, by using a PP2A-specific phosphopeptide substrate. Background levels of phosphatase activity were determined by using an immune complex from a cell extract lacking E4orf4, or by addition of okadaic acid, a PP2A inhibitor, to a phosphatase reaction containing immunoprecipitated wt E4orf4. Fig. 2 shows that mutants that bound PP2A at 13- to 14-fold reduced levels (Fig. 2B, lanes R3–2 and 44), were associated with 10-fold reduced levels of phosphatase activity (Fig. 2C). Mutant A3, which didn’t interact with PP2A, did not associate with phosphatase activity above background levels. Other mutants, which bound PP2A at intermediate levels, were associated with similar phosphatase activities, except mutants R1, R2, and 19, which associated with lower phosphatase levels of activity (21–28% wt levels) than would have been expected by their physical interaction with PP2A (51–67% wt levels) (Figs. 2 and 3). Thus, these results support the conclusion that the E4orf4 domain lying between amino acids 78 and 102 is required for the E4orf4–PP2A interaction.
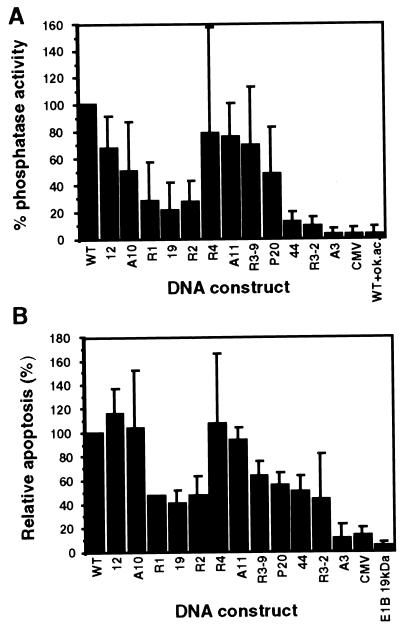
Figure 3.
Induction of apoptosis by E4orf4 mutant proteins. (A) The results from Fig. 2C are shown again, with the mutants arranged by descending intensity of their physical interaction with PP2A. (B) H1299 cells were cotransfected with the cell surface marker (Thy1.2) and the various E4orf4 mutant constructs. Apoptosis was measured by determining the levels of sub-G1 cells appearing in the transfected cell population, stained with the fluorescent Thy1.2-specific antibodies. Background levels were determined either in the absence of E4orf4 (CMV) or in cells transfected with the antiapoptotic adenovirus E1B–19-kDa protein. Apoptosis induced by wt E4orf4 (in 25–35% of the cells, 48 hr posttransfection) was defined as 100%. The average of four experiments is shown.
Induction of Apoptosis by the E4orf4 Mutants.
We have shown that the wt E4orf4 protein induced apoptosis in H1299 cells (5), which lack wt p53 expression. These cells were used further to assay for induction of apoptosis by the mutant E4orf4 proteins, by using flow cytometric analysis. The cells were cotransfected by the Thy1.2 cell surface marker (37) and by the various E4orf4 mutant constructs or an empty vector. After transfection, the cells were stained with a fluorescent antibody that recognizes the surface marker, and with propidium iodide (PI), which stains DNA. Transfected cells (10–30% of the population) were identified by their high fluorescence intensity and were analyzed separately for their DNA content, detected by PI staining. Equal numbers of transfected cells were analyzed for each mutant. Apoptotic cells contain degraded DNA, some of which is lost during the experimental procedure. Thus, apoptotic cells contain less than 2 N DNA content and appear as a “sub-G1” cell population in the fluorescence-activated cell sorter analysis (38). Induction of apoptosis by the various E4orf4 mutants was analyzed by quantitating the sub-G1 fraction in the transfected cell population, and the results are shown in Fig. 3B. Background levels of apoptosis induction were determined by the fraction of sub-G1 cells induced by the empty CMV vector or by the antiapoptotic E1B 19-kDa protein (39). To facilitate easy comparison between the ability of each mutant to bind PP2A and to induce apoptosis, the mutants are arranged in Fig. 3 by descending order of the strength of their physical interaction with PP2A, as predicted by coimmunoprecipitation assays. Fig. 3A shows phosphatase activity levels associated with the mutants, and Fig. 3B demonstrates their ability to induce apoptosis. For most mutants, the abilities to associate with PP2A activity and to induce apoptosis were highly correlated. Mutant A3, which didn’t bind PP2A, did not induce apoptosis above background levels. All other mutants induced from intermediate to high levels of apoptosis. For example, mutants R1, R2, and 19, which were associated with lower levels of phosphatase activity (21–28% wt levels), manifested lower levels of apoptosis (40–50% wt levels). Several mutants that were associated with high levels of PP2A activity induced high levels of apoptosis. Only two mutants, R3–2 and 44, deviated to some extent from the linear correlation between PP2A binding and induction of apoptosis. They bound an active PP2A at very low levels (9–12% wt levels, Fig. 3A), but induced apoptosis to 43–50% of wt levels. These results suggest that an association between E4orf4 and an active PP2A may be required for induction of apoptosis, but even low levels of interaction are sufficient for initiating this pathway.
PP2A Complexes Including the B55 Subunit Are Required for Induction of Apoptosis by E4orf4.
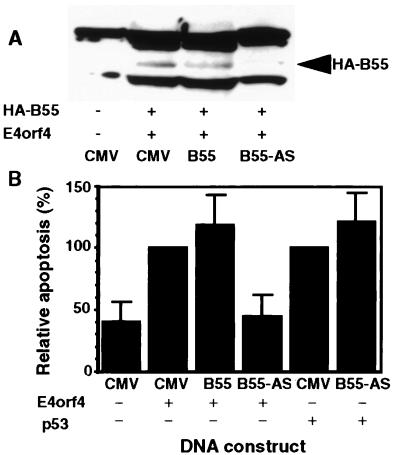
To further examine whether PP2A is required for induction of apoptosis by E4orf4, we measured the effect of an antisense construct of PP2A-B55 on expression of the PP2A-B55 subunit and on induction of apoptosis. The PP2A-B55 subunit was chosen for this assay because it mediates the interaction between E4orf4 and PP2A (8). To measure the effect of the antisense PP2A-B55 construct on expression of PP2A-B55 in the cells, a plasmid encoding HA-PP2A-B55 was cotransfected into H1299 cells with the sense or antisense PP2A-B55 constructs. The level of HA-PP2A-B55 proteins in transfected cells was detected by Western blots. As seen in Fig. 4A, the antisense construct effectively reduced expression of HA-PP2A-B55 in the transfected cells whereas the sense construct did not. Cell killing was measured by counting 4′,6-diamidino-2-phenylindole dihydrochloride-stained apoptotic nuclei in the transfected cell population, identified by the presence of cotransfected green fluorescent protein (5). Fig. 4B demonstrates that the antisense construct inhibited apoptosis induced by E4orf4, whereas the sense construct slightly increased cell killing. The antisense construct did not inhibit p53-induced apoptosis, indicating that PP2A is specifically required for E4orf4-induced apoptosis. These results further support our conclusion that the interaction of E4orf4 with PP2A is required for induction of apoptosis by the viral protein.
Figure 4.
Expression of antisense PP2A-B55 inhibits induction of apoptosis by E4orf4. (A) Plasmids expressing HA-tagged PP2A-B55 and E4orf4 were cotransfected into H1299 cells with the empty CMV vector (CMV), or with a plasmid expressing PP2A-B55 in the sense (B55), or in the antisense (B55-AS) orientation. Expression of HA-PP2A-B55 was detected by a Western blot. (B) H1299 cells were transfected as in A, but the transfection mix contained a plasmid expressing green fluorescent protein and not the HA-tagged PP2A-B55. pCMV-p53 was used in some experiments instead of E4orf4. The number of apoptotic cells was determined 48 hr posttransfection by counting 4′,6-diamidino-2-phenylindole dihydrochloride-stained apoptotic nuclei in the transfected cell population. One hundred transfected cells were counted in each experiment. Apoptosis induced by E4orf4 or p53 was defined as 100%. The average of four experiments is shown.
Oncogene Expression Sensitizes Primary Cells to Killing by E4orf4 Protein.
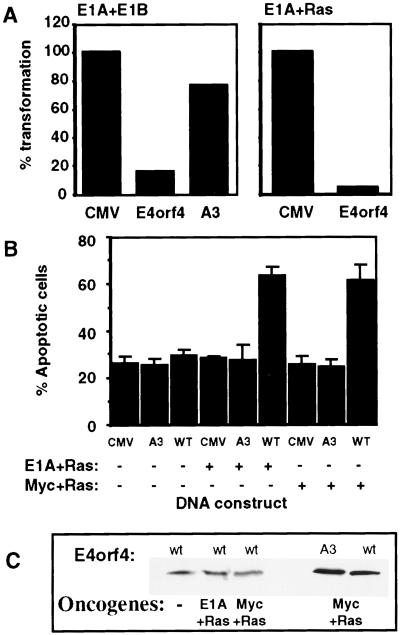
Because E4orf4 induces apoptosis in transformed cells and reduces colony formation by transformed cells (5), we asked whether oncogenic transformation of primary cells by various combinations of oncogenes could be prevented by E4orf4. Baby rat kidney (BRK) cells were cotransfected with the adenovirus oncogenes E1A and E1B or with E1A and activated Ras, together with E4orf4 or mutant A3, and resulting foci of morphologically transformed cells were counted. Fig. 5A shows one of a series of experiments, in which wt E4orf4 reduced the number of foci obtained with the various oncogene combinations by 6.7- to 33-fold, whereas mutant A3 reduced the foci count by 1.3- to 2.2-fold. The other mutants reduced transformation of BRK cells to intermediate levels (data not shown). To test whether the inhibition of oncogenic transformation was the result of a general toxic effect, or whether E4orf4 killed transformed cells specifically, we examined whether E4orf4 kills primary cells in the presence or absence of oncogenes. Primary cell cultures were transfected with E4orf4, with mutant A3, or with the empty CMV vector, either with a combination of oncogenes, or without them. Apoptosis was measured by counting apoptotic nuclei in the transfected cell population. Fig. 5B shows that E4orf4 induced high levels of apoptosis in the presence of E1A plus activated Ras, or Myc plus Ras, compared with its low toxicity in the absence of the oncogenes. Similar results were obtained in the presence of E1A plus E1B (data not shown). Increased E4orf4 activity in the presence of oncogenes did not result from enhanced levels of E4orf4 protein (Fig. 5C) or from enhanced expression of E1A or Myc (results not shown). Mutant A3 did not induce apoptosis even in the presence of oncogenes (Fig. 5B), although it was expressed as well as wt E4orf4 (Fig. 5C). Thus, expression of oncogenes in primary cells sensitized them to killing by E4orf4, leading to E4orf4-induced inhibition of cellular transformation. Furthermore, the abilities to induce apoptosis and to inhibit oncogenic transformation were retained by the same set of E4orf4 mutants and correlated with the ability of the mutant E4orf4 proteins to interact with PP2A.
Figure 5.
Oncogenes sensitize primary cells to E4orf4-induced apoptosis. (A) Baby rat kidney cells were transfected with combinations of oncogenes (E1A plus E1B or E1A plus activated Ras) and with the empty CMV vector or the vector expressing wt E4orf4 or mutant A3. The number of foci obtained in the presence of the empty vector was defined as 100% transformation. (B) The primary cell cultures were transfected as in A, and a green fluorescent protein-expressing plasmid was added to the transfection mix for detection of transfected cells. Apoptosis was measured as in Fig. 4. The average of four experiments is shown. (C) Proteins extracted from transfected cells were chromatographed on 15% SDS gels and the blot was stained with E4orf4-specific antibodies.
DISCUSSION
The results described in this report reveal two important aspects of E4orf4-induced apoptosis. First, the interaction between E4orf4 and PP2A apparently is required for induction of apoptosis by E4orf4, and second, E4orf4 induces apoptosis much more efficiently in oncogenically transformed cells, as compared with untransformed cells, thus leading to a specific elimination of transformed cells.
The first conclusion is supported by two lines of evidence. Initially, a mutation analysis of E4orf4 indicated that a correlation existed between the ability of a mutant E4orf4 protein to bind PP2A and its ability to induce apoptosis. The interaction with PP2A was measured by two types of assays, detection by Western blots of the physical presence of PP2A-C subunit in E4orf4-containing immune complexes and measurement of the level of phosphatase activity associated with the same immune complexes. Both types of assays indicated that a region between amino acids 78 and 102 was important for the binding of PP2A to E4orf4. Mutations in this region (R3–2, A3, and 44) caused a marked reduction in the ability of the protein to interact with PP2A and to be associated with a phosphatase activity measured toward a PP2A-specific phosphopeptide substrate (Fig. 2). In addition, regions at the amino terminus of E4orf4 (mutations R1 and R2 at amino acids 9–10 and 21–23) and at the carboxyl terminus (mutation 19 at amino acid 105) seemed to affect the phosphatase activity of E4orf4-associated PP2A (21–28% wt levels) more than they influenced the physical interaction (51–67% wt levels). Thus, these regions may affect the phosphatase activity once PP2A is bound to E4orf4. For most mutants, a direct correlation was found between the ability to associate with an active PP2A and the ability to induce apoptosis (Fig. 3). A mutant that could not interact with PP2A at all (A3) lost its ability to induce apoptosis, and several mutants, which associated with 3.5- to 5-fold decreased PP2A activity (R1, R2, and 19), retained a reduced ability to induce apoptosis (40–50% wt levels). However, two mutants retaining 8.3- to 11-fold reduced ability to bind an active PP2A (R3–2 and 44) still could induce apoptosis to 43–50% of wt levels. Thus, although the interaction with an active PP2A may be required for induction of apoptosis, it seems that even low levels or short durations of interaction are sufficient to initiate the pathway. The results further suggest the possibility that additional E4orf4-associated factors may cooperate in facilitating E4orf4-induced apoptosis.
The second line of evidence indicating that PP2A is required for E4orf4-induced apoptosis involves experiments with a plasmid expressing PP2A-B55 subunit in an antisense orientation. Introduction of this plasmid into cells led to reduced expression of the endogenous B55 subunit, as represented in the transfected cells by an HA-tagged PP2A-B55 protein (Fig. 4A). Furthermore, including the antisense construct in the transfection mix resulted in a dramatic reduction of E4orf4-induced apoptosis, whereas PP2A-B55 in the sense orientation slightly increased apoptosis (Fig. 4B). Lower levels of the antisense construct reduced E4orf4-induced apoptosis to intermediate levels (results not shown). Thus, in the absence of the PP2A-B55 subunit, E4orf4 cannot induce apoptosis. Expression of the PP2A-B55 antisense construct did not cause a decrease in p53-induced apoptosis, indicating its specific role in the E4orf4 apoptotic pathway. Our results support the conclusion that the interaction with PP2A is required for initiating the E4orf4 apoptotic pathway, although it may not be sufficient. Future studies will reveal the relevant PP2A substrates whose altered phosphorylation triggers this pathway.
Many of the E4orf4 mutant proteins, containing alterations in different regions of the protein, manifested an impaired ability to interact with PP2A and to induce apoptosis, compared with the wt protein. Furthermore, even small deletions introduced into the ends of the E4orf4 sequence resulted in expression of an unstable protein, which could not be detected in cell extracts. These observations suggest that the conformation of the entire protein is important to its stability and function. In addition, fusion of some E4orf4 mutants to a glutathione S-transferase moiety abolished their ability to interact with PP2A, whereas the same native mutant proteins retained this ability (data not shown). These results further support the conclusion that the functional E4orf4 protein is highly sensitive to any changes in its conformation. As a result, caution should be exercised when using epitope-tagged constructs of wt or E4orf4 mutant proteins.
The second major finding described in this manuscript is that E4orf4 kills oncogene-expressing cells much more efficiently than untransformed cells (Fig. 5B). As a result, E4orf4 inhibits oncogenic transformation by various combinations of oncogenes, such as E1A plus E1B, E1A plus Ras (Fig. 5A), and Myc plus Ras (data not shown). It has been reported that E1A sensitizes cells to drug-induced apoptosis by activating caspase-9 in E1A-transformed cells (40). It was suggested that the E1A-induced proapoptotic signal is uncoupled from the apoptotic machinery, and that the added drug restores the link. The mechanism by which E4orf4 interacts with the oncogene-induced latent apoptotic state currently is not understood. However, caspase-independent mechanisms also may be involved in E4orf4-induced apoptosis, because it has been reported that E4orf4 induces apoptosis in Chinese hamster ovary cells by a caspase-independent mechanism (6).
Our results indicate that E4orf4 may have an important therapeutic potential. A high frequency of human cancers are p53 deficient, and conventional therapy of these types of cancer was found to have a poor prognosis, because of their resistance to DNA damage-activated apoptosis. Hence, discovering alternative therapies for p53-deficient tumors will be highly beneficial. E4orf4, which induces p53-independent apoptosis and kills transformed cells preferentially, is thus a candidate for use in cancer gene therapy.
Acknowledgments
We thank I. S. Y. Chen for the BScxThy1.2 plasmid, D. Büchner for technical assistance, and S. Lavi for her thoughtful comments on the manuscript. This work was supported by grants from the Israel Science Foundation, the Israel Cancer Research Fund, the Israel Cancer Association, and Internal Technion Funds.
ABBREVIATIONS
- AP-1
activator protein 1
- PP2A
protein phosphatase 2A
- wt
wild type
- HA
hemagglutinin
- CMV
cytomegalovirus
References
- 1.Müller U, Roberts M P, Engel D A, Doerfler W, Shenk T. Genes Dev. 1989;3:1991–2002. doi: 10.1101/gad.3.12a.1991. [DOI] [PubMed] [Google Scholar]
- 2.Müller U, Kleinberger T, Shenk T. J Virol. 1992;66:5867–5878. doi: 10.1128/jvi.66.10.5867-5878.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bondesson M, Ohman K, Mannervik M, Fan S, Akusjarvi G. J Virol. 1996;70:3844–3851. doi: 10.1128/jvi.70.6.3844-3851.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kanopka A, Muhlemann O, Petersen-Mahrt S, Estmer C, Ohrmalm C, Akusjarvi G. Nature (London) 1998;393:185–187. doi: 10.1038/30277. [DOI] [PubMed] [Google Scholar]
- 5.Shtrichman R, Kleinberger T. J Virol. 1998;72:2975–2982. doi: 10.1128/jvi.72.4.2975-2982.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lavoie J N, Nguyen M, Marcellus R C, Branton P E, Shore G C. J Cell Biol. 1998;140:637–645. doi: 10.1083/jcb.140.3.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marcellus R C, Lavoie J N, Boivin D, Shore G C, Ketner G, Branton P E. J Virol. 1998;72:7144–7153. doi: 10.1128/jvi.72.9.7144-7153.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kleinberger T, Shenk T. J Virol. 1993;67:7556–7560. doi: 10.1128/jvi.67.12.7556-7560.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mumby M C, Walter G. Physiol Rev. 1993;73:673–699. doi: 10.1152/physrev.1993.73.4.673. [DOI] [PubMed] [Google Scholar]
- 10.Kamibayashi C, Estes R, Lickteig R L, Yang S-I, Craft C, Mumby M. J Biol Chem. 1994;269:20139–20148. [PubMed] [Google Scholar]
- 11.McCright B, Virshup D M. J Biol Chem. 1995;270:26123–26128. doi: 10.1074/jbc.270.44.26123. [DOI] [PubMed] [Google Scholar]
- 12.Csortos C, Zolnierowicz S, Bako E, Durbin S D, DePaoli-Roach A A. J Biol Chem. 1996;271:2578–2588. doi: 10.1074/jbc.271.5.2578. [DOI] [PubMed] [Google Scholar]
- 13.Mumby M. Semin Cancer Biol. 1995;6:229–237. doi: 10.1006/scbi.1995.0030. [DOI] [PubMed] [Google Scholar]
- 14.Cryns V, Yuan J. Genes Dev. 1998;12:1551–1570. doi: 10.1101/gad.12.11.1551. [DOI] [PubMed] [Google Scholar]
- 15.Gjertsen B T, Doskeland S O. Biochim Biophys Acta. 1995;1269:187–199. doi: 10.1016/0167-4889(95)00117-b. [DOI] [PubMed] [Google Scholar]
- 16.Yan Y, Shay J W, Wright W E, Mumby M C. J Biol Chem. 1997;272:15220–15226. doi: 10.1074/jbc.272.24.15220. [DOI] [PubMed] [Google Scholar]
- 17.Song Q, Lavin M. Biochem Biophys Res Commun. 1993;190:47–55. doi: 10.1006/bbrc.1993.1009. [DOI] [PubMed] [Google Scholar]
- 18.Morana S J, Wolf C M, Reynolds J E, Brown M K, Eastman A. J Biol Chem. 1996;271:18263–18271. doi: 10.1074/jbc.271.30.18263. [DOI] [PubMed] [Google Scholar]
- 19.Santoro M F, Annand R R, Robertson M M, Peng Y W, Brady M J, Mankovich J A, Hackett M C, Ghayur T, Walter G, Wong W W, Giegel D A. J Biol Chem. 1998;273:13119–13128. doi: 10.1074/jbc.273.21.13119. [DOI] [PubMed] [Google Scholar]
- 20.Deng X, Ito T, Carr B, Mumby M, May W S. J Biol Chem. 1998;273:34157–34163. doi: 10.1074/jbc.273.51.34157. [DOI] [PubMed] [Google Scholar]
- 21.Hinds P, Finlay C A, Quartin R S, Baker S J, Fearon E R, Vogelstein B, Levine A J. Cell Growth Differ. 1990;1:571–580. [PubMed] [Google Scholar]
- 22.Morgenstern J P, Land H. Nucleic Acids Res. 1990;18:3587–3596. doi: 10.1093/nar/18.12.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pallas D C, Weller W, Jaspers S, Miller T B, Lane W S, Roberts T M. J Virol. 1992;66:886–893. doi: 10.1128/jvi.66.2.886-893.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jowett J B M, Planelles V, Poon B, Shah N P, Chen M L, Chen I S Y. J Virol. 1995;69:6304–6313. doi: 10.1128/jvi.69.10.6304-6313.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Logan J, Pilder S, Shenk T. Cancer Cells. 1984;2:527–532. [Google Scholar]
- 26.Neill S D, Hemstrom C, Virtanen A, Nevins J R. Proc Natl Acad Sci USA. 1990;87:2008–2012. doi: 10.1073/pnas.87.5.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shih C, Weinberg R A. Cell. 1982;29:161–169. doi: 10.1016/0092-8674(82)90100-3. [DOI] [PubMed] [Google Scholar]
- 28.Graham F L, Smiley J, Russel W C, Nairn R. J Gen Virol. 1977;36:59–72. doi: 10.1099/0022-1317-36-1-59. [DOI] [PubMed] [Google Scholar]
- 29.Mitsudomi T, Steinberg S M, Nau M M, Carbone D, D’Amico D, Bodner S, Oie H K, Linnoila R I, Mulshine J L, Minna J D, Gazdar A F. Oncogene. 1992;7:171–180. [PubMed] [Google Scholar]
- 30.Leung D W, Chen E, Goeddel D V. Technique. 1989;1:11–15. [Google Scholar]
- 31.Weiss R, S, Gold M O, Hannes V, Javier R T. J Virol. 1997;71:4385–4394. doi: 10.1128/jvi.71.6.4385-4394.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Graham G, Van der Eb A J. Virology. 1973;52:456–457. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- 33.Harlow E, Lane D. Antibodies: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1988. [Google Scholar]
- 34.Deana A, Pinna L A. Biochim Biophys Acta. 1988;968:179–185. doi: 10.1016/0167-4889(88)90006-7. [DOI] [PubMed] [Google Scholar]
- 35.Baykov A A, Evtushenko O A, Avaeva S M. Anal Biochem. 1988;171:266–270. doi: 10.1016/0003-2697(88)90484-8. [DOI] [PubMed] [Google Scholar]
- 36.Nevels M, Rubenwolf S, Spruss T, Wolf H, Dobner T. Proc Natl Acad Sci USA. 1997;94:1206–1211. doi: 10.1073/pnas.94.4.1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stewart S A, Poon B, Jowett J B M, Chen I S Y. J Virol. 1997;71:5579–5592. doi: 10.1128/jvi.71.7.5579-5592.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Darzynkiewicz Z, Bruno S, Delbino G, Gorczyca W, Hotz M A, Lassota P, Traganos F. Cytometry. 1992;13:795–808. doi: 10.1002/cyto.990130802. [DOI] [PubMed] [Google Scholar]
- 39.White E. Genes Dev. 1996;10:1–15. doi: 10.1101/gad.10.1.1. [DOI] [PubMed] [Google Scholar]
- 40.Fearnhead H O, Rodriguez J, Govek E E, Guo W, Kobayashi R, Hannon G, Lazebnik Y A. Proc Natl Acad Sci USA. 1998;95:13664–13669. doi: 10.1073/pnas.95.23.13664. [DOI] [PMC free article] [PubMed] [Google Scholar]