Abstract
We have characterized and cloned newly isolated lectins from hemolymph plasma of the horseshoe crab Tachypleus tridentatus, which we named tachylectins 5A and 5B (TLs-5). TLs-5 agglutinated all types of human erythrocytes and Gram-positive and Gram-negative bacteria. TLs-5 specifically recognize acetyl group-containing substances including noncarbohydrates; the acetyl group is required and is sufficient for recognition. TLs-5 enhanced the antimicrobial activity of a horseshoe crab-derived big defensin. cDNA sequences of TLs-5 indicated that they consist of a short N-terminal Cys-containing segment and a C-terminal fibrinogen-like domain with the highest sequence identity (51%) to that of mammalian ficolins. TLs-5, however, lack the collagenous domain found in a kind of “bouquet arrangement” of ficolins and collectins. Electron microscopy revealed that TLs-5 form two- to four-bladed propeller structures. The horseshoe crab is equipped with a unique functional homologue of vertebrate fibrinogen, coagulogen, as the target protein of the clotting cascade. Our observations clearly show that the horseshoe crab has fibrinogen-related molecules in hemolymph plasma and that they function as nonself-recognizing lectins. An ancestor of fibrinogen may have functioned as a nonself-recognizing protein.
Innate immunity is phylogenetically older than acquired immunity and is present in all multicellular organisms. With this immunity, unique polysaccharides on pathogens are detected through pattern recognition (1, 2). In mammals, collectins and ficolins characterized by the domain organization of a short N-terminal segment, a collagen-like domain, and a C-terminal lectin domain play important roles in innate immunity (3–7). In the horseshoe crab Tachypleus tridentatus, a major defense system is carried by hemolymph that contains granular hemocytes comprising 99% of the total hemocytes (8). The granular hemocytes are filled with two populations of secretory granules, named large and small granules (9). The granules selectively store defense molecules, such as clotting factors, a clottable protein coagulogen, protease inhibitors, lectins, and antimicrobial substances (10). The hemocytes are highly sensitive to lipopolysaccharides (LPS), and various defense molecules stored are secreted by exocytosis in response to the stimulation of LPS. Four types of lectins have been identified in hemocytes, named numerically tachylectin (TL)-1 to -4 (10). TL-1 interacts with Gram-negative bacteria probably through 2-keto-3-deoxyoctonate, one of the constituents of LPS (11). TL-2 binds to d-GlcNAc or d-GalNAc and recognizes staphylococcal lipoteichoic acids and Gram-negative bacteria-derived LPS (12–14). TL-3 (14) and TL-4 (15) specifically bind to S type LPS found in several Gram-negative bacteria with a certain sugar moiety on O-specific polysaccharides. The hemolymph plasma has been also reported to exhibit hemagglutinin activity (10). However, much less is known of plasma-derived lectins. Plasma-derived lectins of horseshoe crabs have been partially purified by using affinity chromatography of bovine submaxillary gland mucin-agarose (16), chitin (17), and GalNAc-agarose (18). Interestingly, all of the lectins contained in these fractions appear to have specificity for the N-acetyl group of carbohydrates. We have found that the hemagglutinin activity in the hemolymph plasma is adsorbed mostly to an N-acetyl group-immobilized resin. We have now identified plasma-derived lectins, named TL-5A and TL-5B (TLs-5), with structural similarity to vertebrate fibrinogens.
MATERIALS AND METHODS
Preparation of N-Acetyl Group-Immobilized Resin.
Toyopearl AF-Amino-650M resin (Tosoh, Tokyo) (10 g) was washed twice with distilled water and mixed with 8 ml of 0.2 M sodium acetate and 4 ml of acetic anhydride and then incubated on ice for 30 min. After incubation, 4 ml of acetic anhydride was added to the mixture, and the incubation continued for further 30 min. The resin was washed several times with distilled water and 1 M NaOH and then was washed with the equilibration buffer for chromatography.
Hemagglutination Activity, Bacterial Agglutination, and Antimicrobial Activity.
Hemagglutinating activity (12), bacterial agglutinating activity (11), and antimicrobial activity (11) were determined as described, except that the assay buffers contained 5 mM CaCl2. The following bacteria were used for agglutinating assay and antimicrobial activity: Staphylococcus aureus 209P, Staphylococcus saprophyticus KD, Micrococcus luteus, Enterococcus hirae, Escherichia coli B, and Escherichia coli K12.
SDS/PAGE and Immunoblotting.
SDS/PAGE was performed in 12.5% slab gels according to the method of Laemmli (19). The gels were stained with Coomassie Brilliant Blue R-250. For immunoblotting, tissues prepared from an adult male of T. tridentatus were dissolved in the sampling buffer of SDS/PAGE. Insoluble materials were removed by centrifugation. Gels were transferred to nitrocellulose membranes by electroblotting. The membranes were treated with the anti-TL-5A or -5B antibody and incubated with horseradish peroxidase-conjugated goat anti-rabbit IgG. Immunoreactive proteins attached to membranes were visualized by using Enhanced Chemiluminescence (ECL; Amersham Pharmacia).
Peptide Sequencing and Amino Acid Analyses.
When the intact TL-5A was subjected to amino acid sequence analysis, no phenylthiohydantoin-amino acid was obtained, suggesting a blocked N terminus. The analysis of TL-5A treated with pyroglutaminase yielded Asn-Lys-Glu-Leu-Cys-Asp-Val-, indicating the N-terminal pyroglutamic acid. The N-terminal sequence of TL-5B was identified to be Asn-Val-His-His-His-Ala-Cys-. The S-pyridylethylated TLs-5 were digested, respectively, with lysylendopeptidase, trypsin, and chymotrypsin and they were also cleaved chemically with CNBr. The resulting peptides were separated by reverse-phase HPLC. Amino acid sequence analysis was performed by using a gas-phase sequencer, model 473A or 477A (Applied Biosystems). For amino acid analysis, samples were hydrolyzed in 6 M HCl or 3 M mercaptoethanesulfonic acid in evacuated and sealed tubes at 110°C for 24, 48, and 72 h. The hydrolysates were analyzed by using a Hitachi L-8500 amino acid analyzer.
Screening of cDNA Library.
The degenerate nucleotide sequences of the primers used for reverse transcription–PCR were based on amino acid sequences of the peptides derived from chymotrypsin digestion: TL-5A, D-M-E-T-D-G for sense and L-P-Q-V-E-M for antisense; TL-5B, E-D-F-Y-Q-W for sense and D-N-D-K-W-E for antisense. Sense and antisense nucleotides were synthesized with an EcoRI site at the 5′ end. RNA from heart (for TL-5A) and RNA from hemocytes (for TL-5B) were used as the templates for PCR. The products of the PCR were subcloned into plasmid Bluescript II SK (Stratagene) for sequence analysis and served as probes for screening heart and hemocyte λZipLox cDNA libraries. Nucleotide sequences on both strands were determined as described (14).
Genomic Southern Blot Analyses of TLs-5.
Genomic DNA (10 μg each) was digested with several restriction enzymes, subjected to agarose gel electrophoresis, and transferred to nitrocellulose membranes. The membranes were probed with the same radiolabeled specific probes used for screening of the cDNA libraries, under high-stringency conditions (6× SSPE/0.5% SDS at 50°C).
Homology Search.
A computer-assisted homology search was made by using the DNA Data Bank of Japan (DDBJ) homology search system and programs fasta Version 3.0 and blast Version 1.4.9.
RESULTS AND DISCUSSION
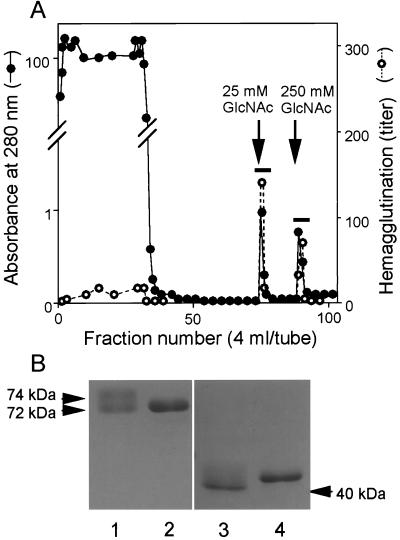
The acetyl group was coupled with a noncarbohydrate resin TOYOPEARL, a hydrophilic vinyl polymer, to avoid nonspecific adsorption of galactose-binding proteins in hemolymph. When hemolymph plasma was applied to the N-acetyl group-immobilized resin, about two thirds of the total hemagglutinating activity bound to the resin (Fig. 1A). Two protein peaks with hemagglutinating activity were obtained by stepwise elution, using different concentrations of GlcNAc. The first lectin eluted by 25 mM GlcNAc gave a smear band of 72–74 kDa on SDS/PAGE, under nonreducing conditions (Fig. 1B, lane 1). In contrast, the second lectin eluted by 250 mM GlcNAc gave a single band of 72 kDa (Fig. 1B, lane 2). After reduction, both protein bands, respectively, shifted to a smear band of 40–42 kDa (Fig. 1B, lane 3) and a single band of 41 kDa (Fig. 1B, lane 4), suggesting that each hemagglutinin consists of two identical subunits connected by disulfide linkage(s). We named these isolated proteins TL-5A and -5B for first and second lectins, respectively. The extinction coefficients at 280 nm for 1% solution were calculated from data on amino acid analysis, 34.1 for TL-5A and 29.2 for TL-5B. About 4 mg of TL-5A and about 3 mg of TL-5B could be purified reproducibly from 500 ml of hemolymph plasma.
Figure 1.
Purification of TLs-5 by using the N-acetyl group-immobilized resin. (A) Hemolymph plasma (500 ml) was applied to the resin (5 ml), equilibrated with 20 mM Tris⋅HCl (pH 8.0) containing 10 mM CaCl2 and washed extensively with the same buffer containing 0.5 M NaCl. Two protein peaks with hemagglutinating activity were obtained by stepwise elution of 25 mM GlcNAc and 250 mM GlcNAc in the equilibration buffer. No proteins were further eluted by extensive washing with the buffer containing 6 M guanidine⋅HCl. The fractions indicated by bars were pooled and dialyzed against 20 mM Tris·HCl (pH 8.0) containing 0.15 M NaCl and 5 mM CaCl2. (B) SDS/PAGE of purified TLs-5 under nonreducing (lanes 1 and 2) and reducing (lanes 3 and 4) conditions.
TLs-5 agglutinated all types of human erythrocytes, indicating that the primary recognition substance is not a blood group antigen (Table 1). TL-5A had stronger hemagglutinating activity than did TL-5B. The minimum agglutination concentration of TL-5A for A type erythrocytes is 400-fold lower than that (1.6 μg/ml) of hemocyte-derived TL-2 (12). The hemagglutinating activities of TLs-5 were completely inhibited by 5 mM EDTA, and this inhibition was overcome by adding an excess amount of CaCl2.
Table 1.
Hemagglutinating activity of TLs-5
| Erythrocyte | Minimum agglutination concentration, μg/ml
|
|
|---|---|---|
| TL-5A | TL-5B | |
| Human A type | 0.004 | 0.077 |
| B type | 0.008 | 0.27 |
| O type | 0.004 | 0.077 |
Mono- and disaccharides containing the acetyl group inhibited the hemagglutination at minimum inhibitory concentrations ranging from 0.6 to 12.5 mM (Table 2), N-acetylneuraminic acid being the most effective. N-Acetyl derivatives of amino acids and other acetyl group-containing substances, including acetylsalicylic acid, acetylcholine, and acetyl CoA were also effective competitors. Furthermore, one of the simplest chemicals containing the acetyl group, sodium acetate (CH3COONa) or acetamide (CH3CONH2), strongly inhibited the activity even at millimolar levels. Propionamide and benzamide also inhibited the hemagglutination, but 4- to 8-fold higher concentrations were required. On the other hand, sodium formate (HCOONa), as well as methyl-α-glucoside and methyl-β-glucoside, were without effect. These results indicate that the methyl group is important but not sufficient for this recognition. The acetyl group is therefore a necessary and sufficient condition. Rough LPS from S. minnesota R595(Re) (20) and E. coli J5 (21) had no effect on the activity, probably because they do not contain acetyl derivatives.
Table 2.
Inhibition of hemagglutinating activity of TLs-5 by various compounds
| Component | Minimum inhibitory concentration
|
|||
|---|---|---|---|---|
| TL-5A | TL-5B | |||
| mM | μg/ml | mM | μg/ml | |
| N-acetyl-d-glucosamine | 1.6 | 1.6 | ||
| N-acetyl-d-galactosamine | 3.1 | 3.1 | ||
| N-acetyl-d-mannosamine | 1.6 | 1.6 | ||
| N-acetyllactosamine | 6.3 | 3.1 | ||
| N-acetylallolactosamine | 1.6 | 1.6 | ||
| N-acetylneuraminic acid | 0.6 | 0.6 | ||
| N-acetylmuramic acid | 12.5 | 6.3 | ||
| N-acetylglycine | 6.3 | 12.5 | ||
| N-acetylglutamic acid | 1.6 | 1.6 | ||
| Nɛ-acetyllysine | 12.5 | 12.5 | ||
| Sodium acetate | 1.3 | 1.3 | ||
| Acetamide | 3.1 | 3.1 | ||
| Propionamide | 12.5 | 12.5 | ||
| Benzamide | 26.3 | 26.3 | ||
| Acetylsalicylic acid | 2.3 | 4.6 | ||
| Acetylcholine | 1.3 | 1.3 | ||
| Acetyl CoA | 1.6 | 1.6 | ||
| Lithium pyruvate | NI | 25.0 | ||
| Colominic acid | 18.9 | 18.9 | ||
| Fetuin | 25.0 | 25.0 | ||
| Lipoteichoic acid (S. aureus) | 25.0 | 12.5 | ||
| LPS | ||||
| E. coli O111B4 (smooth) | 100.0 | 100.0 | ||
| S. minnesota (smooth) | 12.5 | 50.0 | ||
| E. coli EH100 (Ra) | 100.0 | 100.0 | ||
NI, not inhibited at 50 mM.Noninhibitory substances tested: Monosaccharides (50 mM): d-glucose, d-glucosamine, d-galactose, d-galactosamine, d-mannose, l-fucose, methyl-α-glucoside, methyl-β-glucoside, 2-keto-3-deoxyoctonate. Oligo- and polysaccarides (50 mM or 100 μg/ml): sucrose, maltose, lactose, chondroitin sulfate A, chondroitin sulfate B, chondroitin sulfate C, hyaluronic acid, laminarin. LPS (100 μg/ml): S. minnesota R595(Re), E. coli J5(Rc). Others (50 mM): glycine, glutamic acid, sulfatide, sodium formate.
TLs-5 also agglutinated Gram-negative and Gram-positive bacteria, and the activity was more efficient for Gram-negative than for Gram-positive bacteria (Table 3). As in the case of hemagglutinating activity, TL-5A exhibited stronger bacterial agglutinating activity than did TL-5B, particularly against Gram-positive bacteria. TLs-5 at 20 μg/ml had no antimicrobial activity against these bacteria.
Table 3.
Bacterial agglutinating activity of TLs-5
| Bacteria | Minimum agglutination concentration, μg/ml
|
|
|---|---|---|
| TL-5A | TL-5B | |
| Gram-negative | ||
| E. coli B | 0.08 | 0.11 |
| E. coli K12 | 0.04 | 0.05 |
| Gram-positive | ||
| S. aureus 209P | 1.2 | 26.8 |
| S. saprophyticus KD | 1.6 | 15.1 |
| Enterococcus hirae | 0.3 | 26.8 |
| Micrococcus luteus | 2.4 | 26.8 |
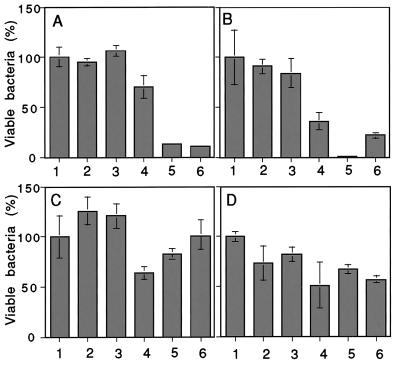
We reported elsewhere that tachycitin and big defensin, antimicrobial polypeptides derived from horseshoe crab hemocytes, had a synergistic effect on antimicrobial activity (22). TLs-5 also enhanced the antimicrobial activity of big defensin against Gram-positive bacteria (Fig. 2 A and B). Interestingly, TLs-5 did not enhance antimicrobial activity against Gram-negative bacteria (Fig. 2 C and D), although agglutination was more efficient against Gram-negative than Gram-positive bacteria. TLs-5 probably interact with big defensin to carry it efficiently to the surface of bacteria. Big defensin possesses two functional domains, the N-terminal antibacterial domain against Gram-positive bacteria and the C-terminal antibacterial domain against Gram-negative bacteria (23). TLs-5 may bind to the C-terminal domain of big defensin, resulting in blockage of the antibacterial activity against Gram-negative bacteria. Hemocyte-derived TL-1 (11) and TL-2 (12) had no such a synergistic effect on the antimicrobial activity of big defensin.
Figure 2.
A synergistic effect of TLs-5 on antimicrobial activity of big defensin. (A) Enterococcus hirae; (B) Staphylococcus aureus 209P; (C) Escherichia coli B; (D) Escherichia coli K12. The bar graph represents the means ± SE of the three measurements. Big defensin (2.5 μg/ml) was mixed with bacteria in the presence or absence of TLs-5 (1 μg/ml). Bar 1, no big defensin and no TLs-5; bar 2, TL-5A; bar 3, TL-5B; bar 4, big defensin; bar 5, big defensin + TL-5A; bar 6, big defensin + TL-5B.
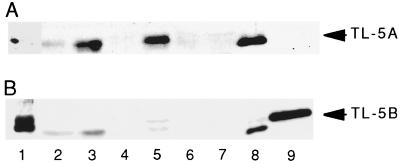
To investigate tissue-specific expression of TLs-5, immunoblotting was done by using polyclonal antibodies. TL-5A was detected in heart and intestine and faintly in hepatopancreas but not in hemocytes, stomach, nervous tissue, and skeletal muscle (Fig. 3A). In contrast, TL-5B was detectable only in hemocytes (Fig. 3B).
Figure 3.
Tissue-specific expression patterns of TLs-5. (A) The membrane was probed with polyclonal antibody against TL-5A. (B) The membrane was probed with polyclonal antibody against TL-5B. The anti-TL-5B antibody cross-reacts with TL-5A. Lane 1, hemocytes; lane 2, hepatopancreas; lane 3, heart; lane 4, stomach; lane 5, intestine; lane 6, nervous tissue; lane 7, skeletal muscle; lane 8, purified TL-5A; lane 9, purified TL-5B.
cDNA clones for TLs-5 were isolated, and nucleotide sequences were determined. Screening a heart cDNA library with the TL-5A-specific probe yielded 4 positive clones, and screening a hemocyte cDNA library with the TL-5B-specific probe yielded 13 clones. The four nucleotide sequences for TL-5A (980–1,176 nt) agreed precisely, with no discrepancies. In the case of TL-5B, the nucleotide sequences of 12 of 13 clones (1,058–1,473 nt) agreed precisely. The remaining one clone for TL-5B had one nucleotide replacement, which replaced Pro-38 with Ser.
The ORF for TL-5A encoded for a mature protein of 269 aa with a signal sequence of 23 residues consisting of a typical hydrophobic core (see the GenBank Database, accession nos. AB024737 and AB024738). The ORF for TL-5B encoded for a mature protein of 289 aa with a signal sequence of 27 residues. As shown in Fig. 4, the deduced amino acid sequences indicated that the two lectins were homologous. The overall sequence identity between TL-5A and TL-5B was 45.1%. TLs-5 contained four putative N-linked glycosylation sites in each deduced sequence (underlined in Fig. 4). The N-glycosidase F treatment resulted in higher mobility bands on SDS/PAGE, under reducing conditions, 36-kDa and 38-kDa bands for TL-5A and a 38-kDa band for TL-5B, indicating that both lectins contain the N-linked sugar(s). By amino acid analysis, galactosamine was detected in addition to glucosamine in hydrolysates of TLs-5, suggesting that O-linked sugar(s) is also linked to TLs-5. Protein analyses showed that the N terminus of TL-5A is pyroglutamic acid. TL-5B, but not TL-5A, contained an integrin-binding sequence of –Arg-Gly-Asp– at positions 112–114.
Figure 4.
Alignment of amino acid sequences of TLs-5, human l-ficolin/P35 (HFCL), and the C-terminal domains of human fibrinogen β- (HFBE) and γ- (HFGA) chains. In the nucleotide sequence, the N-terminal amino acid of TL-5B was aspartic acid; nevertheless, asparagine was identified by amino acid sequence analysis. This discrepancy cannot be clearly explained, but there may be several genes coding isoproteins for TL-5B. The bold letters represent residues identical to TLs-5.
A homology search revealed significant sequence similarity of the C-terminal regions of TLs-5 with those of vertebrate fibrinogens and their related proteins (24), consisting of 210–250 aa. Pairwise comparisons of the C-terminal domains of TLs-5 (Cys-49–Asn-269 in TL-5A and Cys-68–Lys-289 in TL-5B) with the 37 fibrinogen-like domains were made, and normalized alignment scores (NAS) (25) were calculated (Table 4). Based on the relationship between lengths of aligned sequences and NAS, the domains with NAS over ≈250 points are homologous. NAS for fibrinogen β- (340–374) and γ- (279–368) chains derived from various species were sufficiently high to be homologous. In contrast, fibrinogen α-chain had only 15–17% identity with low NAS of 38–83. Gene duplications of individual genes for fibrinogen chains occurred 1,000 million years ago for α-chain and 600 million years ago for β- and γ-chains (26). The low sequence identity of TLs-5 to fibrinogen α-chain and the high sequence identity to β- and γ-chains suggest that the gene duplication of TLs-5 also occurred about 600 million years ago.
Table 4.
Pairwise comparisons of fibrinogen-related sequences with TLs-5
| Protein (accession no.) | Normalized alignment score (% identity)
|
|
|---|---|---|
| TL-5A | TL-5B | |
| Fibrinogen α-chain | ||
| Human (J00128) | 38 (17.2) | 83 (13.7) |
| Rana (U44830) | 83 (15.0) | 50 (15.4) |
| Fibrinogen β-chain | ||
| Human (P02675) | 360 (38.9) | 376 (38.9) |
| Rat (P14480) | 347 (37.3) | 376 (38.9) |
| Bovine (P02676) | 340 (36.9) | 378 (39.3) |
| Chicken (Q02020) | 358 (38.9) | 381 (39.2) |
| Lamprey (P02678) | 374 (40.1) | 372 (38.7) |
| Fibrinogen γ-chain | ||
| Human (P02679) | 336 (36.7) | 354 (38.3) |
| Rat (P02680) | 318 (34.6) | 336 (34.0) |
| Bovine (P12799) | 327 (35.4) | 338 (37.1) |
| Xenopus (P17634) | 336 (36.5) | 369 (38.4) |
| Rana (U44829) | 279 (33.8) | 346 (37.3) |
| Lamprey (P04115) | 338 (38.3) | 396 (39.3) |
| Ficolin | ||
| Human M (D83920) | 474 (47.7) | 505 (50.6) |
| Human L (D49353) | 452 (45.6) | 490 (49.2) |
| Hakata antigen (D88587) | 351 (37.2) | 408 (41.0) |
| Mouse A (AB007813) | 443 (44.7) | 495 (49.7) |
| Mouse B (AF063217) | 453 (45.2) | 515 (50.8) |
| Pig α (A47172) | 429 (44.4) | 505 (50.6) |
| Pig β (B47172) | 445 (45.0) | 497 (50.9) |
| Tenascin | ||
| Human (P24821) | 388 (41.3) | 392 (42.7) |
| Mouse (JQ1322) | 416 (41.9) | 398 (41.1) |
| Pig (S19694) | 416 (42.9) | 396 (42.1) |
| Pig homologue (S28170) | 384 (40.1) | 405 (41.6) |
| Chicken (P10039) | 393 (41.7) | 390 (41.4) |
| Chicken restrictin (JH0675) | 435 (44.8) | 437 (44.9) |
| Chicken cytotactin (J04519) | 399 (44.4) | 395 (40.7) |
| Others | ||
| Human fibrinogen-related protein (JN0596) | 399 (45.1) | 426 (46.0) |
| Human pT49 (S47273) | 374 (41.8) | 399 (41.2) |
| Human CDT6 (Y16132) | 401 (40.1) | 444 (48.9) |
| Human XB gene (P22105) | 375 (39.8) | 396 (41.0) |
| Mouse fibrinogen-like protein (S78773) | 392 (43.6) | 394 (40.8) |
| Mouse c T-cell-specific protein (P12804) | 396 (44.0) | 410 (43.2) |
| Sea cucumber fibrinogen-related protein (P19477) | 376 (40.4) | 392 (42.3) |
| Slug sialic acid binding lectin (AF060450) | 343 (35.0) | 411 (39.8) |
| Snail fibrinogen-related protein (U82479) | 338 (39.1) | 284 (35.6) |
| Drosophila scabrous (P21520) | 312 (31.9) | 327 (31.7) |
| Tachylectin-5A | — | 511 (50.4) |
| Tachylectin-5B | 511 (50.4) | — |
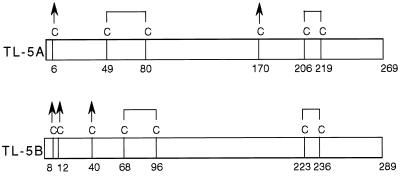
The fibrinogen-like domain of mammalian ficolins showed strikingly sequence identity of up to 51%, and their NAS were almost equivalent to the score between TL-5A and TL-5B. Interestingly, a collagenous domain found in ficolins is missing in the corresponding regions of TLs-5 (Fig. 4). Hemagglutinating activity of TLs-5 is Ca2+-dependent, and fibrin polymerization and crosslinking are also Ca2+-dependent processes. Asp-318 and Asp-320 in the fibrinogen γ-chain have been found to provide their oxygen atoms to coordinate the calcium ion (24, 27), which are conserved in TL-5A (Asp-198 and Asp-200) and TL-5B (Asp-215 and Asp-217). TL-5A and TL-5B contained six and seven cysteine residues, respectively. Based on the disulfide linkages in β- and γ-chains of human fibrinogen, Cys-49–Cys-80 and Cys-206–Cys-219 in TL-5A and Cys68-Cys96 and Cys223-Cys236 in TL-5B could form intramolecular disulfide bonds (Fig. 5). The minimum subunit of 72 kDa of TL-5A or TL-5B is possibly a homodimer linked by interchain disulfide bond(s), and if so, then several of the remaining cysteine residues are involved in interchain disulfide linkages.
Figure 5.
Schematic diagrams of the inter- and intrachain disulfide bonds of TLs-5. Arrows indicate cysteine residues that are possibly involved in interchain disulfide linkages. The disulfide bonds are predicted according to those in human fibrinogen (35).
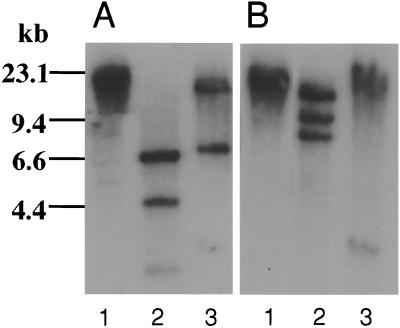
The genes of TLs-5 were examined by Southern blot analysis of the specific cDNA probes used for the screening (Fig. 6). The genomic DNA was completely digested by several restriction enzymes. The TL-5A probe hybridized with multiple bands in HindIII and XhoI digest, and the TL-5B probe did so with multiple bands in XhoI and KpnI digest. Because these enzymes do not digest respective cDNAs, TL-5A and TL-5B are suggested to be encoded by a single gene with at least two introns, or by multiple genes.
Figure 6.
Genomic Southern blot analyses of TLs-5. (A) For TL-5A: lane 1, BamHI; lane 2, HindIII; lane 3, XhoI. (B) For TL-5B: lane 1, BamHI; lane 2, XhoI; lane 3, KpnI.
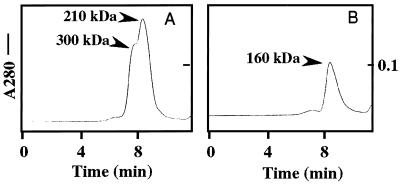
The apparent molecular masses of TLs-5 in solution were determined by using gel filtration. TL-5A showed a major peak of 210 kDa with a shoulder peak of 300 kDa (Fig. 7A), corresponding to hexameric and octameric oligomers, respectively, based on the disulfide-linked homodimer with 72 kDa on SDS/PAGE. On the other hand, TL-5B had a single peak of 160 kDa corresponding to a tetrameric oligomer (Fig. 7B).
Figure 7.
The apparent molecular masses of TLs-5 in solution. (A) Gel filtration for TL-5A. (B) Gel filtration for TL-5B. Gel filtration was carried out on a Bio-Silect TM SEC400–5 column (Bio-Rad Laboratories), equilibrated with 20 mM Tris⋅HCl (pH 7.5) containing 0.15 M NaCl and 5 mM CaCl2, by using a FPLC system (Amersham Pharmacia). Proteins eluted were monitored at 280 nm.
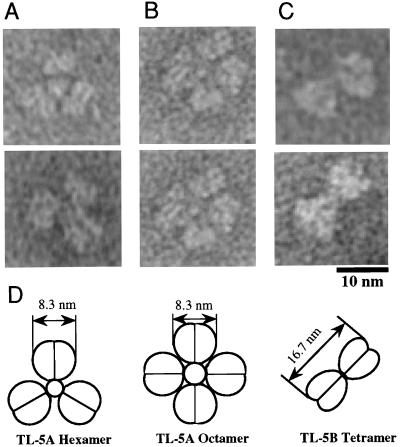
Electron microscopy of negatively stained TLs-5 provided higher resolution images of their oligomeric structures. TL-5A formed a three- or four-bladed propeller structure, and TL-5B formed a two-bladed propeller structure (Fig. 8). Assuming that each blade corresponds to the disulfide-linked dimer, the molecular masses estimated based on these structural images are consistent with the apparent molecular masses of TLs-5 determined by gel filtration. If the ligand-binding site(s) is located in each blade of TLs-5, TL-5A could have more avidity against polymeric ligands than TL-5B. This may explain why TL-5A exhibits higher hemagglutinating and bacterial agglutinating activities than does TL-5B. The small globe at the center of the propeller structures is likely to be a bundle domain of the propeller blades. The small globe was not observed for every propeller structure on electron micrographs, which means that the bundle domain likely projects at one side of the structures.
Figure 8.
Electron micrographs of negatively stained TLs-5. (A) A hexameric structure of TL-5A. (B) An octameric structure of TL-5A. (C) A tetrameric structure of TL-5B. (D) Schematic models of the oligomeric structures of TLs-5. Electron microscopy was performed by using the JEM200EX electron microscope (JEOL, Tokyo) by negative staining with 0.5% uranyl acetate. The length measurements of the protein molecules were made on micrographs printed at ×360,000 magnification.
Ficolins are composed of an N-terminal cysteine-containing oligomerization segment followed by a collagen-like region and a C-terminal fibrinogen-like globular domain to form a “bundle-of-tulips” or “bouquet” overall structure, like mammalian C1q and collectins (3–7, 28). Ficolins and collectins play important roles in host defense in nonself recognition by binding to carbohydrates on microorganisms and enhancing the neutrophil opsonic activity (29, 30). Human l-ficolin/P35 interacts with GlcNAc-conjugated BSA (29) and adsorbs to a nonsugar substance, Tris-derivatized CNBr-Sepharose (31). We found that human l-ficolin/P35 also binds to the N-acetyl group-immobilized resin, under the same conditions used for TLs-5 (data not shown), suggesting that l-ficolin/P35 also recognizes the acetyl group on the surface of pathogens. In contrast, Hakata antigen (32) and a C-terminal globular domain of human fibrinogen generated by plasmin digest, fragment D (33), did not bind to this resin. In light of the overall structural similarity of TLs-5 to these mammalian lectins involved in innate immunity and the tissue-specific localization of TLs-5 at hemolymph plasma and intestine (Fig. 3), where self and nonself substances meet, we propose that TLs-5 must be the primary lectins recognizing invading microbes in the front line of the defense system of horseshoe crab.
It has been suggested that nonvertebrates may have fibrinogen, perhaps present inside blood cells (26). The horseshoe crab is equipped with a unique functional homologue of vertebrate fibrinogen, coagulogen, as the target protein of the clotting cascade (10). However, recent crystallographic analysis indicated that coagulogen is a structural homologue of nerve growth factor (34). Our observations show here that the horseshoe crab has fibrinogen-related molecules in hemolymph plasma and that they function as nonself-recognizing lectins. An ancestor of fibrinogen-like proteins may have functioned as a nonself-recognizing protein.
Acknowledgments
We thank Drs. M. Matsushita and T. Fujita (Fukushima Medical College) for providing human l-ficolin/P35, Dr. N. Hamasaki (Kyushu University) for Hakata antigen, Dr. M. Matsuda (Jichi Medical School) for fragment D, W. Kamada for technical assistance, and M. Ohara for helpful comments on this manuscript. This work was supported by a Grant-in-Aid for scientific research from Ministry of Education, Science, Sports and Culture of Japan (S.K.) and Core Research for Evolutional Science and Technology of Japan Science and Technology Corporation (S.K.).
ABBREVIATIONS
- TL
tachylectin
- LPS
lipopolysaccharides
- NAS
normalized alignment scores
- TLs-5
TL-5A and TL-5B
Footnotes
References
- 1.Janeway C A., Jr Cold Spring Harbor Symp Quant Biol. 1989;54:1–13. [Google Scholar]
- 2.Medzhitov R, Janeway C A., Jr Cell. 1997;91:295–298. doi: 10.1016/s0092-8674(00)80412-2. [DOI] [PubMed] [Google Scholar]
- 3.Weis W I, Drickamer K. Structure (London) 1994;2:1227–1240. doi: 10.1016/S0969-2126(94)00124-3. [DOI] [PubMed] [Google Scholar]
- 4.Turner M W. Immunol Today. 1996;17:532–540. doi: 10.1016/0167-5699(96)10062-1. [DOI] [PubMed] [Google Scholar]
- 5.Ichijo H, Hellman U, Wernstedt C, Gonez L J, Claesson-Welsh L, Heldin C-H, Miyazono K. J Biol Chem. 1993;268:14505–14513. [PubMed] [Google Scholar]
- 6.Lu J, Le Y. Immunobiology. 1998;199:190–199. doi: 10.1016/S0171-2985(98)80026-0. [DOI] [PubMed] [Google Scholar]
- 7.Hansen S, Holmskov U. Immunobiology. 1998;199:165–189. doi: 10.1016/S0171-2985(98)80025-9. [DOI] [PubMed] [Google Scholar]
- 8.Toh Y, Mizutani A, Tokunaga F, Muta T, Iwanaga S. Cell Tissue Res. 1991;266:137–147. [Google Scholar]
- 9.Shigenaga T, Takayenoki Y, Kawasaki S, Seki N, Muta T, Toh Y, Ito A, Iwanaga S. J Biochem. 1993;114:307–316. doi: 10.1093/oxfordjournals.jbchem.a124173. [DOI] [PubMed] [Google Scholar]
- 10.Iwanaga S, Kawabata S, Muta T. J Biochem. 1998;123:1–15. doi: 10.1093/oxfordjournals.jbchem.a021894. [DOI] [PubMed] [Google Scholar]
- 11.Saito T, Kawabata S, Hirata M, Iwanaga S. J Biol Chem. 1995;270:14493–14499. doi: 10.1074/jbc.270.24.14493. [DOI] [PubMed] [Google Scholar]
- 12.Okino N, Kawabata S, Saito T, Hirata M, Takagi T, Iwanaga S. J Biol Chem. 1995;270:31008–31015. doi: 10.1074/jbc.270.52.31008. [DOI] [PubMed] [Google Scholar]
- 13.Beisel H-G, Kawabata S, Iwanaga S, Huber R, Bode W. EMBO J. 1999;18:2313–2322. doi: 10.1093/emboj/18.9.2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Inamori K, Saito T, Iwaki D, Nagira T, Iwanaga S, Arisaka F, Kawabata S. J Biol Chem. 1999;274:3272–3278. doi: 10.1074/jbc.274.6.3272. [DOI] [PubMed] [Google Scholar]
- 15.Saito T, Hatada M, Iwanaga S, Kawabata S. J Biol Chem. 1997;272:30703–30708. doi: 10.1074/jbc.272.49.30703. [DOI] [PubMed] [Google Scholar]
- 16.Shishikura F, Sekiguchi K. J Biochem. 1983;93:1539–1546. doi: 10.1093/oxfordjournals.jbchem.a134292. [DOI] [PubMed] [Google Scholar]
- 17.Matsumoto I, Yamaguchi H, Seno N, Shibata Y, Okuyama T. In: Proceedings of the Second International Conference on Chitin and Chitosan. Hirano S, Tokura S, editors. Tottori: The Japanese Society of Chitin and Chitosan; 1982. pp. pp.165–170. [Google Scholar]
- 18.Shimizu S, Ito M, Takahashi N, Niwa M. In: Biomedical Applications of the Horseshoe crab (Limulidae) Cohen E, editor. New York: Liss; 1979. pp. 625–639. [Google Scholar]
- 19.Laemmli U K. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 20.Holst O, Brade H. In: Chemical Structure of the Core Region of Lipopolysaccharides. Morrison D C, Ryan J L, editors. Boca Ranton, FL: CRC Press; 1992. pp. 135–170. [Google Scholar]
- 21.Müller-Loennies S, Holst O, Brade H. Eur J Biochem. 1994;224:751–760. doi: 10.1111/j.1432-1033.1994.t01-1-00751.x. [DOI] [PubMed] [Google Scholar]
- 22.Kawabata S, Nagayama R, Hirata M, Shigenaga T, Lal Agarwara K, Saito T, Cho J, Nakajima H, Takagi T, Iwanaga S. J Biochem. 1996;120:1253–1260. doi: 10.1093/oxfordjournals.jbchem.a021549. [DOI] [PubMed] [Google Scholar]
- 23.Saito T, Kawabata S, Shigenaga T, Takayenoki Y, Cho J, Nakajima H, Hirata M, Iwanaga S. J Biochem. 1995;117:1131–1137. doi: 10.1093/oxfordjournals.jbchem.a124818. [DOI] [PubMed] [Google Scholar]
- 24.Doolittle R F. Protein Sci. 1992;1:1563–1577. doi: 10.1002/pro.5560011204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Doolittle R F. Of Urfs and Orfs: A Primer on How to Analyze Derived Amino Acid Sequences. Mill Valley, CA: University Science Books; 1987. pp. 10–15. [Google Scholar]
- 26.Doolittle R F. Ann NY Acad Sci. 1983;408:13–27. doi: 10.1111/j.1749-6632.1983.tb23231.x. [DOI] [PubMed] [Google Scholar]
- 27.Yee V C, Pratt K P, Cote H C F, Trong I L, Chung D W, Davie E W, Stenkamp R E, Teller D C. Structure (London) 1996;15:125–138. doi: 10.1016/s0969-2126(97)00171-8. [DOI] [PubMed] [Google Scholar]
- 28.Ohashi T, Erickson H P. J Biol Chem. 1997;272:14220–14226. doi: 10.1074/jbc.272.22.14220. [DOI] [PubMed] [Google Scholar]
- 29.Matsushita M, Endo Y, Taira S, Sato Y, Fujita T, Ichikawa N, Nakata M, Mizuochi T. J Biol Chem. 1996;271:2448–2454. doi: 10.1074/jbc.271.5.2448. [DOI] [PubMed] [Google Scholar]
- 30.Lu J, Le Y, Kon O L, Chan J, Lee S H. Immunology. 1996;89:289–294. doi: 10.1046/j.1365-2567.1996.d01-732.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Le Y, Kon O L, Lu J. FEBS Lett. 1998;425:367–370. doi: 10.1016/s0014-5793(98)00267-1. [DOI] [PubMed] [Google Scholar]
- 32.Sugimoto R, Yae Y, Akaiwa M, Kitajima S, Shibata Y, Sato H, Hirata J, Okochi K, Izuhara K, Hamasaki N. J Biol Chem. 1998;273:20721–20727. doi: 10.1074/jbc.273.33.20721. [DOI] [PubMed] [Google Scholar]
- 33.Spraggon G, Everse S J, Doolittle R F. Nature (London) 1997;389:455–462. doi: 10.1038/38947. [DOI] [PubMed] [Google Scholar]
- 34.Bergner A, Oganessyan V, Muta T, Iwanaga S, Typke D, Huber R, Bode W. EMBO J. 1996;15:6789–6797. [PMC free article] [PubMed] [Google Scholar]
- 35.Bouma H, III, Takagi T, Doolittle R F. Thromb Res. 1978;13:557–562. doi: 10.1016/0049-3848(78)90142-1. [DOI] [PubMed] [Google Scholar]